Abstract
Monogenic diabetes refers to diabetes mellitus (DM) caused by a mutation in a single gene and accounts for approximately 1%–5% of diabetes. Correct diagnosis is clinically critical for certain types of monogenic diabetes, since the appropriate treatment is determined by the etiology of the disease (e.g., oral sulfonylurea treatment of HNF1A/HNF4A-diabetes vs. insulin injections in type 1 diabetes). However, achieving a correct diagnosis requires genetic testing, and the overlapping of the clinical features of monogenic diabetes with those of type 1 and type 2 diabetes has frequently led to misdiagnosis. Improvements in sequencing technology are increasing opportunities to diagnose monogenic diabetes, but challenges remain. In this Review, we describe the types of monogenic diabetes, including common and uncommon types of maturity-onset diabetes of the young, multiple causes of neonatal DM, and syndromic diabetes such as Wolfram syndrome and lipodystrophy. We also review methods of prioritizing patients undergoing genetic testing, and highlight existing challenges facing sequence data interpretation that can be addressed by forming collaborations of expertise and by pooling cases.
Introduction
Monogenic diabetes is caused by a single defect in one of over 40 genes (1, 2). Since maturity-onset diabetes of the young (MODY) was named by Fajans for the type 2 diabetes–like presentation in young people with an autosomal dominant pattern of inheritance (3, 4), our understanding of phenotypic and genetic heterogeneity in monogenic diabetes has increased. The major monogenic diabetes categories are MODY, neonatal diabetes mellitus (NDM), and syndromic diabetes (5). Misdiagnosis is frequent because of overlapping of phenotypes with type 1 diabetes (T1D), such as young onset and leanness, and with type 2 diabetes (T2D), such as preserved β cell function and family history. Tailored treatment of some monogenic diabetes depends on the disease’s underlying etiology — e.g., oral sulfonylurea treatment of HNF1A/HNF4A-MODY — and requires genetic testing to diagnose. Here we will describe monogenic diabetes types, etiologies, diagnosis, management, and strategies to improve diagnosis.
Monogenic versus polygenic diabetes
Monogenic and polygenic diabetes are traditionally considered distinct, with monogenic diabetes resulting from one highly penetrant variant in one gene in a given individual and polygenic diabetes resulting from the contribution of several variants with smaller effects in the context of environmental/lifestyle factors. In T1D, autoimmune dysfunction is the prominent mechanism, with variation in the major histocompatibility locus and other genomic factors combining with apparent environmental triggers to result in β cell loss and diabetes. In monogenic diabetes, highly penetrant variants, mostly causing extremely impaired β cell development and insulin secretion, cause diabetes regardless of other risk factors. T2D, sometimes considered a diagnosis of exclusion, is a heterogeneous group of disorders involving smaller genetic effects on multiple mechanisms, including insulin secretion and insulin sensitivity, combining with environmental and lifestyle factors, mostly impacting insulin sensitivity. While this distinction is important both scientifically and clinically, emerging studies of the genetic architecture of diabetes reveal more of a spectrum with respect to the penetrance of genetic variants and their relative role in diabetes. For example, the HNF4A variant p.R114W, found in 0.02% of non-Finnish Europeans, has been shown to be overrepresented in patients with MODY (OR = 30.4 vs. public variant databases) but to have a distinct clinical phenotype (including lack of sulfonylurea response) and much lower penetrance than other HNF4A MODY mutations (54% vs. 71% by age 30; ref. 6). In Mexican Americans, HNF1A variant p.E508K (NM_000545.8, rs483353044) was associated with T2D with a much greater effect size (OR = 4.2) than most polygenic T2D variants, with diabetic carriers and noncarriers having similar onset age and BMI (7). T2D polygenic risk scores have also shown evidence of modifying age at diagnosis of monogenic diabetes (8). Finally, while lack of features of either autoimmunity or obesity/metabolic syndrome raises the likelihood of monogenic diabetes, these features can coexist with monogenic diabetes — particularly obesity, given its high prevalence especially in youth. In the Treatment Options for Diabetes in Adolescents and Youth (TODAY) clinical trial, in which overweight or obesity was required for enrollment of newly diagnosed youth with T2D, at least 4.5% were identified as having MODY. Those with HNF4A-MODY had poor response to metformin, representing a previously missed opportunity for optimal treatment (9). In summary, monogenic and polygenic forms of diabetes exist along more of a continuum than previously appreciated. Therefore, knowledge about monogenic diabetes not only provides opportunities for etiology-based treatment of the minority of individuals with highly penetrant variants, but also informs broader understanding of diabetes etiology.
Types of monogenic diabetes
Maturity-onset diabetes of the young
MODY comprises most monogenic diabetes cases, with classical characteristics of young diagnosis age, family history of diabetes in an autosomal dominant pattern of transmission, and insulin independence, with some types having additional features (Table 1). While 14 genes have now been designated as MODY genes in OMIM and/or the literature, three of these (BLK, PAX4, and KLF11) have been proposed for elimination based on a recent study (10) (see Table 1 for the remaining 11 along with RFX6, recently proposed as an additional MODY gene; ref. 11). Variants in GCK, HNF1A, and HNF4A are responsible for most MODY cases, followed by HNF1B (12). Given the known genetic etiology of most MODY cases and the increased frequency of pediatric T2D due to increased childhood overweight and obesity prevalence, it has been suggested that this term be abandoned in favor of terms describing the etiology of the type of diabetes, such as transcription factor diabetes for MODY caused by mutations in the transcription factor genes HNF1A, HNF4A, HNF1B, and others (13). Moreover, it can be argued that any diabetes designation is unsuitable for the usually benign condition of heterozygous GCK deficiency, which is characterized by only mildly elevated glucose levels often not reaching the diabetic range and, more to the point, generally does not lead to diabetic micro- and macrovascular complications (14).
Table 1. Genetic causes of maturity-onset diabetes of the young.
Common types of MODY-classified monogenic diabetes
HNF1A-MODY and HNF4A-MODY are caused by variants in genes encoding HNF1 homeobox A and hepatic nuclear factor 4α, respectively. These transcription factors play essential roles in transcription of genes related to β cell development and insulin secretion. HNF1A variants decrease expression of HNF1A target genes (15). Among patients diagnosed with diabetes, HNF1A-MODY is the most common MODY. To date, over 400 HNF1A variants and 100 HNF4A variants have been discovered from MODY families (16).
HNF1A/HNF4A-MODY is usually diagnosed in adolescence or early adulthood. Compared with T2D, HNF1A-MODY and HNF4A-MODY occur at younger ages with lower BMI, lower hemoglobin A1c and triglycerides, and similar risk for microvascular complications. Approximately 50% of patients with HNF4A-MODY are macrosomic, which is attributed paradoxically to transient neonatal hyperinsulinemic hypoglycemia at birth (17). Hyperinsulinemic hypoglycemia was also recently observed in some patients with HNF1A-MODY (18).
Individuals with HNF1A- and HNF4A-diabetes have increased sensitivity to sulfonylureas, an insulin-stimulating class of drug (19, 20), such that low doses are effective, and more typical T2D doses cause hypoglycemia. Sulfonylureas bind to the subunit of the KATP channel to depolarize the β cell and release insulin. In a randomized clinical trial, low doses of sulfonylureas (e.g., 20–40 mg gliclazide daily) produced better glucose control than metformin in HNF1A- and HNF4A-MODY (21). In an observational study, most patients with presumed T1D who were subsequently found to have HNF1A-diabetes gained glycemic control when treatment was changed from insulin to sulfonylureas (19). Glucagon-like peptide-1 receptor agonist monotherapy (22) or sulfonylurea in combination with dipeptidyl peptidase-4 inhibitor (23) was recently demonstrated to achieve good glycemic control in HNF1A-MODY with reduced or no hypoglycemia, suggesting possible utility as a first-line HNF1A/HNF4A-diabetes treatment.
GCK encodes glucokinase, an enzyme catalyzing glucose phosphorylation at glycolysis initiation. GCK is a pancreatic β cell glucose sensor; genetic defects change the glucose-stimulated insulin secretion threshold (24, 25). In the United Kingdom, the prevalence of GCK-hyperglycemia was estimated at 0.1% among White Europeans (26) — higher than that of HNF1A-diabetes because the lack of symptoms keeps many cases from coming to medical attention. Further studies are needed in other populations. GCK-hyperglycemia has limited phenotypic heterogeneity; most patients have lifelong mild, persistent, and asymptomatic fasting hyperglycemia within the prediabetes range (27), with hemoglobin A1c values not exceeding 7.5% (60 mmol/mol; ref. 28), though some have glucose levels that meet the diabetes mellitus (DM) criteria, and a few have T2D and related complications, likely due to additional genetic and environmental risk factors (29, 30). Glucose levels are resistant to lowering by insulin or oral agents. Moreover, since GCK-hyperglycemia does not appear to be associated with significant microvascular and macrovascular diabetes complications (14, 31), patients with GCK-hyperglycemia usually do not require glucose-lowering medication, except possibly during pregnancy. Maternal GCK mutations increase risk for macrosomia and associated perinatal complications similarly to gestational or pre-gestational diabetes of any type owing to the excess insulin secretion in response to a hyperglycemic intrauterine environment. Fetal GCK mutations decrease birthweight as a result of poor insulin response. A paternally inherited fetal mutation places the fetus at risk for low birthweight in a normoglycemic intrauterine environment. A maternal mutation creates a hyperglycemic intrauterine environment for fetal insulin secretion needed for normal growth of a GCK-deficient fetus, and thus attempts at normalizing maternal glucose may result in harm. In pregnant women with GCK mutations, it is recommended that, at minimum, fetal growth be monitored by serial ultrasound to guide treatment, but it is ideal to know the fetal mutation status early in pregnancy (32). A noninvasive technique for determining fetal GCK mutation status from cell-free DNA in maternal circulation is being developed that will enable women with a mutation-positive fetus to be discharged from high-risk antenatal care (33).
HNF1B variants are estimated to account for less than 1% of MODY (34). Patients with HNF1B defects may exhibit early-onset DM only; diabetes with renal, pancreas, or liver phenotypes (renal cysts and diabetes [RCAD] syndrome); or other features with or without diabetes, such as neurodevelopmental disorders (35, 36) and hypomagnesemia. HNF1B genotype-phenotype correlation is currently unclear, with clinical heterogeneity even among family members with the same variant. However, renal outcome as measured by estimated glomerular filtration rate has been reported to be better in deletion versus nondeletion variants (36, 37); this is hypothesized to result from a dominant-negative effect (38, 39). Some HNF1B-MODY initially responds to sulfonylurea or repaglinide (36) but may ultimately require insulin.
Rare MODY-classified monogenic diabetes types
ATP-sensitive potassium channel (KATP channel) diabetes.
Pathogenic variants in ABCC8 and KCNJ11, the genes encoding sulfonylurea receptor 1 (SUR1) and the inward rectifier potassium channel 11 (Kir6.2), subunits of the ATP-sensitive potassium channel (KATP channel) found in β cells (Figure 1), are common causes of NDM (either permanent or transient; see below) but also can occasionally cause diabetes with later childhood or young adult onset (sometimes referred to as MODY12 [ref. 40] and MODY13 [ref. 41], respectively). KATP diabetes is discussed further in the NDM section below.
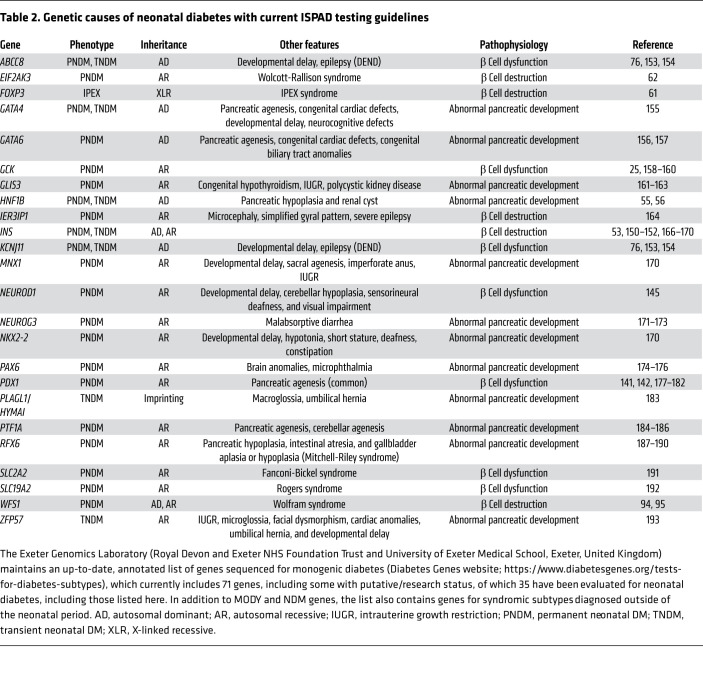
Figure 1. Schematic representation of glucose-induced insulin secretion and MODY-associated genes.
The pancreatic KATP channel directly regulates insulin secretion. It is a hetero-octamer formed by four subunits of the inward rectifier potassium channel 11 (Kir6.2, encoded by KCNJ11) and four sulfonylurea receptor 1 (SUR1, encoded by ABCC8) subunits. Glucose enters the β cell and glucokinase phosphorylates glucose to glucose-6-phosphate, which further breaks down in the glycolysis and citric acid cycle to produce ATP. The increased ATP/MgATP ratio leads to the closure of the KATP channel and causes depolarization of the β cell membrane and subsequent activation of voltage-gated calcium channels. Calcium flows into the cell through activated voltage-gated calcium channel and triggers the insulin to be released from the β cell. Transcription factors (HNF1A, HNF4A, HNF1B, NEUROD1, PDX1, and RFX6) constitute a network that regulates the expression of insulin and β cell development and proliferation. The MODY-associated genes are labeled in red.
The prominent and rarer types of MODY and their genetic and clinical features are summarized in Table 1. Emerging findings obtained through next-generation sequencing (NGS) to identify new causes of MODY have suggested potential roles of APPL1 (42) and PCBD1 (43) in MODY.
Neonatal DM
NDM is defined as diabetes diagnosed within the first 6 months of age and can be either permanent (PNDM) or transient (TNDM). Clinical features of NDM also include intrauterine growth retardation, failure to thrive, polyuria, and severe dehydration (44, 45). Depending on the genetic etiology, some patients can also have birth defects and neurological disorders (46). It affects 1 in 90,000 to 260,000 live births (47, 48), 50% being PNDM and 50% being TNDM (44).
The diabetes phenotype in TNDM results from inadequate insulin production presenting at the first week of life and resolving by 18 months (44), but 50% of patients relapse during early adulthood (46). Approximately 60%–70% of TNDM is caused by overexpression of paternally expressed imprinted genes on chromosome 6q24 (hereafter referred to as 6q24-TNDM) resulting from paternally inherited duplications or paternal disomy for the region or chromosome (both copies inherited from the father; ref. 49). The remaining cases mostly result from mutations in KATP channels, KCNJ11 (50) and ABCC8 (51), which tend to be functionally less severe than those causing PNDM (52). There are also rare occurrences attributed to mutations in INS (encoding the insulin precursor molecule preproinsulin; refs. 51, 53, 54), HNF1B (55, 56), and other genes (Table 2). It remains undetermined why only some TNDM patients relapse later, but theories about β cell function and the development of insulin resistance at puberty represent possible explanations (57).
Table 2. Genetic causes of neonatal diabetes with current ISPAD testing guidelines.
Some of the same genes implicated in TNDM, including ABCC8, KCNJ11, and INS, also have variants that more commonly cause PNDM. Homozygous or compound heterozygous inactivating GCK mutations cause PNDM (ref. 25 and Table 2). PNDM can also be part of IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked) syndrome, caused by variants in the FOXP3 gene; Wolcott-Rallison syndrome, caused by variants in EIF2AK3; and others (58). Unlike more common types of PNDM, the PNDM of IPEX syndrome (and a few other rare types of monogenic diabetes; refs. 59, 60) is autoimmune, as FOXP3 is crucial in maintaining regulatory T cells’ normal function of inhibiting proliferation and cytokine production of other T cells (61). PNDM in Wolcott-Rallison syndrome is possibly due to increased pancreatic β cell apoptosis that is regulated by EIF2AK3 (62). Genes implicated in NDM and associated phenotypes are listed in Table 2. Since a genetic cause has been identified in only 82% of patients with NDM (63), the search continues through exome sequencing, most recently implicating YIPF5 in autosomal recessive neonatal diabetes and microcephaly (64).
It is recommended that patients diagnosed with diabetes in the first 6 months of life obtain immediate genetic testing to identify the subtype, since T1D is extremely rare in this subgroup. Approximately 80%–85% of NDM cases have an identifiable genetic cause (63), half of these being KATP-diabetes caused by KCNJ11 or ABCC8 mutations, treatable with high-dose sulfonylureas rather than insulin (50, 65, 66). The benefit of identifying patients with KATP-diabetes is thus considerable, and many studies have attempted to establish genotype-phenotype correlation (67) to facilitate the prediction of patients’ clinical courses based on genetic data.
NDM caused by pathogenic variants in KATP channels.
Activating KATP channel gene variants cause NDM by decreasing ATP’s ability to achieve channel closure in multiple ways (68). Whether variants will cause PNDM or TNDM (or, rarely, MODY) is determined in part by the functional severity of the mutation as well as which gene is involved, with KCNJ11 variants mainly associated with PNDM and most ABCC8 variants linked to TNDM (51, 69). Diabetes severity could be partially explained by the extent to which the variant impacts ATP sensitivity (70); however, the same variant in one family could cause both NDM and MODY in different patients (e.g., the KCNJ11 C42R variant; ref. 71), suggesting that other mechanisms influence the development of clinical presentation. Loss-of-function (LOF) mutations in both genes cause an increase in insulin secretion and present as congenital hyperinsulinemic hypoglycemia when found in the homozygous or compound heterozygous state (72, 73) and when dominant LOF mutations are found in the heterozygous state. In addition, paternally inherited recessive LOF mutations in combination with somatic loss of maternal 11p15.5 chromosomal region cause focal hyperinsulinism (74).
KATP-NDM is autosomal-dominantly inherited but often (60%–84%) arises de novo (75, 76). Some individuals with NDM have neurological features in addition to DM (77), as KATP channels are expressed in other tissues, including muscle and brain. Common KATP-NDM features include muscle weakness, developmental delay, and early-onset epilepsy (DEND syndrome), while those with intermediate DEND (iDEND) syndrome do not have epilepsy (78). Treatment of KATP-NDM caused by either KCNJ11 or ABCC8 variants with high-dose sulfonylureas has proven safe and effective for both short-term and long-term glycemic control and may resolve CNS features (79–82). Ninety percent of KATP-NDM patients could switch from insulin therapy to sulfonylurea successfully (79, 83), with mutation severity (77, 84, 85) and diabetes duration before the transition (86) predicting the likelihood of success. For patients who cannot completely transfer to sulfonylurea, combining insulin and sulfonylurea has shown favorable results (87).
6q24-TNDM.
Although patients with 6q24-TNDM always present with growth retardation and hyperglycemia during the neonatal period, different etiologies, including paternal uniparental disomy, partial duplication of paternal origin, or a methylation defect of maternal origin on 6q24, all lead to the overexpression of PLAG1 and HYMAI, encoding a zinc finger protein (ZFP) and long noncoding RNA, respectively. In other cases, ZFP57 variants cause hypomethylation of multiple imprinted loci, including at the 6q24 locus. The treatment for the first onset of diabetes is insulin, and many are treated with insulin during remission, while some are successfully treated with sulfonylureas or a combination of sulfonylureas and insulin (88, 89). Compared with KATP-TNDM patients, patients with 6q24-TNDM were observed to have lower birthweight and earlier presentation (90). Some patients with 6q24-TNDM may also experience hyperinsulinemic hypoglycemia following diabetes remission (91). Certain congenital abnormalities, such as macroglossia, are characteristic of 6q24-TNDM and thus could help to distinguish this type of TNDM from other types in considering testing strategies.
Syndromic diabetes
In addition to RCAD syndrome due to HNF1B variants as described above, other monogenic syndromes include DM as one of the clinical features. We describe the best-characterized of these syndromes below.
Wolfram syndrome.
Two types of Wolfram syndrome (WS) corresponding to two causative genes have been identified to date. Wolfram syndrome 1 (WS1), characterized by diabetes insipidus, DM, optic atrophy, and deafness, is a rare autosomal recessive disease caused by variants in wolframin ER transmembrane glycoprotein (WFS1). Severe cases with dominant heterozygous variants are also reported (92). Often, patients’ first manifestation is DM at an average age of 6 years. Though most WS1 patients require daily insulin as therapy, the high morbidity and mortality rates as well as low average age of death make an accurate and timely diagnosis essential. Recently, a presentation similar to WS1 in many WFS1 mutation–negative patients was linked to variants in CDGSH iron sulfur domain 2 (CISD2) and thus named Wolfram syndrome 2 (WS2). Clinical features of WS2 resemble those of WS1 but without diabetes insipidus and with the addition of peptic ulcer bleeding and defective platelet aggregation (93). In addition, there are some WFS1 mutations that cause isolated diabetes with significantly reduced penetrance or nonpenetrance for other WS-related features (94, 95).
Insulin resistance due to insulin receptor defects.
Genetic defects in the insulin receptor gene (INSR) result in several insulin resistance syndromes, which are distinguished from typical insulin resistance not only by their severity but by normal lipid profiles because the etiology is directly due to defects in insulin receptor signaling rather than obesity and its sequelae (96). The most common type is type A insulin resistance syndrome, which has autosomal dominant and autosomal recessive forms. Type A insulin resistance syndrome affects predominantly nonobese females and presents with extreme insulin resistance, acanthosis nigricans, hirsutism, and polycystic ovarian disease (97, 98). Rabson-Mendenhall syndrome (RMS) is an intermediate form of insulin resistance with autosomal recessive inheritance. Patients with RMS have clinical features of extreme insulin resistance, acanthosis nigricans, hirsutism, dental precocity, thick nails, pineal hyperplasia, genital enlargement in both males and females, abdominal distension, and other distinctive dysmorphic features (99, 100). The most severe form is Donohue syndrome (DS), an autosomal recessive disorder in which patients present with failure to thrive, severe hyperinsulinemia, and fasting hypoglycemia. Patients with DS seldom survive infancy (101). LOF variants in the fibronectin type III (FnIII) domain are proposed to be associated with more severe DS, and there are genotype-phenotype and structure-phenotype correlations of INSR variants (102).
Lipodystrophy.
Monogenic lipodystrophy is a group of diseases featuring a complete or partial lack of adipose tissue and adipose tissue–derived hormones, which results in insulin resistance and other metabolic complications. Unlike insulin receptor defects, the lack of adipose tissue in lipodystrophy leads to dyslipidemia and insulin resistance due to spillover of fat into ectopic areas, paradoxically similar to the consequences of obesity (96). Based on the loss of adipose tissue, this disease can be divided into congenital generalized lipodystrophy (CGL) and familial partial lipodystrophy (FPLD). CGL is an autosomal recessive disease; pathogenic variants in genes encoding 1-acylglycerol-3-phosphate O-acyltransferase 2 (AGPAT2) and Berardinelli-Seip congenital lipodystrophy 2 (BSCL2) account for most CGL cases, with rare cases being caused by pathogenic variants in CAV1 and PTRF. CGL patients show common features, such as generalized lipodystrophy, muscular appearance, DM, and dyslipidemia; however, patients with pathogenic BSCL2 variants display lower serum leptin levels than patients with pathogenic AGPAT2 variants (103) but a higher rate of developing intellectual disability (104). The majority of FPLD cases are caused by pathogenic variants in lamin A/C (LMNA) or PPARγ (PPARG), and there are also other rarer forms caused by pathogenic variants in PLIN1, AKT2, LIPE, CIDEC, and PCYT1A. Body fat deficiency in FPLD is found on limbs, buttocks, and hips. Patients with pathogenic variants of either LMNA or PPARG appear to benefit similarly from leptin replacement therapy with metreleptin (105) in terms of improved glycemia and cardiometabolic outcomes.
Mitochondrial diabetes.
Mitochondrial diabetes, also known as maternally inherited diabetes and deafness (MIDD), is caused by pathogenic variants in mitochondrial DNA, mostly tRNA variant m.3243A>G. Patients often present with diabetes in adulthood, but a greater proportion of mutated mitochondrial genomes in the affected tissues is associated with a younger age of diagnosis of diabetes in some studies (106). The hearing loss associated with m.3243A>G is bilateral, sensorineural, and progressive, typically preceding the diagnosis of diabetes (107, 108). Other clinical features such as macular pattern dystrophy, nephropathy, and neurological symptoms are more common in rarer forms of mitochondrial diabetes than the classical form (109). The penetrance of mitochondrial diabetes is estimated to be nearly 100% by the age of 70 years. The disease etiology determined that patients have impaired insulin secretion, and insulin treatment is eventually required for most patients. The effects of other treatments, such as coenzyme Q10 and PPARγ agonists, were only evaluated in single cases, thus requiring caution for application. To better screen patients suspected to have mitochondrial diabetes, clinical features including diabetes and hearing loss on the maternal side are key. Tian et al. established a mitochondrial diabetes score system with good performance (100% sensitivity, 69.9% specificity) to select patients diagnosed with T2D for genetic testing in a Chinese cohort (110), although this system needs validation in other populations.
Challenges in identifying and diagnosing monogenic diabetes
The broad application of personalized medicine to patients with monogenic diabetes faces challenges in two aspects: detecting patients suspected of having monogenic diabetes to pursue etiology-based therapies and accurately interpreting sequence variants of monogenic diabetes genes.
Monogenic diabetes detection methods
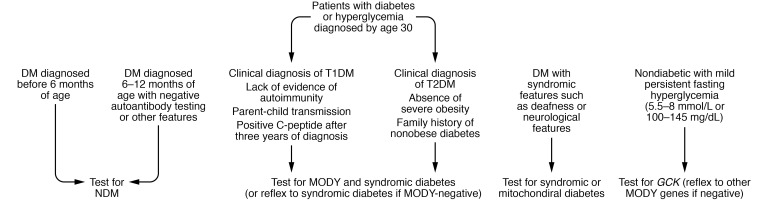
At present, practical guidelines for systematic screening for monogenic diabetes have been limited. The International Society for Pediatric and Adolescent Diabetes (ISPAD) has recommended testing for NDM in all patients diagnosed with diabetes before the age of 6 months as well as in patients diagnosed with diabetes before the age of 12 months with negative islet antibodies. This recommendation not only has the potential to dramatically improve care at the individual level when KATP-diabetes is diagnosed but has been shown to be cost-effective in this population (111). However, adult and pediatric T1D and T2D populations, which also include misdiagnosed patients with monogenic diabetes (112), are more challenging to screen routinely for MODY (111) and can be challenging especially for clinicians with limited experience diagnosing MODY. More complex screening criteria based on age of onset, family history, endogenous insulin secretion, nonobesity, and absence of pancreatic autoantibodies are needed to achieve cost-effectiveness and an ideal balance of sensitivity and specificity (113–115). The American Diabetes Association (ADA) recommends some scenarios for considering testing individuals who do not fit into the T1D or T2D classifications (2). A proposed algorithm to increase sensitivity is shown in Figure 2. Clinicians are referred to the primary source (116) as well as current ADA (2) and ISPAD guidelines (1) for further guidance; additional development is needed and is ongoing in this area.
Figure 2. Proposed diagnostic algorithm for monogenic diabetes.
Though the majority of patients diagnosed between 6 and 12 months have T1D, NDM can exist in these patients; genetic testing should be considered if they test negative for autoantibody, have extrapancreatic features, or have unusual family history (1, 2). High prevalence of MODY was observed in C-peptide–positive T2D diagnosed before 30 years regardless of metabolic syndrome status (116).
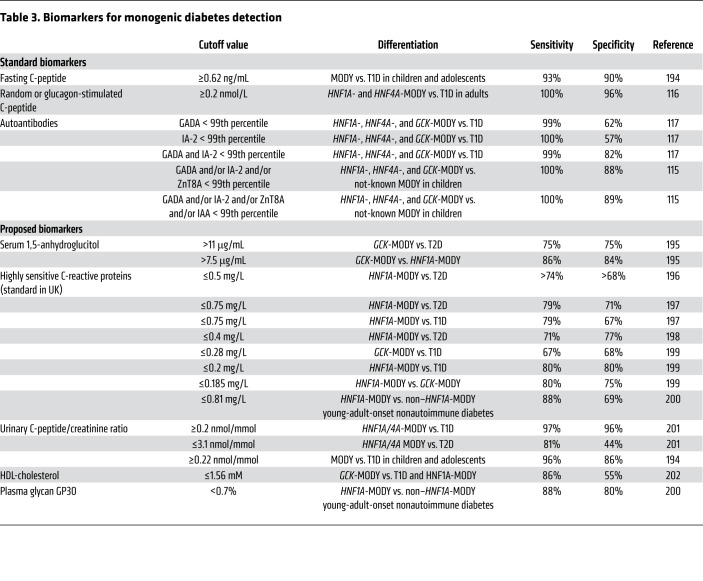
Biomarkers or derived scores avoid reliance on clinical judgments and arbitrary cutoffs and establish a quantitative evaluation that could be validated and replicated across cohorts. The Swedish Better Diabetes Diagnosis (BDD) study showed that absence of glutamic acid decarboxylase (GAD), islet antigen-2, zinc transporter 8 antibodies, and insulin autoantibodies could be a good discriminator, since in this study, MODY patients were only identified from the antibody-negative group, and 15% of antibody-negative patients had MODY (115). However, other studies have shown that 1%–2% of patients diagnosed with MODY are GAD antibody positive (117), reducing the antibody’s sensitivity as a screen. Meanwhile, the types of autoantibodies tested on each patient may vary depending on the clinic; thus, using negative antibodies as a screening method may not be practical without standardization. Table 3 summarizes published biomarkers other than pancreatic antibodies that have been utilized to distinguish monogenic diabetes subtypes from T1D or T2D. Limited by the low prevalence of monogenic diabetes, these biomarkers were developed in selected populations to differentiate the most common types of MODY.
Table 3. Biomarkers for monogenic diabetes detection.
In addition to biomarkers, Shields et al. established a MODY calculator predicting the possibility of testing positive for MODY given a set of common clinical criteria (118). In the initial cohort of White European patients who were diagnosed before the age of 35, the cutoff of probability at 40% yields sensitivity of 96% and specificity of 91% in differentiating MODY from T2D, and yields 87% sensitivity and 88% specificity for MODY versus T1D. Validations in other cohorts with different ethnic backgrounds show variable outcomes, suggesting room for improvement, including the need for a more ethnically diverse reference database.
Selection of method and genes for testing
Previously, molecular diagnosis of monogenic diabetes was usually performed through Sanger sequencing of one or several common-cause genes based on clinical suspicion. With the development of NGS, all known monogenic diabetes genes or even a patient’s whole exome can be analyzed simultaneously. Targeted panels typically include all the MODY genes, or at least the most common ones, as well as the NDM and syndromic forms of diabetes genes. There are both advantages and disadvantages to using NGS gene panels. The low price of massively parallel sequencing enables the analysis of additional genes that were reported to be associated with syndromic forms of diabetes. This is useful because patients with syndromic forms of diabetes may lack or appear to lack the clinical features that would lead to testing of a single syndromic gene (119). However, it is important that diagnostic panels not include genes with weak or disputed associations with monogenic diabetes, or, if they are included for surveillance purposes, that they not be reported (120). The yields of these panels will not only facilitate molecular diagnosis but also add rare or novel variants to the knowledge base for future studies. Sanger confirmation is sometimes needed after variant discovery in NGS panels, though increasingly less so except in difficult regions of the genome. Regardless of methodology, it is becoming increasingly clear that evaluating only exonic regions will overlook some causal variants, as variants in the noncoding regulatory and deep intronic regions and 5′- and 3′-UTRs have also been implicated in monogenic diabetes (16, 121).
Searching for monogenic diabetes using exome or genome sequencing enables novel gene discovery and also requires caution. The coverage of exome sequencing may not be complete, leading to the risk of false negatives (122, 123). In addition, as exome or genome sequencing could discover variants that are potentially important to health or reproduction but are unrelated to the clinical indication, the reporting of such secondary findings must be addressed, with consideration of the recommendations of organizations such as the American College of Medical Genetics and Genomics (ACMG; ref. 124). With these caveats, this approach can serve as a powerful tool for searching for candidate genes in patients with monogenic diabetes for whom variants in known genes have not been found (125).
Variant classification
Key to diagnosing monogenic diabetes and other genetic conditions is not only identifying the variant but also distinguishing disease-causing variants from normal variation. Previous approaches to determine whether a variant identified in a patient was disease-causing involved sequencing a group of matched controls (usually 100–200 people) to assess the variant’s presence in the general population. This approach was limited because the sample size was too small to rule out population prevalence being too high for the disease; e.g., HNF1A-diabetes has an estimated population prevalence of 1 in 10,000. Moreover, the extent to which rare but benign genetic variation existed in the population studied was not known and was thus probably underestimated. As NGS has begun to boom, the problem of large quantities of genetic data for interpretation has arisen for genetic diseases in general. The genetic and phenotypic heterogeneity of monogenic diabetes, and its overlapping features with T1D and T2D, together increase the difficulty of interpreting the pathogenicity of variants found in patients suspected to have monogenic diabetes. On the other hand, NGS emergence has led to the availability of exome and genome sequences of over 100,000 individuals of diverse ancestries in the gnomAD database, dramatically improving the ability to assess variant frequency in the general population. Additional resources have emerged, including computational predictive tools (126–128), and other sources of data, including phenotype specificity, familial segregation, and functional studies, are also used. However, there is subjectivity in assigning pathogenicity to variants, and in the early 2010s, a lack of consistency of variant interpretation across laboratories became apparent.
In 2015, the ACMG and the Association for Molecular Pathology (AMP) jointly published a consensus recommendation on standards and guidelines for clinical genomic variant interpretation (129). The guidelines were developed through data sharing by a large number of American Board of Medical Genetics and Genomics–certified clinical molecular geneticists and pathologists from Clinical Laboratory Improvement Amendment/College of American Pathologists–accredited laboratories. The recommendations suggested that variants could be assigned to a five-tier system of classification: (a) pathogenic, (b) likely pathogenic, (c) uncertain significance, (d) likely benign, or (e) benign. The proposed sets of criteria include population data, computational and predictive data, clinical data, functional data (in vitro studies), and pedigree segregation. Each criterion is weighted by different levels of strength based on observed evidence and combined with other collected criteria to reach a conclusion. Since the publication of the initial ACMG/AMP guidelines, additional refinements have been published to improve rigor, including recommendations for evaluating the strength of evidence for LOF (130), standards for assessing functional studies (131), and application of a Bayesian quantitative point system (132).
Value of establishing gene-specific rules
The aim of the ACMG/AMP guidelines is to provide a universal set of criteria for interpreting variants for Mendelian disease. Additionally, each gene-disease pair requires further specification to reflect the specific disease frequency, clinical features, and genotype-phenotype relationships. In 2013, the Clinical Genome (ClinGen) Resource was founded by the National Human Genome Research Institute to serve as a knowledge base that defines gene-disease relationships, curate variants of genetic disease using a standardized approach, and distribute information about the variants to researchers and clinicians. Since then, dozens of expert panels and working groups have been formed to examine specific gene or disease groups for determining clinical significance and constructing gene-specific standardizations. The Monogenic Diabetes Expert Panel (MDEP), established in 2017, brings together experts and data to adapt the ACMG/AMP variant interpretation guidelines for monogenic diabetes genes and classify variants using these gene-specific rules, thereby improving the accuracy of variant classification in these genes and in turn improving the ability to accurately diagnose monogenic diabetes (133).
Value of data sharing
The establishing of guidelines is fundamental to standardized and concordant interpretation of monogenic diabetes gene variants. This process calls for expertise in endocrinology, molecular genetic testing, genetic counseling, and biochemistry. To reach the full potential of precision medicine in monogenic diabetes, centralization of case-level data is important. For instance, when the variant being evaluated is not observed in the general population but is observed in affected individuals, a higher number of occurrences leads to a higher level of evidence supporting pathogenicity. However, the uncommonness of monogenic diabetes often makes it difficult for individual laboratories to acquire enough cases. By pooling case data, expert panels can achieve levels of case-based evidence for pathogenicity not possible for any single laboratory or clinic.
Value of functional evidence
Well-established functional studies on variants boost the understanding of disease mechanisms and provide evidence supporting or disputing the pathogenicity of the variants. Studies have shown that functional analyses clarify variant interpretation in HNF1A-MODY variants, especially when family segregation data or phenotype data are not available (134). Caution is needed in using these data, because not all functional assays reflect the disease mechanism and not all variants impact the function in the same way. Full inspection of the consequences of a variant may require multiple assays to reach a conclusion (135). Systematic validation and statistical quantification of the level of strength of pathogenicity or benignity in functional assays are recommended (131). This approach encourages high-throughput mutation screenings, such as saturation mutagenesis (136) and systematic functional profiling of variants identified in the population (137, 138), which consist of pathogenic and benign variants. The MDEP is currently developing standards for evaluating evidence from luciferase assays for transactivation, which assess transcriptional activity of HNF1A and HNF4A variants, along with assays of DNA binding activity and protein expression (138, 139). For GCK variants, similar work is focused on the relative activity index of glucokinase as a measure of enzyme kinetic characteristics (140). In the longer term, multiplexed assays of variant effect (MAVEs) could provide comprehensive catalogs of allelic effects that can be interrogated to aid variant interpretation. This approach is particularly well suited for transcription factors such as HNF1A. It is important to note that functional evidence does not single-handedly implicate a variant in disease; the functional data must be evaluated in concert with the population and clinical data to make a pathogenicity determination.
Conclusion
Accurate genetic diagnosis of monogenic diabetes is crucial for patients, since it helps optimize treatment, especially for some patients switching from insulin or metformin to low-dose sulfonylureas (HNF1A-MODY and HNF4A-MODY) or no treatment (GCK-MODY) or from insulin to high-dose sulfonylureas (KATP-diabetes). Additionally, accurate monogenic diabetes diagnosis leads to better familial risk management and clinical course prediction. Advancement in genetic testing technology has increased the capacity of genetic diagnosis while decreasing sequencing cost. However, until we can offer genetic testing to every patient with diabetes, prioritizing patients with high suspicion of monogenic diabetes through assessment of their biomarker profiles or probability score is more practical. Monogenic diabetes provides an example of translating research findings into clinical practice that improves diagnosis and quality of life. Multidisciplinary expert collaboration and case sharing combined with incorporation of basic science into sequence variant interpretation will lead to improved diagnosis. Establishing clear guidelines for evaluating the causality of individual variants by this process is essential for widespread diagnosis of monogenic diabetes; more broadly, routine incorporation of emerging genomic data into the care of diabetes and disease in general is needed to realize the full potential of personalized and precision medicine. And as we celebrate the 100th anniversary of insulin’s discovery, it seems fitting to now celebrate and disseminate our more recently discovered ability to identify individuals who can make their own insulin once they have received the appropriate genomic diagnosis and treatment.
Acknowledgments
This work was supported by NIH grants U24-HD093486 and U01-HG007775 (to TIP). ALG is a Wellcome Senior Fellow in Basic Biomedical Science and is supported by the Wellcome Trust (095101, 200837) and the NIH (U01-DK105535, U01-DK085545, P30-DK116074).
Version 1. 02/01/2021
Electronic publication
Footnotes
Conflict of interest: ALG’s spouse is an employee of Genentech and holds stock and stock options in Roche. TIP receives research and salary support from the Regeneron Genetics Center and Regeneron Pharmaceuticals.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(3):e142244.https://doi.org/10.1172/JCI142244.
Contributor Information
Haichen Zhang, Email: Haichen.Zhang@som.umaryland.edu.
Kevin Colclough, Email: kevin.colclough@nhs.net.
References
- 1.Hattersley AT, et al. ISPAD clinical practice consensus guidelines 2018: the diagnosis and management of monogenic diabetes in children and adolescents. Pediatr Diabetes. 2018;19(suppl 27):47–63. doi: 10.1111/pedi.12772. [DOI] [PubMed] [Google Scholar]
- 2.American Diabetes Association. 2. Classification and diagnosis of diabetes: Standards of Medical Care in Diabetes—2020. Diabetes Care. 2020;43(suppl 1):S14–S31. doi: 10.2337/dc20-S002. [DOI] [PubMed] [Google Scholar]
- 3.Tattersall RB. Mild familial diabetes with dominant inheritance. Q J Med. 1974;43(170):339–357. [PubMed] [Google Scholar]
- 4.Fajans SS, et al. The Banting Memorial Lecture 1978. Clinical and etiologic heterogeneity of idiopathic diabetes mellitus. Diabetes. 1978;27(11):1112–1125. doi: 10.2337/diab.27.11.1112. [DOI] [PubMed] [Google Scholar]
- 5. Weiss RE, Refetoff S, eds. Genetic Diagnosis of Endocrine Disorders. Academic Press; 2016. [Google Scholar]
- 6.Laver TW, et al. The common p.R114W HNF4A mutation causes a distinct clinical subtype of monogenic diabetes. Diabetes. 2016;65(10):3212–3217. doi: 10.2337/db16-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sigma Type 2 Diabetes Consortium. et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311(22):2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lango Allen H, et al. Polygenic risk variants for type 2 diabetes susceptibility modify age at diagnosis in monogenic HNF1A diabetes. Diabetes. 2010;59(1):266–271. doi: 10.2337/db09-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinberger JW, et al. Monogenic diabetes in overweight and obese youth diagnosed with type 2 diabetes: the TODAY clinical trial. Genet Med. 2018;20(6):583–590. doi: 10.1038/gim.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. doi: 10.1111/dme.2_13570. Laver T, et al. Redefining the pathogenicity of Maturity Onset Diabetes of the Young (MODY) genes: BLK, PAX4 and KLF11 do not cause MODY. Paper presented at: Diabetes UK Professional Conference 2018; March 14–16, 2018; London ExCeL, London, UK. Accessed Jan 8, 2020. [DOI]
- 11.Patel KA, et al. Heterozygous RFX6 protein truncating variants are associated with MODY with reduced penetrance. Nat Commun. 2017;8(1):888. doi: 10.1038/s41467-017-00895-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thanabalasingham G, Owen KR. Diagnosis and management of maturity onset diabetes of the young (MODY) BMJ. 2011;343:d6044. doi: 10.1136/bmj.d6044. [DOI] [PubMed] [Google Scholar]
- 13.Murphy R, et al. Clinical implications of a molecular genetic classification of monogenic beta-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4(4):200–213. doi: 10.1038/ncpendmet0778. [DOI] [PubMed] [Google Scholar]
- 14.Steele AM, et al. Prevalence of vascular complications among patients with glucokinase mutations and prolonged, mild hyperglycemia. JAMA. 2014;311(3):279–286. doi: 10.1001/jama.2013.283980. [DOI] [PubMed] [Google Scholar]
- 15.Haliyur R, et al. Human islets expressing HNF1A variant have defective β cell transcriptional regulatory networks. J Clin Invest. 2019;129(1):246–251. doi: 10.1172/JCI121994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colclough K, et al. Mutations in the genes encoding the transcription factors hepatocyte nuclear factor 1α and 4α in maturity-onset diabetes of the young and hyperinsulinemic hypoglycemia. Hum Mutat. 2013;34(5):669–685. doi: 10.1002/humu.22279. [DOI] [PubMed] [Google Scholar]
- 17.Pearson ER, et al. Macrosomia and hyperinsulinaemic hypoglycaemia in patients with heterozygous mutations in the HNF4A gene. PLoS Med. 2007;4(4):e118. doi: 10.1371/journal.pmed.0040118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yau D, et al. Congenital hyperinsulinism due to mutations in HNF1A. Eur J Med Genet. 2020;63(6):103928. doi: 10.1016/j.ejmg.2020.103928. [DOI] [PubMed] [Google Scholar]
- 19.Shepherd M, et al. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med. 2009;26(4):437–441. doi: 10.1111/j.1464-5491.2009.02690.x. [DOI] [PubMed] [Google Scholar]
- 20.Pearson ER, et al. Sensitivity to sulphonylureas in patients with hepatocyte nuclear factor-1alpha gene mutations: evidence for pharmacogenetics in diabetes. Diabet Med. 2000;17(7):543–545. doi: 10.1046/j.1464-5491.2000.00305.x. [DOI] [PubMed] [Google Scholar]
- 21.Pearson ER, et al. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362(9392):1275–1281. doi: 10.1016/S0140-6736(03)14571-0. [DOI] [PubMed] [Google Scholar]
- 22.Fantasia KL, Steenkamp DW. Optimal glycemic control in a patient with HNF1A MODY with GLP-1 RA monotherapy: implications for future therapy. J Endocr Soc. 2019;3(12):2286–2289. doi: 10.1210/js.2019-00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen AS, et al. Efficacy and safety of glimepiride with or without linagliptin treatment in patients with HNF1A diabetes (maturity-onset diabetes of the young type 3): a randomized, double-blinded, placebo-controlled, crossover trial (GLIMLINA) Diabetes Care. 2020;43(9):2025–2033. doi: 10.2337/dc20-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrne MM, et al. Insulin secretory abnormalities in subjects with hyperglycemia due to glucokinase mutations. J Clin Invest. 1994;93(3):1120–1130. doi: 10.1172/JCI117064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osbak KK, et al. Update on mutations in glucokinase (GCK), which cause maturity-onset diabetes of the young, permanent neonatal diabetes, and hyperinsulinemic hypoglycemia. Hum Mutat. 2009;30(11):1512–1526. doi: 10.1002/humu.21110. [DOI] [PubMed] [Google Scholar]
- 26.Chakera AJ, et al. The 0.1% of the population with glucokinase monogenic diabetes can be recognized by clinical characteristics in pregnancy: the Atlantic Diabetes in Pregnancy cohort. Diabetes Care. 2014;37(5):1230–1236. doi: 10.2337/dc13-2248. [DOI] [PubMed] [Google Scholar]
- 27.Carmody D, et al. GCK-MODY in the US National Monogenic Diabetes Registry: frequently misdiagnosed and unnecessarily treated. Acta Diabetol. 2016;53(5):703–708. doi: 10.1007/s00592-016-0859-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stride A, et al. Cross-sectional and longitudinal studies suggest pharmacological treatment used in patients with glucokinase mutations does not alter glycaemia. Diabetologia. 2014;57(1):54–56. doi: 10.1007/s00125-013-3075-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wedrychowicz A, et al. Phenotype heterogeneity in glucokinase-maturity-onset diabetes of the young (GCK-MODY) patients. J Clin Res Pediatr Endocrinol. 2017;9(3):246–252. doi: 10.4274/jcrpe.4461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuesta-Munoz AL, et al. Clinical heterogeneity in monogenic diabetes caused by mutations in the glucokinase gene (GCK-MODY) Diabetes Care. 2010;33(2):290–292. doi: 10.2337/dc09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szopa M, et al. Prevalence of retinopathy in adult patients with GCK-MODY and HNF1A-MODY. Exp Clin Endocrinol Diabetes. 2015;123(9):524–528. doi: 10.1055/s-0035-1559605. [DOI] [PubMed] [Google Scholar]
- 32.Hattersley AT, et al. Mutations in the glucokinase gene of the fetus result in reduced birth weight. Nat Genet. 1998;19(3):268–270. doi: 10.1038/953. [DOI] [PubMed] [Google Scholar]
- 33.Caswell RC, et al. Noninvasive fetal genotyping by droplet digital PCR to identify maternally inherited monogenic diabetes variants. Clin Chem. 2020;66(7):958–965. doi: 10.1093/clinchem/hvaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edghill EL, et al. HNF1B deletions in patients with young-onset diabetes but no known renal disease. Diabet Med. 2013;30(1):114–117. doi: 10.1111/j.1464-5491.2012.03709.x. [DOI] [PubMed] [Google Scholar]
- 35.Dubois-Laforgue D, et al. Intellectual disability in patients with MODY due to hepatocyte nuclear factor 1B (HNF1B) molecular defects. Diabetes Metab. 2017;43(1):89–92. doi: 10.1016/j.diabet.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Dubois-Laforgue D, et al. Diabetes, associated clinical spectrum, long-term prognosis, and genotype/phenotype correlations in 201 adult patients with hepatocyte nuclear factor 1B (HNF1B) molecular defects. Diabetes Care. 2017;40(11):1436–1443. doi: 10.2337/dc16-2462. [DOI] [PubMed] [Google Scholar]
- 37.Clissold RL, et al. Chromosome 17q12 microdeletions but not intragenic HNF1B mutations link developmental kidney disease and psychiatric disorder. Kidney Int. 2016;90(1):203–211. doi: 10.1016/j.kint.2016.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomura H, et al. Loss-of-function and dominant-negative mechanisms associated with hepatocyte nuclear factor-1beta mutations in familial type 2 diabetes mellitus. J Biol Chem. 1999;274(19):12975–12978. doi: 10.1074/jbc.274.19.12975. [DOI] [PubMed] [Google Scholar]
- 39.Lindner TH, et al. A novel syndrome of diabetes mellitus, renal dysfunction and genital malformation associated with a partial deletion of the pseudo-POU domain of hepatocyte nuclear factor-1beta. Hum Mol Genet. 1999;8(11):2001–2008. doi: 10.1093/hmg/8.11.2001. [DOI] [PubMed] [Google Scholar]
- 40.Bowman P, et al. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia. 2012;55(1):123–127. doi: 10.1007/s00125-011-2319-x. [DOI] [PubMed] [Google Scholar]
- 41.Bonnefond A, et al. Whole-exome sequencing and high throughput genotyping identified KCNJ11 as the thirteenth MODY gene. PLoS One. 2012;7(6):e37423. doi: 10.1371/journal.pone.0037423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prudente S, et al. Loss-of-function mutations in APPL1 in familial diabetes mellitus. Am J Hum Genet. 2015;97(1):177–185. doi: 10.1016/j.ajhg.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simaite D, et al. Recessive mutations in PCBD1 cause a new type of early-onset diabetes. Diabetes. 2014;63(10):3557–3564. doi: 10.2337/db13-1784. [DOI] [PubMed] [Google Scholar]
- 44.Polak M, Cave H. Neonatal diabetes mellitus: a disease linked to multiple mechanisms. Orphanet J Rare Dis. 2007;2:12. doi: 10.1186/1750-1172-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aguilar-Bryan L, Bryan J. Neonatal diabetes mellitus. Endocr Rev. 2008;29(3):265–291. doi: 10.1210/er.2007-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Busiah K, et al. Neuropsychological dysfunction and developmental defects associated with genetic changes in infants with neonatal diabetes mellitus: a prospective cohort study [corrected] Lancet Diabetes Endocrinol. 2013;1(3):199–207. doi: 10.1016/S2213-8587(13)70059-7. [DOI] [PubMed] [Google Scholar]
- 47.Iafusco D, et al. Minimal incidence of neonatal/infancy onset diabetes in Italy is 1:90,000 live births. Acta Diabetol. 2012;49(5):405–408. doi: 10.1007/s00592-011-0331-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Slingerland AS, et al. Referral rates for diagnostic testing support an incidence of permanent neonatal diabetes in three European countries of at least 1 in 260,000 live births. Diabetologia. 2009;52(8):1683–1685. doi: 10.1007/s00125-009-1416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Temple IK, Shield JP. 6q24 transient neonatal diabetes. Rev Endocr Metab Disord. 2010;11(3):199–204. doi: 10.1007/s11154-010-9150-4. [DOI] [PubMed] [Google Scholar]
- 50.Gloyn AL, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. doi: 10.1056/NEJMoa032922. [DOI] [PubMed] [Google Scholar]
- 51.Babenko AP, et al. Activating mutations in the ABCC8 gene in neonatal diabetes mellitus. N Engl J Med. 2006;355(5):456–466. doi: 10.1056/NEJMoa055068. [DOI] [PubMed] [Google Scholar]
- 52.Gloyn AL, et al. Relapsing diabetes can result from moderately activating mutations in KCNJ11. Hum Mol Genet. 2005;14(7):925–934. doi: 10.1093/hmg/ddi086. [DOI] [PubMed] [Google Scholar]
- 53.Garin I, et al. Recessive mutations in the INS gene result in neonatal diabetes through reduced insulin biosynthesis. Proc Natl Acad Sci U S A. 2010;107(7):3105–3110. doi: 10.1073/pnas.0910533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnefond A, et al. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. J Biol Chem. 2011;286(32):28414–28424. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edghill EL, et al. Hepatocyte nuclear factor-1β mutations cause neonatal diabetes and intrauterine growth retardation: support for a critical role of HNF-1β in human pancreatic development. Diabet Med. 2006;23(12):1301–1306. doi: 10.1111/j.1464-5491.2006.01999.x. [DOI] [PubMed] [Google Scholar]
- 56.Yorifuji T, et al. Neonatal diabetes mellitus and neonatal polycystic, dysplastic kidneys: phenotypically discordant recurrence of a mutation in the hepatocyte nuclear factor-1β gene due to germline mosaicism. J Clin Endocrinol Metab. 2004;89(6):2905–2908. doi: 10.1210/jc.2003-031828. [DOI] [PubMed] [Google Scholar]
- 57.Kelsey MM, Zeitler PS. Insulin resistance of puberty. Curr Diab Rep. 2016;16(7):64. doi: 10.1007/s11892-016-0751-5. [DOI] [PubMed] [Google Scholar]
- 58.Rubio-Cabezas O, et al. Permanent neonatal diabetes mellitus—the importance of diabetes differential diagnosis in neonates and infants. Eur J Clin Invest. 2011;41(3):323–333. doi: 10.1111/j.1365-2362.2010.02409.x. [DOI] [PubMed] [Google Scholar]
- 59.Johnson MB, et al. Monogenic autoimmune diseases of the endocrine system. Lancet Diabetes Endocrinol. 2016;4(10):862–872. doi: 10.1016/S2213-8587(16)30095-X. [DOI] [PubMed] [Google Scholar]
- 60.Johnson MB, et al. A type 1 diabetes genetic risk score can discriminate monogenic autoimmunity with diabetes from early-onset clustering of polygenic autoimmunity with diabetes. Diabetologia. 2018;61(4):862–869. doi: 10.1007/s00125-018-4551-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bacchetta R, et al. From IPEX syndrome to FOXP3 mutation: a lesson on immune dysregulation. Ann N Y Acad Sci. 2018;1417(1):5–22. doi: 10.1111/nyas.13011. [DOI] [PubMed] [Google Scholar]
- 62.Rubio-Cabezas O, et al. Wolcott-Rallison syndrome is the most common genetic cause of permanent neonatal diabetes in consanguineous families. J Clin Endocrinol Metab. 2009;94(11):4162–4170. doi: 10.1210/jc.2009-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Franco E, et al. The effect of early, comprehensive genomic testing on clinical care in neonatal diabetes: an international cohort study. Lancet. 2015;386(9997):957–963. doi: 10.1016/S0140-6736(15)60098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De Franco E, et al. YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J Clin Invest. 2020;130(12):6338–6353. doi: 10.1172/JCI141455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vaxillaire M, et al. Kir6.2 mutations are a common cause of permanent neonatal diabetes in a large cohort of French patients. Diabetes. 2004;53(10):2719–2722. doi: 10.2337/diabetes.53.10.2719. [DOI] [PubMed] [Google Scholar]
- 66.Flanagan SE, et al. Mutations in KCNJ11, which encodes Kir6.2, are a common cause of diabetes diagnosed in the first 6 months of life, with the phenotype determined by genotype. Diabetologia. 2006;49(6):1190–1197. doi: 10.1007/s00125-006-0246-z. [DOI] [PubMed] [Google Scholar]
- 67.Pipatpolkai T, et al. New insights into KATP channel gene mutations and neonatal diabetes mellitus. Nat Rev Endocrinol. 2020;16(7):378–393. doi: 10.1038/s41574-020-0351-y. [DOI] [PubMed] [Google Scholar]
- 68.Ashcroft FM, et al. Neonatal diabetes and the KATP channel: from mutation to therapy. Trends Endocrinol Metab. 2017;28(5):377–387. doi: 10.1016/j.tem.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patch AM, et al. Mutations in the ABCC8 gene encoding the SUR1 subunit of the KATP channel cause transient neonatal diabetes, permanent neonatal diabetes or permanent diabetes diagnosed outside the neonatal period. Diabetes Obes Metab. 2007;9(suppl 2):28–39. doi: 10.1111/j.1463-1326.2007.00772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lang V, Light PE. The molecular mechanisms and pharmacotherapy of ATP-sensitive potassium channel gene mutations underlying neonatal diabetes. Pharmgenomics Pers Med. 2010;3:145–161. doi: 10.2147/PGPM.S6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yorifuji T, et al. The C42R mutation in the Kir6.2 (KCNJ11) gene as a cause of transient neonatal diabetes, childhood diabetes, or later-onset, apparently type 2 diabetes mellitus. J Clin Endocrinol Metab. 2005;90(6):3174–3178. doi: 10.1210/jc.2005-0096. [DOI] [PubMed] [Google Scholar]
- 72.Aguilar-Bryan L, et al. Cloning of the beta cell high-affinity sulfonylurea receptor: a regulator of insulin secretion. Science. 1995;268(5209):423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 73.Thomas P, et al. Mutation of the pancreatic islet inward rectifier Kir6.2 also leads to familial persistent hyperinsulinemic hypoglycemia of infancy. Hum Mol Genet. 1996;5(11):1809–1812. doi: 10.1093/hmg/5.11.1809. [DOI] [PubMed] [Google Scholar]
- 74.Glaser B, et al. Hyperinsulinism caused by paternal-specific inheritance of a recessive mutation in the sulfonylurea-receptor gene. Diabetes. 1999;48(8):1652–1657. doi: 10.2337/diabetes.48.8.1652. [DOI] [PubMed] [Google Scholar]
- 75.Slingerland AS, Hattersley AT. Mutations in the Kir6.2 subunit of the KATP channel and permanent neonatal diabetes: new insights and new treatment. Ann Med. 2005;37(3):186–195. doi: 10.1080/07853890510007287. [DOI] [PubMed] [Google Scholar]
- 76.Flanagan SE, et al. Update of mutations in the genes encoding the pancreatic beta-cell K(ATP) channel subunits Kir6.2 (KCNJ11) and sulfonylurea receptor 1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2009;30(2):170–180. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 77.Proks P, et al. Molecular basis of Kir6.2 mutations associated with neonatal diabetes or neonatal diabetes plus neurological features. Proc Natl Acad Sci U S A. 2004;101(50):17539–17544. doi: 10.1073/pnas.0404756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hattersley AT, Ashcroft FM. Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new scientific insights, and new therapy. Diabetes. 2005;54(9):2503–2513. doi: 10.2337/diabetes.54.9.2503. [DOI] [PubMed] [Google Scholar]
- 79.Pearson ER, et al. Switching from insulin to oral sulfonylureas in patients with diabetes due to Kir6.2 mutations. N Engl J Med. 2006;355(5):467–477. doi: 10.1056/NEJMoa061759. [DOI] [PubMed] [Google Scholar]
- 80.Bowman P, et al. Effectiveness and safety of long-term treatment with sulfonylureas in patients with neonatal diabetes due to KCNJ11 mutations: an international cohort study. Lancet Diabetes Endocrinol. 2018;6(8):637–646. doi: 10.1016/S2213-8587(18)30106-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mlynarski W, et al. Sulfonylurea improves CNS function in a case of intermediate DEND syndrome caused by a mutation in KCNJ11. Nat Clin Pract Neurol. 2007;3(11):640–645. doi: 10.1038/ncpneuro0640. [DOI] [PubMed] [Google Scholar]
- 82.Slingerland AS, et al. Sulphonylurea therapy improves cognition in a patient with the V59M KCNJ11 mutation. Diabet Med. 2008;25(3):277–281. doi: 10.1111/j.1464-5491.2007.02373.x. [DOI] [PubMed] [Google Scholar]
- 83.Rafiq M, et al. Effective treatment with oral sulfonylureas in patients with diabetes due to sulfonylurea receptor 1 (SUR1) mutations. Diabetes Care. 2008;31(2):204–209. doi: 10.2337/dc07-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Masia R, et al. An ATP-binding mutation (G334D) in KCNJ11 is associated with a sulfonylurea-insensitive form of developmental delay, epilepsy, and neonatal diabetes. Diabetes. 2007;56(2):328–336. doi: 10.2337/db06-1275. [DOI] [PubMed] [Google Scholar]
- 85.Sumnik Z, et al. Sulphonylurea treatment does not improve psychomotor development in children with KCNJ11 mutations causing permanent neonatal diabetes mellitus accompanied by developmental delay and epilepsy (DEND syndrome) Diabet Med. 2007;24(10):1176–1178. doi: 10.1111/j.1464-5491.2007.02228.x. [DOI] [PubMed] [Google Scholar]
- 86.Babiker T, et al. Successful transfer to sulfonylureas in KCNJ11 neonatal diabetes is determined by the mutation and duration of diabetes. Diabetologia. 2016;59(6):1162–1166. doi: 10.1007/s00125-016-3921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Misra S, et al. Permanent neonatal diabetes: combining sulfonylureas with insulin may be an effective treatment. Diabet Med. 2018;30(2):170–180. doi: 10.1111/dme.13758. [DOI] [PubMed] [Google Scholar]
- 88.Fu JL, et al. Relapsed 6q24-related transient neonatal diabetes mellitus successfully treated with sulfonylurea. Chin Med J (Engl) 2019;132(7):846–848. doi: 10.1097/CM9.0000000000000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Carmody D, et al. Role of noninsulin therapies alone or in combination in chromosome 6q24-related transient neonatal diabetes: sulfonylurea improves but does not always normalize insulin secretion. Diabetes Care. 2015;38(6):e86–87. doi: 10.2337/dc14-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Docherty LE, et al. Clinical presentation of 6q24 transient neonatal diabetes mellitus (6q24 TNDM) and genotype-phenotype correlation in an international cohort of patients. Diabetologia. 2013;56(4):758–762. doi: 10.1007/s00125-013-2832-1. [DOI] [PubMed] [Google Scholar]
- 91.Flanagan SE, et al. Hypoglycaemia following diabetes remission in patients with 6q24 methylation defects: expanding the clinical phenotype. Diabetologia. 2013;56(1):218–221. doi: 10.1007/s00125-012-2766-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.De Franco E, et al. Dominant ER stress-inducing WFS1 mutations underlie a genetic syndrome of neonatal/infancy-onset diabetes, congenital sensorineural deafness, and congenital cataracts. Diabetes. 2017;66(7):2044–2053. doi: 10.2337/db16-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rigoli L, Di Bella C. Wolfram syndrome 1 and Wolfram syndrome 2. Curr Opin Pediatr. 2012;24(4):512–517. doi: 10.1097/MOP.0b013e328354ccdf. [DOI] [PubMed] [Google Scholar]
- 94.Bansal V, et al. Identification of a missense variant in the WFS1 gene that causes a mild form of Wolfram syndrome and is associated with risk for type 2 diabetes in Ashkenazi Jewish individuals. Diabetologia. 2018;61(10):2180–2188. doi: 10.1007/s00125-018-4690-3. [DOI] [PubMed] [Google Scholar]
- 95.Bonnycastle LL, et al. Autosomal dominant diabetes arising from a Wolfram syndrome 1 mutation. Diabetes. 2013;62(11):3943–3950. doi: 10.2337/db13-0571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Semple RK, et al. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. J Clin Invest. 2009;119(2):315–322. doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.O’Rahilly S, Moller DE. Mutant insulin receptors in syndromes of insulin resistance. Clin Endocrinol (Oxf) 1992;36(2):121–132. doi: 10.1111/j.1365-2265.1992.tb00945.x. [DOI] [PubMed] [Google Scholar]
- 98.Hegele RA. Monogenic forms of insulin resistance: apertures that expose the common metabolic syndrome. Trends Endocrinol Metab. 2003;14(8):371–377. doi: 10.1016/S1043-2760(03)00142-5. [DOI] [PubMed] [Google Scholar]
- 99.al-Gazali LI, et al. A syndrome of insulin resistance resembling leprechaunism in five sibs of consanguineous parents. J Med Genet. 1993;30(6):470–475. doi: 10.1136/jmg.30.6.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kadowaki H, et al. Four mutant alleles of the insulin receptor gene associated with genetic syndromes of extreme insulin resistance. Biochem Biophys Res Commun. 1997;237(3):516–520. doi: 10.1006/bbrc.1997.7181. [DOI] [PubMed] [Google Scholar]
- 101.Kirkwood A, et al. Donohue syndrome: a review of literature, case series, and anesthetic considerations. Paediatr Anaesth. 2018;28(1):23–27. doi: 10.1111/pan.13273. [DOI] [PubMed] [Google Scholar]
- 102.Hosoe J, et al. Structural basis and genotype-phenotype correlations of INSR mutations causing severe insulin resistance. Diabetes. 2017;66(10):2713–2723. doi: 10.2337/db17-0301. [DOI] [PubMed] [Google Scholar]
- 103.Agarwal AK, et al. Phenotypic and genetic heterogeneity in congenital generalized lipodystrophy. J Clin Endocrinol Metab. 2003;88(10):4840–4847. doi: 10.1210/jc.2003-030855. [DOI] [PubMed] [Google Scholar]
- 104.Van Maldergem L, et al. Genotype-phenotype relationships in Berardinelli-Seip congenital lipodystrophy. J Med Genet. 2002;39(10):722–733. doi: 10.1136/jmg.39.10.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sekizkardes H, et al. Efficacy of metreleptin treatment in familial partial lipodystrophy due to PPARG vs LMNA pathogenic variants. J Clin Endocrinol Metab. 2019;104(8):3068–3076. doi: 10.1210/jc.2018-02787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ohkubo K, et al. Mitochondrial gene mutations in the tRNA(Leu(UUR)) region and diabetes: prevalence and clinical phenotypes in Japan. Clin Chem. 2001;47(9):1641–1648. doi: 10.1093/clinchem/47.9.1641. [DOI] [PubMed] [Google Scholar]
- 107.Uimonen S, et al. Hearing impairment in patients with 3243A-->G mtDNA mutation: phenotype and rate of progression. Hum Genet. 2001;108(4):284–289. doi: 10.1007/s004390100475. [DOI] [PubMed] [Google Scholar]
- 108.Murphy R, et al. Clinical features, diagnosis and management of maternally inherited diabetes and deafness (MIDD) associated with the 3243A>G mitochondrial point mutation. Diabet Med. 2008;25(4):383–399. doi: 10.1111/j.1464-5491.2008.02359.x. [DOI] [PubMed] [Google Scholar]
- 109.Decoux-Poullot AG, et al. Clinical phenotype of mitochondrial diabetes due to rare mitochondrial DNA mutations. Ann Endocrinol (Paris) 2020;81(2-3):68–77. doi: 10.1016/j.ando.2020.04.007. [DOI] [PubMed] [Google Scholar]
- 110.Tian LH, et al. A screening approach for mitochondrial tRNALeu(UUR) A3243G mutation in a hospital-based population with diabetes. Chin Med J (Engl) 2018;131(9):1117–1119. doi: 10.4103/0366-6999.230729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Naylor R. Economics of genetic testing for diabetes. Curr Diab Rep. 2019;19(5):23. doi: 10.1007/s11892-019-1140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Todd J, et al. Monogenic diabetes in the progress for diabetes genetics in youth (ProDiGY) collaboration. Diabetes. 2018;18(8):57. doi: 10.2337/db18-268-OR. [DOI] [Google Scholar]
- 113.Naylor RN, et al. Cost-effectiveness of MODY genetic testing: translating genomic advances into practical health applications. Diabetes Care. 2014;37(1):202–209. doi: 10.2337/dc13-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Thomas ER, et al. Diagnosis of monogenic diabetes: 10-year experience in a large multi-ethnic diabetes center. J Diabetes Investig. 2016;7(3):332–337. doi: 10.1111/jdi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carlsson A, et al. Absence of islet autoantibodies and modestly raised glucose values at diabetes diagnosis should lead to testing for MODY: lessons from a 5-year pediatric swedish national cohort study. Diabetes Care. 2020;43(1):82–89. doi: 10.2337/dc19-0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thanabalasingham G, et al. Systematic assessment of etiology in adults with a clinical diagnosis of young-onset type 2 diabetes is a successful strategy for identifying maturity-onset diabetes of the young. Diabetes Care. 2012;35(6):1206–1212. doi: 10.2337/dc11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.McDonald TJ, et al. Islet autoantibodies can discriminate maturity-onset diabetes of the young (MODY) from Type 1 diabetes. Diabet Med. 2011;28(9):1028–1033. doi: 10.1111/j.1464-5491.2011.03287.x. [DOI] [PubMed] [Google Scholar]
- 118.Shields BM, et al. The development and validation of a clinical prediction model to determine the probability of MODY in patients with young-onset diabetes. Diabetologia. 2012;55(5):1265–1272. doi: 10.1007/s00125-011-2418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ellard S, et al. Improved genetic testing for monogenic diabetes using targeted next-generation sequencing. Diabetologia. 2013;56(9):1958–1963. doi: 10.1007/s00125-013-2962-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ellard S, et al. Prediction algorithms: pitfalls in interpreting genetic variants of autosomal dominant monogenic diabetes. J Clin Invest. 2020;130(1):14–16. doi: 10.1172/JCI133516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Carmody D, et al. Continued lessons from the INS gene: an intronic mutation causing diabetes through a novel mechanism. J Med Genet. 2015;52(9):612–616. doi: 10.1136/jmedgenet-2015-103220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kwak SH, et al. Clinical whole exome sequencing in early onset diabetes patients. Diabetes Res Clin Pract. 2016;122:71–77. doi: 10.1016/j.diabres.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 123.Johansson S, et al. Exome sequencing and genetic testing for MODY. PLoS One. 2012;7(5):e38050. doi: 10.1371/journal.pone.0038050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kalia SS, et al. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 125.Shim YJ, et al. Identification of candidate gene variants in Korean MODY Families by whole-exome sequencing. Horm Res Paediatr. 2015;83(4):242–251. doi: 10.1159/000368657. [DOI] [PubMed] [Google Scholar]
- 126.Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31(13):3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ioannidis NM, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99(4):877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Abou Tayoun AN, et al. Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat. 2018;39(11):1517–1524. doi: 10.1002/humu.23626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brnich SE, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med. 2019;12(1):3. doi: 10.1186/s13073-019-0690-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tavtigian SV, et al. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum Mutat. doi: 10.1002/humu.24088. doi: 10.1002/humu.24088. [published online July 26, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rivera-Muñoz EA, et al. ClinGen Variant Curation Expert Panel experiences and standardized processes for disease and gene-level specification of the ACMG/AMP guidelines for sequence variant interpretation. Hum Mutat. 2018;39(11):1614–1622. doi: 10.1002/humu.23645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Malikova J, et al. Functional analyses of HNF1A-MODY variants refine the interpretation of identified sequence variants. J Clin Endocrinol Metab. 2020;105(4):dgaa051. doi: 10.1210/clinem/dgaa051. [DOI] [PubMed] [Google Scholar]
- 135.Raimondo A, et al. Phenotypic severity of homozygous GCK mutations causing neonatal or childhood-onset diabetes is primarily mediated through effects on protein stability. Hum Mol Genet. 2014;23(24):6432–6440. doi: 10.1093/hmg/ddu360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Majithia AR, et al. Prospective functional classification of all possible missense variants in PPARG. Nat Genet. 2016;48(12):1570–1575. doi: 10.1038/ng.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Thormaehlen AS, et al. Systematic cell-based phenotyping of missense alleles empowers rare variant association studies: a case for LDLR and myocardial infarction. PLoS Genet. 2015;11(2):e1004855. doi: 10.1371/journal.pgen.1004855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Althari S, et al. Unsupervised clustering of missense variants in HNF1A using multidimensional functional data aids clinical interpretation. Am J Hum Genet. 2020;107(4):670–682. doi: 10.1016/j.ajhg.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Najmi LA, et al. Functional investigations of HNF1A identify rare variants as risk factors for type 2 diabetes in the general population. Diabetes. 2017;66(2):335–346. doi: 10.2337/db16-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Gloyn AL, et al. Glucokinase and the regulation of blood sugar. In: Matschinsky FM, Magnuson MA, eds. Glucokinase and Glycemic Disease: From Basics to Novel Therapeutics. Karger; 2004:92–109. [Google Scholar]
- 141.Stoffers DA, et al. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat Genet. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 142.Fajans SS, et al. Obesity and hyperinsulinemia in a family with pancreatic agenesis and MODY caused by the IPF1 mutation Pro63fsX60. Transl Res. 2010;156(1):7–14. doi: 10.1016/j.trsl.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Caetano LA, et al. PDX1 -MODY and dorsal pancreatic agenesis: new phenotype of a rare disease. Clin Genet. 2018;93(2):382–386. doi: 10.1111/cge.13044. [DOI] [PubMed] [Google Scholar]
- 144.Yu H, et al. Identification of a novel mutation site in maturity‑onset diabetes of the young in a Chinese family by whole‑exome sequencing. Mol Med Rep. 2019;20(3):2373–2380. doi: 10.3892/mmr.2019.10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Horikawa Y, Enya M. Genetic dissection and clinical features of MODY6 (NEUROD1-MODY) Curr Diab Rep. 2019;19(3):12. doi: 10.1007/s11892-019-1130-9. [DOI] [PubMed] [Google Scholar]
- 146.Raeder H, et al. Pancreatic lipomatosis is a structural marker in nondiabetic children with mutations in carboxyl-ester lipase. Diabetes. 2007;56(2):444–449. doi: 10.2337/db06-0859. [DOI] [PubMed] [Google Scholar]
- 147.Raeder H, et al. Mutations in the CEL VNTR cause a syndrome of diabetes and pancreatic exocrine dysfunction. Nat Genet. 2006;38(1):54–62. doi: 10.1038/ng1708. [DOI] [PubMed] [Google Scholar]
- 148.Vesterhus M, et al. Pancreatic function in carboxyl-ester lipase knockout mice. Pancreatology. 2010;10(4):467–476. doi: 10.1159/000266284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Liu M, et al. INS-gene mutations: from genetics and beta cell biology to clinical disease. Mol Aspects Med. 2015;42:3–18. doi: 10.1016/j.mam.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Stoy J, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci U S A. 2007;104(38):15040–15044. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Edghill EL, et al. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes. 2008;57(4):1034–1042. doi: 10.2337/db07-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Molven A, et al. Mutations in the insulin gene can cause MODY and autoantibody-negative type 1 diabetes. Diabetes. 2008;57(4):1131–1135. doi: 10.2337/db07-1467. [DOI] [PubMed] [Google Scholar]
- 153.Gloyn AL, et al. Mutations in the genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2006;27(3):220–231. doi: 10.1002/humu.20292. [DOI] [PubMed] [Google Scholar]
- 154.De Franco E, et al. Update of variants identified in the pancreatic beta-cell KATP channel genes KCNJ11 and ABCC8 in individuals with congenital hyperinsulinism and diabetes. Hum Mutat. 2020;41(5):884–905. doi: 10.1002/humu.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Shaw-Smith C, et al. GATA4 mutations are a cause of neonatal and childhood-onset diabetes. Diabetes. 2014;63(8):2888–2894. doi: 10.2337/db14-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Allen HL, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet. 2011;44(1):20–22. doi: 10.1038/ng.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.De Franco E, et al. GATA6 mutations cause a broad phenotypic spectrum of diabetes from pancreatic agenesis to adult-onset diabetes without exocrine insufficiency. Diabetes. 2013;62(3):993–997. doi: 10.2337/db12-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Borowiec M, et al. Phenotype variability and neonatal diabetes in a large family with heterozygous mutation of the glucokinase gene. Acta Diabetol. 2011;48(3):203–208. doi: 10.1007/s00592-011-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bennett K, et al. Four novel cases of permanent neonatal diabetes mellitus caused by homozygous mutations in the glucokinase gene. Pediatr Diabetes. 2011;12(3 pt 1):192–196. doi: 10.1111/j.1399-5448.2010.00683.x. [DOI] [PubMed] [Google Scholar]
- 160.Esquiaveto-Aun AM, et al. A new compound heterozygosis for inactivating mutations in the glucokinase gene as cause of permanent neonatal diabetes mellitus (PNDM) in double-first cousins. Diabetol Metab Syndr. 2015;7:101. doi: 10.1186/s13098-015-0101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wen X, Yang Y. Emerging roles of GLIS3 in neonatal diabetes, type 1 and type 2 diabetes. J Mol Endocrinol. 2017;58(2):R73–R85. doi: 10.1530/JME-16-0232. [DOI] [PubMed] [Google Scholar]
- 162.Alghamdi KA, et al. Extended clinical features associated with novel Glis3 mutation: a case report. BMC Endocr Disord. 2017;17(1):14. doi: 10.1186/s12902-017-0160-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fu C, et al. Mutation screening of the GLIS3 gene in a cohort of 592 Chinese patients with congenital hypothyroidism. Clin Chim Acta. 2018;476:38–43. doi: 10.1016/j.cca.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 164.Poulton CJ, et al. Microcephaly with simplified gyration, epilepsy, and infantile diabetes linked to inappropriate apoptosis of neural progenitors. Am J Hum Genet. 2011;89(2):265–276. doi: 10.1016/j.ajhg.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Polak M, et al. Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early infancy: a report from the French ND (Neonatal Diabetes) Study Group. Diabetes. 2008;57(4):1115–1119. doi: 10.2337/db07-1358. [DOI] [PubMed] [Google Scholar]
- 166.Colombo C, et al. Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Invest. 2008;118(6):2148–2156. doi: 10.1172/JCI33777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bonfanti R, et al. Insulin gene mutations as cause of diabetes in children negative for five type 1 diabetes autoantibodies. Diabetes Care. 2009;32(1):123–125. doi: 10.2337/dc08-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rubio-Cabezas O, et al. Testing for monogenic diabetes among children and adolescents with antibody-negative clinically defined Type 1 diabetes. Diabet Med. 2009;26(10):1070–1074. doi: 10.1111/j.1464-5491.2009.02812.x. [DOI] [PubMed] [Google Scholar]
- 169.Porksen S, et al. Disease progression and search for monogenic diabetes among children with new onset type 1 diabetes negative for ICA, GAD- and IA-2 antibodies. BMC Endocr Disord. 2010;10:16. doi: 10.1186/1472-6823-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Flanagan SE, et al. Analysis of transcription factors key for mouse pancreatic development establishes NKX2-2 and MNX1 mutations as causes of neonatal diabetes in man. Cell Metab. 2014;19(1):146–154. doi: 10.1016/j.cmet.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rubio-Cabezas O, et al. Permanent neonatal diabetes and enteric anendocrinosis associated with biallelic mutations in NEUROG3. Diabetes. 2011;60(4):1349–1353. doi: 10.2337/db10-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pinney SE, et al. Neonatal diabetes and congenital malabsorptive diarrhea attributable to a novel mutation in the human neurogenin-3 gene coding sequence. J Clin Endocrinol Metab. 2011;96(7):1960–1965. doi: 10.1210/jc.2011-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang J, et al. Mutant neurogenin-3 in congenital malabsorptive diarrhea. N Engl J Med. 2006;355(3):270–280. doi: 10.1056/NEJMoa054288. [DOI] [PubMed] [Google Scholar]
- 174.Yasuda T, et al. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes. 2002;51(1):224–230. doi: 10.2337/diabetes.51.1.224. [DOI] [PubMed] [Google Scholar]
- 175.Wen JH, et al. Paired box 6 (PAX6) regulates glucose metabolism via proinsulin processing mediated by prohormone convertase 1/3 (PC1/3) Diabetologia. 2009;52(3):504–513. doi: 10.1007/s00125-008-1210-x. [DOI] [PubMed] [Google Scholar]
- 176.Solomon BD, et al. Compound heterozygosity for mutations in PAX6 in a patient with complex brain anomaly, neonatal diabetes mellitus, and microophthalmia. Am J Med Genet A. 2009;149A(11):2543–2546. doi: 10.1002/ajmg.a.33081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stoffers DA, et al. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat Genet. 1997;15(1):106–110. doi: 10.1038/ng0197-106. [DOI] [PubMed] [Google Scholar]
- 178.Wright NM, et al. Permanent neonatal diabetes mellitus and pancreatic exocrine insufficiency resulting from congenital pancreatic agenesis. Am J Dis Child. 1993;147(6):607–609. doi: 10.1001/archpedi.1993.02160300013005. [DOI] [PubMed] [Google Scholar]
- 179.Thomas IH, et al. Neonatal diabetes mellitus with pancreatic agenesis in an infant with homozygous IPF-1 Pro63fsX60 mutation. Pediatr Diabetes. 2009;10(7):492–496. doi: 10.1111/j.1399-5448.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schwitzgebel VM, et al. Agenesis of human pancreas due to decreased half-life of insulin promoter factor 1. J Clin Endocrinol Metab. 2003;88(9):4398–4406. doi: 10.1210/jc.2003-030046. [DOI] [PubMed] [Google Scholar]
- 181.Nicolino M, et al. A novel hypomorphic PDX1 mutation responsible for permanent neonatal diabetes with subclinical exocrine deficiency. Diabetes. 2010;59(3):733–740. doi: 10.2337/db09-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Edghill EL, et al. Sequencing PDX1 (insulin promoter factor 1) in 1788 UK individuals found 5% had a low frequency coding variant, but these variants are not associated with Type 2 diabetes. Diabet Med. 2011;28(6):681–684. doi: 10.1111/j.1464-5491.2011.03269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Temple IK, et al. Transient neonatal diabetes: widening the understanding of the etiopathogenesis of diabetes. Diabetes. 2000;49(8):1359–1366. doi: 10.2337/diabetes.49.8.1359. [DOI] [PubMed] [Google Scholar]
- 184.Sellick GS, et al. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat Genet. 2004;36(12):1301–1305. doi: 10.1038/ng1475. [DOI] [PubMed] [Google Scholar]
- 185.Hoveyda N, et al. Neonatal diabetes mellitus and cerebellar hypoplasia/agenesis: report of a new recessive syndrome. J Med Genet. 1999;36(9):700–704. [PMC free article] [PubMed] [Google Scholar]
- 186.Tutak E, et al. A Turkish newborn infant with cerebellar agenesis/neonatal diabetes mellitus and PTF1A mutation. Genet Couns. 2009;20(2):147–152. [PubMed] [Google Scholar]
- 187.Mitchell J, et al. Neonatal diabetes, with hypoplastic pancreas, intestinal atresia and gall bladder hypoplasia: search for the aetiology of a new autosomal recessive syndrome. Diabetologia. 2004;47(12):2160–2167. doi: 10.1007/s00125-004-1576-3. [DOI] [PubMed] [Google Scholar]
- 188.Chappell L, et al. A further example of a distinctive autosomal recessive syndrome comprising neonatal diabetes mellitus, intestinal atresias and gall bladder agenesis. Am J Med Genet A. 2008;146A(13):1713–1717. doi: 10.1002/ajmg.a.32304. [DOI] [PubMed] [Google Scholar]
- 189.Martinovici D, et al. Neonatal hemochromatosis and Martinez-Frias syndrome of intestinal atresia and diabetes mellitus in a consanguineous newborn. Eur J Med Genet. 2010;53(1):25–28. doi: 10.1016/j.ejmg.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 190.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463(7282):775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sansbury FH, et al. SLC2A2 mutations can cause neonatal diabetes, suggesting GLUT2 may have a role in human insulin secretion. Diabetologia. 2012;55(9):2381–2385. doi: 10.1007/s00125-012-2595-0. [DOI] [PubMed] [Google Scholar]
- 192.Shaw-Smith C, et al. Recessive SLC19A2 mutations are a cause of neonatal diabetes mellitus in thiamine-responsive megaloblastic anaemia. Pediatr Diabetes. 2012;13(4):314–321. doi: 10.1111/j.1399-5448.2012.00855.x. [DOI] [PubMed] [Google Scholar]
- 193.Touati A, et al. Transient neonatal diabetes mellitus and hypomethylation at additional imprinted loci: novel ZFP57 mutation and review on the literature. Acta Diabetol. 2019;56(3):301–307. doi: 10.1007/s00592-018-1239-3. [DOI] [PubMed] [Google Scholar]
- 194.Yilmaz Agladioglu S, et al. Urinary C-Peptide/creatinine ratio can distinguish maturity-onset diabetes of the young from type 1 diabetes in children and adolescents: a single-center experience. Horm Res Paediatr. 2015;84(1):54–61. doi: 10.1159/000375410. [DOI] [PubMed] [Google Scholar]
- 195.Pal A, et al. Evaluation of serum 1, 5 anhydroglucitol levels as a clinical test to differentiate subtypes of diabetes. Diabetes Care. 2010;33(2):252–257. doi: 10.2337/dc09-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Thanabalasingham G, et al. A large multi-centre European study validates high-sensitivity C-reactive protein (hsCRP) as a clinical biomarker for the diagnosis of diabetes subtypes. Diabetologia. 2011;54(11):2801–2810. doi: 10.1007/s00125-011-2261-y. [DOI] [PubMed] [Google Scholar]
- 197.McDonald TJ, et al. High-sensitivity CRP discriminates HNF1A-MODY from other subtypes of diabetes. Diabetes Care. 2011;34(8):1860–1862. doi: 10.2337/dc11-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Owen KR, et al. Assessment of high-sensitivity C-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1A mutations. Diabetes Care. 2010;33(9):1919–1924. doi: 10.2337/dc10-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Fu J, et al. Using clinical indices to distinguish MODY2 (GCK Mutation) and MODY3 (HNF1A Mutation) from Type 1 diabetes in a young Chinese population. Diabetes Ther. 2019;10(4):1381–1390. doi: 10.1007/s13300-019-0647-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Juszczak A, et al. Plasma fucosylated glycans and c-reactive protein as biomarkers of HNF1A-MODY in young adult-onset nonautoimmune diabetes. Diabetes Care. 2019;42(1):17–26. doi: 10.2337/dc18-0422. [DOI] [PubMed] [Google Scholar]
- 201.Besser RE, et al. Urinary C-peptide creatinine ratio is a practical outpatient tool for identifying hepatocyte nuclear factor 1-α/hepatocyte nuclear factor 4-α maturity-onset diabetes of the young from long-duration type 1 diabetes. Diabetes Care. 2011;34(2):286–291. doi: 10.2337/dc10-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fendler W, et al. HDL cholesterol as a diagnostic tool for clinical differentiation of GCK-MODY from HNF1A-MODY and type 1 diabetes in children and young adults. Clin Endocrinol (Oxf) 2011;75(3):321–327. doi: 10.1111/j.1365-2265.2011.04052.x. [DOI] [PubMed] [Google Scholar]