Abstract
Background
Severe acute respiratory syndrome caused by novel coronavirus 2 (SARS-CoV-2) emerged in Wuhan (China) in December 2019. Here we evaluated a panel of biomarkers to phenotype patients and to define the role of immuno-inflammatory mediators as biomarkers of severity.
Materials and methods
Serum samples were obtained from 24 COVID-19 patients on admission to hospital, before any treatment or infusion of intravenous steroids or invasive ventilation. KL-6 IL-6 and C-peptide were measured by chemiluminescent enzyme immunoassay. IL-6 assay was validated for accuracy and precision. The validity of variables used to distinguish severe from mild-to-moderate patients was assessed by areas under curves (AUC) of the receiver operating characteristic (ROC) and logistic regression was performed to combine parameters of the two groups.
Results
In the severe group, IL-6, CRP and KL-6 concentrations were significantly higher than in mild-to-moderate patients. KL-6, IL-6 and CRP concentrations were directly correlated with each other. ROC curve analysis of the logistic regression model including IL-6, KL-6 and CRP showed the best performance with an AUC of 0.95.
Conclusions
Besides corroborating previous reports of over-expression of IL-6 in severe COVID-19 patients requiring mechanical ventilation, analytical determination of other mediators showed that IL-6 concentrations were correlated with those of KL-6 and CRP. The combination of these three prognostic bioindicators made it possible to distinguish severe COVID-19 patients with poor prognosis from mild-to-moderate patients.
Keywords: IL-6, KL-6, Biomarkers, COVID-19
Abbreviations: IL, interleukin; KL-6, Krebs von den Lungen; CRP, c-reactive protein; LDH, lactate dehydrogenase
1. Introduction
Severe acute respiratory syndrome caused by new coronavirus 2 (SARS-CoV-2) emerged in Wuhan (China) in December 2019 [1], [2]. In a few months, the etiological agent spread worldwide and was declared pandemic by the World Health Organization (WHO) in March 2020 [3], [4]. The severe pneumonia-associated respiratory syndrome caused by the new coronavirus shows different clinical phenotypes, varying from mild-moderate to severe and critical with acute respiratory distress syndrome, requiring hospitalization and mechanical ventilation [5], [6], [44]. In general, male patients over 65 years with chronic lung disease and/or metabolic disorders such as arterial hypertension, obesity and diabetes show higher risk of severe disease [7], [8], [9], [49]. Although the physiopathology of this extremely contagious disease is not fully understood, sepsis is considered a critical illness prototype for coronavirus 2 (COVID-19) pathogenesis, since severe cases are associated with immune disorder, lymphopenia (mainly CD4 and B cells) and hyper-cytokinaemia, elevated serum levels of IL-6 and IL-10 being predictive of severe course [10], [11], [12]. A recent study reported high levels of IL-6, IL-8 and TNF in peripheral blood of COVID-19 patients, contributing to hyperinflammation [46], [13].
Aberrant host immune responses and especially inflammatory cytokine storms contribute to alveolar exudation and lung damage [52], [51]. Pleiotropic Th2 cytokines, including IL-6, mediate the transition from the acute to the chronic phase of infective processes [11], [14] and their number in critical patients is about an order of magnitude greater than in mild COVID-19 patients [9], [15], [16]. Significant dysregulation of serum levels of coagulation factors and anti-fibrinolytic components have been observed in subjects with high levels of IL-6, suggesting a link to COVID-19 pathophysiology [17], [18], [50]. Thus, clinical trials of different monoclonal antibodies (such as JAK-STAT moAbs) that block many inflammatory cytokines (including IL6) were recently approved as an effective treatment to improve prognosis in severe COVID-19 patients [10], [19]. Different commercial immunoassays are available to analyse serum concentrations of IL-6 for clinical and research purposes [20].
Metabolic disorders, such as diabetes, in COVID-19 patients, have been associated with higher mortality, especially in older people. These comorbidities can modify host immunity and disease progression [21]. Obesity has also been associated with a negative prognosis: adipocytokines have an impact on metabolic, oncological and rheumatological disorders [22] and it was recently suggested that adipocyte dysfunction may lead to a specific immune environment predisposing obese patients to respiratory failure during COVID-19 [23]. Different studies aiming to assist clinicians in early identification of severe and critical patients have tested combinations of immune-mediators, including inflammatory cytokines, against age, comorbidities and clinical features [24].
The aim of the present study was to evaluate a panel of biomarkers including IL-6, KL-6, C-peptide, CRP, LDH, glycemia and pancreatic amylase. These molecules were analysed to phenotype our population of COVID-19 patients and to define a possible role of immuno-inflammatory mediators as biomarkers of severity.
2. Methods
2.1. Study population
Forty-one patients with COVID-19 (27 males (65.8%), 64.6 ± 18.4 years), hospitalized at Siena University Hospital in March and April 2020, and 30 healthy controls (18 males (60%), 59 ± 9.8 years) were enrolled consecutively in the study. They underwent clinical and radiological evaluation and were divided into mild-to-moderate and severe groups (the latter requiring intubation and invasive mechanical ventilation). Signs, symptoms, radiological and immunological features and serum concentrations of inflammatory biomarkers were entered in a database.
Serum samples were obtained from 24 of the patients on admission to hospital, before any treatment or infusion of intravenous steroids or invasive ventilation, and from the 30 healthy controls. Serum aliquots were stored at −80 °C until assay. All patients gave their written informed consent to the study that was approved by our local ethics committee (BIOBANCA-MIU-2010).
2.2. KL-6 and C-peptide assay
Assay of serum Krebs von den Lungen-6 (sKL-6) is based on agglutination of sialylated carbohydrate antigen with KL-6 mAb reagent (Fujirebio Europe, UK). Concentrations (expressed in U/ml) were determined by measuring changes in absorbance as described in previous papers [25], [26]. Test reagents for C-peptide were from Fujirebio Inc. (Tokyo, Japan). This biomolecule was measured by the Lumipulse G600 II system. The limit of detection and limit of quantification were 0.001 nmol/L for C-peptide. The manufacturer’s reference interval was 0.06–0.24 ng/ml. The tests were conducted according to the manufacturer's instructions.
2.3. IL-6: Quality control of analytical determinations and comparison of results
Serum concentrations of IL-6 were determined with an automatic biochemical analyzer (Cobas 8000 e602, Roche, Mannheim, Germany) and detected by electro-chemiluminescent immunoassay (ECLIA). Serum samples were also analysed by Lumipulse G600 II (Fujirebio, Japan).
Precision was evaluated according to CLSI EP5-A3 guidelines [27] using two concentrations of reagent control serum assays: Precicontrol multimarkers-1 (PCMM1) (Roche, Germany) (median 40.8 (range 32.2–49.4) pg/mL and PCMM2 (median 247 (range 195–299) pg/mL. Control serum assays were performed in duplicate, twice a day for 20 consecutive days. Linearity was evaluated according to CLSI EP6-A guidelines [25]. Four concentrations of IL-6 calibrators from the IL-6 kit (Fujirebio Inc.) were used. To establish the regression equation, the measurements were repeated four times for each concentration. To compare methods, we measured 54 serum samples from COVID-19 patients and healthy subjects. Comparisons were performed according to CLSI EP9-A3 guidelines [26].
2.4. Other markers
C-reactive protein IV (CRP) was measured by immunoturbidimetric test; α-amylase pancreatic protein (AMY-P) and lipase colorimetric (LIPC) by a colorimetric test; lactate dehydrogenase (LDH), glucose HK 3 (GLU-3), alanine aminotransferase and aspartate (ALT and AST, respectively) by an UV test. Control and calibrator reagents were supplied by Roche/Hitachi, Mannheim, Germany and analysed with a Cobas 8000 e602-c702 automatic biochemical analyzer (Roche/Hitachi, Mannheim, Germany).
2.5. Statistical analysis
The results are reported as means ± SD or as medians and inter quartiles (25th and 75th percentiles) for continuous variables. The Shapiro-Wilk test showed that the data did not have a normal distribution. To evaluate precision, the coefficient of variation was calculated. To evaluate linearity, the coefficient of determination (R2) was determined by logistic regression. Bias between systems was calculated by Bland-Altman analysis.
One-way ANOVA non parametric test (Kruskal-Wallis test) and Dunn test were therefore performed for multiple comparisons. The chi-squared test was applied to categorical variables. The validity of variables used to distinguish severe from mild-to-moderate patients was assessed by areas under (AUC) receiver operating characteristic (ROC) curves and a logistic regression was performed to compare parameters between the two groups. Sensitivity, specificity, and positive and negative predictive values (PPV and NPV, respectively) were calculated for cut-offs of the different variables. The Youden index (J = max [sensitivity + specificity − 1]) was used to establish the best cut-offs. Spearman’s rank correlation coefficients were assessed for relationships between variables in each group. Statistical analysis and graphic representation of the data were performed by GraphPad Prism 8.0 software.
3. Results
3.1. Study population
The main characteristics of the COVID-19 population, including demographic data and IL-6 levels, are reported in Table 1 . Other markers, together with clinical and immunological findings are reported in Table 2 . There was a prevalence of males in both groups: 78.5% and 80% in mild-moderate and severe patients, respectively. Bilateral diffuse pneumonia was detected in 60% of the severe group and 57.2% of mild-moderate patients. Regarding symptoms, 8 out of 14 (58%) mild-moderate patients and 6 out of 10 (60%) severe patients showed at least two symptoms at onset. Seven out of 24 patients were without comorbidities. In particular, the severe group included four patients with arterial hypertension, one with diabetes, one with heart failure, one with lung disease and four with cancer (one breast cancer, one melanoma, one leukemia and one adenoma). Five mild-to-moderate patients showed arterial hypertension, four dyslipidaemia, three heart failure, two lung diseases (one asthma and one COPD) and one stroke.
Table 1.
Demographic data and IL-6 concentrations in healthy controls, Mild to moderate and severe groups.
| Healthy controls (n = 30) | Mild to Moderate (n = 14) | Severe (n = 10) | P value | |
|---|---|---|---|---|
| Age (m ± SD) | 59 ± 9.8 | 62.2 ± 15.6 | 65.2 ± 8 | ns |
| Gender (M:F) | 18/12 | 11/3 | 8/2 | ns |
| Smoking Habits (never/current/former) | 12/3/15 | 6/4/4 | 3/1/7 | ns |
| IL-6 (pg/ml) (median (IQR)) | 19.2 (16.7–22.8) | 32.4 (11.8–143.5) | 333.9 (66.3–843.2) | 0.0004 |
Table 2.
Clinical, immunological and radiological data of COVID-19 groups.
| Mild to moderate group (n = 14) | Severe group (n = 10) | P value | |
|---|---|---|---|
| Symptoms | |||
| Fever | 13 | 8 | ns |
| Cough | 4 | 4 | ns |
| Vomit | 1 | 0 | ns |
| Weakness | 1 | 1 | ns |
| Dyspnoea | 12 | 6 | 0.04 |
| Comorbidities | |||
| yes/no | 10/4 | 7/3 | ns |
| KL-6 (U/ml) (median (IQR)) | 320 (226.3–927.8) | 903 (333.8–1956) | 0.035 |
| CRP (mg/l) (median (IQR)) | 3.5 (2.2–5) | 5.4 (4.5–12.8) | 0.03 |
| LDH (U/l) (median (IQR)) | 256 (227.3–329) | 342.5 (290–585) | ns |
| Glycemia (mg/dl) (median (IQR)) | 98.7 (75–113.5) | 109 (103–125) | ns |
| Lipase (U/l) (median (IQR)) | 21 (18–25) | 23 (15–29) | ns |
| Pancreatic Amylase (U/l) (median (IQR)) | 28.7 (15–41.5) | 29.5 (17–35) | ns |
| AST (U/l)(median (IQR)) | 22.5 (14–27) | 23.25 (12–49.5) | ns |
| ALT (U/l) (median (IQR)) | 14.2 (10–18) | 20.5 (12–26.5) | ns |
| C-peptide (ng/ml) (median (IQR)) | 1.72 (1–1.93) | 2.6 (1.73–2.8) | ns |
| Blood count | |||
| RBC (cell/mm3) | 4.3 ± 0.9 | 4.5 ± 0.5 | ns |
| WBC (cell/mm3) | 4.9 ± 1.6 | 7.6 ± 6 | ns |
| PLT (cell/mm3) | 278 ± 156 | 183.7 ± 62 | ns |
| Leucocytes counts | |||
| Lymphocytes (×103/ml) | 18.2 ± 11 | 13.4 ± 6.4 | 0.04 |
| Neutrophils (×103/ml) | 74 ± 13 | 77 ± 8 | ns |
| Eosinophils (×103/ml) | 0.05 ± 0.1 | 0.25 ± 0.7 | ns |
| Monocytes (×103/ml) | 7.4 ± 2.7 | 8.8 ± 9 | ns |
| Basophils (×103/ml) | 0.22 ± 0.2 | 0.15 ± 0.13 | ns |
| Lymphocytes subsets | |||
| CD4(%) | 44.4 ± 7.8 | 44.6 ± 13 | ns |
| CD8(%) | 22 ± 8.7 | 31 ± 18 | ns |
| CD19(%) | 18.1 ± 7 | 12.6 ± 9.3 | 0.03 |
| NK(%) | 13 ± 8 | 10 ± 6.9 | ns |
| Chest X ray | |||
| (monolateral/bilateral/bilateral diffused) | 2/4/8 | 0/4/6 | ns |
3.2. IL-6 assay and analytical validation
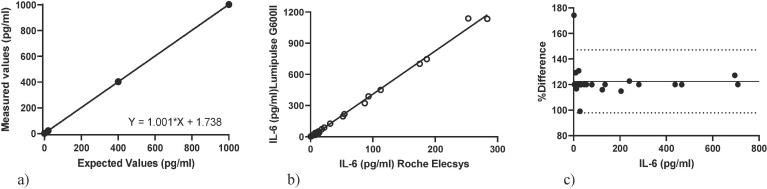
The precision of IL-6 analytical determinations (CV%) for PCMM1 and PCMM2 reagent concentrations was 1.49% for the lower and 0.52% for the higher, respectively. The results of the linearity evaluation for IL-6 are shown in Fig. 1 a. The regression equation between the expected and the measured value was Y = 1001 * X + 1738. The coefficient of determination (R2) (range: 0–1000 pg/ml) of the regression analysis was 0.999. IL-6 concentrations were higher when measured by Lumipulse G600 II than by the Roche Elecsys system in COVID-19 patients (62.4 (21.4–371) vs 16.13 (5.5–91.5) pg/ml, p < 0.0001). R2 between Lumipulse G600 II and Roche Elecsys results for IL-6 was 0.98 (p < 0.001) (Fig. 1b).
Fig. 1.
. (a) Linearity of IL-6 in the Lumipulse G600II system. (b) Method comparison of the two technology. (c) Assay validation Bland-Altman plots. Dotted lined indicates 95% limits of agreement.
Bland–Altman difference analysis revealed a mean bias of 122.5 ± 12.5 (95% limits of agreement 97.9–147.1) for IL-6 between the Lumipulse G600 II and the Roche Elecsys systems (Fig. 1c).
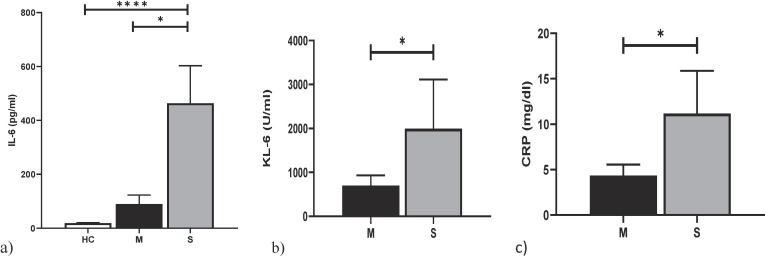
In the severe group, IL-6 concentrations were significantly higher than in mild-to-moderate patients (p = 0.009) and healthy controls (p = 0.0004) (Fig. 2 a). Serum concentrations of IL-6 in mild to moderate patients were not significantly different from those of controls (median 32.4 and 19.2 pg/ml, p = 0.76).
Fig. 2.
. (a) comparison analysis of IL-6 between Healthy controls (HC), Mild-to-moderate and Severe Covid-19 Patients. (b–c) comparison analysis of KL-6 and CRP between Mild-to-moderate and Severe Covid-19 Patients. *p < 0.05, **p < 0.01, ***p < 0.001, *** p < 0.0001.
3.3. Other serum biomarkers
Serum concentrations of KL-6 were correlated with IL-6 concentrations (r = 0.43; p = 0.04). They were higher in severe than in mild to moderate patients (p = 0.035) (Fig. 2b), and showed similar concentrations in healthy and mild-moderate patients (221, 226.3 and 903 U/ml for controls, mild-moderate and severe patients, respectively). CRP levels were significantly higher in severe than in mild to moderate patients (p = 0.03) (Fig. 2c). No significant differences in C-peptide concentrations were found between the two groups of COVID-19 patients (p > 0.05), although levels were above the normal range suggested by the manufacturer. Our population only included a single type 2 diabetes patient. Serum concentrations of CRP were correlated with KL-6 (r = 0.51; p = 0.04) and were significantly higher in severe than in mild-to-moderate patients (p = 0.03). In turn, CRP concentrations were very significantly correlated with LDH (r = 0.71; p = 0.002), without significant differences in LDH concentrations in the two groups (p = 0.10).
3.4. Immunological markers
Comparison of groups showed lower concentrations of peripheral lymphocytes in severe than in mild to moderate patients (13.4 ± 6.4 and 18.2 ± 11 respectively, p = 0.04). The same trend was found for CD19 percentages which were 18.1 ± 7 and 12.6 ± 9.3, respectively (p = 0.03). No other significant differences in immunological data were found. Interestingly, peripheral concentrations of platelets showed an indirect correlation with KL-6 and CD4 percentages (r = −0.56, p = 0.04 and r = −0.63, p = 0.01, respectively), in contrast with a direct correlation between platelet count and CD8 percentages (r = 0.53, p = 0.03).
3.5. Panel of biomarkers
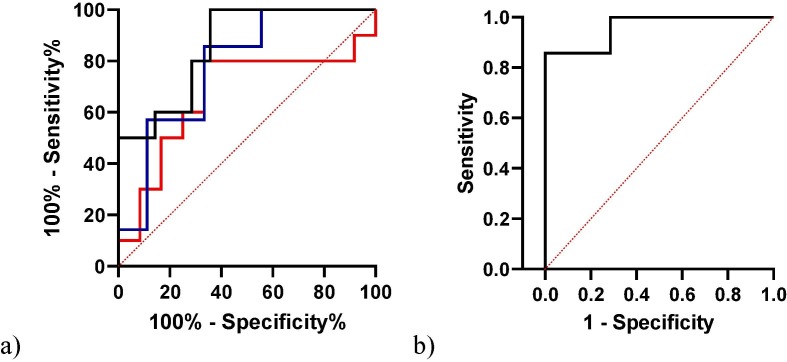
The ROC analysis of IL-6 data distinguished COVID-19 patients from healthy subjects, with an area under the curve of 0.78 (95% CI 0.63–94, p = 0.009) and a best cut-off value of 27.3 pg/ml (75% sensitivity, 100% specificity). ROC analysis also distinguished mild to moderate from severe patients. Serum IL-6 showed the best performance with an area under the curve of 0.85 (95% CI 0.79–1, p = 0.003) and best cut-off value of 62.45 pg/ml (80% sensitivity, 71% specificity) (Fig. 3 a). In the logistic regression, severe patients were tested as dependent variable against KL-6, CRP, LDH, C-peptide and IL-6 as independent variables. ROC curve analysis combining IL-6, KL-6 and CRP proved to have the best performance, showing an AUC of 0.95 (95% CI 0.86–1; NPP(%) 85.7, PPP(%) 85.7, p = 0.004) (Fig. 3b).
Fig. 3.
. (a) ROC curve analysis of CRP (blue), KL-6 (Red) and IL-6 (black) between mild to moderate and severe Covid-19 patients. (b) Combination model of KL-6, IL-6 and CRP.
4. Discussion
This monocentric retrospective study examined the clinical and laboratory characteristics of a population of patients with COVID-19 admitted to an Italian university hospital.
Combinations of biomarkers were compared in mild-to-moderate versus severe patients in search of a panel of biomarkers with prognostic value. We also evaluated the analytical performance of an IL-6 assay kit based on chemiluminescent enzyme immunoassay, which was confirmed to be good: cut-off values discriminating healthy subjects from COVID-19 patients were 7.0 and 27.3 pg/ml for Cobas 6000 and Lumipulse G600 II, respectively.
Analysis of IL-6 and other inflammatory biomarkers in serum of COVID-19 patients showed higher levels of IL-6, KL-6 and CRP in patients admitted to the ICU and requiring intubation and mechanical ventilation due to diffuse interstitial pneumonia than in mild-to-moderate patients. IL-6 is involved in cytokine storms and huge increases in its concentrations have been reported in COVID-19 patients at high risk of death [28], [29], [30]. An increase in inflammatory mediators, such as CRP and IL-6, in the course of the disease may help identify patients with poor prognosis, requiring rapid clinical intervention [31], [32]. Wang et al. reported high CRP concentrations in the early stages of SARS-CoV-2 infection and documented a positive correlation between CRP levels and the severity of lung damage detected by HRCT (i.e. CRP emerged as a prognostic biomarker) [33].
Interestingly, Zhu Z et al. compared clinical and immune-inflammatory parameters in non-severe and severe patients, reporting IL-6, CRP and hypertension as independent predictors of COVID-19 severity [34]. To our knowledge, ours is the first study investigating the combination of KL-6, IL-6 and CRP concentrations in a population of COVID patients. KL-6 is a mucin-like glycoprotein widely studied in interstitial lung diseases [35], [36], [47], [48] and its concentrations in peripheral blood can reflect severe interstitial lung damage, epithelial lung alterations and regenerative processes secondary to SARS-CoV-2 infection [37]. Serum KL-6 concentrations were determined in a Japanese population of COVID-19 patients treated with ECMO due to severe lung involvement, and were in the same range as those of our cohort [38]. Other authors recently analysed serum concentrations of KL-6 in COVID-19 patients in relation to COVID-19-related acute respiratory distress syndrome, and suggested KL-6 as a differential prognostic biomarker [39], [40], [45].
Xue M et al. found KL-6 levels in severe patients significantly upregulated with respect to mild patients, supporting the idea that this mediator reflects the level of lung injury and inflammation [41]. In line with these findings, the same trend was confirmed in our albeit limited population. No data is currently available on C-peptide in COVID-19 patients. The findings of the present study suggest for the first time that inflammatory status induced by SARS-CoV-2 may be associated with insulin resistance.
Platelet count also showed a correlation with immunological data, in particular with CD4 and CD8 percentages and KL-6 concentrations. Recent papers report that platelet percentages are depressed in very severe patients [42], [43]. In particular, Tien R et al. showed that critical patients had low lymphocyte and platelet counts and elevated CRP and IL-6 concentrations. Our data was perfectly in line with these findings and for the first time, also demonstrates a correlation between IL and 6 and KL-6 concentrations.
Although there is much data in the literature on over-expression of IL-6 in severe COVID-19 patients requiring mechanical ventilation, besides corroborating previous reports, our results suggest that a panel of three prognostic bioindicators can improve recognition of severe forms. The combination of KL-6, IL-6 and CRP made it possible to distinguish severe COVID-19 patients with poor prognosis from mild-to-moderate patients.
CRediT authorship contribution statement
L. Bergantini: Conceptualization, Methodology, Software. E. Bargagli: Conceptualization, Methodology, Software. M. d’Alessandro: Data curation, Writing - original draft. R.M. Refini: Data curation, Writing - original draft. P. Cameli: Data curation, Writing - original draft. L. Galasso: Visualization, Investigation. C. Scapellato: Visualization, Investigation. F. Montagnani: Supervision. S. Scolletta: Supervision. F. Franchi: Supervision. S. Valente: Supervision. D. Bennett: Software, Validation. G. Sebastiani: Software, Validation. B. Frediani: Writing - review & editing. F. Dotta: Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu S., Wang W., Wang Y., Litvinova M., Luo K., Ren L., et al. Infectivity, susceptibility, and risk factors associated with SARS-CoV-2 transmission under intensive contact tracing in Hunan, China. MedRxiv Prepr. Serv. Health Sci. 2020 doi: 10.1038/s41467-021-21710-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahn D.-G., Shin H.-J., Kim M.-H., Lee S., Kim H.-S., Myoung J., et al. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19) J. Microbiol. Biotechnol. 2020;30(3):313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’alessandro M., Bennett D., Montagnani F., Cameli P., Perrone A., Bergantini L., et al. Peripheral lymphocyte subset monitoring in COVID19 patients: a prospective Italian real-life case series. Minerva Med. 2020 [Google Scholar]
- 5.Kirtipal N., Bharadwaj S., Kang S.G. From SARS to SARS-CoV-2, insights on structure, pathogenicity and immunity aspects of pandemic human coronaviruses. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect Dis. 2020;13:104502. doi: 10.1016/j.meegid.2020.104502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: the anger of inflammation. Cytokine. 2020;1(133):155151. doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lake M.A. What we know so far: COVID-19 current clinical knowledge and research. Clin. Med. Lond. Engl. 2020;20(2):124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.F. Wang, H. Hou, Y. Luo, G. Tang, S. Wu, M. Huang, et al., The laboratory tests and host immunity of COVID-19 patients with different severity of illness, JCI Insight [Internet], 5(10) (2020). May 21 (cited 2020 Jun 8). Available from: https://insight.jci.org/articles/view/137799. [DOI] [PMC free article] [PubMed]
- 9.Wan S., Yi Q., Fan S., Lv J., Zhang X., Guo L., et al. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID-19) infected patients. Br. J. Haematol. 2020 doi: 10.1111/bjh.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.E.J. Giamarellos-Bourboulis, M.G. Netea, N. Rovina, K. Akinosoglou, A. Antoniadou, N. Antonakos, et al., Complex immune dysregulation in COVID-19 patients with severe respiratory failure, Cell Host Microbe (Internet) (2020) (cited 2020 Apr 22). Available from: https://linkinghub.elsevier.com/retrieve/pii/S1931312820302365. [DOI] [PMC free article] [PubMed]
- 11.Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;28 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noroozi R., Branicki W., Pyrc K., Łabaj P.P., Pospiech E., Taheri M., et al. Altered cytokine levels and immune responses in patients with SARS-CoV-2 infection and related conditions. Cytokine. 2020;1(133):155143. doi: 10.1016/j.cyto.2020.155143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahn R., Schmidt T., Golestani K., Mossberg A., Gullstrand B., Bengtsson A.A., et al. Mismatch between circulating cytokines and spontaneous cytokine production by leukocytes in hyperinflammatory COVID-19. J. Leukoc. Biol. 2020;13 doi: 10.1002/JLB.5COVBCR0720-310RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka T., Kishimoto T. Targeting interleukin-6: all the way to treat autoimmune and inflammatory diseases. Int. J. Biol. Sci. 2012;8(9):1227–1236. doi: 10.7150/ijbs.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., et al. Detectable serum SARS-CoV-2 viral load (RNAaemia) is closely correlated with drastically elevated interleukin 6 (IL-6) level in critically ill COVID-19 patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H.-J. Deng, Q.-X. Long, B.-Z. Liu, J.-H. Ren, P. Liao, J.-F. Qiu, et al., Cytokine biomarkers of COVID-19 3 Jun 2020. medRxiv 2020.05.31.20118315.
- 17.D’Alessandro A., Thomas T., Dzieciatkowska M., Hill R.C., Francis R.O., Hudson K.E., et al. Serum proteomics in COVID-19 patients: altered coagulation and complement status as a function of IL-6 level. J. Proteome Res. 2020;14 doi: 10.1021/acs.jproteome.0c00365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.X. Hou, X. Zhang, X. Wu, M. Lu, D. Wang, M. Xu, et al., Serum protein profiling reveals a landscape of inflammation and immune signaling in early-stage COVID-19 infection, Mol. Cell. Proteomics MCP, 11 Aug 2020. [DOI] [PMC free article] [PubMed]
- 19.J. Meletiadis, S. Tsiodras, P. Tsirigotis, Interleukin-6 blocking vs. JAK-STAT inhibition for prevention of lung injury in patients with COVID-19, Infect. Dis. Ther., 12 Aug 2020. [DOI] [PMC free article] [PubMed]
- 20.Glady L., Lavaux T., Charchour R., Lacorte J.-M., Lessinger J.-M. Interleukin-6 chemiluminescent immunoassay on Lumipulse G600 II: analytical evaluation and comparison with three other laboratory analyzers. Clin. Chem. Lab. Med. 2020 doi: 10.1515/cclm-2019-1145. [DOI] [PubMed] [Google Scholar]
- 21.Kim M.K., Jeon J.H., Kim S.W., Moon J.S., Cho N.H., Han E., et al. The Clinical characteristics and outcomes of patients with moderate-to-severe coronavirus disease 2019 infection and diabetes in Daegu, South Korea. Diab. Metab. J. 2020 doi: 10.4093/dmj.2020.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maximus P.S., Al Achkar Z., Hamid P.F., Hasnain S.S., Peralta C.A. Adipocytokines: are they the theory of everything? Cytokine. 2020;1(133):155144. doi: 10.1016/j.cyto.2020.155144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Méry G., Epaulard O., Borel A.-L., Toussaint B., Le Gouellec A. COVID-19: underlying Adipokine storm and angiotensin 1–7 umbrella. Front. Immunol. 2020;11:1714. doi: 10.3389/fimmu.2020.01714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X., Yu M.-Q., Shen Q., Wang L.-Z., Yan R.-D., Zhang M.-Y., et al. Analysis of inflammatory parameters and disease severity for 88 hospitalized COVID-19 patients in Wuhan. China Int. J. Med. Sci. 2020;17(13):2052–2062. doi: 10.7150/ijms.47935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tholen D.W., Kroll M., Astles J.R. CLSI; Wayne, Pa., U.S.A: 2003. Evaluation of the Linearity of Quantitative Measurement Procedures a Statistical Approach; Approved Guideline. [Google Scholar]
- 26.Budd J.R. CLSI; Wayne, PA: 2013. Clinical and Laboratory Standards Institute. Measurement Procedure Comparison and Bias Estimation Using Patient Samples: Approved Guideline. [Google Scholar]
- 27.McEnroe R.J., Durham A.P., Goldford M.D., Kondratovich M.V., Lababidi S., Magari R., et al. Clinical Laboratory Standards Institute; Wayne: 2014. Evaluation of Precision of Quantitative Measurement Procedure; Approved Guideline. [Google Scholar]
- 28.Zhang C., Wu Z., Li J.-W., Zhao H., Wang G.-Q. The cytokine release syndrome (CRS) of severe COVID-19 and Interleukin-6 receptor (IL-6R) antagonist Tocilizumab may be the key to reduce the mortality. Int. J. Antimicrob. Agents. 2020;29:105954. doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pedersen S.F., Ho Y.-C. SARS-CoV-2: a storm is raging. J. Clin. Invest. 2020;13 doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagunas-Rangel F.A., Chávez-Valencia V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020;16 doi: 10.1002/jmv.25900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terpos E., Ntanasis-Stathopoulos I., Elalamy I., Kastritis E., Sergentanis T.N., Politou M., et al. Hematological findings and complications of COVID-19. Am. J. Hematol. 2020 doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou Y., Han T., Chen J., Hou C., Hua L., He S., et al. Clinical and autoimmune characteristics of severe and critical cases of COVID-19. Clin. Transl. Sci. 2020 doi: 10.1111/cts.12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang L. C-reactive protein levels in the early stage of COVID-19. Med. Mal. Infect. 2020 doi: 10.1016/j.medmal.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Z., Cai T., Fan L., Lou K., Hua X., Huang Z., et al. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int. J. Infect. Dis. 2020;1(95):332–339. doi: 10.1016/j.ijid.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.d’Alessandro M., Bergantini L., Cameli P., Vietri L., Lanzarone N., Alonzi V., et al. Krebs von den Lungen-6 as a biomarker for disease severity assessment in interstitial lung disease: a comprehensive review. Biomark Med. 2020;14(8):665–674. doi: 10.2217/bmm-2019-0545. [DOI] [PubMed] [Google Scholar]
- 36.d’Alessandro M., Carleo A., Cameli P., Bergantini L., Perrone A., Vietri L., et al. BAL biomarkers’ panel for differential diagnosis of interstitial lung diseases. Clin. Exp. Med. 2020;20(2):207–216. doi: 10.1007/s10238-020-00608-5. [DOI] [PubMed] [Google Scholar]
- 37.d’Alessandro M., Cameli P., Refini R.M., Bergantini L., Alonzi V., Lanzarone N., et al. Serum KL-6 concentrations as a novel biomarker of severe COVID19. J. Med. Virol. 2020;29 doi: 10.1002/jmv.26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Japan ECMsOnet for COVID-19 Nationwide system to centralize decisions around ECMO use for severe COVID-19 pneumonia in Japan (Special Correspondence) J. Intensive Care. 2020;8:29. doi: 10.1186/s40560-020-00445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.H. Nakamura, K. Miyagi, M. Otsuki, Y. Higure, N. Nishiyama, T. Kinjo, et al., Serum KL‐6 can distinguish between different phenotypes of severe COVID‐19, J. Med. Virol. (Internet), 2020 (cited 2020 Aug 17). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7361808/. [DOI] [PMC free article] [PubMed]
- 40.d’Alessandro M., Cameli P., Bergantini L., Franchi F., Scolletta S., Bargagli E. Serum concentrations of Krebs von den Lungen-6 in different COVID-19 phenotypes. J. Med. Virol. 2020;14 doi: 10.1002/jmv.26431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue M., Zheng P., Bian X., Huang Z., Huang H., Zeng Y., et al. Exploration and correlation analysis of changes in Krebs von den Lungen-6 levels in COVID-19 patients with different types in China. Biosci. Trends. 2020 doi: 10.5582/bst.2020.03197. [DOI] [PubMed] [Google Scholar]
- 42.Tian R., Wu W., Wang C., Pang H., Zhang Z., Xu H., et al. Clinical characteristics and survival analysis in critical and non-critical patients with COVID-19 in Wuhan, China: a single-center retrospective case control study. Sci. Rep. 2020;10(1):17524. doi: 10.1038/s41598-020-74465-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rampotas A., Pavord S. Platelet aggregates, a marker of severe COVID-19 disease. J Clin Pathol. 2020;16 doi: 10.1136/jclinpath-2020-206933. [DOI] [PubMed] [Google Scholar]
- 44.Daga S. Employing a systematic approach to biobanking and analyzing clinical and genetic data for advancing COVID-19 research. Eur. J. Hum. Genet. 2021 doi: 10.1038/s41431-020-00793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.d’Alessandro M. Serial KL-6 measurements in COVID-19 patients. Int. Emerg Med. 2021 doi: 10.1007/s11739-020-02614-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benetti E. Clinical and molecular characterization of COVID-19 hospitalized patients. PLoS One. 2020 doi: 10.1371/journal.pone.0242534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.d’Alessandro M. Serum KL-6 levels in pulmonary Langerhans’ cell histiocytosis. Eur. J. Clin. Invest. 2020 doi: 10.1111/eci.13242. [DOI] [PubMed] [Google Scholar]
- 48.Bergantini I. Serial KL-6 analysis in patients with idiopathic pulmonary fibrosis treated with nintedanib. Respir. Investig. 2019 doi: 10.1016/j.resinv.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Landi C. A system biology study of BALF from patients affected by idiopathic pulmonary fibrosis (IPF) and healthy controls. Proteomics Clin. Appl. 2014 doi: 10.1002/prca.201400001. [DOI] [PubMed] [Google Scholar]
- 50.Bargagli E. Serum analysis of coagulation factors in IPF and NSIP. Inflammation. 2014 doi: 10.1007/s10753-013-9706-z. [DOI] [PubMed] [Google Scholar]
- 51.Vietri I. Serum amyloid A: A potential biomarker of lung disorders. Respir. Invest. 2020 doi: 10.1016/j.resinv.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 52.Castelli V., et al. Cytokine Storm in COVID-19: “When You Come Out of the Storm, You Won’t Be the Same Person Who Walked in”. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.02132. [DOI] [PMC free article] [PubMed] [Google Scholar]