Abstract
Humans have been infected with Mycobacterium tuberculosis (Mtb) for thousands of years. While tuberculosis (TB), one of the deadliest infectious diseases, is caused by uncontrolled Mtb infection, over 90% of presumed infected individuals remain asymptomatic and contain Mtb in a latent TB infection (LTBI) without ever developing disease, and some may clear the infection. A small number of heavily Mtb-exposed individuals appear to resist developing traditional LTBI. Because Mtb has mechanisms for intracellular survival and immune evasion, successful control involves all of the arms of the immune system. Here, we focus on immune responses to Mtb in humans and nonhuman primates and discuss new concepts and outline major knowledge gaps in our understanding of LTBI, ranging from the earliest events of exposure and infection to success or failure of Mtb control.
Introduction
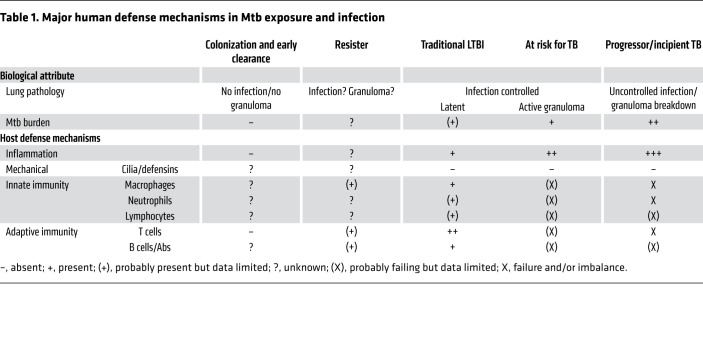
Mycobacterium tuberculosis (Mtb), a bacterium transmitted through respiratory droplets, is one of the most successful human pathogens. With approximately 10 million cases and 1.45 million associated deaths per year, tuberculosis (TB), which is caused by uncontrolled Mtb infection, is the world’s most lethal infectious disease next to COVID-19 (1). Failure of TB control programs and the lack of a highly efficacious vaccine against TB have refocused attention on the earliest events in TB pathogenesis — the acquisition and control of Mtb bacilli in the human lung. Because of its ability to infect and survive in macrophages (reviewed in ref. 2), Mtb can persist and cause, in most individuals, a clinically inapparent infection referred to as latent TB infection (LTBI) (reviewed in ref. 3). However, TB and LTBI are not binary classifications but rather terms comprising a heterogeneous spectrum (reviewed in ref. 4). Our inability to detect persistent/latent Mtb bacilli makes it impossible to determine who among those presumed infected and asymptomatic have cleared the bacilli (5), remain latently infected, or will progress to uncontrolled infection/TB (Table 1). Instead, we rely on a detectable cellular immune response to Mtb antigens in the form of a positive tuberculin skin test (TST) and/or blood-based IFN-γ release assay (IGRA) as surrogates for presumed LTBI (3, 6–8). Therefore, LTBI is an operational and not a pathogenetic definition.
Table 1. Major human defense mechanisms in Mtb exposure and infection.
Since a quarter of the world’s population is estimated to have LTBI, there is a large reservoir from which TB can emerge to fuel its worldwide pandemic (9). Understanding all of the immune components that result in LTBI or resistance to it, and in the continued control or possibly clearance of Mtb, is critical for insights into protective immunity to Mtb and for determining who is at risk of developing TB (10). Genetic studies indicate that Mtb may have coevolved with humans for more than 6000 years, which likely contributed to its success in intracellular survival and escape from innate and adaptive immune mechanisms (10–13). The bacterial pathogenesis, evolution, and strain diversity of Mtb have been extensively reviewed elsewhere (10–14). Based on human and nonhuman primate (NHP) studies, we here focus on new concepts and point out major knowledge gaps in efforts to understand the complexity of immune responses in LTBI.
Models for human LTBI
Animal models have provided insight into essential mechanisms of TB pathogenesis, but few reflect the heterogeneity of human responses to Mtb, particularly during the early events of control and containment in the airways (refs. 15, 16, and reviewed in refs. 17–19). NHPs, especially macaques, have been invaluable models for Mtb infection of the lung. They display the full spectrum of host responses and clinical manifestations that most closely resemble those in humans (reviewed in refs. 20, 21). Macaques differ in their susceptibility to Mtb — around 90% of rhesus and 60% of cynomolgus macaques develop TB after low-dose airway infection (20–22). Both macaque models are being used to study TB pathogenesis and TB vaccine responses, and provide important insights into T and B cell–mediated correlates and mechanisms of protection against Mtb and its progression to TB in the setting of immunosuppression (e.g., SIV infection) and T and B cell depletion (reviewed in refs. 17–21; refs. 23–26). The cynomolgus model, owing to its higher rate of Mtb control, is more suitable for investigation of the earliest events in the lung leading to granuloma development, and LTBI or progression to TB (15–18, 20–22). With sophisticated imaging, systems immunology, and computational modeling approaches (27), NHP models will continue to enhance our understanding of pathogenesis in human TB and LTBI.
Human granuloma models allow for analyses of early host-pathogen interactions during Mtb infection (reviewed in ref. 28). They bring together cells such as mononuclear phagocytes, lymphocytes, fibroblasts, and epithelial cells, and allow investigation of the impact of different human immune components on early granuloma formation. Mycobacterial growth inhibition assays are another tool for in vitro/ex vivo assessment of immune responses to Mtb in humans (reviewed in ref. 29). While these in vitro systems have limitations, such as short infection duration, limited cell type diversity, and inability to model kinetics of immune cell recruitment, these models likely will continue to become more sophisticated and contribute to our understanding of human granuloma formation.
Development and spectrum of LTBI
Based on animal studies, after inhalation some Mtb bacilli reach distal alveolar spaces where they are engulfed by alveolar macrophages, resident dendritic cells (DCs), and/or recruited mononuclear phagocytes (reviewed in refs. 18, 30). Infected cells travel to local lymphoid tissues (e.g., bronchus-associated lymphoid tissue or mediastinal lymph nodes) where Mtb antigens are processed and presented by DCs to initiate an adaptive immune response. In most, this results in pulmonary granuloma formation, which controls or eliminates Mtb (reviewed in refs. 4, 18). Failure of adaptive, mostly cell-mediated immune responses to control Mtb, as seen for example in newborns and advanced HIV disease, results in direct progression from infection to pulmonary or disseminated TB (reviewed in refs. 31–33).
Studies in macaques have expanded our understanding of immune mechanisms in LTBI (reviewed in refs. 17–21). These studies show that granulomas can be initiated by a single bacillus, are heterogeneous, and develop independent trajectories, with some becoming sterile, some containing small numbers of Mtb, and others progressing with necrosis and uncontrolled bacterial growth either naturally or when immune suppression is applied. NHP studies have also helped establish that controlled granulomas consist of a core of macrophages and neutrophils/polymorphonuclear cells surrounded by T and B cells expressing a balanced panel of proinflammatory (e.g., IFN-γ, IL-17, TNF-α) and antiinflammatory (e.g., IL-10, TGF-β) cytokines (reviewed in ref. 4), and that concurrent Mtb infection is protective against a secondary Mtb challenge (34). Understanding the differences between granulomas that control and those that do not control Mtb is a critical area of research.
In most individuals who are not overtly immune-compromised, adaptive immune responses control Mtb growth, primarily through T cells, which, through secretion of cytokines such as IFN-γ and TNF-α and cytolytic function, promote the ability of macrophages to control the growth of Mtb (reviewed in refs. 18, 35). The majority (about 90%) of these individuals do not progress from infection to disease (reviewed in refs. 3, 4). Evidence that they have been exposed to Mtb and are likely infected stems from their positive TST and/or IGRA, in which case they meet the criteria for having LTBI (3, 6–8). The TST is based on a delayed-type hypersensitivity response to a mixture of 100–200 denatured Mtb proteins and peptides, referred to as purified protein derivative (PPD). Because many proteins in PPD are also found in other mycobacteria, including the current TB vaccine strain M. bovis bacillus Calmette-Guérin (BCG) (36), responses to PPD may not be Mtb specific. The more Mtb-specific blood-based IGRAs measure CD4+ T cell responses to peptides from Mtb-specific proteins, such as ESAT6, CFP10, and TB10.4, which are not generated by most nontuberculous mycobacteria and BCG.
Epidemiologic and cohort studies indicate that the risk of progression from LTBI to disease is around 5%–10% and is greatest in the first 1–2 years after TST/IGRA conversion (37–40). This observation suggests that in recent TST/IGRA converters, progression from infection to disease reflects poor control of the initial Mtb infection, allowing continued slow Mtb replication until the uncontrolled infection becomes clinically apparent. In children, very high versus low IGRA responses can differentiate risk of progression to TB, but the magnitude of response is of less value in adults (41, 42) and does not reflect mycobacterial burden or state of protective immune activation in LTBI. Some individuals progress from LTBI to TB years later, but estimates of rates vary widely (reviewed in ref. 43). Epidemiologic studies on the impact of immunosuppression (e.g., HIV infection, anti-TNF therapy, and organ or bone marrow transplantation) on people with LTBI estimate that only a minority develop TB (reviewed in ref. 5). Because progression is seen in non–TB-endemic settings where the risk for Mtb reinfection is low, these data suggest that those who progressed harbored viable Mtb whereas those who did not may have cleared the bacilli. Biomarker studies are making inroads into determining who is at risk for progression from LTBI to TB (reviewed in ref. 44), but prospective validation studies are needed to determine the ability of these biomarkers to estimate Mtb exposure and infection, size of mycobacterial burden, and level of protective immunity.
While some people with heavy Mtb exposure appear to resist what we define as LTBI (reviewed in ref. 45 and discussed below), many individuals with LTBI who progress to TB do not have an obvious acquired immunodeficiency or risk factor, suggesting potential undefined genetic risk factors. Higher rates of TB in monozygotic than in dizygotic twins provided evidence for a role for human genetics (46). Furthermore, Mendelian susceptibility to mycobacterial disease (MSMD) has defined the importance of the IFN-γ/Stat1/IL-12 axis for host defenses against mycobacteria, including Mtb (47). However, genetic association studies have yet to directly link a gene, locus, or gene network with a specific mechanism to explain resistance or susceptibility to TB (reviewed in ref. 48). New data indicate genetic variations associated with TST conversion in Brazilian TB household contacts (49), but more studies focusing on earlier phases of TB pathogenesis, including susceptibility to Mtb infection and development of LTBI, are needed.
In TB-endemic settings the vast majority of people with LTBI are unable to pinpoint a recent Mtb exposure, remain well, and do not progress to TB. The term LTBI implies that small numbers of “latent” but viable Mtb bacilli are contained in granulomas and can reactivate to cause TB (reviewed in ref. 3). Because it is not possible to detect latent bacilli in vivo yet, we cannot parse individuals with LTBI into those harboring “latent” Mtb and those who may have cleared the bacilli. However, we know that for most the cellular immune response to Mtb that defines LTBI reflects control of exposure to and/or infection with Mtb (Table 1; and reviewed in ref. 33). We further know that LTBI comprises a spectrum of host immune responses, likely influencing the potential clearance or degree of persistent Mtb burden (reviewed in ref. 4). While not measuring Mtb directly, studies using PET-CT can provide insight into this spectrum of immune activation and its correlation with Mtb control or progression to TB. Improved understanding of all of the immune components that result in resistance, clearance, or maintenance of LTBI will enhance our insights into protective immunity to Mtb.
Resistance to traditional LTBI
In TB endemic settings or environments with heavy Mtb exposure (e.g., sharing a berthing compartment at sea with an individual with pulmonary TB), some people remain TST and/or IGRA negative (reviewed in ref. 45). Recent studies from Uganda, India, and Indonesia have extended these earlier observations of individuals who appear to resist the development of “traditional” LTBI despite extensive Mtb exposure (50–54). We estimate that 5%–10% of adult TB household contacts in a TB-endemic urban environment such as Kampala, Uganda remain TST/IGRA negative and clinically well after prolonged follow-up (53). Furthermore, approximately 10% of South African miners, who may have the highest Mtb exposure in the world, remain TST negative after years of working in the mines (55).
The lack of a traditional LTBI response in heavily Mtb-exposed people raises several interesting immunopathogenesis questions. Do individuals who resist Mtb infection (resisters) have a unique respiratory mucosal immune response that clears Mtb from airways before it reaches the alveolus? Are innate or trained macrophages able to control Mtb without help from T cells? Do resisters have an alternative T cell response not measured by TST/IGRA that clears and/or controls Mtb? Is there a role for protective B cell responses? Is there a role for genetics? As with traditional LTBI, the inability to detect Mtb does not allow us to determine whether and which resisters could be latently infected or may have cleared Mtb (5, 56). Understanding the host response and immune mechanism(s) of these LTBI resisters may identify novel protective immune responses to Mtb.
Based on cohort studies of Ugandan household contacts who were highly exposed to Mtb but remained TST and IGRA negative during an almost decade-long follow-up period, we have evidence for differences in both innate and adaptive immune responses (52, 56). Monocyte-derived macrophages from resisters and people with LTBI differed in gene expression and metabolic programs in response to Mtb, suggesting their contribution to resistance to a traditional LTBI response (57, 58). In addition, we found non–IFN-γ T cell responses to the Mtb-specific proteins ESAT6 and CFP10 in resisters, while their overall T cell responses revealed normal IFN-γ responses (56). These non–IFN-γ T cell responses were associated with Mtb-specific antibody profiles and characteristics, indicating that resisters were Mtb-exposed. Among Indonesian TB household contacts, those resisting Mtb infection had evidence for trained immune responses (59). Importantly, while the cohort was small, there was no evidence that resisters were at increased risk of progression to TB, i.e., their immune responses were adequate to control their exposure to aerosolized Mtb.
Based on these data, we believe that these Ugandan resisters may have developed an alternative form of LTBI. Resisters might have enhanced macrophage capacity to control Mtb, due to either trained immunity or genetic factors, and less need for an expansive T cell response. Alternatively, resisters may have a unique combination of B and T cell responses that help macrophages control Mtb. Some elements of these resisters’ immune responses likely are also present in subsets of people with traditional LTBI. Studies of the immune responses of well-characterized resisters from various settings may provide insights into alternative mechanisms of protection against Mtb, TB host-directed therapies, and approaches to vaccine development.
T cells and LTBI
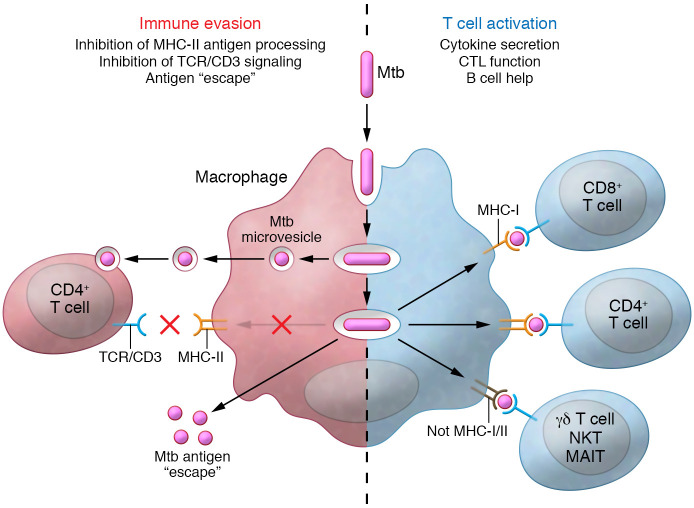
T cells are critical for successful containment of Mtb by macrophages in granulomas (Figure 1), and many T cell subsets respond to a wide range of Mtb antigens. These subsets can broadly be defined as classical MHC-restricted T cells and donor-unrestricted T cells (DURTs), with the former responding to wide-ranging Mtb peptides (reviewed in ref. 60) and the latter to a restricted set of mostly nonprotein antigens (reviewed in ref. 61). HIV-induced CD4+ T cell depletion and its association with TB risk provide the strongest evidence for the dominant role of CD4+ T cells in controlling Mtb (reviewed in ref. 33). Murine MHC-II knockout and NHP CD4+ T cell depletion studies further support this central role of MHC-II–restricted CD4+ T cells (reviewed in ref. 35). Polyfunctional CD4+ T cells expressing IFN-γ, TNF-α, and IL-2 are associated with protective responses (62), and effector/memory CD4+ T cells responsive to Mtb antigens are found in the bronchoalveolar lavage fluid of people with LTBI (63). In addition, CD4+ Treg and Th17 responses to Mtb are found in LTBI, but their role in controlling Mtb infection is less clear.
Figure 1. Evasion of T cell recognition versus T cell activation by Mtb-infected antigen-presenting cells.
The paradox of the T cell response to Mtb is that, on the one hand, Mtb antigens, when appropriately processed by an activated antigen-presenting cell, elicit a broad T cell response in a person with LTBI. This involves many T cell subsets responding to a wide range of antigens. These Mtb-activated T cells secrete predominantly Th1 cytokines and chemokines, possess cytotoxic T lymphocyte (CTL) function, and can provide help to B cells. On the other hand, Mtb harbored by macrophages can use a variety of mechanisms to interfere with T cell recognition. These mechanisms have primarily been identified for CD4+ T cells and include inhibition of MHC-II antigen processing, antigen escape, and inhibition of T cell receptor–CD3 signaling by Mtb glycolipids, but some may also apply to other T cell subsets. MAIT, mucosal-associated invariant T cell.
Mtb-activated human CD4+ T cells help macrophages control intracellular mycobacteria through secretion of cytokines and cytotoxic T lymphocyte function (ref. 64 and reviewed in refs. 35, 65). In addition to these direct effector roles, CD4+ T cell subsets also provide important helper functions for other immune cells involved in LTBI, including help for CD8+ T cell and DURT expansion, and for antibody production by B cells (reviewed in refs. 66, 67). While the central role of CD4+ T cells in LTBI and protection against TB is well established, the key Mtb antigens recognized by protective T cells have not been identified. CD4+ T cells (and CD8+ T cells; see below) from people with LTBI demonstrate broad reactivity to Mtb peptides (60, 68), but only a limited number of antigens are recognized by most individuals with LTBI. Antigens expressed by MHC molecules on Mtb-infected cells remain largely unknown. Identifying these antigens is essential to define the key protective T cells for LTBI.
MHC-I–restricted CD8+ T cells responsive to Mtb are found in peripheral blood and in the bronchoalveolar lavage fluid of humans and NHPs with LTBI (ref. 69 and reviewed in refs. 19, 35, 70). Recent computational modeling studies based on LTBI in NHP data suggest that multifunctional CD8+ T cells have a central role in preventing Mtb dissemination (27). Furthermore, DURTs that respond to Mtb may have a role in innate responses to Mtb (reviewed in refs. 35, 61). These include γ-δ T cells, CD1d-restricted natural killer (NK) T cells, and mucosal-associated invariant T cells. However, while DURTs are biologically intriguing, their specific roles in protecting against Mtb still need to be characterized. Despite our broad knowledge of cell-mediated immunity in LTBI, many knowledge gaps remain: (a) What T cell protective antigens are most relevant to the recognition of infected cells? (b) Does Mtb’s ability to inhibit antigen processing limit antigens presented by infected cells? (c) How do BCG vaccination and exposure to environmental mycobacteria modulate T cell responses after Mtb infection? (d) How do CD8+ T cells contribute to protective immunity in LTBI? (e) How do the granuloma milieu and architecture impact T cell function?
Mtb’s evasion of T cell recognition
Elegant cell biology and functional studies have defined a number of molecular mechanisms used by Mtb to resist innate immune mechanisms in macrophages and DCs, including disruption of progression to phagolysosome fusion, and resisting of killing by superoxide, autophagy, and apoptosis (reviewed in refs. 30, 35, 65, 71, 72). Mtb can also indirectly and directly interfere with recognition of infected cells by CD4+ T cells (Figure 1). For example, Mtb lipoproteins can activate TLR2 signaling in macrophages, which inhibits IFN-γ–driven expression of MHC-II molecules (73); Mtb’s secreted protein EsxH can interfere with CD4+ T cell activation (74); and Mtb-infected DCs can export antigens to uninfected cells, thereby limiting their antigen presentation to and activation of CD4+ T cells (75).
Mtb resides in macrophage phagosomes, which resemble an endosomal recycling compartment that traffics molecules and bacterial vesicles. Release of bacterial microvesicles allows Mtb products, which include lipids, proteins, and glycolipids such as lipoarabinomannan (LAM), to reach T cells in the proximity of infected cells (76–78). Exposure of CD4+ T cells to LAM or LAM-containing microvesicles inhibits proximal T cell receptor–CD3 signaling, which induces GRAIL (gene regulating anergy in lymphocytes), rendering LAM-exposed CD4+ T cells anergic (79). Similar inhibitory mechanisms are likely applicable to CD8+ T cells, and DURTs, since they all rely on CD3 for activation. Despite these known direct and indirect mechanisms of Mtb interference with T cell recognition of infected cells, questions remain: (a) Do these evasion mechanisms impact non-CD4+ T cells? (b) During which stages of Mtb infection and disease do they affect the immune response? (c) Which of these different T cell evasion mechanisms dominates, and at what stage of Mtb pathogenesis in vivo?
Antibodies and B cells in LTBI
Antibodies may contribute to long-term Mtb control in LTBI (reviewed in refs. 80–84). Serum IgG from individuals exposed to or latently infected with Mtb can be protective in vitro and in vivo against Mtb (85–87). Mtb resisters carry IgM against ESAT6 and CFP10 and other Mtb antigens and have class-switched IgG antibody responses, suggesting a role in these persistently TST/IGRA-negative but heavily Mtb-exposed individuals (56). In contrast, few studies support a protective role for anti-Mtb antibodies from TB patients (88).
Antibodies can bind mycobacterial surface molecules and interact with Fc receptors (FcRs) on phagocytes (reviewed in refs. 80–84). While binding to surface molecules can activate complement and prevent bacterial adhesion and invasion of host cells, subclasses or isotypes and their distinct Fc glycosylation profiles can influence FcR-mediated effects, including inflammatory versus noninflammatory responses. Through FcγR, mycobacterial multi- and single-antigen-specific polyclonal IgG from asymptomatic Mtb-exposed and infected people can enhance Mtb phagocytosis and growth inhibition, and antibody-dependent cellular cytotoxicity (85, 87, 89). Enhanced cytotoxic responses mediated mostly by FcγRIIIa (CD16) and NK cells were also observed in LTBI (87, 90). These data demonstrate the important interplay between antibodies and the innate immune system in LTBI.
The range of mycobacterial antigens targeted by protective antibodies remains poorly understood. Transfer studies with murine IgG or IgA monoclonal antibodies (mAbs) in Mtb-infected mice suggest that antibodies targeting the surface glycan arabinomannan (AM), the glycolipid lipoarabinomannan (LAM), the surface protein heparin-binding hemagglutinin (HBHA), the heat shock protein HspX, and the 38-kDa adhesion protein PstS1 might be protective (reviewed in ref. 91). Vaccination with AM and antigen 85 followed by passive transfer of antibodies was moderately protective against Mtb in mice (92). In humans, antibodies against antigen 85 and AM/LAM appear to be protective (85, 93, 94), but experimental data with human mAbs remain scarce.
Attempts to identify significantly different antigen-specific antibody responses in LTBI versus TB are ongoing, but have provided few conclusions to date (95, 96). In both NHPs and humans, antibody responses to Mtb are heterogeneous (85, 97–101), likely because of granuloma heterogeneity (reviewed in ref. 19), large numbers of differentially expressed Mtb antigens (102), and/or prior exposure to BCG and/or nontuberculous mycobacteria (85, 100, 103). This heterogeneity contributes to the challenges of delineating specific protective antibodies against Mtb.
A limited number of functional human multi- or single-antigen-specific polyclonal antibody studies have been performed (56, 85–88, 104). Protective ex vivo efficacy was reversed when total serum IgG from asymptomatic health care workers was preabsorbed with Mtb (86). We found that anti-AM IgG isolated from high-titer asymptomatic TST-positive individuals was protective in vitro and in vivo (85). In line with serum anti-AM IgG studies from an adult BCG vaccination trial (100), our data further suggested the importance of targeting specific glycan epitopes within AM and support the protective role of IgG to certain Mtb surface antigens and epitopes.
Efforts to generate human mAbs against Mtb-specific antigens/epitopes are ongoing (105, 106), which will help define the roles of variable and Fc domains. Human mAb isotypes against LAM and HBHA generated from plasmablasts and memory B cells of TB patients and Mtb-exposed health care workers demonstrated different effector functions (105); IgG enhanced and IgA inhibited Mtb uptake by human lung epithelial cells and macrophages, irrespective of the target, although neither differences in FcR expressions between these cell types nor effects on intracellular Mtb growth were taken into consideration.
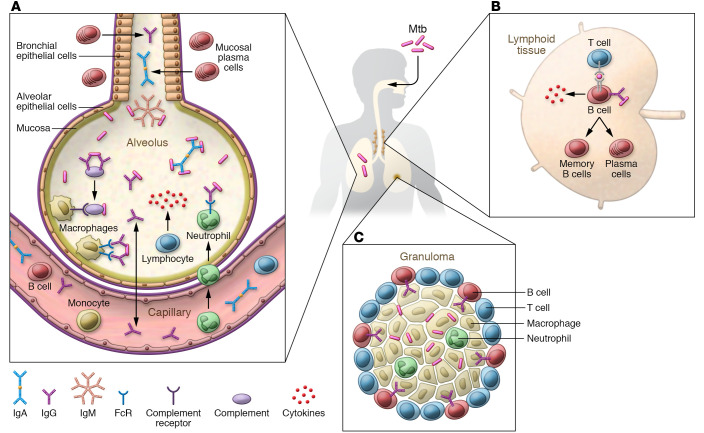
Antibodies in the airways could serve as a first line of defense against inhaled Mtb (Figure 2). For example, secretory IgA could bind to Mtb antigens and thereby prevent Mtb adhesion to and infection of airway cells, while in parallel facilitating elimination of Mtb via mucociliary clearance. Passive transfer studies support a protective role of poly- and monoclonal IgG and IgA against Mtb in the airways (reviewed in refs. 83, 91; refs. 85, 86). Polyfunctional Th17 cells, IL-10, and increased airway IgA levels were associated with protection against Mtb in NHPs mucosally vaccinated with BCG (107), and mucosal vaccination of mice and NHPs with the MTBVAC vaccine indicated a role of mucosal secretory antibodies against Mtb (108). The role of antibodies after intravenous BCG, shown to be more protective than airway vaccination, remains to be determined (109).
Figure 2. Potential protective roles of antibodies and B cells in the lung during both initial Mtb exposure and LTBI.
(A) Antibody isotypes (IgA, IgG, and IgM) could impact Mtb in the lower airways through binding, opsonization, complement activation, and FcR-mediated enhanced phagocytosis and intracellular growth reduction by phagocytes. (B) B cells located in germinal centers of lymphoid tissues could control infection through (i) enhancing antigen presentation to T cells; (ii) production of helper cytokines for T cells; and (iii) generation of antibodies that could modulate innate and adaptive immune responses. (C) Both the presence of B cells and the pro- and antiinflammatory capacities of antibodies could influence the formation of functional granulomas and thereby contribute to the control of Mtb.
Antibodies also can synergize with T cells in controlling Mtb (92), and, in addition to being influenced by T cells (reviewed in ref. 66), B cells may regulate T cell and cytokine responses during Mtb infection, thereby influencing inflammation and granuloma formation (reviewed in refs. 110, 111). B cells are present in the granulomatous lesions of Mtb-infected mice, non-human primates, and humans. Although inconsistent results of murine studies have led to controversy regarding the protective effects of B cells in Mtb infection (reviewed in ref. 110), recent data show an association of smaller lung B cell follicles with increased Mtb susceptibility in male versus female mice (112), and NHP studies support the beneficial effects of B cells in the lung. Despite a lack of difference in outcome between B cell–depleted and nondepleted Mtb-infected cynomolgus macaques, B cell depletion influenced local T cell and cytokine responses, resulting in increased Mtb burden at the granuloma level (113). Expanded B cell follicles in the lungs of Mtb- and SIV-coinfected rhesus macaques were also associated with lack of progression to TB (24).
In humans, household contacts with LTBI and TB patients were shown to have atypical B cell phenotypes associated with a compromised T cell response, which, in TB patients, resolved after antituberculous treatment (114). These atypical B cells showed diminished proliferation and immunoglobulin and cytokine production, supporting their lack of function in TB. Circulating naive B cells are reduced in LTBI, possibly as a result of sequestration at the site of infection (90). B cells form prominent aggregates in the lungs of Mtb-infected humans, NHPs, and mice (24, 115–118). Nevertheless, because of the conflicting associations with disease outcome, the role of these lung B cell aggregates remains to be determined.
Overall, many questions remain regarding the roles of antibodies and B cells in the defense against Mtb: (a) What are the critical antigens in antibody-mediated immunity against Mtb? (b) How do epitope specificity and Fc glycosylation influence success and failure of Mtb control? (c) What are the protective roles and mechanisms of IgG, IgA, and IgM during Mtb exposure and infection? (d) Do antibodies have direct effects on Mtb? (e) What are the essential interactions between the humoral and other immune arms in the defense against Mtb? (f) What role do B cells and pulmonary B cell aggregates have in Mtb infection? A better understanding of these roles will inform immunotherapy and TB vaccine development.
Innate immune responses and LTBI
Innate immune cells, both lymphoid and myeloid, have a central role in the host response to Mtb (reviewed in refs. 30, 35, 61, 119). Recent studies have expanded our understanding of the range of innate cells, such as the myeloid-derived polymorphonuclear cells (PMNs) and innate lymphoid cells and DURTs (discussed above), involved in responses to Mtb and influencing the complexity of macrophage responses. Nevertheless, whereas the centrality of macrophages as nidus and site of Mtb control in LTBI is well established, the role of other innate cells in LTBI is less clear. In vivo innate responses upon Mtb infection can only be studied in experimental animals, and in vitro studies of macrophage functions are primarily performed with bone marrow–derived macrophages from C57BL/6 mice, considered resistant to Mtb infection, and with human blood monocyte-derived macrophages and macrophage cell lines. Where results from these studies fit in the spectrum from Mtb exposure to LTBI and TB in humans is not straightforward.
As a facultative intracellular pathogen in macrophages, Mtb depends on phagocytosis for host cell entry. Thus, the receptor repertoire of these cells defines infectivity and shapes downstream host responses. Upon inhalation, Mtb trapped in the alveolar surfactant phospholipid layer can be bound by surfactant proteins A and D for indirect phagocytosis by alveolar macrophages, a defense mechanism deficient in the elderly (120). Once phagocytosed, Mtb proliferates in macrophages by interfering with phagosome maturation through cell wall glycolipids (72, 121, 122). The mycobacterial phagosome communicates dynamically with endosomes and delivers mycobacterial antigens into the lysosomal degradation pathway for antigen processing. During phagocytosis, Mtb also triggers a set of pattern recognition receptors, which induce both proinflammatory (IL-1, IL-12/-23, TNF-α, and type I IFNs) and antiinflammatory (IL-10) responses (reviewed in refs. 123–125). Mtb cell wall glycolipids interacting with C-type lectins can switch a proinflammatory to an antiinflammatory IL-10 response (122, 126). Alveolar macrophages exhibit a predominantly antiinflammatory M2 phenotype, which Mtb can use to establish its intracellular niche (reviewed in refs. 72, 127).
Alveolar macrophages also transport Mtb into the bronchus-associated lymphoid tissue, where, in LTBI, they transfer antigens to DCs to trigger adaptive T cell responses that help control Mtb growth (reviewed in ref. 128). Recent studies suggest that group 3 innate lymphoid cells (ILC3s) are involved in Mtb control (129). These cells were associated with enhanced alveolar macrophage recruitment in the lungs of Mtb-infected mice and, when depleted, reduced bacterial control. In TB patients, ILC3 accumulated in the lungs and were depleted in the blood with normalization after TB treatment (129), but their role in LTBI remains to be determined. High levels of circulating NK cells in LTBI may also play a role in controlling Mtb during LTBI, which is further supported by the observation that NK cell levels are low in TB and return to baseline after TB treatment (90).
In LTBI, immune activation by IFN-γ, TNF-α, and autocrine IL-15 (probably reinforced by vitamin D3) can enhance Mtb control by accelerating phagosome maturation, production of microbicidal effectors, augmented glycolysis, and induced autophagy (130–132). The relevance of autophagy as an anti-Mtb effector of activated macrophage remains to be determined (133).
Prior pathogen exposure can train innate immunity. For example, BCG vaccination can epigenetically prime NK cells and monocytes/macrophages for a more focused secondary response (reviewed in ref. 134). Distinct innate immune cell and cytokine responses in Indonesian TB household contacts support a role for trained immunity in early clearance of Mtb in humans (59). In mice infected intravenously with BCG or Mtb, IFN-γ was found to be an important factor in regulating macrophage trained immunity by enhancing myelopoiesis and expansion of lineage–cKit+Sca1+ (LKS) bone marrow stem cells (135, 136). Mycobacterial interaction with LKS leads to innate imprinting of myeloid cells by altering their epigenetic profile, thereby rendering mature macrophages more effective against Mtb and likely contributing to trained immunity in LTBI (137).
The role(s) of PMNs in Mtb pathogenesis is an active area of research. In Mtb-resistant mice, numbers of infected PMNs are only transiently increased following infection (30). In contrast, susceptible mouse strains such as C3HeB/FeJ mice and NOS2- or Atg5-knockout mice had PMN infiltrates associated with exacerbation of necrotic granulomas and earlier death due to higher Mtb loads (133, 138, 139). These latter data indicate that NOS2 and Atg5 are essential to restrict Mtb growth, likely through interference with PMN influx and associated pathology. Recent data from Mtb-infected mice further suggest that long-lived PMNs can accumulate in the lungs and serve as an intracellular niche for Mtb growth and persistence (140).
Necrotic PMN-laden granulomas in susceptible mice share features with those found in infected NHPs and in TB patients, where PMNs represent the dominant Mtb-infected cell population (141, 142). The pro- and antiinflammatory cytokine profiles of PMNs in Mtb-infected NHP granulomas suggest that the cells have an important immunoregulatory role. The abundance of PMNs in human and NHP TB lesions, together with a PMN-associated transcriptomic signature in PBMCs of TB patients (143), and enhanced PMN-driven inflammation in TB patients with type 1 diabetes (144, 145), links PMNs with disease, rather than LTBI. However, it is not known whether PMNs drive disease progression, or whether they are attracted to granulomas as a result of failed Mtb control. In NHPs, PMNs are part of stable Mtb granulomas, and uptake of infected PMNs by DCs facilitates T cell priming in mice (146), suggesting a protective role. It therefore remains unclear whether PMNs, with the right balance of inflammatory effects, contribute to Mtb control after initial exposure and in LTBI.
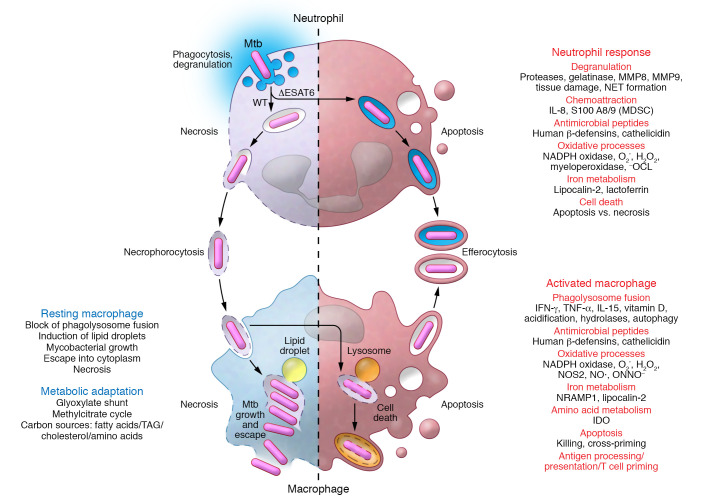
In vitro, virulent Mtb strains drive PMNs quickly into necrotic cell death (147, 148). Necrotic Mtb-infected PMNs release neutrophil extracellular traps as an antimicrobial effector but do not kill Mtb. Instead, clearance of necrotic Mtb-infected PMNs by macrophages promotes mycobacterial growth in these more sustainable host cells. Subsequently, infected macrophages also succumb to necrotic cell death and release mycobacteria to infect new phagocytes, thereby continuing the infectious cycle. IL-8 from infected PMNs and macrophages feeds an influx of PMNs and sustains a cycle of host cell necrosis, necrophorocytosis (phagocytic removal of necrotic cellular debris), and bacterial growth in TB lesions (reviewed in ref. 149).
Mtb-triggered PMN necrosis requires myeloperoxidase-derived (MPO-derived) reactive oxygen species. Inhibition of MPO rescues infected PMNs from necrosis and restores the macrophage’s ability to control Mtb upon efferocytosis of infected but apoptotic PMNs (148). Therefore, MPO and other factors associated with PMN-driven pathology may represent intriguing targets for host-directed therapy for TB, shifting the balance back toward LTBI (reviewed in refs. 35, 149–152). Yet only interactions between infected resting macrophages and PMNs have been studied (Figure 3). Thus, the impact of macrophage activation for dealing with infected PMNs remains to be determined (72). Overall, many questions on the role of innate cells in LTBI remain, including the role of trained immunity, macrophage heterogeneity and activation in granulomas, Mtb’s metabolic state, and the protective versus detrimental role of PMNs.
Figure 3. Interactions between infected PMNs and macrophages can determine the balance of exacerbating versus protective host responses in LTBI.
Wild-type but not attenuated Mtb drives PMNs into necrotic cell death in a myeloperoxidase-dependent manner. Uptake of Mtb together with necrotic PMNs by resting macrophages further promotes mycobacterial propagation and macrophage necrosis, releasing Mtb for another round of intracellular replication and macrophage death. In contrast, immune activation in LTBI equips macrophages with a potent antimicrobial armamentarium to control and possibly eliminate Mtb. MDSC, myeloid-derived suppressor cells; NET, neutrophil extracellular traps; –OCL, hypochlorite.
Conclusions
In most Mtb-infected individuals, LTBI is established through finely regulated immune responses. Summarizing known facts and important areas of LTBI research in humans and NHPs, we have pointed out critical gaps in understanding how the immune system protects against or controls Mtb. While the interaction between activated macrophages and CD4+ T cells is central for Mtb control in LTBI, recent discoveries reveal a more complex picture with roles for genetic factors, other T cell subsets, innate lymphoid cells, B cells and antibodies, trained immunity, and possibly more. Some host defenses may promote excessive inflammation, and, if not regulated properly, exacerbate pathology and facilitate progression to disease and Mtb transmission. LTBI and variants thereof, as seen in resisters, rely on both innate and adaptive immunity. The goal of parsing LTBI is to identify the immune mechanisms of the more than 90% who successfully control Mtb versus the few at risk for disease. Given our inability to distinguish who harbors dead versus live bacilli, and determine Mtb burden, LTBI remains an operational definition, hampering the triaging of care to those at greatest risk for progression to TB. Given the difficulty in identifying and preventing acute exposure and infection with Mtb in humans, animal and careful observational human studies are needed to determine the essential local immune responses necessary for elimination or long-term control of this wily pathogen with its plethora of immune evasion mechanisms.
Acknowledgments
This work was supported in part by funds from the NIH/National Institute of Allergy and Infectious Diseases to JMA (AI146329, AI127173, and AI117927) and to WHB (AI125642, AI124348, AI124348, AI147319, and contract 75N93019C00071), and by grants from the Leibniz Research Alliance INFECTIONS ´21, Leibniz Science Campus Evolung, the German Science Foundation (IRTG 1911; Scha 514 5-1), and the Ministry of Education and Research (German Center for Infection Research, TB-Sequel) to UES.
Version 1. 02/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(3):e136222.https://doi.org/10.1172/JCI136222.
Contributor Information
W. Henry Boom, Email: whb@case.edu.
Jacqueline M. Achkar, Email: jacqueline.achkar@einstein.yu.edu.
References
- 1. WHO. Global Tuberculosis Report 2019. World Health Organization; 2019. [Google Scholar]
- 2.Huang L, et al. Mycobacterium tuberculosis: bacterial fitness within the host macrophage. Microbiol Spectr. 2019;7(2):10.1128/microbiolspec.BAI-0001–2019. doi: 10.1128/microbiolspec.BAI-0001-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Getahun H, et al. Latent Mycobacterium tuberculosis infection. N Engl J Med. 2015;372(22):2127–2135. doi: 10.1056/NEJMra1405427. [DOI] [PubMed] [Google Scholar]
- 4.Lin PL, Flynn JL. The end of the binary era: revisiting the spectrum of tuberculosis. J Immunol. 2018;201(9):2541–2548. doi: 10.4049/jimmunol.1800993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr MA, et al. Is Mycobacterium tuberculosis infection life long? BMJ. 2019;367:l5770. doi: 10.1136/bmj.l5770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO. Latent Tuberculosis Infection: Updated and Consolidated Guidelines for Programmatic Management. Geneva: World Health Organization; 2018. License: CC BY-NC-SA 3.0 IGO. [PubMed] [Google Scholar]
- 7.Lewinsohn DM, et al. Official American Thoracic Society/Infectious Diseases Society of America/Centers for Disease Control and Prevention Clinical Practice Guidelines: Diagnosis of Tuberculosis in Adults and Children. Clin Infect Dis. 2017;64(2):111–115. doi: 10.1093/cid/ciw778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mack U, et al. LTBI: latent tuberculosis infection or lasting immune responses to M. tuberculosis? A TBNET consensus statement. Eur Respir J. 2009;33(5):956–973. doi: 10.1183/09031936.00120908. [DOI] [PubMed] [Google Scholar]
- 9.Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 2016;13(10):e1002152. doi: 10.1371/journal.pmed.1002152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagneux S. Ecology and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(4):202–213. doi: 10.1038/nrmicro.2018.8. [DOI] [PubMed] [Google Scholar]
- 11.Orgeur M, Brosch R. Evolution of virulence in the Mycobacterium tuberculosis complex. Curr Opin Microbiol. 2018;41:68–75. doi: 10.1016/j.mib.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 12.Menardo F, et al. The molecular clock of Mycobacterium tuberculosis. PLoS Pathog. 2019;15(9):e1008067. doi: 10.1371/journal.ppat.1008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saelens JW, et al. Mycobacterial evolution intersects with host tolerance. Front Immunol. 2019;10:528. doi: 10.3389/fimmu.2019.00528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrt S, et al. Metabolic principles of persistence and pathogenicity in Mycobacterium tuberculosis. Nat Rev Microbiol. 2018;16(8):496–507. doi: 10.1038/s41579-018-0013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin PL, et al. Quantitative comparison of active and latent tuberculosis in the cynomolgus macaque model. Infect Immun. 2009;77(10):4631–4642. doi: 10.1128/IAI.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coleman MT, et al. Early changes by (18)Fluorodeoxyglucose positron emission tomography coregistered with computed tomography predict outcome after Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2014;82(6):2400–2404. doi: 10.1128/IAI.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bucsan AN, et al. The current state of animal models and genomic approaches towards identifying and validating molecular determinants of Mycobacterium tuberculosis infection and tuberculosis disease. Pathog Dis. 2019;77(4):ftz037. doi: 10.1093/femspd/ftz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sia JK, Rengarajan J. Immunology of Mycobacterium tuberculosis infections. Microbiol Spectr. 2019;7(4):10.1128/microbiolspec.GPP3-0022–2018. doi: 10.1128/microbiolspec.gpp3-0022-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cadena AM, et al. Heterogeneity in tuberculosis. Nat Rev Immunol. 2017;17(11):691–702. doi: 10.1038/nri.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pena JC, Ho WZ. Monkey models of tuberculosis: lessons learned. Infect Immun. 2015;83(3):852–862. doi: 10.1128/IAI.02850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scanga CA, Flynn JL. Modeling tuberculosis in nonhuman primates. Cold Spring Harb Perspect Med. 2014;4(12):a018564. doi: 10.1101/cshperspect.a018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiello P, et al. Rhesus macaques are more susceptible to progressive tuberculosis than cynomolgus macaques: a quantitative comparison. Infect Immun. 2018;86(2):e00505–17. doi: 10.1128/IAI.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucsan AN, et al. Mechanisms of reactivation of latent tuberculosis infection due to SIV coinfection. J Clin Invest. 2019;129(12):5254–5260. doi: 10.1172/JCI125810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foreman TW, et al. CD4+ T-cell-independent mechanisms suppress reactivation of latent tuberculosis in a macaque model of HIV coinfection. Proc Natl Acad Sci U S A. 2016;113(38):E5636–E5644. doi: 10.1073/pnas.1611987113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diedrich CR, et al. SIV and Mycobacterium tuberculosis synergy within the granuloma accelerates the reactivation pattern of latent tuberculosis. PLoS Pathog. 2020;16(7):e1008413. doi: 10.1371/journal.ppat.1008413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin PL, Flynn JL. CD8 T cells and Mycobacterium tuberculosis infection. Semin Immunopathol. 2015;37(3):239–249. doi: 10.1007/s00281-015-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wessler T, et al. A computational model tracks whole-lung Mycobacterium tuberculosis infection and predicts factors that inhibit dissemination. PLoS Comput Biol. 2020;16(5):e1007280. doi: 10.1371/journal.pcbi.1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elkington P, et al. In vitro granuloma models of tuberculosis: potential and challenges. J Infect Dis. 2019;219(12):1858–1866. doi: 10.1093/infdis/jiz020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner R, et al. In vitro mycobacterial growth inhibition assays: a tool for the assessment of protective immunity and evaluation of tuberculosis vaccine efficacy. Vaccine. 2016;34(39):4656–4665. doi: 10.1016/j.vaccine.2016.07.058. [DOI] [PubMed] [Google Scholar]
- 30.Srivastava S, et al. Beyond macrophages: the diversity of mononuclear cells in tuberculosis. Immunol Rev. 2014;262(1):179–192. doi: 10.1111/imr.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tornheim JA, Dooley KE. Tuberculosis associated with HIV infection. Microbiol Spectr. 2017;5(1):TNMI7-0028-2016. doi: 10.1128/microbiolspec.TNMI7-0028-2016. [DOI] [PubMed] [Google Scholar]
- 32.Boisson-Dupuis S, et al. Inherited and acquired immunodeficiencies underlying tuberculosis in childhood. Immunol Rev. 2015;264(1):103–120. doi: 10.1111/imr.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jasenosky LD, et al. T cells and adaptive immunity to Mycobacterium tuberculosis in humans. Immunol Rev. 2015;264(1):74–87. doi: 10.1111/imr.12274. [DOI] [PubMed] [Google Scholar]
- 34.Cadena AM, et al. Concurrent infection with Mycobacterium tuberculosis confers robust protection against secondary infection in macaques. PLoS Pathog. 2018;14(10):e1007305. doi: 10.1371/journal.ppat.1007305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayer-Barber KD, Barber DL. Innate and adaptive cellular immune responses to Mycobacterium tuberculosis infection. Cold Spring Harb Perspect Med. 2015;5(12):a018424. doi: 10.1101/cshperspect.a018424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho YS, et al. Deciphering the proteome of the in vivo diagnostic reagent “purified protein derivative” from Mycobacterium tuberculosis. Proteomics. 2012;12(7):979–991. doi: 10.1002/pmic.201100544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reichler MR, et al. Risk factors for tuberculosis and effect of preventive therapy among close contacts of persons with infectious tuberculosis. Clin Infect Dis. 2020;70(8):1562–1572. doi: 10.1093/cid/ciz438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloot R, et al. Risk of tuberculosis after recent exposure. A 10-year follow-up study of contacts in Amsterdam. Am J Respir Crit Care Med. 2014;190(9):1044–1052. doi: 10.1164/rccm.201406-1159OC. [DOI] [PubMed] [Google Scholar]
- 39.Reichler MR, et al. Risk and timing of tuberculosis among close contacts of persons with infectious tuberculosis. J Infect Dis. 2018;218(6):1000–1008. doi: 10.1093/infdis/jiy265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fox GJ, et al. Household-contact investigation for detection of tuberculosis in Vietnam. N Engl J Med. 2018;378(3):221–229. doi: 10.1056/NEJMoa1700209. [DOI] [PubMed] [Google Scholar]
- 41.Andrews JR, et al. Serial QuantiFERON testing and tuberculosis disease risk among young children: an observational cohort study. Lancet Respir Med. 2017;5(4):282–290. doi: 10.1016/S2213-2600(17)30060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gupta RK, et al. Quantitative IFN-γ release assay and tuberculin skin test results to predict incident tuberculosis. A prospective cohort study. Am J Respir Crit Care Med. 2020;201(8):984–991. doi: 10.1164/rccm.201905-0969OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Menzies NA, et al. Progression from latent infection to active disease in dynamic tuberculosis transmission models: a systematic review of the validity of modelling assumptions. Lancet Infect Dis. 2018;18(8):e228–e238. doi: 10.1016/S1473-3099(18)30134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmed M, et al. Immune correlates of tuberculosis disease and risk translate across species. Sci Transl Med. 2020;12(528):eaay0233. doi: 10.1126/scitranslmed.aay0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons JD, et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat Rev Immunol. 2018;18(9):575–589. doi: 10.1038/s41577-018-0025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Comstock GW. Tuberculosis in twins: a re-analysis of the Prophit survey. Am Rev Respir Dis. 1978;117(4):621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 47.Bustamante J, et al. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 2014;26(6):454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McHenry ML, et al. Genetics and evolution of tuberculosis pathogenesis: New perspectives and approaches. Infect Genet Evol. 2020;81:104204. doi: 10.1016/j.meegid.2020.104204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cubillos-Angulo JM, et al. Polymorphisms in interferon pathway genes and risk of Mycobacterium tuberculosis infection in contacts of tuberculosis cases in Brazil. Int J Infect Dis. 2020;92:21–28. doi: 10.1016/j.ijid.2019.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma N, et al. Clinical and epidemiological characteristics of individuals resistant to M. tuberculosis infection in a longitudinal TB household contact study in Kampala, Uganda. BMC Infect Dis. 2014;14:352. doi: 10.1186/1471-2334-14-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mave V, et al. Infection free “resisters” among household contacts of adult pulmonary tuberculosis. PLoS One. 2019;14(7):e0218034. doi: 10.1371/journal.pone.0218034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stein CM, et al. Long-term stability of resistance to latent Mycobacterium tuberculosis infection in highly exposed tuberculosis household contacts in Kampala, Uganda. Clin Infect Dis. 2019;68(10):1705–1712. doi: 10.1093/cid/ciy751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stein CM, et al. Resistance and susceptibility to Mycobacterium tuberculosis infection and disease in tuberculosis households in Kampala, Uganda. Am J Epidemiol. 2018;187(7):1477–1489. doi: 10.1093/aje/kwx380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verrall AJ, et al. Early clearance of Mycobacterium tuberculosis: the INFECT case contact cohort study in Indonesia. J Infect Dis. 2020;221(8):1351–1360. doi: 10.1093/infdis/jiz168. [DOI] [PubMed] [Google Scholar]
- 55.Hanifa Y, et al. Prevalence of latent tuberculosis infection among gold miners in South Africa. Int J Tuberc Lung Dis. 2009;13(1):39–46. [PubMed] [Google Scholar]
- 56.Lu LL, et al. IFN-gamma-independent immune markers of Mycobacterium tuberculosis exposure. Nat Med. 2019;25(6):977–987. doi: 10.1038/s41591-019-0441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seshadri C, et al. Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection. PLoS One. 2017;12(4):e0175844. doi: 10.1371/journal.pone.0175844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Simmons JD, et al. Monocyte metabolic programs are associated with resistance to TST/IGRA conversion. Poster presented at: Keystone Symposia, Tuberculosis: Immunity and Immune Evasion; January 16–20, 2020; Santa Fe, NM. [Google Scholar]
- 59.Verrall AJ, et al. Early clearance of Mycobacterium tuberculosis is associated with increased innate immune responses. J Infect Dis. 2020;221(8):1342–1350. doi: 10.1093/infdis/jiz147. [DOI] [PubMed] [Google Scholar]
- 60.Lindestam Arlehamn CS, et al. Antigens for CD4 and CD8 T cells in tuberculosis. Cold Spring Harb Perspect Med. 2014;4(7):a018465. doi: 10.1101/cshperspect.a018465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Joosten SA, et al. Harnessing donor unrestricted T-cells for new vaccines against tuberculosis. Vaccine. 2019;37(23):3022–3030. doi: 10.1016/j.vaccine.2019.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Harari A, et al. Dominant TNF-α+ Mycobacterium tuberculosis-specific CD4+ T cell responses discriminate between latent infection and active disease. Nat Med. 2011;17(3):372–376. doi: 10.1038/nm.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Silver RF, et al. Recruitment of antigen-specific Th1-like responses to the human lung following bronchoscopic segmental challenge with purified protein derivative of Mycobacterium tuberculosis. Am J Respir Cell Mol Biol. 2003;29(1):117–123. doi: 10.1165/rcmb.4840. [DOI] [PubMed] [Google Scholar]
- 64.Tsukaguchi K, et al. CD4+ α β T cell and γ δ T cell responses to Mycobacterium tuberculosis. Similarities and differences in Ag recognition, cytotoxic effector function, and cytokine production. J Immunol. 1995;154(4):1786–1796. [PubMed] [Google Scholar]
- 65.Ernst JD. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe. 2018;24(1):34–42. doi: 10.1016/j.chom.2018.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Crotty S. A brief history of T cell help to B cells. Nat Rev Immunol. 2015;15(3):185–189. doi: 10.1038/nri3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laidlaw BJ, et al. The multifaceted role of CD4(+) T cells in CD8(+) T cell memory. Nat Rev Immunol. 2016;16(2):102–111. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang JD, et al. Mycobacterium tuberculosis-specific CD4+ and CD8+ T cells differ in their capacity to recognize infected macrophages. PLoS Pathog. 2018;14(5):e1007060. doi: 10.1371/journal.ppat.1007060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tan JS, et al. Human alveolar T lymphocyte responses to Mycobacterium tuberculosis antigens: role for CD4+ and CD8+ cytotoxic T cells and relative resistance of alveolar macrophages to lysis. J Immunol. 1997;159(1):290–297. [PubMed] [Google Scholar]
- 70.Lewinsohn DA, et al. Comprehensive definition of human immunodominant CD8 antigens in tuberculosis. NPJ Vaccines. 2017;2:8. doi: 10.1038/s41541-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.VanderVen BC, et al. The minimal unit of infection: Mycobacterium tuberculosis in the macrophage. Microbiol Spectr. 2016;4(6):10.1128/microbiolspec.TBTB2-0025–2016. doi: 10.1128/microbiolspec.TBTB2-0025-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunol Rev. 2015;264(1):182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harding CV, Boom WH. Regulation of antigen presentation by Mycobacterium tuberculosis: a role for Toll-like receptors. Nat Rev Microbiol. 2010;8(4):296–307. doi: 10.1038/nrmicro2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Portal-Celhay C, et al. Mycobacterium tuberculosis EsxH inhibits ESCRT-dependent CD4+ T-cell activation. Nat Microbiol. 2016;2:16232. doi: 10.1038/nmicrobiol.2016.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Srivastava S, et al. Antigen export reduces antigen presentation and limits T cell control of M. tuberculosis. Cell Host Microbe. 2016;19(1):44–54. doi: 10.1016/j.chom.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Athman JJ, et al. Bacterial membrane vesicles mediate the release of Mycobacterium tuberculosis lipoglycans and lipoproteins from infected macrophages. J Immunol. 2015;195(3):1044–1053. doi: 10.4049/jimmunol.1402894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sande OJ, et al. Mannose-capped lipoarabinomannan from Mycobacterium tuberculosis induces CD4+ T cell anergy via GRAIL. J Immunol. 2016;196(2):691–702. doi: 10.4049/jimmunol.1500710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Prados-Rosales R, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121(4):1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Athman JJ, et al. Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J Immunol. 2017;198(5):2028–2037. doi: 10.4049/jimmunol.1601199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Achkar JM, et al. B cells and antibodies in the defense against Mycobacterium tuberculosis infection. Immunol Rev. 2015;264(1):167–181. doi: 10.1111/imr.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobs AJ, et al. Antibodies and tuberculosis. Tuberculosis (Edinb) 2016;101:102–113. doi: 10.1016/j.tube.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li H, Javid B. Antibodies and tuberculosis: finally coming of age? Nat Rev Immunol. 2018;18(9):591–596. doi: 10.1038/s41577-018-0028-0. [DOI] [PubMed] [Google Scholar]
- 83.Tran AC, et al. Emerging themes for the role of antibodies in tuberculosis. Immune Netw. 2019;19(4):e24. doi: 10.4110/in.2019.19.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawahara JY, et al. A case for antibodies as mechanistic correlates of immunity in tuberculosis. Front Immunol. 2019;10:996. doi: 10.3389/fimmu.2019.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen T, et al. Capsular glycan recognition provides antibody-mediated immunity against tuberculosis. J Clin Invest. 2020;130(4):1808-1822. doi: 10.1172/JCI128459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li H, et al. Latently and uninfected healthcare workers exposed to TB make protective antibodies against Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2017;114(19):5023–5028. doi: 10.1073/pnas.1611776114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu LL, et al. A functional role for antibodies in tuberculosis. Cell. 2016;167(2):433–443.e14. doi: 10.1016/j.cell.2016.08.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Shea MK, et al. Immunological correlates of mycobacterial growth inhibition describe a spectrum of tuberculosis infection. Sci Rep. 2018;8(1):14480. doi: 10.1038/s41598-018-32755-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu LL, et al. Antibody Fc glycosylation discriminates between latent and active tuberculosis. J Infect Dis. 2020;222(12):2093–2102. doi: 10.1093/infdis/jiz643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Roy Chowdhury R, et al. A multi-cohort study of the immune factors associated with M. tuberculosis infection outcomes. Nature. 2018;560(7720):644–648. doi: 10.1038/s41586-018-0439-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Achkar JM, Casadevall A. Antibody-mediated immunity against tuberculosis: implications for vaccine development. Cell Host Microbe. 2013;13(3):250–262. doi: 10.1016/j.chom.2013.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Prados-Rosales R, et al. Enhanced control of Mycobacterium tuberculosis extrapulmonary dissemination in mice by an arabinomannan-protein conjugate vaccine. PLoS Pathog. 2017;13(3):e1006250. doi: 10.1371/journal.ppat.1006250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Costello AM, et al. Does antibody to mycobacterial antigens, including lipoarabinomannan, limit dissemination in childhood tuberculosis? Trans R Soc Trop Med Hyg. 1992;86(6):686–692. doi: 10.1016/0035-9203(92)90192-F. [DOI] [PubMed] [Google Scholar]
- 94.Fletcher HA, et al. T-cell activation is an immune correlate of risk in BCG vaccinated infants. Nat Commun. 2016;7:11290. doi: 10.1038/ncomms11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kimuda SG, et al. Humoral responses to Rv1733c, Rv0081, Rv1735c, and Rv1737c DosR regulon-encoded proteins of Mycobacterium tuberculosis in individuals with latent tuberculosis infection. J Immunol Res. 2017;2017:1593143. doi: 10.1155/2017/1593143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Coppola M, et al. Differences in IgG responses against infection phase related Mycobacterium tuberculosis (Mtb) specific antigens in individuals exposed or not to Mtb correlate with control of TB infection and progression. Tuberculosis (Edinb) 2017;106:25–32. doi: 10.1016/j.tube.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 97.Kunnath-Velayudhan S, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107(33):14703–14708. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kunnath-Velayudhan S, Gennaro ML. Immunodiagnosis of tuberculosis: a dynamic view of biomarker discovery. Clin Microbiol Rev. 2011;24(4):792–805. doi: 10.1128/CMR.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kunnath-Velayudhan S, et al. Proteome-scale antibody responses and outcome of Mycobacterium tuberculosis infection in nonhuman primates and in tuberculosis patients. J Infect Dis. 2012;206(5):697–705. doi: 10.1093/infdis/jis421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen T, et al. Association of human antibodies to arabinomannan with enhanced Mycobacterial opsonophagocytosis and intracellular growth reduction. J Infect Dis. 2016;214(2):300–310. doi: 10.1093/infdis/jiw141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Song L, et al. Identification of antibody targets for tuberculosis serology using high-density nucleic acid programmable protein arrays. Mol Cell Proteomics. 2017;16(4 suppl 1):S277–S289. doi: 10.1074/mcp.M116.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee BY, Horwitz MA. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J Clin Invest. 1995;96(1):245–249. doi: 10.1172/JCI118028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shah JA, et al. Nontuberculous Mycobacteria and heterologous immunity to tuberculosis. J Infect Dis. 2019;220(7):1091–1098. doi: 10.1093/infdis/jiz285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Valliere S, et al. Enhancement of innate and cell-mediated immunity by antimycobacterial antibodies. Infect Immun. 2005;73(10):6711–6720. doi: 10.1128/IAI.73.10.6711-6720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zimmermann N, et al. Human isotype-dependent inhibitory antibody responses against Mycobacterium tuberculosis. EMBO Mol Med. 2016;8(11):1325–1339. doi: 10.15252/emmm.201606330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Choudhary A, et al. Characterization of the antigenic heterogeneity of lipoarabinomannan, the major surface glycolipid of Mycobacterium tuberculosis, and complexity of antibody specificities toward this antigen. J Immunol. 2018;200(9):3053–3066. doi: 10.4049/jimmunol.1701673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dijkman K, et al. Prevention of tuberculosis infection and disease by local BCG in repeatedly exposed rhesus macaques. Nat Med. 2019;25(2):255–262. doi: 10.1038/s41591-018-0319-9. [DOI] [PubMed] [Google Scholar]
- 108.Aguilo N, et al. Respiratory immunization with a whole cell inactivated vaccine induces functional mucosal immunoglobulins against tuberculosis in mice and non-human primates. Front Microbiol. 2020;11:1339. doi: 10.3389/fmicb.2020.01339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Darrah PA, et al. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature. 2020;577(7788):95–102. doi: 10.1038/s41586-019-1817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chan J, et al. The role of B cells and humoral immunity in Mycobacterium tuberculosis infection. Semin Immunol. 2014;26(6):588–600. doi: 10.1016/j.smim.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Loxton AG. B cells and their regulatory functions during tuberculosis: latency and active disease. Mol Immunol. 2019;111:145–151. doi: 10.1016/j.molimm.2019.04.012. [DOI] [PubMed] [Google Scholar]
- 112.Hertz D, et al. Increased male susceptibility to Mycobacterium tuberculosis infection is associated with smaller B cell follicles in the lungs. Sci Rep. 2020;10(1):5142. doi: 10.1038/s41598-020-61503-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Phuah J, et al. Effects of B Cell depletion on early Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2016;84(5):1301–1311. doi: 10.1128/IAI.00083-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Joosten SA, et al. Patients with tuberculosis have a dysfunctional circulating B-cell compartment, which normalizes following successful treatment. PLoS Pathog. 2016;12(6):e1005687. doi: 10.1371/journal.ppat.1005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Linge I, et al. B-lymphocytes forming follicle-like structures in the lung tissue of tuberculosis-infected mice: dynamics, phenotypes and functional activity. Tuberculosis (Edinb) 2017;102:16–23. doi: 10.1016/j.tube.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 116.Maglione PJ, et al. B cells moderate inflammatory progression and enhance bacterial containment upon pulmonary challenge with Mycobacterium tuberculosis. J Immunol. 2007;178(11):7222–7234. doi: 10.4049/jimmunol.178.11.7222. [DOI] [PubMed] [Google Scholar]
- 117.Phuah JY, et al. Activated B cells in the granulomas of nonhuman primates infected with Mycobacterium tuberculosis. Am J Pathol. 2012;181(2):508–514. doi: 10.1016/j.ajpath.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Torrado E, et al. Differential and site specific impact of B cells in the protective immune response to Mycobacterium tuberculosis in the mouse. PLoS One. 2013;8(4):e61681. doi: 10.1371/journal.pone.0061681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dallenga T, Schaible UE. Neutrophils in tuberculosis—first line of defence or booster of disease and targets for host-directed therapy? Pathog Dis. 2016;74(3):ftw012. doi: 10.1093/femspd/ftw012. [DOI] [PubMed] [Google Scholar]
- 120.Moliva JI, et al. The lung mucosa environment in the elderly increases host susceptibility to Mycobacterium tuberculosis infection. J Infect Dis. 2019;220(3):514–523. doi: 10.1093/infdis/jiz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Patin EC, et al. Trehalose dimycolate interferes with FcγR-mediated phagosome maturation through Mincle, SHP-1 and FcγRIIB signalling. PLoS One. 2017;12(4):e0174973. doi: 10.1371/journal.pone.0174973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Turner J, Torrelles JB. Mannose-capped lipoarabinomannan in Mycobacterium tuberculosis pathogenesis. Pathog Dis. 2018;76(4):fty026. doi: 10.1093/femspd/fty026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ishikawa E, et al. Recognition of mycobacterial lipids by immune receptors. Trends Immunol. 2017;38(1):66–76. doi: 10.1016/j.it.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 124.Stamm CE, et al. Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol Rev. 2015;264(1):204–219. doi: 10.1111/imr.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Patin EC, et al. Macrophage inducible C-type lectin as a multifunctional player in immunity. Front Immunol. 2017;8:861. doi: 10.3389/fimmu.2017.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Patin EC, et al. Mincle-mediated anti-inflammatory IL-10 response counter-regulates IL-12 in vitro. Innate Immun. 2016;22(3):181–185. doi: 10.1177/1753425916636671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Russell DG, et al. Immunometabolism at the interface between macrophages and pathogens. Nat Rev Immunol. 2019;19(5):291–304. doi: 10.1038/s41577-019-0124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Randall TD. Bronchus-associated lymphoid tissue (BALT) structure and function. Adv Immunol. 2010;107:187–241. doi: 10.1016/B978-0-12-381300-8.00007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ardain A, et al. Group 3 innate lymphoid cells mediate early protective immunity against tuberculosis. Nature. 2019;570(7762):528–532. doi: 10.1038/s41586-019-1276-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hackett EE, et al. Mycobacterium tuberculosis limits host glycolysis and IL-1β by restriction of PFK-M via MicroRNA-21. Cell Rep. 2020;30(1):124–136.e4. doi: 10.1016/j.celrep.2019.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bloom BR, Modlin RL. Mechanisms of defense against intracellular pathogens mediated by human macrophages. Microbiol Spectr. 2016;4(3):10.1128/microbiolspec.MCHD-0006-2015. doi: 10.1128/microbiolspec.MCHD-0006-2015. [DOI] [PubMed] [Google Scholar]
- 132.Fabri M, et al. Vitamin D is required for IFN-gamma-mediated antimicrobial activity of human macrophages. Sci Transl Med. 2011;3(104):104ra2. doi: 10.1126/scitranslmed.3003045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kimmey JM, et al. Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature. 2015;528(7583):565–569. doi: 10.1038/nature16451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Koeken V, et al. Trained innate immunity and resistance to Mycobacterium tuberculosis infection. Clin Microbiol Infect. 2019;25(12):1468–1472. doi: 10.1016/j.cmi.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 135.Reece ST, et al. Mycobacterium tuberculosis-infected hematopoietic stem and progenitor cells unable to express inducible nitric oxide synthase propagate tuberculosis in mice. J Infect Dis. 2018;217(10):1667–1671. doi: 10.1093/infdis/jiy041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tornack J, et al. Human and mouse hematopoietic stem cells are a depot for dormant Mycobacterium tuberculosis. PLoS One. 2017;12(1):e0169119. doi: 10.1371/journal.pone.0169119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kaufmann E, et al. BCG educates hematopoietic stem cells to generate protective innate immunity against tuberculosis. Cell. 2018;172(1-2):176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 138.Mishra BB, et al. Nitric oxide prevents a pathogen-permissive granulocytic inflammation during tuberculosis. Nat Microbiol. 2017;2:17072. doi: 10.1038/nmicrobiol.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yeremeev V, et al. Neutrophils exacerbate tuberculosis infection in genetically susceptible mice. Tuberculosis (Edinb) 2015;95(4):447–451. doi: 10.1016/j.tube.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 140.Lovewell RR, et al. Granulocytes act as a niche for Mycobacterium tuberculosis growth. Mucosal Immunol. 2021;14(1):229–241. doi: 10.1038/s41385-020-0300-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Eum SY, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137(1):122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gideon HP, et al. Neutrophils express pro- and anti-inflammatory cytokines in granulomas from Mycobacterium tuberculosis-infected cynomolgus macaques. Mucosal Immunol. 2019;12(6):1370–1381. doi: 10.1038/s41385-019-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Berry MP, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Prada-Medina CA, et al. Systems immunology of diabetes-tuberculosis comorbidity reveals signatures of disease complications. Sci Rep. 2017;7(1):1999. doi: 10.1038/s41598-017-01767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kumar NP, et al. Persistent inflammation during anti-tuberculosis treatment with diabetes comorbidity. Elife. 2019;8:e46477. doi: 10.7554/eLife.46477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Blomgran R, Ernst JD. Lung neutrophils facilitate activation of naive antigen-specific CD4+ T cells during Mycobacterium tuberculosis infection. J Immunol. 2011;186(12):7110–7119. doi: 10.4049/jimmunol.1100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Corleis B, et al. Escape of Mycobacterium tuberculosis from oxidative killing by neutrophils. Cell Microbiol. 2012;14(7):1109–1121. doi: 10.1111/j.1462-5822.2012.01783.x. [DOI] [PubMed] [Google Scholar]
- 148.Dallenga T, et al. M. tuberculosis-induced necrosis of infected neutrophils promotes bacterial growth following phagocytosis by macrophages. Cell Host Microbe. 2017;22(4):519–530.e3. doi: 10.1016/j.chom.2017.09.003. [DOI] [PubMed] [Google Scholar]
- 149.Dallenga T, et al. Targeting neutrophils for host-directed therapy to treat tuberculosis. Int J Med Microbiol. 2018;308(1):142–147. doi: 10.1016/j.ijmm.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 150.Dorhoi A, Kaufmann SH. Pathology and immune reactivity: understanding multidimensionality in pulmonary tuberculosis. Semin Immunopathol. 2016;38(2):153–166. doi: 10.1007/s00281-015-0531-3. [DOI] [PubMed] [Google Scholar]
- 151.Schaible UE, et al. Strategies to improve vaccine efficacy against tuberculosis by targeting innate immunity. Front Immunol. 2017;8:1755. doi: 10.3389/fimmu.2017.01755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mayer-Barber KD, Sher A. Cytokine and lipid mediator networks in tuberculosis. Immunol Rev. 2015;264(1):264–275. doi: 10.1111/imr.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]