Graphical abstract
Keywords: ATA and its Alkaline earth metal complexes, Synthesis, Spectroscopy, Virtual study in treating COVID-19
Abstract
The salt of Aurintricarboxylic acid (ATA) was utilized in this study to synthesize new alkaline earth metal ion complexes. The analytical results proposed the isolation of mononuclear (Sr+2&Ba+2) and binuclear complexes (Mg+2&Ca+2). These complexes were analyzed by available analytical and spectral techniques. The tetrahedral geometry was suggested for all complexes (SP3) through bidentate binding mode of ligand with each central atom. UV–Vis spectra reveal the influence of L → M charge transfer and the estimated optical band gap mostly appeared close to that for known semiconductors. XRD, SEM and TEM studies were executed for new complexes and reflects the nano-crystallinity and homogeneous morphology. The structural forms of ATA and its complexes were optimized by DFT/B3LYP under 6-31G and LANL2DZ basis sets. The output files (log, chk &fchk) were visualized on program screen and according to numbering scheme, many physical features were obtained. It is worthy to note that, a virtual simulation for the inhibition affinity towards COVID-19 proteins as proactive study before the actual application, was done for ATA and its complexes. This was done in addition to drugs currently applied in curing (Hydroxychloroquine & Lopinavir), for comparison and recommendation. Drug-likeness parameters were obtained to evaluate the optimal pharmacokinetics to ensure efficacy. Furthermore, simulated inhibition for COVID-19 cell-growth, was conducted by MOE-docking module. The negative allosteric binding mode represents good inhibitory behavior of ATA, Ba(II)-ATA complex and Lopinavir only. All interaction outcomes of Hydroxychloroquine drug reflect unsuitability of this drug in treating COVID-19. On the other hand, there is optimism for ATA and Lopinvir behaviors in controlling COVID-19 proliferation.
1. Introduction
It is unfortunate now that the Corona pandemic has claimed tens of thousands of human lives, which put researchers in a race against time to save humanity from this pandemic virus, each research in its own or available way. The main obstacle is that until now there is no way to prevent this virus, but only the medications used reduce the symptoms associated with it. Although previously promising treatments have been found for the Corona family discovered in recent decades and are now being tested versus COVID-19, but studies have not yet been completed and all drugs are still in the field of experiments. Such as, Lopinavir drug that displayed effectiveness against viruses as HIV and SARS-CoV-2. Also, Hydroxychloroquine is currently used to treat malaria, autoimmune lupus and can prevent viruses. The World Health Organization (WHO) urges researchers to investigate and search for an effective drug as soon as possible. Consequently, the area of diligence of researchers will remain open for a not short period of time until a treatment for this devastating virus is reached, and this is what we have done in our research. Aurintricarboxylic acid (ATA) is a polymeric carboxylated triphenyl methane derivative [1] which known by its variable biological efficiency and action modes. Also, ATA is enzymatic inhibitor due to its binding with nucleic acid, DNA and RNA polymerases, nucleases, DNA topoisomerase and reverse transcriptase (RT) [2]. Moreover, ATA also has been reported as inhibitor for protein synthesis through blocking RNA association with ribosomes, which disrupts cellular signaling pathways by different mechanisms [3]. Previously we synthesized aurintricarboxylic acid complexes from VO(II), Cr(III), Mn(II) and Fe(III) ions, which fully characterized by analytical, spectral and theoretical tools [4]. It was discovered that ATA compound has the ability to inhibit influenza virus by reducing its ability to reproduce in cultured cells after infection [5]. The activity of ATA compound or its analogues is observed in reducing viral infection when used immediately after infection. So, this compound is considered a drug that is strongly recommended as anti-infectious diseases, including Corona virus [6]. In addition to, calcium plays multiple roles inside living cells and often plays the role of the messenger that sends signals between plasma membrane and intracellular systems [7], [8]. Several studies have confirmed the role of magnesium in the effectiveness of DNA induced by antibacterial drugs [9]. A series of mononuclear or binuclear complexes from alkaline earth metals were previously prepared [10], [11] and showed superiority as antimicrobial drugs [12]. Proceeding from this, we decided to work hard in this research over a salt of Aurintricarboxylic acid (ATA) that is a known inhibitor for influenza virus and also mimics some antiviral drugs. Then was used to prepare its alkaline earth metal ion complexes, as a try to raise its activity by adding such metal ions, particularly Ca(II) and Mg(II). After completing the accurate description of complexes, a comparison was done computationally versus two drugs currently used (Hydroxychloroquine & Lopinavir) to treat Corona virus pandemic (COVID-19) and finally we will present a recommendation of the best one.
2. Methodology
2.1. Reagents
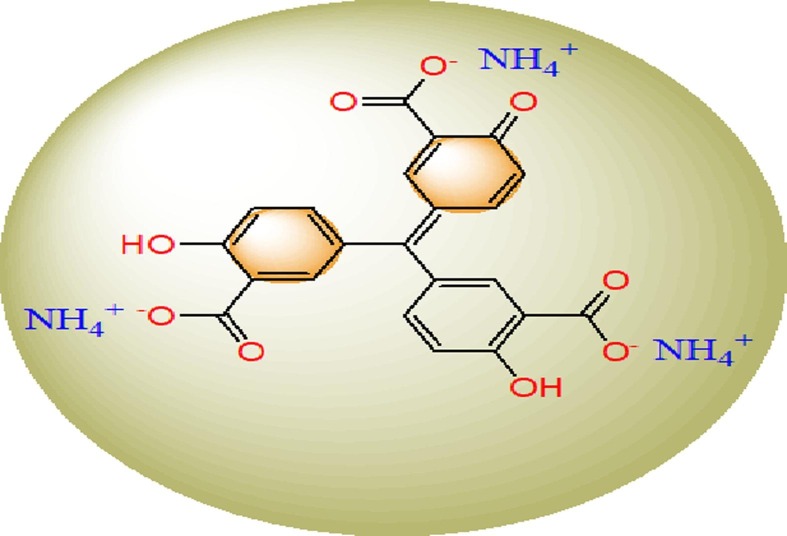
Aurintricarboxylic acid ammonium salt (ATA) (Scheme 1 ), MaCl2, CaCl2, SrCl2 and BaCl2 were the compounds used to prepare complexes, which obtained from Merck as BDH (British Drug Houses Company). Furthermore, Dimethyl sulfoxide (DMSO) and EtOH solvents were obtained from Sigma & Aldrich and have spectroscopic nature then used without any pre-treatment.
Scheme.
1. Aurintricarboxylic acid ammonium salt (ATA) ligand.
2.2. Synthesis of alkaline earth metal ion complexes
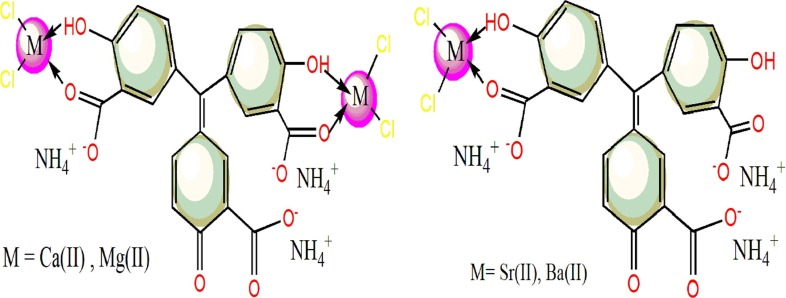
A simple strategy was utilized to prepare the targeted alkaline earth metal ion complexes (Scheme 1S). By using, 1:2 M ratio from ATA ligand (dark red) and each colorless metal salt that was mixed in EtOH/H2O solvent (3:1). O.947 g (2 mmol) of ATA ligand was dissolved in 10 mL bi-distilled H2O and then mixed with each salt such as, MgCl2 (0.381 g), CaCl2 (0.444 g), SrCl2 (0.634 g) or BaCl2 (0.833 g), which dissolved in 30 mL EtOH. A colored precipitates were appeared during reflux at 75 °C up to 3 h, then filtered off and dried in desiccators over CaCl2. The yields of Sr(II) and Ba(II) complexes around 70%, while the yields of Ca(II) and Mg(II) complexes up to 53%.
2.3. Equipments used
All instruments used in this study were presented as pictures, for simplicity (Scheme 2S), also the characterization of each instrument was typed below the picture. While, the working conditions will be addressed during discussion. The metal content percentage was evaluated complexometrically [Ca(II)&Mg(II)] or by flam atomic absorption [Ba(II) & Sr(II)]. Whereas, the chloride percentage was assessed gravimetrically [13].
2.4. Computational studies
-
A)
Geometry optimization
Standard software (Gaussian 09) [14] was used to configure the geometries to reach the best atomic distribution within the ligand and its complexes. DFT method was the suitable one, which applied under valence double-zeta polarized basis set and correlation-consistent basis set (6-31G & LANL2DZ). Three functional files as log, chk and chkf (formulated chk file by Gaussian-prog) were exported and containing all quantum calculations. Furthermore, other theoretical indices were estimated using known simple equations which depend on frontier energy gaps (ΔE) [15].
-
B)
Effective virtual screening against COVID-19 virus
-
i)
Drug-likeness assay
This computation was executed by Swiss ADME link and then after orientation steps, important pharmacokinetics were estimated. This aims to find out how the behavior of studied compound closely related to pharmacological behavior [16].
-
ii)
Simulated inhibition for COVID-19 proliferation
The most recent certified program used to evaluate the therapeutic behavior of suggested drugs is MOE module (Molecular Operating Environmental, Vs. 2014–2018), which applied here. This in-silico study was used over ATA ligand and its alkaline metal ion complexes in comparing to antiviral drugs that currently used in treating COVID-19 (Hydroxychloroquine & Lopinavir). On the other side two significant COVID-19 cell-proteins were selected for this test as 6lu7and 6lzg proteins in co-crystal form (Scheme 3S) [17]. After that, many configuration steps must be done before starting the docking process. First of all, each tested compound must be optimized through energy minimization then render atomic charges which followed by adjusting potential energy. Other adjustment was executed by MMFF94x force field and consequently a new database was built for the configured compound as MDB file [18]. The other instillation part concerned with protein was started by adding hydrogen atoms over assigned receptors. Then connecting receptor types, fixing potential energy and looking for binding sites and dummies [18]. Accordingly, choose the targeted MDB file and starting the docking process, which consumed variable times that cover 30 poses for each docking process. Some of these poses may clash protein helix and must be excluded completely based on London dG-scoring function, which advanced twice times by triangle matcher [19]. The inhibition activity was ranked for true docking pathways (H-bond length ≤ 3.5 Å) based on data exported from each process.
3. Results and discussion
3.1. General properties
The used ATA ligand that a known oxygen-rich or multi-donor compound, may behave by variable ways depending on metal properties [4]. The elemental analysis obtained for complexes (Table 1 ), suggests either mononuclear with Sr(II) & Ba(II) ions or binuclear complexes with Mg(II) & Ca(II) ions. This variation may be supported by higher tendency of Mg(II) and Ca(II) towards complexation due to their smaller sizes than the other alkaline earth metal ions (Sr(II) & Ba(II)). Also, the molar conductivity measurements for 1 mmol (in DMSO) showed the ligand retained the electrolytic feature (124.45 Ohm-1cm2mol−1) after complexation, but decrease slightly (103.91–120.13Ohm-1cm2mol−1) [20]. This result reflects unchanged charges over coordination sphere of the complexes, which asserts on the covalence presence of chloride within the sphere.
Table 1.
Analytical and physical properties of ATA and its alkaline earth metal ion complexes.
| Compound Empirical formula | M.P.,°C | % Calculated (Found.) |
|||||
|---|---|---|---|---|---|---|---|
| C | H | N | Cl | M | |||
| 1)ATA(C22H23N3O9)(473.43) | 220 | 55.81(55.62) | 4.90(4.66) | 8.88(9.23) | --- | 124.45 | |
| 2)[Mg2Cl4(C22H23N3O9)](663.85) | ˃300 | 39.80(39.30) | 3.49(3.22) | 6.33(6.61) | 21.36(21.33) | 7.32(7.33) | 103.91 |
| 3)[Ca2Cl4(C22H23N3O9)]H2O(713.42) | ˃300 | 37.04(37.31) | 3.53(3.79) | 5.89(6.13) | 19.88(19.71) | 11.24(11.09) | 112.48 |
| 4)[SrCl2(C22H23N3O9)]H2O(649.97) | ˃300 | 40.65(40.23) | 3.88(3.59) | 6.46(6.67) | 10.91(10.78) | 13.48(13.36) | 119.67 |
| 5)[BaCl2(C22H23N3O9)](681.67) | ˃300 | 38.76(38.66) | 3.40(3.16) | 6.16(6.40) | 10.40(10.49) | 20.15(20.20) | 120.13 |
3.2. Vibrational modes
IR spectroscopy was used to identify the mode of ATA binding towards central metal ions inside the complexes. The comparative view for the obtained complexes spectra (Fig. 1 S) with that of corresponding ligand (ATA), reveals the changes on functional-group vibrations (Table 2 ). As known, the symmetric distribution of function groups (CO, OH &COO–) within ATA structure, offers the same opportunity for their coordination. So, the judgment on coordination seems difficult without quantum study. In this case, the computational data for free ligand is essential to judge on the coordination mode perfectly. Estimated charges of oxygen atoms over C(15) = O(18), C(3)-O(10)H and C(25)-O(28)H groups were 0.02604. −0.12569 and −0.12880, respectively. This values over the high coordination availability for OH groups than the carbonyl which includes oxygen atom appeared positively charged instead of negativity. This may be due to the contribution of C = O in effective resonance inside quinonoid structure. In addition, monitoring the vibration within COO– group through ν(C = O)ac&ν(C-O)ac beside the vibration of aromatic C = O and OH groups, the coordination mode was simply proposed. Whereas, proposing the coordination mode must coincide with the number of coordinating central-atoms which evaluated analytically. Thus, the ligand behaves as bidentate within mononuclear complexes (Sr+2&Ba+2), while as a tetradentate within binuclear complexes (Mg+2&Ca+2). This is based on observable shift (to lower wavenumber) for carbonyl vibration ν(C = O)ac and OH vibrations (ν & δ), which agrees with their coordination either as bidentate or tetradentate mode [21]. While, more or less unshift of ν(C-O)ac vibration (1270 cm−1), points to its ruling out from coordination. The broadness around 1700–1850 cm−1 is due to intensive intraligand H-bonding, which expected with presence of three ionic bond within ligand structure. In addition to, the vibration or rotation of water molecule presented in Ca(II) and Sr(II) complexes, was appeared at ≈ 3420, 820 and 690 cm−1 and assigned for ν(OH), ρr(H2O) and ρw(H2O), respectively [22], [23]. Finally, the metallic bond vibration (M−O) was appeared around 508–562 cm−1 range.
Fig. 1.
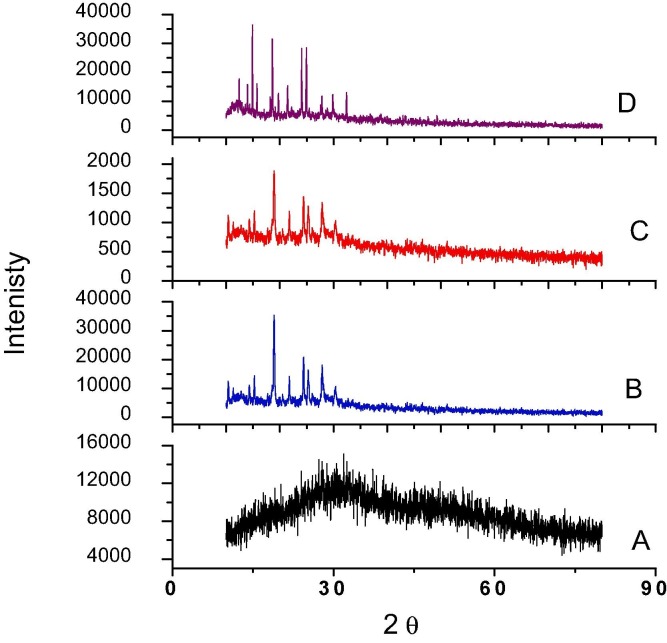
XRD patterns for studied Mg(II), Ca(II), Sr(II) and Ba(II) complexes (A-D).
Table 2.
IR spectra bands (cm−1) of ATA and its alkaline earth metal ion complexes.
| Compound | υs(NH4+) | υ(OH) | δ(OH) | (C = O) | υ(C-O)ac | ρr(H2O) | ρw(H2O) | υ(M−O) |
|---|---|---|---|---|---|---|---|---|
| 1)ATA(C22H23N3O9) | 3430, 3230, 1446 | 3480 | 1380 | 1582, 1662 | 1270 | --- | --- | --- |
| 2)[Mg2Cl4(C22H23N3O9)] | 3327, 3213, 1440 | 3440 | 1365 | 1565, 1624 | 1267 | --- | --- | 520 |
| 3)[Ca2Cl4(C22H23N3O9)]H2O | 3338, 3231, 1434 | 3469 | 1392 | 1565, 1630 | 1250 | 817 | 692 | 562 |
| 4)[SrCl2(C22H23N3O9)]H2O | 3214, 3017, 1446 | 3467 | 1399 | 1570; 1660 | 1256 | 823 | 680 | 526 |
| 5)[BaCl2(C22H23N3O9)] | 3380, 3237, 1434 | 3467 | 1380 | 1580; 1657 | 1149 | --- | --- | 508 |
1H NMR analysis was executed for the complexes, but they did not dissolved in available solvents (DMSO‑d 6 & CDCl3). I have added the figure for Mg(II) complex in supplementary file (Fig. 2 S) as example, for assurance. The figure includes signals of TMS (Reference tetramethylsilane) at δ 0, DMSO‑d 6 δ ~ 2.5 & 3.0 and CDCl3 at δ ~ 7.5 ppm. So, there is no significant signals appeared.
Fig. 2.
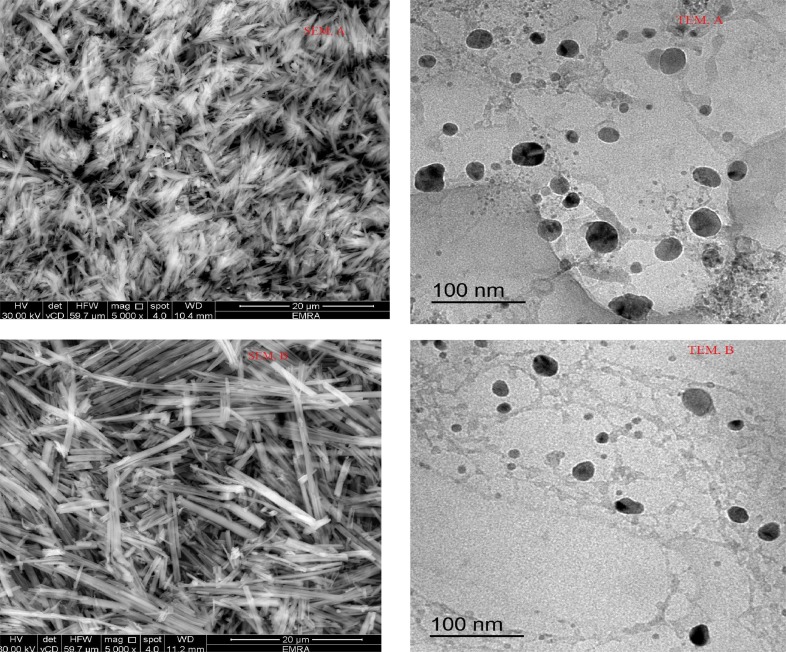
SEM & TEM for Mg(II), A and Ca(II), B complexes of ATA ligand.
3.3. Electronic transition and optical band gap (Eg)
UV–Vis spectra was scanned for the ligand and its alkaline earth metal ion complexes (Fig. 3 S), to assess the changes in intraligand transitions as well as charge transfer bands. Although, the absence of any ligand field transition with such metal ions, but the comparison between absorption bands, may offer a good information. Commonly, the complexation has a great impact on the whole electronic transition within the ligand, especially on ligand field transitions that are very sensitive with coordination [24]. Hypochromic effect was clearly observed with π → π*and n → π* transitions in complexes (Table 3 ). Furthermore, significant charge transfer transition from ligand to metal effects on the color of complexes which must be colorless as known for the compounds of these metals. The intensive colors appeared for these complexes (reddish brown or red), refer to effective ligand → metal charge transfer (LMCT) [7]. The most favorable geometry for such s-block elements, was the tetrahedral structure by SP3 hybridization (Scheme 2 ). The optical band gap (Eg, eV) which considered as the lowest energy of photon essential to excite electron and leaves its position as a positive hole although electrostatic attraction. Accordingly, the lower the band gap value, the easier the electronic transition within flexible electron cloud, which favored in semiconductors or fluorescence [25]. This gap can be calculated for studied compounds based on absorption spectra and applying the following relations;
Fig. 3.
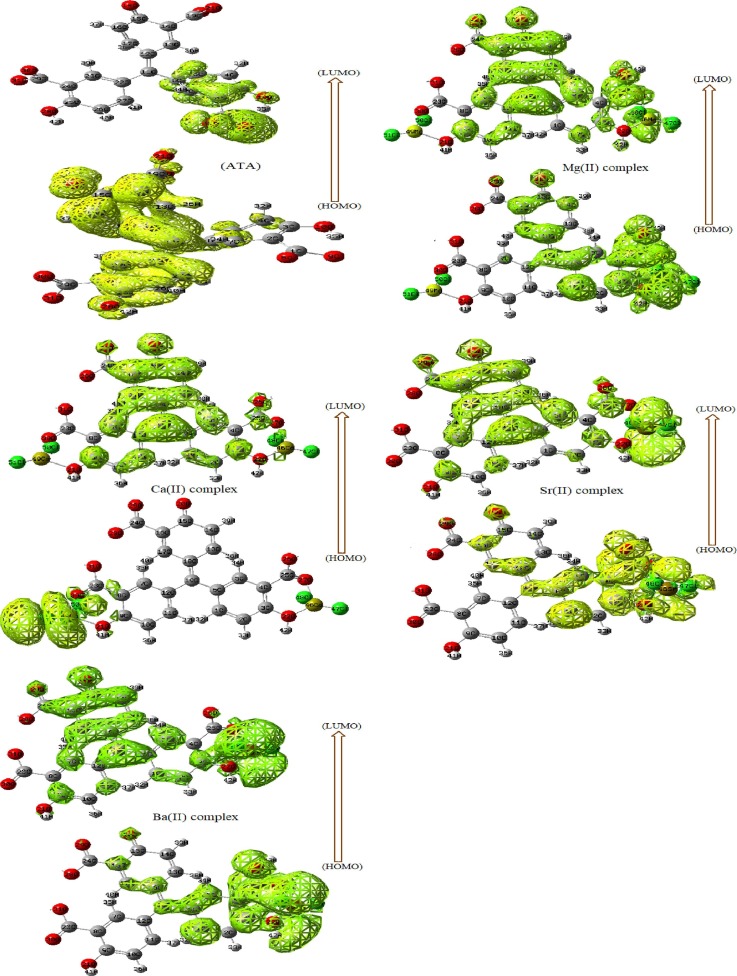
Frontier level maps for the ligand, Mg(II), Ca(II), Sr(II) and Ba(II) complexes(A-E).
Table 3.
Electronic transition bands in ATA and its alkaline earth metal ion complexes.
| Complexes | Color | Intraligand and CT bands (nm); assignment |
|---|---|---|
| 1)ATA(C22H23N3O9) | Red | 559.5 (n → π*), 524.5 (n → σ*), 376.0, 329.4, 294.3(π → π*) |
| 2)[Mg2Cl4(C22H23N3O9)] | Reddish Brown | 546.9, 469.8, 312.5, 265.8 |
| 3)[Ca2Cl4(C22H23N3O9)]H2O | Brown | 551.4, 467.5, 319.2, 267.2 |
| 4)[SrCl2(C22H23N3O9)]H2O | Red | 553.3, 472.7, 319.2, 270.8 |
| 5)[BaCl2(C22H23N3O9)] | Brown | 490.7, 372.8, 286.84 |
Scheme 2.
The coordination spheres of alkaline earth metal ion complexes from ATA ligand.
i) α = 1/d ln A (1), where d is the cell-width. ii) αhυ = A (hυ-Eg)m (2), which representing a relationship between absorption coefficient and optical band gap (Eg). Then and after aggregating the two relations (1&2), Eg values can be estimated [25]. Where m is 0.5 or 2, for direct or indirect transition, respectively and A is the independent constant of energy. The α values from relation 1, were used to obtain (αhυ)2, so the relation can be drawn against hυ (Fig. 4 S). A parallel straight line extended to meet x-axis, the value of band gap can be obtained (eV). The lower values were recorded with Mg(II) and Ba(II) complexes beside ATA ligand as 2.131, 3.028 and 3.33 eV, respectively. These values are promising due to they agree perfectly with that know for common semiconductors (0.8–3.6 eV) [26].
Fig. 4.
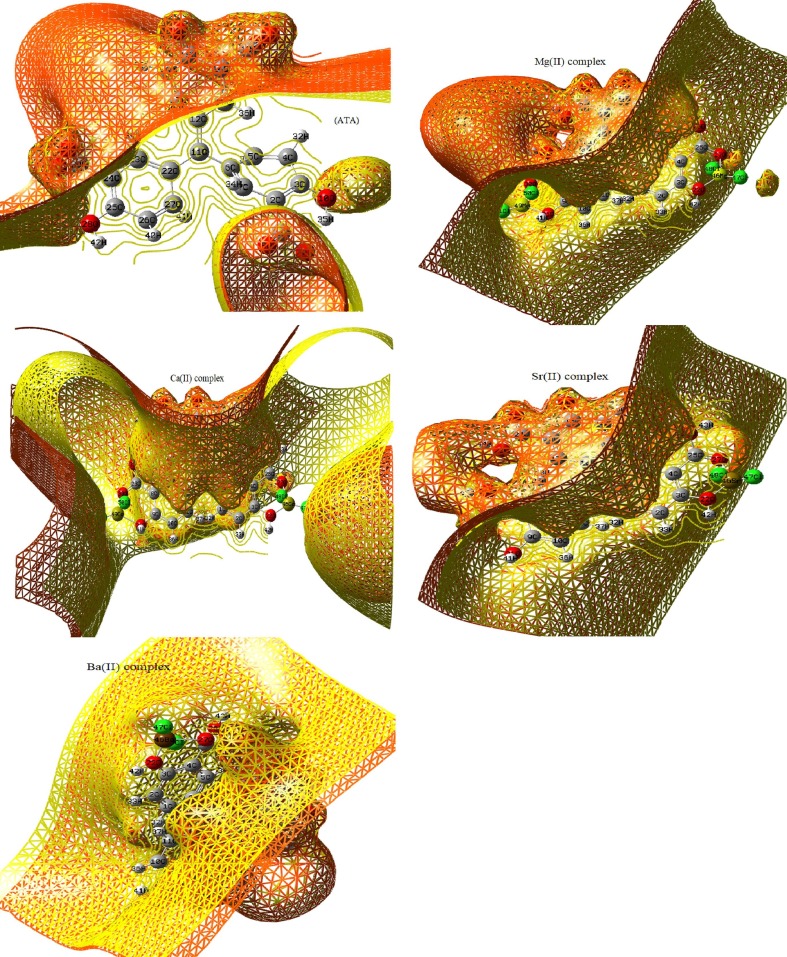
Iso-surface with array plots for ATA, Mg(II), Ca(II), Sr(II) and Ba(II) complexes(A-E).
3.4. Thermogravimetric analysis
TGA study was used to evaluate the degree of stability or resistance of new complexes for pyrolysis. The obtained thermo-degradation curves (Fig. 5 S) represent the stability of Mg(II) and Ba(II) complexes, while the other two complexes are thermally unstable. The instability of Ca(II) and Sr(II) complexes, is only due to presence of crystal water molecule attached outer the coordination sphere. Three degradation steps were recorded with all complexes except Sr(II) complex in which the TGA curve divided into four steps till 620 °C. The hypothetical degradation paths were proposed and tabulated (Table 1S). Most complexes displayed high stability during successive degradation till ≈ 800 °C, which appeared from high residual percentage (except Sr(II) complex). This may refer to the strength of bonds surround the central atoms, particularly with binuclear complexes.
Fig. 5.
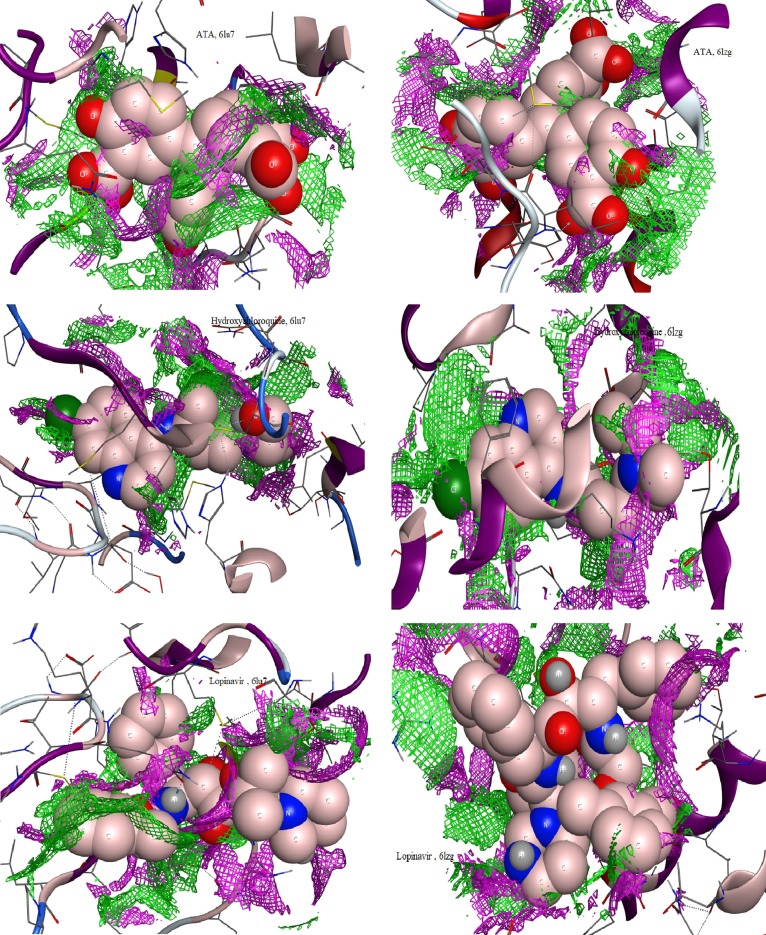
Contact preferences recorded with ATA ligand and suggested drugs towards COVID-19 protein (6lu7 & 6lzg).
3.5. XRD, SEM and TEM analyses
XRD scanning was done for all tested complexes by Cu/Kα1 radiation-source at 2θ =10˚<2θ < 90˚ range (Fig. 1). Applying known method, the diffraction patterns achieved reveal the purity of synthesizes from starting materials (have known patterns in literature) [27]. Also, crystal-lattice dynamic reflects nano-crystalline feature for all complexes except Mg(II) one. Consequently and by using FWHM method, crystallite parameters were estimated by Bragg and Scherrer equations [28] for all complexes except the amorphous one (Mg+2). Such parameters as intensity, 2θ, d-spacing, particulate-size, crystal strain(ε) and dislocation density (δ) were calculated and tabulated (Table 4 ). The data showed two complexes sizes close to be in nanometer range (87.91 &102.62 nm) and their crystals having low imperfections according to the values of ε & δ parameters [29]. Moreover, SEM and TEM images that obtained by applying high-energy beam of electrons either to scan or go through thin samples, respectively (Fig. 2 &6S). The images of two types, were appeared by high-quality which facilitates investigating surface morphology, microstructure, crystallographic information and grain boundaries [30]. Three-dimensional SEM images for the complexes appeared very similar, which indicates extreme symmetry of structures. As well as, the crystalline feature of particles was obviously appeared as needle-particulate shape. Furthermore, TEM images appeared highly clear and homogeneous that exhibited small particles with few aggregation (black spots). The purity and crystallinity were basically suggested and further confirmed from these images (Table 5 ).
Table 4.
Crystal parameters for nano-crystalline complexes by using FWHM method.
| Compounds | Size (Å) | 2θ | Intensity | d-spacing (Å) | ε | δ(Å−2) | FWHM |
|---|---|---|---|---|---|---|---|
| 1)[Ca2Cl4(C22H23N3O9)]H2O | 10.262 | 18.59 | 35,266 | 4.769 | 0.056 | 0.0095 | 0.143 |
| 2)[SrCl2(C22H23N3O9)]H2O | 8.791 | 18.88 | 1857 | 4.696 | 0.064 | 0.0129 | 0.167 |
| 3)[BaCl2(C22H23N3O9)] | 14.90 | 14.54 | 35,652 | 6.087 | 0.049 | 0.0045 | 0.098 |
Table 5.
Drug-likeness parameters for ATA ligand, its alkaline earth metal ion complexes and two drugs.
| Properties | ATA | Mg(II) complex | Ca(II) complex | Sr(II) complex | Ba(II) complex | Hydroxyl-chloroquine | Lopinavir |
|---|---|---|---|---|---|---|---|
| TPSA (Å2) | 127.92 | 167.90 | 159.82 | 168.11 | 145.17 | 48.39 | 120.00 |
| Log Po/w | 1.31 | 1.54 | 1.58 | 1.40 | 1.47 | 3.29 | 4.06 |
| GI absorption | High | Low | Low | Low | Low | High | High |
| BBB permeant | No | No | No | No | No | Yes | No |
| Synthetic accessibility(SA) | 3.45 | 4.30 | 4.31 | 4.34 | 4.31 | 2.82 | 5.71 |
| Log Kp (skin permeation) | −5.79 cm/s | −5.02 cm/s | −5.21 cm/s | −5.79 cm/s | −6.10 cm/s | −5.81 cm/s | −6.12 cm/s |
3.6. Computation studies
3.6.1. Geometry optimization
i) General properties
Using DFT method under 6-31G and LANL2DZ basis sets, ATA ligand and its alkaline earth metal ion complexes were structurally optimized (Fig. 7S). Each optimization process yielded three files (log, chk & fchk) that summarized all computational characteristics, which presented over program screen according to numbering scheme.
Accordingly, HOMO and LUMO frontiers were firstly drawn (Fig. 3) over new cubic contour, to see the default distribution of electron cloud over the whole molecule. As seen, the HOMO frontier was distributed perfectly over ATA structure as well as in complexes, which particularly condensed around central atoms. On the other side, the LUMO frontier was specifically focused on certain side except Ba(II) complex was completely covered.
Moreover, other physical parameters (Table 2S) as electronegativity (χ), absolute-softness (σ), electrophilicity (ω), global hardness (η), global softness (S) and chemical potential (µ), were estimated depending on frontier energy gap (Δ = ELUMO-EHOMO) [15]. The following remarks can be summarized regarding the data calculated;
-
iii)
Softness characteristic was appeared with all compounds except Sr(II) complex has the lower value, which points to their flexibility in biological behaviors.
-
iv)
Also, electrophilicity (ω) appeared high with ATA ligand, which points to its high ability to accept electrons.
-
v)
The lower dipole moment values (Ddebye) of ATA ligand and its Ca(II) complex, reflect the reduced magnitude of electrostatic polarity over the molecule which increase its lipophilic feature [31], [32].
-
vi)
Lower heat of formation (E, a.u.) values of Mg(II) and Ca(II) complexes, reflect their high stability.
-
vii)
Electrostatic and iso-surface maps
Electrostatic potential maps (MEP) (Fig. 8S) were established for all optimized structures through fchk file and after exporting a new cubic contour. These maps differentiate electrophilic, nucleophilic and zero potential features by using blue, red and green colors, respectively [33]. MEP of free ATA ligand, exhibited high nucleophilic feature of COO– groups as well as OH groups. This offer their high priority in coordination towards the metal ions. On the other side, the complexes maps exhibited notable disappearance of nucleophilic feature for coordinating groups (C = O & OH) due to the formation of coordinate bonds.
Moreover, the iso-surface with array plots maps (Fig. 4), were also established over new cubic contour over all structures. By which, the electron density was measured on several points over the surface-grid and these points must be connected to reach the iso-surface shape. As seen, a widespread yellow lines (inner contour) were observed as well as outer contour lines (red), particularly with free ATA ligand. This reflects presence of high unsaturated surface boundary in the ligand, which able to acquire electrons from surrounding [34]. This feature is preferable in the interaction with biological systems.
3.6.2. Effective virtual screening against COVID-19 virus
-
i)
Drug-likeness assay
Designing an effective drug to face this pandemic needs many supportive studies that focus on proactive studies before the actual application in the laboratory. This is should not be done on any proposed drug easily, but it is better to be done on a drug that is expected to succeed before actual application and this is what we care about here. The proposed drug should have optimal pharmacokinetics that evaluate the absence of blood brain barrier (BBB) penetration to ensure safety, as a first step. A new tool (Swiss ADME) has been developed to assess fundamental physicochemical parameters [35]. Such parameters were TPSA parameter (topological polar surface area), Partition coefficient (Log P o/w), Synthetic accessibility (SA), gastrointestinal absorption (GI), blood–brain barrier (BBB) and skin permeation (Log K p), to obtain desirability score of drug-likeness attribute (Table 5, Fig. 9S). The proposed antiviral ligand (ATA) and its complexes were screened virtually versus two medicines that currently tried as Hydroxychloroquine and Lopinavir, to put a sincere proposal that adheres to scientific honesty. The results of the studied compounds may allow the prospective design when it retains the desired features in the likeness of drug and its ability to inhibit COVID-cell after successful penetration for sebaceous wall. Accordingly, TPSA parameter, which considered a measurable for surface polarity, appeared lower to preferable extent with the two drugs as well as ATA ligand. Partition coefficient (Log P o/w) values of ATA and its complexes are indicator for their promising biological activity in general. This points to their preferable lipophilicity within cell lipid. The gastrointestinal absorption (GI), BBB permeant and skin permeation (Log K p) values agree with the previous proposal except the unfavorable feature of Hydroxychloroquine in BBB feature. The synthetic accessibility (SA) parameter (1–10 range) appeared very good with ATA and Hydroxychloroquine. Finally from this introductory assay, there is optimism for ATA and Lopinavir behavior as antivirals, so we will do more in-depth study which is the following.
-
ii)
Simulated inhibition for COVID-19 proliferation
To study the binding affinity of ATA and its alkaline earth metal ion complexes with COVID-19 proteins (6lu7 & 6lzg), the molecular dynamic simulation at top scoring poses was performed by MOE module. Where, 6lu7 protein is a crystal structure of COVID-19 main protease, while 6lzg protein is the crystal structure of novel Coronavirus spike receptor-binding domain complexed with its receptor ACE2 (Scheme 3S).
From a comparative point of view, this simulation was also conducted on Hydroxychloroquine and Lopinavir as the drugs currently applied in curing. The maps of interaction fingerprint (Figs. 10S-12S) and contact preferences (Fig. 5) as well as the interaction affinity parameters (Tables 6 & 3S), were exported to evaluate the negative allosteric binding mode [36].
Table 6.
Interaction data for new compounds or two reference drugs with COVID-19 cell-proteins.
| Compounds | Proteins | ligand | Receptor | Interaction | Distance(Å) | E (Kcal/mol) | S(energy score) |
|---|---|---|---|---|---|---|---|
| 1)ATA | |||||||
| 6lu7 | O20 O9 O30 O31 |
SD MET 165 (A) ND2 ASN 142 (A) N GLY 143 (A) OG SER 144 (A) |
H-donor H-acceptor H-acceptor H-acceptor |
3.32 3.13 2.99 2.75 |
−0.5 –2.7 –3.1 –1.8 |
−6.7357 | |
| 6lzg | O9 O9 O30 O9 O9 O30 |
NH1 ARG 219 (A) NH2 ARG 219 (A) NZ LYS 94 (A) NH1 ARG 219 (A) NH2 ARG 219 (A) NZ LYS 94 (A) |
H-acceptor H-acceptor H-acceptor ionic ionic ionic |
3.26 2.93 2.95 3.26 2.9 2.95 |
−1.7 –2.3 –12.6 –3.0 –4.9 –4.8 |
−5.8931 | |
| 2)Mg(II)complex | 6lu7 | O26 O31 O26 O31 |
OH TYR 37 (A) NZ LYS 88 (A) NZ LYS 88 (A) NZ LYS 88 (A) |
H-acceptor ionic ionic ionic |
3.16 2.93 3.06 2.93 |
−1.2 –4.9 –4.1 –5.0 |
−1.8707 |
| 6lzg | Mg41 | OD1 ASP 38 (A) | ionic | 2.27 | −11.7 | 0.8686 | |
| 3)Ca(II)complex | 6lu7 | O26 | NZ LYS 61 (A) | ionic | 2.92 | −5.0 | −0.5276 |
| 6lzg | Cl46 O31 Ca44 |
NZ LYS 441 (A) NZ LYS 441 (A) OD1 ASP 367 (A) |
H-acceptor ionic ionic |
2.87 2.87 2.46 |
−2.7 –5.4 –9.3 |
3.1528 | |
| 4)Sr(II)complex | |||||||
| 6lu7 | O31 | SG CYS 145 (A) | H-donor | 3.41 | −1.1 | −2.6354 | |
| 6lzg | C25 Sr42 |
NZ LYS 234 (A) OE2 GLU 483 (A) |
Ionic ionic |
3.11 2.51 |
−3.8 –8.7 |
−4.9474 | |
| 5)Ba(II) complex | 6lu7 | O28 O30 |
SD MET 165 (A) CA ARG 188 (A) |
H-donor H-acceptor |
3.46 3.26 |
−1.7 –0.7 |
−6.0648 |
| 6lzg | O30 O30 O28 O30 Ba42 |
NH1 ARG 403 (B) OH TYR 505 (B) NH2 ARG 393 (A) NH1 ARG 403 (B) NE ARG 408 (B) |
H-acceptor H-acceptor ionic ionic ionic |
2.93 2.92 3.21 2.93 3.34 |
−4.4 –2.3 –3.2 –5.0 –2.6 |
−5.5695 | |
| Hydroxychloroquine drug | 6lu7 | --- | --- | --- | --- | --- | −5.2454 |
| 6lzg | O23 6-ring 6-ring |
OD1 ASP 367 (A) CB ILE 291 (A) CB ILE 291 (A) |
H-donor pi-H pi-H |
2.97 3.91 4.59 |
−3.5 –0.7 –0.6 |
−6.0729 | |
| Lopinavir drug | 6lu7 | O36 6-ring 6-ring |
NZ LYS 5 (A) NZ LYS 137 (A) CB SER 284 (A) |
H-acceptor pi-cation pi-H |
2.87 4.08 4.33 |
−5.1 –0.7 –0.7 |
−6.258 |
| 6lzg | N3 6-ring |
NH1 ARG 403 (B) NE2 GLN 96 (A) |
H-acceptor pi-H | 3.23 4.27 |
−2.5 –1.3 |
−7.6147 |
Accordingly the following information were aggregated from interaction patterns;
-
i)
The solvent contact as a doted proximity contour surround the molecule, which appeared perfectly covered ATA ligand, Hydroxychloroquine and Lopinavir than the other compounds.
-
ii)
The doted arrows that represent the H-bonding between ligand sites and amino acid residues, showed polar (pink circles) and nonpolar (green circles) receptors from the kind of side chain acceptor or donor and basic backbone acceptor. Such were recorded basically with ATA and its complexes towards COVID-19 proteins. In addition to, the metal ion complexes exceeded by electrostatic interaction through O-.
-
iii)
Hydroxychloroquine showed OH– side chain donor and 6-ring interaction towards 6lzg protein only [5].
-
iv)
Lopinavir showed a polar side chain acceptor (through C = O or N) and 6-ring contact, which appeared intensively with the two COVID-19 proteins.
The effective H-bonding cooperation between the interacting sites towards negative allosteric sites, were summarized from interaction parameters (Table 6) as follow;
-
i)
Significant inhibition activity was recorded with ATA ligand, Ba(II) complex as well as Lopinavir drug towards COVID-19 cell-proteins. This is based on their high scoring energy values recorded (Table 6), which seem close to each other's. While, Hydroxychloroquine comes in the second order after them. The activity of other complexes, was appeared moderate to nil.
-
ii)
Regarding ATA ligand binding, the interaction was directed towards MET165, ASN142, GLY143, SEP144, ARG219 and LYS94 amino acid residues. While, the complexes bonding was directed towards many residues of Lysine, Glutamate and Arginine.
-
v)
Hydroxychloroquine bonding was directed towards Asp367 and lle291 residues, while, Lopinavir bonding was directed towards Lys137, Ser284, ArgB403 and GlnA96 residues. With respect to true docking paths (bond length ≤ 3.5 Å), the inhibition features were obtained [37].
-
vi)
The contact preferences (Fig. 5) of ATA and two handled drugs, exhibited well penetration of ATA and Lopinavir within amino acid residues, which indicates perfect interaction.
-
vii)
The energy content appeared lower with ATA interaction poses till reach −12.6 Kcal/mol which reflects stability than Lopinavir patterns that reach to −5.0 Kcal/mol only.
The inhibition outcomes exclude the effectiveness of Hydroxychloroquine drug in treatment of COVID-19 virus, while Lopinavir as well as our proposed drug (ATA) may be the best in treating Corona pandemic.
-
iii)
Binding affinity calculations
To indicate the binding affinity of new compounds as well as the antiviral drugs, we estimated some essential parameters (Table 3S). Total energy, electrostatic energy, heat of formation, ionization potential, sum of atomic polariazability and log P (GCUT, 0/3), were the targeted parameters, to confirm previous suggestions. The inhibitory binding of ATA towards COVID-19 proteins, is absolutely evident in comparison with the two drugs (Hydroxychloroquine & Lopinavir), through the highly reduced total energy with highly exothermic interaction. Also, it's lower ionization potential and atomic polariazability, evidence high solubility in fats, which facilitates biological handling after penetrating cell-membrane. Finally, high minimization of partition coefficient (log P) for ATA and two drugs give them best opportunity for miscibility with lipids and contact directly with biological systems. But the contact features are strongly affected by other conditions, which may be improved as in ATA and Lopinavir [6].
4. Conclusions
Aurintricarboxylic acid (ATA) and its new alkaline earth metal ion complexes, were interested in this study for computerized treatment towards COVID-19 pandemic. The synthesized complexes were proposed either 1:1 or 1:2 (L: M) molar ratio within tetrahedral configuration. This is depending on spectral (IR, UV–Vis, XRD, SEM&TEM) analytical and conformational analyses. The main objective was dealt with approved ways (drug-likeness & docking) to reach the closest perception of compounds behavior compared to two drugs currently applied towards Corona pandemic. The outcomes of inhibition affinity towards COVID-19, reveal the promising behavior of Lopinavir as well as ATA compound. On the other side, ineffectiveness was observed for Hydroxychloroquine drug which unfortunately be used now extensively.
5. Recommendation
This medication implemented Hydroxychloroquine is not the best in treating the emerging Corona pandemic. But Lopinavir and our proposed drug (ATA), may be the best and I invite specialists to try them practically that we did not have the facilities to do that by ourselves.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
Taif University Researches Supporting Project number (TURSP-2020/01), Taif University, Taif, Saudi Arabia.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.inoche.2021.108472.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Michal H., Rachel B., Avraham K. Endocrinology. 2002;143:837. [Google Scholar]
- 2.Kadhier A.F. J. Kerbala Univ. 2009;7:190. [Google Scholar]
- 3.Skidmore A.F., Beebee T.J. Biochem. J. 1989;263:73. doi: 10.1042/bj2630073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saad F.A., Elghalban M.G., Al-Fahemi J.H., Yarkandy N., El-Metwaly N.M., Abou-Melha K.S., Al-Hazmi G.A., Saleh K.A. J. Therm. Anal. Calorim. 2017;128:1565–1578. [Google Scholar]
- 5.Obrecht A.S., Urban N., Schaefer M., Röse A., Kless A., Meents J.E., Lampert A., Abdelrahman A., Müller C.E., Schmalzing G., Hausmann R. Neuropharmacology. 2019;158 doi: 10.1016/j.neuropharm.2019.107749. [DOI] [PubMed] [Google Scholar]
- 6.Kuban-Jankowska A., Sahu K.K., Gorska M., Niedzialkowski P., Tuszynski J.A., Ossowski T., Wozniak M. World J. Microbiol. Biotechnol. 2016;32:163. doi: 10.1007/s11274-016-2123-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Refat M.S., El-Metwaly N.M. J. Mol. Struct. 2011;988:111–118. [Google Scholar]
- 8.Starosta W., Leciejewicz J. J. Coord. Chem. 2009;62:1240. [Google Scholar]
- 9.Chen Z.F., Li B.Q., Xie Y.R., Xiong R.G., You X.Z., Feng X.L. Inorg. Chem. Commun. 2001;4:1346. [Google Scholar]
- 10.Ruben M., Lehn J.M., Müller P. Chem. Soc. Rev. 2006;35:1056. doi: 10.1039/b517267p. [DOI] [PubMed] [Google Scholar]
- 11.El-Megharbel S.M., Hamza R.Z., Refat Spectrochim M.S. Acta A Mol. Biomol. Spectrosc. 2015;135:915–928. doi: 10.1016/j.saa.2014.07.101. [DOI] [PubMed] [Google Scholar]
- 12.Shebl M., Khalil S.M.E., Taha A., Mahdi M.A.N. J. Mol. Struct. 2012;1027:140–149. [Google Scholar]
- 13.Vogel A.I. Longman; London: 1986. Textbook of Quantitative Inorganic Analysis. [Google Scholar]
- 14.R. Dennington, T. Keith, J. Millam, Gauss View, Version 4.1.2, SemichemInc, Shawnee Mission, KS, 2007.
- 15.El-Ayaan U., El-Metwally N.M., Youssef M.M., El Bialy S.A.A. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2007;68:1278–1286. doi: 10.1016/j.saa.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 16.R. Shah, A. Alharbi, A. M. Hameed, F. Saad, R. Zaky, A. M. Khedr, N. El‑Metwaly, J. Inorg. Organomet. Poly. Mat. https://doi.org/10.1007/s10904-020-01505-w.
- 17.H.M Abumelha, J.H Al-Fahemi, I. Althagafi, A.A. Bayazeed, Z.A. Al-Ahmed, A.M. Khedr, N. El-Metwaly, J. Inorg. Organomet. Poly. Mat. https://doi.org/10.1007/s10904-020-01503-y.
- 18.Althagafi I., El-Metwaly N., Farghaly T.A. Molecules. 2019;24:1741. doi: 10.3390/molecules24091741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Hazmi G.A.A., Abou-Melha K.S., El-Metwaly N.M., Althagafi I., Shaaban F., Zaky R. Appl. Organometal. Chem. 2020;34 [Google Scholar]
- 20.Geary W.J. J. Coord. Chem. Rev. 1971;7:81–122. [Google Scholar]
- 21.Saad F.A., Al-Fahemi J.H., El-Ghamry H., Khedr A.M., Elghalban M.G., El-Metwaly N.M. J. Therm. Anal. Calorim. 2018;131:1249–1267. [Google Scholar]
- 22.Abu-Melha K.S., El-Metwally N.M. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2008;70:277–283. doi: 10.1016/j.saa.2007.07.058. [DOI] [PubMed] [Google Scholar]
- 23.El-Megharbel S.M., El-Metwaly N.M., Refat M.S. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2015;149:263–270. doi: 10.1016/j.saa.2015.04.069. [DOI] [PubMed] [Google Scholar]
- 24.Lever A.B.P. Elsevier; Amsterdam: 1986. Inorganic Electronic Spectroscopy. [Google Scholar]
- 25.Mott N.F., Davis E.A. Calendron Press; Oxford: 1979. Electrochemical Process In Non-crystalline Materials. [Google Scholar]
- 26.Karipcin F., Dede B., Caglar Y., Hur D., Ilican S., Caglar M., Sahin Y. Optic Commun. 2007;272:131–137. [Google Scholar]
- 27.Cullity B.D. 2nd edn., Addison-Wesley Inc.; 1993. Elements of X-Ray Diffraction. [Google Scholar]
- 28.Cullity B.D., Stock S.R. Prentice Hall; New Jersey: 2001. Elements of X-Ray Diffraction. [Google Scholar]
- 29.Saad F.A., El-Metwaly N.M., Khedr A.M. J. Inorg. Organomet. Polym. 2019;29:1606–1624. [Google Scholar]
- 30.Refat M.S., El-Metwaly N.M. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011;81:215–256. doi: 10.1016/j.saa.2011.05.108. [DOI] [PubMed] [Google Scholar]
- 31.Al-Hazmi G.A.A., Abou-Melha K.S., El-Metwaly N.M., Althagafi I., Shaaban F., Zaki R. Appl. Organomet. Chem. 2020;34 [Google Scholar]
- 32.Mi H., Xiao G., Chen X. Comput Theor. Chem. 2015;1072:7–14. [Google Scholar]
- 33.Costa R.A., Pitt P.O., Pinheiro M.L.B., Oliveira K.M.T., Salome K.S., Barison A., Costa E.V. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2017;174:94–104. doi: 10.1016/j.saa.2016.11.018. [DOI] [PubMed] [Google Scholar]
- 34.Prasad S.B.B., Naveen S., Ananda Kumar C.S., Lokanath N.K., Raghu A.V., Daraghmeh I., Reddy K.R., Warad I. J. Mol. Struct. 2018;1167:215e226. [Google Scholar]
- 35.Shaaban S., Fasseur D.V., Andreoletti P., Zarrouk A., Richard P., Negm A., Manolikakes G., Jacob C., Malki M.C. Bioorg. Chem. 2018;80:43–56. doi: 10.1016/j.bioorg.2018.05.019. [DOI] [PubMed] [Google Scholar]
- 36.Al-Hazmi G.A.A., Abou-Melha K.S., El-Metwaly N.M., Althagafi I., Shaaban F., Elghalban M.G., El-Gamil M.M. Appl. Organomet. Chem. 2019;34 [Google Scholar]
- 37.Al-Hazmi G.A.A., Abou-Melha K.S., El-Metwaly N.M., Althagafi I., Zaky R., Shaaban F., Inorg J. Organomet. Poly. 2019 doi: 10.1007/s10904-019-01326-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.