Abstract
Remdesivir is a nucleoside analog approved by the US FDA for treatment of COVID-19. Here, we present a 3.9-Å-resolution cryo-EM reconstruction of a remdesivir-stalled RNA-dependent RNA polymerase complex, revealing full incorporation of 3 copies of remdesivir monophosphate (RMP) and a partially incorporated fourth RMP in the active site. The structure reveals that RMP blocks RNA translocation after incorporation of 3 bases following RMP, resulting in delayed chain termination, which can guide the rational design of improved antiviral drugs.
Keywords: remdesivir, SARS-CoV-2, RNA-dependent RNA polymerase, COVID-19 treatment
Graphical abstract
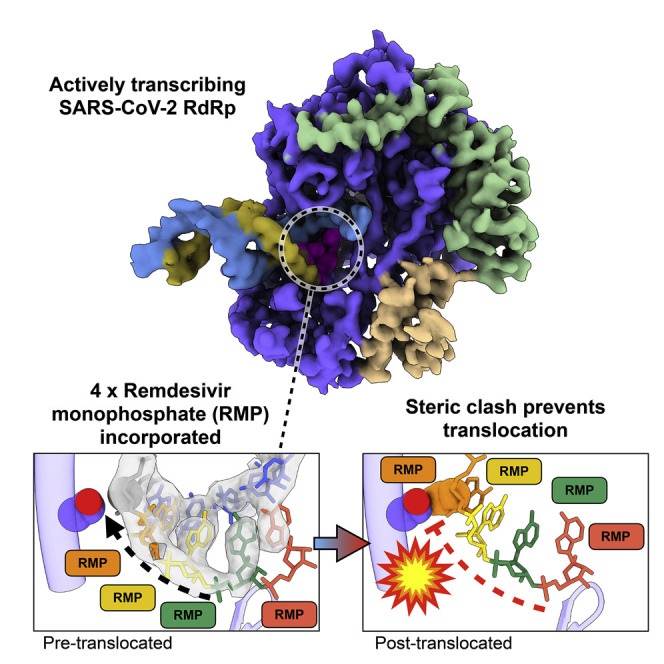
Bravo et al. reveal the structural basis for remdesivir-induced stalling of the SARS-CoV-2 RNA-dependent RNA polymerase. The structure of the stalled RdRp shows that chain translocation is blocked by steric clashes between incorporated remdesivir and the polymerase.
Introduction
Although vaccines appear to be on track to mitigate the coronavirus disease 2019 (COVID-19) pandemic, there is still a need for direct acting antiviral drugs to treat severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) infections when immunization fails and for future coronavirus outbreaks.
Structures of the core SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) complex (consisting of the enzyme nsp12 and the accessory subunits nsp7 and nsp8) have revealed the structural basis for RNA replication (Chen et al., 2020; Hillen et al., 2020; Wang et al., 2020b; Yan et al., 2020, 2021; Yin et al., 2020). However, the mechanism of action of remdesivir, an adenosine analog that was recently approved by the US Foos and Drug Administration (FDA) for COVID-19 treatment, remains to be established (Beigel et al., 2020; Wang et al., 2020a). Kinetic studies have shown that remdesivir triphosphate (RTP) is incorporated rapidly with an efficiency that is 2-fold greater than ATP and is rapidly extended within 1 s, but the polymerase stalls after adding 2 or 3 nt on top of the newly incorporated remdesivir monophosphate (RMP) (Dangerfield et al., 2020; Gordon et al., 2020). Here, we provide the structure of a SARS-CoV-2 polymerase-RNA complex stalled after the incorporation of remedesivir, revealing the underlying mechanistic basis for the inhibition of replication.
Results
Kinetics of remdesivir incorporation and stalling by RdRp
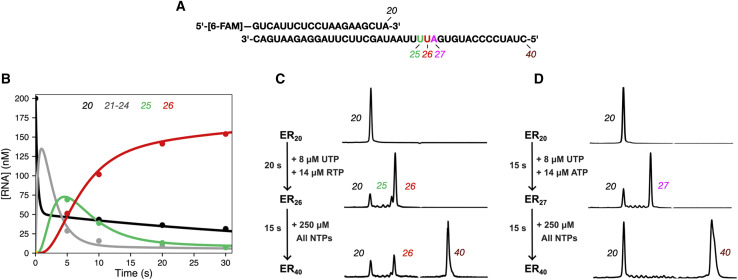
We examined the kinetics of remdesivir incorporation and subsequent stalling of continued RNA replication by incubating the RdRp with a 6-carboxyfluorescein (FAM)-labeled primer:template RNA duplex (Figure 1 A) and then initiating the polymerization by the addition of uracil triphosphate (UTP) and RTP (in the presence of 5 mM Mg2+). After quenching the reaction with EDTA, we resolved and quantified the reaction products at different time points using capillary electrophoresis (CE) by monitoring fluorescence. While we have previously demonstrated the rapid incorporation of 2 UTP and 3 RTP molecules within 1 s (Dangerfield et al., 2020), here, we found that over the course of 20 s, there is a significantly slower incorporation of a fourth RTP (a rate of 0.13 s−1, at the concentrations listed in Figure 1B). Note that we did not observe further addition of UTP on top of the 4 RMPs observed under these conditions, indicating that the RdRp complex has stalled (Figure 1C). CE reveals a mixture of 25- and 26-nt products corresponding to the extended primer with a mixture of 3 (10%) and 4 RMP (90%) molecules incorporated, respectively (Figure 1C). Under the same conditions in the presence of UTP and ATP, we see the extension form a 27-nt product, indicating that premature stalling does not occur with ATP (Figure 1D).
Figure 1.
RdRp stalls after incorporation of 4 remdesivirs
(A) The RNA substrate used in experiments consists of a 20-nt, 5′-[6-FAM]-labeled primer annealed to a 40-nt template. The sequence is from the 3′ end of the SARS-CoV-2 genome, preceding the poly-A tail.
(B) Time course of UTP + RTP incorporation by the RdRp complex. A solution of 3.33 μM nsp12/7/8, 4.7 μM nsp8, and 200 nM FAM-20/40 RNA was mixed with 14 μM RTP and 8 μM UTP to start the reaction. Time points were quenched by the addition of EDTA, and samples were analyzed by capillary electrophoresis. Lengths of various RNA products are given at the top of the panel. The solid lines through the data points are from the fit by simulation in KinTek Explorer.
(C) UTP + RTP product formation and extension. The chromatogram for the zero-time point is given at the top (ER20). Initially, RTP and UTP were added for 20 s (ER26) at the concentrations given in the figure. Then, 250 μM of all NTPs were added for 15 s to allow extension to the full-length product (ER40). On the timescale of the experiment, only a fraction of ER26 is converted to the full-length product (ER40).
(D) UTP + ATP product formation and extension. The experiment was performed as in (C), but with ATP instead of RTP in the first mixing step, allowing extension to the ER27 product. After addition of all of the nucleotides for 15 s in the second mixing step, the ER27 complex is completely converted to the full-length ER40 product.
Following the addition of 250 μM of all 4 nt to the 27-nt product formed with ATP, we observed complete extension to the full-length 40-nt product within 15 s. In contrast, when we add 250 μM of all 4 nucleotides to the RMP-stalled RNA, only 70% of the 26-nt (RTP) primer was fully extended to yield a 40-nt product within 15 s. The apparent extension rate of 0.08 s−1 represents a 4,000-fold inhibition of normal incorporation. While our results are in agreement with previous studies that have shown that RMP incorporation results in stalling after being extended by 3 nucleotides, our data collected with a highly active replicating enzyme show that the inhibition is not absolute and can be overcome by incubation at high nucleotide concentrations.
A physical barrier between remdesivir and RdRp blocks translocation
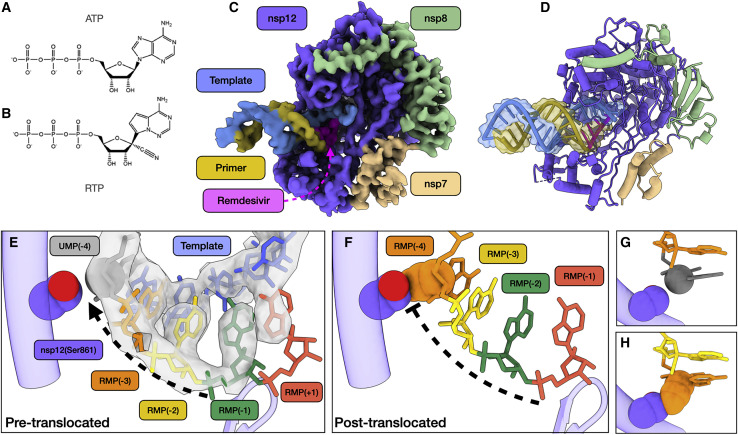
We used conditions that lead to stalling in our kinetic analysis above to prepare samples for cryoelectron microscopy (cryo-EM), allowing us to determine the structure of a bona fide elongation intermediate at a global resolution of 3.9 Å (Figure 2 ). We observed well-resolved primer:template duplex density, allowing de novo modeling of a remdesivir-stalled primer elongation intermediate. Within the active site of the RdRp complex, RMP occupies positions −3, −2, and −1 (i.e., 3, 2, and 1 nucleotides downstream of the active site, respectively). We also observed additional density in the polymerase active site position (position +1). This density corresponds to the nucleobase of a fourth remdesivir, consistent with our kinetic analysis (Figure 1C). This suggests that while the fourth RMP can participate in Watson-Crick base pairing with the template UMP and stacking interactions with neighboring RMP, the sugar-phosphate backbone may be somewhat disordered. Surprisingly, the fourth RMP was retained at the polymerase active site and did not translocate to allow the binding of the next nucleotide (UTP in our sequence). Thus, our data suggest that the polymerase stalls because the translocation step is blocked. We modeled a hypothetical post-translocated state, where the first RMP is moved to the −4 position, which revealed a steric clash between the C1 cyano group of RMP(−4) and the Ser861-Cβ of nsp12 (Figures 2E–2H). This residue provides a physical barrier to slow duplex translocation and may lead to perturbations near or in the active site that slow the rate of chain elongation. Our model provides a structural basis for stalling RdRp after incorporation of the fourth RMP. This is in agreement with previous studies that have demonstrated that mutations of nsp12(Ser861) reduce the inhibitory effect of remdesivir (Gordon et al., 2020; Tchesnokov et al., 2020). Our structure also shows that the cyano group of each of the 4 RMP molecules is readily accommodated at the active site, with no apparent steric interactions with the enzyme. The proposed steric interaction would only occur as the RNA duplex attempts to translocate to allow the binding and incorporation of UTP.
Figure 2.
Structural basis of SARS-CoV-2 RdRp complex stalling by remdesivir
(A) Chemical structures of adenosine triphosphate.
(B) Chemical structure of remdesivir triphosphate.
(C) Cryo-EM reconstruction of remdesivir-stalled RdRp complex.
(D) Corresponding atomic model of remdesivir-stalled RdRp complex.
(E) Close-up view of polymerase active site. Four copies of remdesivir monophosphate (RMP) occupy positions +1 (active site), −1, −2, and −3. Uracil monophosphate (UMP) is in position −4 (gray). C1 carbon of UMP and side chain of nsp12(Ser861) are shown as spheres. Cryo-EM density corresponding to template RNA strand (blue) and primer strand positions +1 through −5 is shown (transparent gray). The active site loop is also shown.
(F) Hypothetical model of post-translocated state, whereby the first copy of RMP is translocated into the −4 polymerase position. The RMP(−4) cyano group and nsp12(Ser861) are shown as spheres and show a steric clash.
(G) Spatial relationship between nucleotide at the −4 position and side chain of nsp12(Ser861) in pre-translocated state.
(H) Spatial relationship between nucleotide at −4 position and side chain of nsp12(Ser861), illustrating the clash between the cyano group of RMP that prevents translocation.
See Table 1 and Figure S1 for cryo-EM data collection, model validation, and analysis.
Discussion
Combined with our prior kinetic analysis, our structure suggests why the SARS-CoV-2 RdRp can rapidly incorporate 3 molecules of RMP, then more slowly incorporate a fourth RMP, which remains in the active site. This demonstrates that the RNA fails to be translocated to open the active site to allow binding of the next nucleotide. Translocation is inhibited by the steric interaction between Ser861 and the cyano group of the first RMP (seen in the structure at the −4 position). Interpretation of the structural data together with the kinetic data define the mechanism of inhibition by remdesivir as a delayed chain extension inhibitor. We propose that the steric block between the cyano group of RMP at −4 slows translocation to inhibit continued polymerization. This inhibition is not absolute, in that the subsequent addition of a high concentration of the 4 nucleotides supports the extension of 70% of the RNA within 20 s.
While there have been multiple structures of SARS-CoV-2 with a single copy of RMP incorporated determined (Kokic et al., 2020; Wang et al., 2020b; Yin et al., 2020), our structure reveals the first insights into a bona fide stalled elongation complex of an actively polymerizing enzyme. Initial structural studies with a single copy of RMP in positions −1 (Wang et al., 2020b) and +1 (Yin et al., 2020) demonstrate that RMP is able to be incorporated into the nascent transcript through Watson-Crick base pairing with template strand uracil bases. However, our structural and kinetic data show that inhibition only occurs after the RMP is buried by three nucleotides. Another recent study (Kokic et al., 2020) determined multiple structures of RdRp in complex with synthetic substrates in which RMP had been incorporated at specific positions. While both that study and ours have identified a clash between RMP(−4) and nsp12(Ser861) as the translocation barrier inhibiting elongation and thus inducing RdRp stalling, our additional analysis reveals that this state represents a kinetically trapped intermediate.
These studies were performed in the absence of the proofreading exonuclease (nsp10/14), and as such, we cannot fully define the fate of remdesivir and subsequent elongation products that will be observed with the complete holoenzyme. Exonuclease selectivity is determined at the polymerization site by stalling of the polymerase to slow the extension of a mismatched base pair, which then serves as a trigger for exonuclease removal by altering the kinetic partitioning between extension versus excision (Donlin et al., 1991). Stalling of the polymerase allows time for the transfer of the primer strand from the polymerase to the exonuclease site. Our data suggest that RMP may be resistant to excision, not because of the inherent properties of RMP, but rather because the subsequent extension after the first RMP incorporation (Dangerfield et al., 2020) is faster than rates of primer strand transfer and excision at the exonuclease site seen with other polymerases (Donlin et al., 1991; Johnson and Johnson, 2001). Thus, we expect the kinetic partitioning to favor extension rather than excision of the first RMP.
The steric clash between the serine and cyano groups of RMP does not completely block polymerization, so over time the RNA is extended (at a net rate of 0.08 s−1) to bury the incorporated RMP in the absence of the proofreading exonuclease. After the addition of 3 nucleotides on top of the first RMP, stalling of the polymerase is expected to lead to exonuclease removal of the 3′ terminal base. According to this kinetic model, the excised base would then be rapidly replaced, and could lead to a steady-state balance with repeated cycles of excision and replacement of the 3′ terminal base pair, effectively blocking further extension while protecting RMP from excision. Remdesivir is 5 times more effective in viral mutants with reduced exonuclease activity (Agostini et al., 2018), so it is clear that the exonuclease reduces the effectiveness of remdesivir. In the absence of the exonuclease, remdesivir could become buried in the genome, which could cause lethal mutagenesis (Shannon et al., 2020). Further studies are required to establish the kinetic parameters and structural features that govern exonuclease proofreading relative to RMP-induced polymerase stalling. Nonetheless, our current data provide a structural basis for RMP-induced stalling, which may guide the development of more effective nucleoside analogs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| BL21 E. coli | New England Biolabs | Cat. # C2530H |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 NSP8 | Dangerfield et al., 2020 | N/A |
| SARS-CoV-2 NSP12/7/8 Complex | Dangerfield et al., 2020 | N/A |
| Remdesivir triphosphate | Gilead Sciences Inc. | GS-443902; CAS: 1355149-45-9 |
| Deposited data | ||
| Coordinates of SARS-CoV-2 nsp7-8-12:template:primer:(RMP4) | This paper | PDB: 7L1F |
| Cryo-EM map of SARS-CoV-2 nsp7-8-12:template:primer:(RMP4) | This paper | EMD-23109 |
| Supplemental information | Mendeley Data | https://doi.org/10.17632/ry9rk6tmcn.1 |
| Oligonucleotides | ||
| RNA Primer: 5′-[FAM]-GUC AUUCUCCUAAGAAGCUA-3′ |
Integrated DNA Technologies | N/A |
| RNA Template: 5′-CUAUCCCC AUGUGAUUUUAAUAGCUU CUUAGGAGAAUGAC-3′ |
Integrated DNA Technologies | N/A |
| Cy3 Internal Standard DNA: 5′-[Cy3]-CCGTGAGTTGGTTG GACGGCTGCGAGGC-3′ |
Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| pcIts,ind+-(NSP8) | Dangerfield et al., 2020 | Deposited to Addgene – 160656 |
| pQE-(NSP12)-pcIts,ind+-(NSP7-NSP8) | Dangerfield et al., 2020 | Deposited to Addgene–160540 |
| pG-Tf2 | Takara Biosciences | Cat. # 3340 |
| Software and algorithms | ||
| KinTek Explorer v10 | KinTek Corp. | https://kintekexplorer.com |
| Gene Mapper v5 | ThermoFisher Scientific | https://www.thermofisher.com/order/catalog/product/4370784#/4370784 |
| SerialEM | Mastronarde, 2005 | https://bio3d.colorado.edu/SerialEM/ |
| Warp | Tegunov and Cramer, 2019 | http://www.warpem.com/warp/# |
| CryoSparc | Punjani et al., 2017 | https://cryosparc.com/ |
| Coot | Emsley and Cowtan, 2004 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot |
| ChimeraX | Goddard et al., 2018 | https://www.cgl.ucsf.edu/chimerax/ |
| DeepEMhancer | Sanchez-Garcia et al., 2020 | https://github.com/rsanchezgarc/deepEMhancer |
| PHENIX | Adams et al., 2010 | https://www.phenix-online.org/documentation/index.html |
| COSMIC2 | Cianfrocco et al., 2017 | https://cosmic-cryoem.org/ |
| eLBOW | Moriarty et al., 2009 | https://www.phenix-online.org/documentation/reference/elbow.html |
| Other | ||
| C-flat CF-2/2 200 mesh grids | Protochips, Inc. | CF-2/2-2Cu-50 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Kenneth A. Johnson (kajohnson@utexas.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The cryo-EM structure and associated atomic coordinates are available at the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD:23109 and PDB: 7L1F, respectively.
Method details
Preparation of protein
The SARS CoV-2 RdRp (nsp12/7/8) was expressed and purified without tags to provide a highly active enzyme for transient kinetic and structural studies using cryo-electron microscopy (cryo-EM). Tag-free nsp12/7/8 complex and nsp8 were expressed in E. coli and purified as described in detail previously (Dangerfield et al., 2020, 2021). A detailed summary of each expression and purification is given below.
Nsp8 was expressed in BL21 E. coli harboring the plasmid pcIts,ind+-(NSP8). Cells were grown in Terrific Broth (2.4% [w/v] yeast extract, 2% [w/v] tryptone, 0.4% [v/v] glycerol, 17 mM KH2PO4, 72 mM K2HPO4) with 100 μg/ml ampicillin for selection. Protein expression was induced by shifting the culture temperature from 30°C to 42°C for 20 minutes, followed by 3 hours at 38°C before harvesting the cells. The cells were lysed by sonication, then the lysate was clarified by centrifugation and separated on the following columns: Q Sepharose-FF, SP Sepharose-FF, HiTrap Blue-FF, Superdex 200. The protein was then dialyzed into Storage Buffer-1 (50 mM Tris-HCl pH 7.5, 0.1 mM EDTA, 20 mM KCl, 1 mM DTT, 50% [v/v] glycerol), aliquoted, and stored at −80°C.
The three peptides for the nsp12/7/8 complex were co-expressed in BL21 E. coli harboring the plasmids pG-Tf2 and pQE-(NSP12)-pcIts,ind+-(NSP7-NSP8). Cells were grown in Terrific Broth with 30 μg/ml kanamycin and 20 μg/ml chloramphenicol for selection at 30°C. Protein expression was induced by addition of tetracycline to 10 ng/ml for 20 minutes, followed by addition of IPTG to 0.5 mM and nalidixic acid to 50 μg/ml, then incubation overnight at 16°C. Harvested cells were lysed by sonication, then the lysate was clarified by centrifugation and separated on the following columns: Q Sepharose-FF, HiTrap Blue-FF, Heparin Sepharose 6-FF, SP Sepharose-FF, Superdex 200. The protein was then dialyzed into Storage Buffer-2 (50 mM Tris-HCl pH 7.5, 0.1 mM EDTA, 50 mM NaCl, 1 mM DTT, 50% [v/v] glycerol), aliquoted, and stored at −80°C.
RNA substrates and other materials
RNA samples were prepared in DEPC treated water (Ambion). RNA substrates were purchased from Integrated DNA technologies with RNase free HPLC purification and resuspended in Annealing Buffer (10 mM Tris-HCl pH 7, 50 mM NaCl, 0.1 mM EDTA). Concentration of each oligo was determined by absorbance at 260 nm using the extinction coefficients 222,360 M-1 cm-1 and 403,100 M-1 cm-1 for the FAM-20 nt primer and the 40 nt template, respectively. The double stranded RNA substrate was prepared by mixing each oligo at a 1:1 molar ratio, heating to 75°C for 3 minutes, then cooling slowly to room temperature over approximately 2 hours. Oligonucleotides were stored at −20°C. UTP was purchased from New England Biolabs. Remdesivir triphosphate (GS-443902) was a kindly provided by Gilead Sciences and concentration was determined by absorbance at 245 nm using the extinction coefficient 24,100 M-1 cm-1 (Dangerfield et al., 2020). Remdesivir triphosphate was stored at −20°C.
Extension reactions and analysis by capillary electrophoresis
All reactions were conducted in Reaction Buffer (40 mM Tris-HCl pH 7, 50 mM NaCl, 5 mM MgCl2, 1 mM DTT) (Dangerfield et al., 2020). Concentrations of enzyme, RNA and nucleotides are given in the legend for Figure 1. (concentrations given are the final concentrations after mixing). Reactions were performed at room temperature (∼25°C) in Reaction Buffer after allowing the RdRp complex and RNA to equilibrate for approximately 30 minutes. Samples were quenched by adding 0.6 M EDTA to a final concentration of 0.4 M. One μL of each sample was diluted into 10 μL of HiDi formamide (ThermoFisher) containing 1 nM Cy3 internal standard DNA oligo (5′-[Cy3]-CCGTGAGTTGGTTGGACGGCTGCGAGGC-3′, purchased from Integrated DNA Technologies) in a 96 well plate. Samples were analyzed on an Applied Biosystems 3130xl Genetic Analyzer instrument equipped with a 36 cm capillary array (ThermoFisher) and nanoPOP-6 polymer (Molecular Cloning Laboratories). The oven temperature was set to 65°C. Before injecting samples, a pre-run electrophoresis step was performed at 15 kV for 3 minutes. Samples were injected at 3.6 kV for 12 s, then the voltage was ramped up to 15 kV over 40, 15 s steps. Fluorescence was monitored for 800 s using the G5 dye set and all peak intensities were within the linear range of the instrument. Peaks were integrated with GeneMapper 5 software (ThermoFisher).
Cryo-EM sample preparation, data collection and processing
Samples were prepared in Reaction Buffer. A solution of 3.33 μM nsp12/7/8, 4.67 μM nsp8, 3.33 μM FAM-20/40 RNA was mixed with 14 μM remdesivir triphosphate (RTP) and 7.5 μM UTP to start the reaction at room temperature (∼25°C) (concentrations of reaction components are given after mixing). The reaction was allowed to proceed for 10-20 s before application to glow discharged holey carbon grids (C-flat 4/2, Protochips Inc.), blotted for 0.5 s with a blot force of 4 and rapidly plunged into liquid ethane using an FEI Vitrobot MarkIV.
Data was collected on an FEI Titan Krios cryo-electron microscope equipped with a K3 Summit direct electron detector (Gatan, Pleasanton, CA). Images were recorded with SerialEM (Mastronarde, 2005), with a pixel size of 1.1Å over a defocus range of −1.5 to −2.5 μm. 2446 movies were recorded at 13.3 electrons/pixel/second for 6 s (80 frames) to give a total dose of 80 electrons/pixel. CTF correction, motion correction and particle picking were performed in real-time using WARP (Tegunov and Cramer, 2019), resulting in 2,340,544 particles, which were uploaded to cryoSPARC v2 (Punjani et al., 2017).
Particles were subjected to multiple rounds of 3D classification in cryoSPARC, and a final set of 116,748 particles was refined to a global resolution of 3.89 Å based on the 0.143 FSC criterion. Map sharpening was performed using DeepEMhancer (Sanchez-Garcia et al., 2020) as implemented in COSMIC2 (Cianfrocco et al., 2017). For modeling, the published structure of nsp7-8-12 (PDB: 7bv2 (Yin et al., 2020)) was used as a starting model. Non-proteinaceous molecules (i.e., RNA, ligands) were removed, and the template:primer duplex was built de novo in Coot (Emsley and Cowtan, 2004). Restraints for RMP were generated using eLBOW (Moriarty et al., 2009)), and structures were subjected to real-space refinement using Phenix (which resulted in a map-to-model FSC of 4.0Å at the 0.5 threshold) (Afonine et al., 2018). Figures were prepared using ChimeraX (Goddard et al., 2018).
Quantification and statistical analysis
Structural statistics are provided in Table 1 .
Table 1.
Cryo-EM data collection and processing statistics
| Data collection and processing | |
| Magnification | 22,500× |
| Voltage (kV) | 300 |
| Electron exposure (e−/Å) | 80 |
| Defocus range (μM) | −1.5 to −2.5 |
| Symmetry imposed | C1 |
| Initial particle images | 2,340,544 |
| Final particle images | 116,748 |
| Map resolution (Å) | 3.89 |
| FSC threshold | 0.143 |
| Map resolution range (Å) | 3.3 to >8 |
| Refinement | |
| Initial model used (PDB code) | PDB: 7BV2 |
| Model resolution (Å) | 3.9 |
| FSC threshold | 0.5 |
| Map sharpening B factor (Å2) | 114.5 |
| Model composition | |
| Nonhydrogen atoms | 8,820 |
| Residues (protein/RNA) | 1,009/35 |
| B factors (Å2), min/max/mean | |
| Protein | 60.2/114.2/92.6 |
| RNA | 81.2/219.3/145.3 |
| RMSDs | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.388 |
| Validation | |
| MolProbity score | 1.66 |
| clashscore | 4.15 |
| Poor rotamers (%) | 0 |
| Ramachandran plot (%) | |
| Favored | 92.58 |
| Allowed | 7.42 |
| Disallowed | 0 |
FSC, Fourier shell correlation.
Acknowledgments
We thank Gilead (assisted by Brian Schultz and Joy Feng) for providing the Remdesivir triphosphate used in this study. This work was supported in part by Welch Foundation grants F-1604 (to K.A.J.) and F-1938 (to D.W.T.), National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) grant R01AI110577 (to K.A.J.), National Institute of General Medical Sciences (NIGMS) of the NIH grants R01114223 (to K.A.J.) and R35GM138348 (to D.W.T.), Army Research Office grant W911NF-15-1-0120 (to D.W.T.), and a Robert J. Kleberg, Jr. and Helen C. Kleberg Foundation Medical Research Award (to D.W.T.). D.W.T is a CPRIT Scholar supported by the Cancer Prevention and Research Institute of Texas (RR160088) and an Army Young Investigator supported by the Army Research Office (W911NF-19-1-0021). D.W.T. is supported by the David Taylor Excellence Fund in Structural Biology made possible with support from Judy and Henry Sauer. Data were collected at the Sauer Structural Biology Lab at the University of Texas at Austin.
Author contributions
J.P.K.B. performed the cryo-EM, structure determination, and modeling. T.L.D. purified the enzyme, performed the kinetic studies, and prepared the samples for cryo-EM. J.P.K.B., T.L.D., D.W.T., and K.A.J. analyzed and interpreted the data and wrote the manuscript. D.W.T. and K.A.J. supervised and secured funding for the studies.
Declaration of interests
The authors declare no competing interests.
Published: January 28, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2021.01.035.
Supplemental information
References
- Afonine P.V., Poon B.K., Read R.J., Sobolev O.V., Terwilliger T.C., Urzhumtsev A., Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., et al. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. MBio. 2018;9:e00221-e18. doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. ACTT-1 Study Group Members Remdesivir for the Treatment of Covid-19 - Final Report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell. 2020;182:1560–1573.e13. doi: 10.1016/j.cell.2020.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cianfrocco M.A., Wong M., Youn C., Wagner R., Leschziner A.E. Practice & Experience in Advanced Research Computing; New Orleans, LA: 2017. COSMIC²: A Science Gateway for Cryo-Electron Microscopy Structure Determination. July 9–13. [Google Scholar]
- Dangerfield T.L., Huang N.Z., Johnson K.A. Remdesivir Is Effective in Combating COVID-19 because It Is a Better Substrate than ATP for the Viral RNA-Dependent RNA Polymerase. iScience. 2020;23:101849. doi: 10.1016/j.isci.2020.101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangerfield T.L., Huang N.Z., Johnson K.A. Expression and purification of tag-free SARS-CoV-2 RNA-dependent RNA polymerase in Escherichia coli. STAR Protoc. 2021 doi: 10.1016/j.xpro.2021.100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin M.J., Patel S.S., Johnson K.A. Kinetic partitioning between the exonuclease and polymerase sites in DNA error correction. Biochemistry. 1991;30:538–546. doi: 10.1021/bi00216a031. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295:6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Johnson A.A., Johnson K.A. Exonuclease proofreading by human mitochondrial DNA polymerase. J. Biol. Chem. 2001;276:38097–38107. doi: 10.1074/jbc.M106046200. [DOI] [PubMed] [Google Scholar]
- Kokic G., Hillen H.S., Tegunov D., Dienemann C., Seitz F., Schmitzova J., Farnung L., Sievert A., Hobartner C., Cramer P. Mechanism of SARS-CoV-2 polymerase inhibition by remdesivir. Nat. Commun. 2020;12:279. doi: 10.1038/s41467-020-20542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Moriarty N.W., Grosse-Kunstleve R.W., Adams P.D. electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr. D Biol. Crystallogr. 2009;65:1074–1080. doi: 10.1107/S0907444909029436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Sanchez-Garcia R., Gomez-Blanco J., Cuervo A., Carazo J.M., Sorzano C.O.S., Vargas J. DeepEMhancer: a deep learning solution for cryo-EM volume post-processing. bioRxiv. 2020 doi: 10.1101/2020.06.12.148296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Selisko B., Le N.T.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., et al. Favipiravir strikes the SARS-CoV-2 at its Achilles heel, the RNA polymerase. bioRxiv. 2020 doi: 10.1101/2020.05.15.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., Gordon C.J., Woolner E., Kocinkova D., Perry J.K., Feng J.Y., Porter D.P., Götte M. Template-dependent inhibition of coronavirus RNA-dependent RNA polymerase by remdesivir reveals a second mechanism of action. J. Biol. Chem. 2020;295:16156–16165. doi: 10.1074/jbc.AC120.015720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D., Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell. 2020;182:417–428.e13. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020;11:5874. doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Ge J., Zheng L., Zhang Y., Gao Y., Wang T., Huang Y., Yang Y., Gao S., Li M., et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell. 2021;184:184–193.e10. doi: 10.1016/j.cell.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Mao C., Luan X., Shen D.D., Shen Q., Su H., Wang X., Zhou F., Zhao W., Gao M., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020;368:1499–1504. doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM structure and associated atomic coordinates are available at the Electron Microscopy Data Bank and Protein Data Bank with accession codes EMD:23109 and PDB: 7L1F, respectively.