Abstract
Clonal expansion of infected CD4+ T cells is a major mechanism of HIV-1 persistence and a barrier to achieving a cure. Potential causes are homeostatic proliferation, effects of HIV-1 integration, and interaction with antigens. Here, we show that it is possible to link antigen responsiveness, the full proviral sequence, the integration site, and the T cell receptor β-chain (TCRβ) sequence to examine the role of recurrent antigenic exposure in maintaining the HIV-1 reservoir. We isolated CMV- and Gag-responding CD4+ T cells from 10 treated individuals. Proviral populations in CMV-responding cells were dominated by large clones, including clones harboring replication-competent proviruses. TCRβ repertoires showed high clonality driven by converging adaptive responses. Although some proviruses were in genes linked to HIV-1 persistence (BACH2, STAT5B, MKL1), the proliferation of infected cells under antigenic stimulation occurred regardless of the site of integration. Paired TCRβ and integration site analysis showed that infection could occur early or late in the course of a clone’s response to antigen and could generate infected cell populations too large to be explained solely by homeostatic proliferation. Together, these findings implicate antigen-driven clonal selection as a major factor in HIV-1 persistence, a finding that will be a difficult challenge to eradication efforts.
Keywords: AIDS/HIV, Immunology
Keywords: Adaptive immunity, Clonal selection, T cells
Introduction
Latently infected CD4+ T cells represent the main barrier to an HIV-1 cure (1–3). Decay of the latent reservoir, defined as the pool of cells containing inducible and replication-competent proviruses, is slow (4, 5), necessitating life-long antiretroviral therapy (ART). Proviral sequencing and integration site analysis have shown that the proliferation of infected cells is a major mechanism of HIV-1 persistence (6–11). Proliferation begins early after transmission (12) for cells carrying both defective and replication-competent proviruses (13–18).
Despite evidence for persistence by division, factors driving the proliferation of infected cells remain unclear. Homeostatic proliferation has been linked to reservoir maintenance and size (19). Stimulation with IL-7, a key mediator of T cell homeostasis and survival (20), allows CD4+ T cell activation and proliferation without latency reversal (21, 22). In addition, retroviral integration into certain genes may affect cell fate by insertional mutagenesis (23). Proviruses integrated into STAT5B and BACH2 are frequently detected in individuals on ART (8, 9, 24) and are clustered within specific introns in the same orientation relative to host gene transcription, suggesting postintegration positive selection. However, selection of specific integration sites has been observed only for a few of the large number of genes in which integration has been detected.
The majority of HIV-1–infected cells are found in central and effector memory CD4+ T cell subsets (19, 25, 26), which rely on highly regulated programs of proliferation and differentiation driven by antigen stimulation (27, 28). Longitudinal analysis of the HIV-1 reservoir during ART shows fluctuating dynamics of infected cells resembling the typical expansion-contraction phases of adaptive immune responses (29). Proviral DNA has been detected in CD4+ T cells specific for herpesviruses (30, 31), Candida albicans (32), influenza (33), tetanus (33, 34), and HIV-1 itself (35). However, it is unclear whether enrichment of HIV-1 DNA in certain antigen-specific cells is due to a higher susceptibility to HIV-1 infection (35) or a higher proliferation of infected cells driven by recurrent antigen exposure (36). Recently, Mendoza et al. showed that CD4+ T cell clones containing either defective or intact proviruses are present among CD4+ T cells responsive to chronic viral antigens (37). To provide definitive evidence for antigen-driven clonal expansion in vivo and assess its contribution to HIV-1 persistence relative to integration site–driven proliferation, we studied the adaptive CD4+ T cell responses to CMV and HIV-1 Gag in individuals on ART.
Results
Accessing HIV-1–infected antigen-responsive CD4+ T cells.
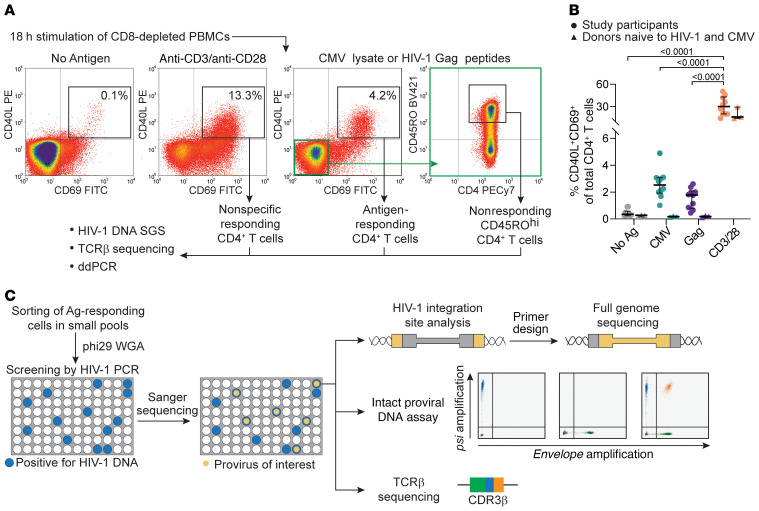
To study antigen-responsive CD4+ T cells infected with HIV-1, we obtained PBMCs from 10 HIV-1–infected adults on suppressive ART for a median of 8 years (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI145254DS1). Cells were depleted of CD8+ T cells and stimulated with CMV or HIV-1 Gag antigens for 18 hours. Because the CMV peptidome is large (213 ORFs) and CMV-specific T cell responses are broad and heterogeneous (38, 39), we used lysates of CMV-infected cells rather than immunodominant proteins (e.g., pp65). To study HIV-1–specific cells, we used overlapping Gag peptides, since responses to class II–restricted Gag epitopes have been well characterized (40). We sorted responding cells on the basis of upregulation of both CD40 ligand (CD40L) and CD69 (Figure 1A). The median frequencies of CMV- and Gag-responding cells among all CD4+ T cells were 2.5% and 1.8%, respectively (Figure 1B), comparable to findings in previous studies (39, 41). For each antigen, we also sorted CD40L and CD69 double-negative cells with high expression of CD45RO to obtain memory populations depleted of responding cells (Figure 1A). PBMCs from HIV–CMV– donors showed no increase in CD40L+CD69+ cells compared with conditions without antigens (Figure 1B and Supplemental Figure 1C). We also sorted CD40L+CD69+ T cells responding to anti-CD3/anti-CD28 beads to obtain a population representing the CD4+ T cell repertoire (Figure 1A). As expected, we observed that a higher proportion of cells became activated in response to anti-CD3/anti-CD28 beads (median of 30%, P < 0.0001), and a lower fraction of these were memory cells (Supplemental Figure 1B).
Figure 1. Experimental approach to study HIV-1–infected, antigen-responding CD4+ T cells.
(A) Experimental design and gating logic to isolate cells responding to stimulation. CD8-depleted PBMCs were stimulated with no antigen, anti-CD3/anti-CD28–conjugated beads, CMV lysates, or HIV-1 Gag peptides. CD4+ T cells upregulating both activation markers CD40L and CD69 were sorted. For CMV and Gag stimulations, nonresponding cells with high CD40RO expression were also isolated (highlighted in green). (B) Frequencies of CD4+ T cells responding to the indicated stimulation. Mean values of all time points are shown for each of 10 participants; horizontal bars show the median and interquartile values. Statistical significance was determined by 1-way ANOVA. (C) Experimental design to characterize the clones of the HIV-1–infected antigen-responding cells. Samples from follow-up time points were processed as in A; responding cells were sorted in small pools and subjected to WGA. Pools containing infected cells were detected by u5-gag or env PCR. Proviruses matching potential clones previously identified by single-genome sequencing were detected by Sanger sequencing. Whole-genome–amplified DNA was then used for integration site analysis, full proviral genome sequencing, the intact proviral DNA assay, and TCRβ sequencing. Ag, antigen.
Initially, DNA from sorted cells was used for single-genome sequencing of HIV-1 proviruses to study the clonality of infected cells, whereas TCRβ sequencing was used to assess the clonality of all sorted cells (see Methods). Subsequently, responding cells from the same donors (Supplemental Figure 1) were sorted in small pools representing limiting dilution with respect to infected cells (Figure 1C). We then used whole-genome amplification (WGA) with phi29 polymerase to generate thousands of copies of cell genomes (Supplemental Figure 3; see complete unedited blots in the supplemental material) (42, 43). PCR on whole-genome–amplified DNA identified HIV-1–positive pools, and then sequencing identified the proviruses of interest for determination of the integration site and the full proviral sequence (Figure 1C).
Identical proviral sequences are common among antigen-specific cells.
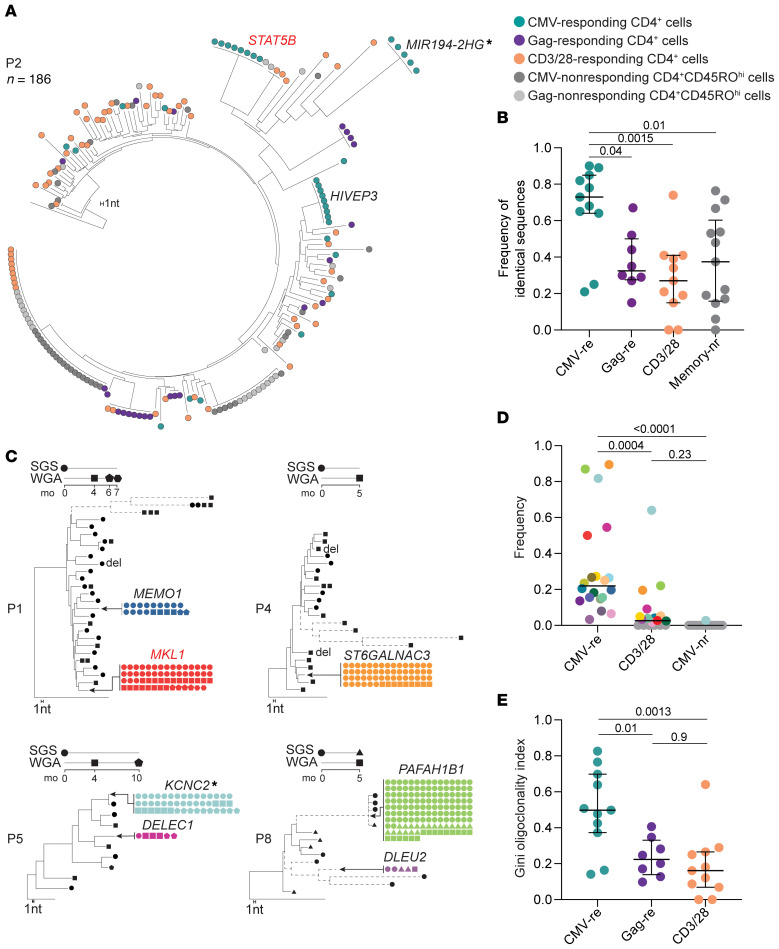
Figure 2A shows a phylogenetic tree of 186 HIV-1 sequences from independent limiting dilution PCRs for a representative participant (P2). We found that higher frequencies of identical sequences (appearing as “rakes” on the tree) were present in both antigen-responding (0.65 and 0.67 for CMV and Gag, respectively) and unrelated memory cells (0.68) compared with cells responding to anti-CD3/anti-CD28 stimulation (0.21), likely reflecting the presence of naive cells in the latter condition. We identified at least 1 set of identical sequences in antigen-responding cells from all participants (10 of 10 for CMV and 8 of 8 for Gag) (Figure 2B, Supplemental Figure 4, and Supplemental Table 2). Integration site analysis proved that most identical proviral sequences were true clones of infected cells (see below). In aggregated single-genome sequences (SGSs) from all participants (n = 1787), a higher fraction of the proviral sequences were identical among CMV-responding cells than among Gag-responding cells, anti-CD3/anti-CD28–responding cells, or memory cells that did not respond to CMV or Gag (Figure 2B). These results link responsiveness to a chronic viral antigen to in vivo proliferation of HIV-1–infected cells. Figure 2C shows clonal proviruses with defined integration sites (see below) dominating HIV-1–infected, CMV-responding cells in 4 participants (P1, P3, P5, P8); we identified these sequences in multiple samples collected up to 10 months apart, demonstrating stable persistence.
Figure 2. HIV-1–infected, CMV-responding cells are enriched in proviral populations generated by clonal expansion.
(A) Representative NJ tree of 186 independent HIV-1 u5-gag DNA SGSs from participant P2, rooted to the HIV-1 subtype C consensus. Sequences from different sorted populations are color coded (see legend). A branch distance of 1 nucleotide is shown on the tree scale. (B) Frequencies of identical proviral sequences within sorted populations for all 10 participants. (C) NJ trees of HIV-1 sequences recovered from CMV-responding CD4+ T cells from 4 participants. Identical sequences are collapsed onto the same branch, and trees are rooted to the HIV-1 subtype B consensus sequence. Dashed branches indicate hypermutated proviruses. Symbols indicate the method and time point used to generate the sequences. Large CMV-specific clones are colored as in D, and the gene containing or closest to the integration site is indicated (see Supplemental Table 3 for detailed integration site data). (D) Dot plot showing increased frequencies of probable clones identified in CMV-responding cells compared with cells responding to anti-CD3/anti-CD28 stimulation or CMV-nonresponding memory cells. Only clones confirmed by integration site or potential clones composed of at least 4 sequences were included. Probable clones are color-coded across stimulation conditions and as in C and Supplemental Figure 4. (E) Dot plot showing higher clonality of proviral populations from CMV-responding cells measured with the Gini coefficient. Horizontal bars show the median and interquartile range. Statistical significance was determined by 1-way ANOVA. CMV-nr, CMV-nonresponding; CMV-re, CMV-responding; Gag-re, Gag-responding; Memory nr, CMV-nonresponding memory cells. Gene symbols next to the tree branches show the genes containing or closest to (indicated by an asterisk) the integration site. Genes previously linked to the persistence of HIV-1–infected cells are highlighted in red
Considering all sets of 4 or more identical sequences to be potential clones, we calculated the frequencies of cells from each potential clone among all HIV-1–infected, CMV-responding cells from each donor. Potential clones had a median frequency of 0.22 (range, 0.03–0.89). In the control population of CMV-nonresponding CD45ROhi cells, the same sequences were either absent or present at a much lower frequency (Figure 2D). In some cases, the relative abundance of these clonal variants was not only dominant among HIV-1–infected, CMV-responding cells, but also among HIV-1–infected cells responding to anti-CD3/anti-CD28 (Supplemental Figure 4), suggesting that in some cases, CMV-specific clones represent the most expanded clones of HIV-1–infected cells. For example, in participant P8, sequences from the provirus integrated in the PAFAH1B1 gene represented 87% of all HIV-1 sequences from CMV-responding cells and 22% of those from cells responding to anti-CD3/anti-CD28 (Figure 2C and Supplemental Figure 4). To compare the clonality of HIV-1–infected cells across conditions, we used the Gini coefficient, a measure of distribution previously used to estimate oligoclonality in human T cell leukemia virus, type 1 (HTLV-1) infection (44). Proviral populations from CMV-responding cells showed significantly higher oligoclonality than did those from Gag- or anti-CD3/anti-CD28–responding cells (Figure 2E). As expected, nonresponding memory cells also contained groups of identical sequences (Figure 2A and Supplemental Figure 4), specific for other unknown antigens and in some cases present at high frequencies.
These results show that clonally expanded proviral populations were present within CMV- and Gag-responding CD4+ T cells; the significantly higher clonality of the infected cells responding to CMV suggests that the chronic immune responses to CMV antigens, characterized by memory inflation (30), contribute to proliferation and maintenance of the HIV-1 reservoir in many infected individuals.
Integration site analysis of HIV-1–infected, antigen-responding clones.
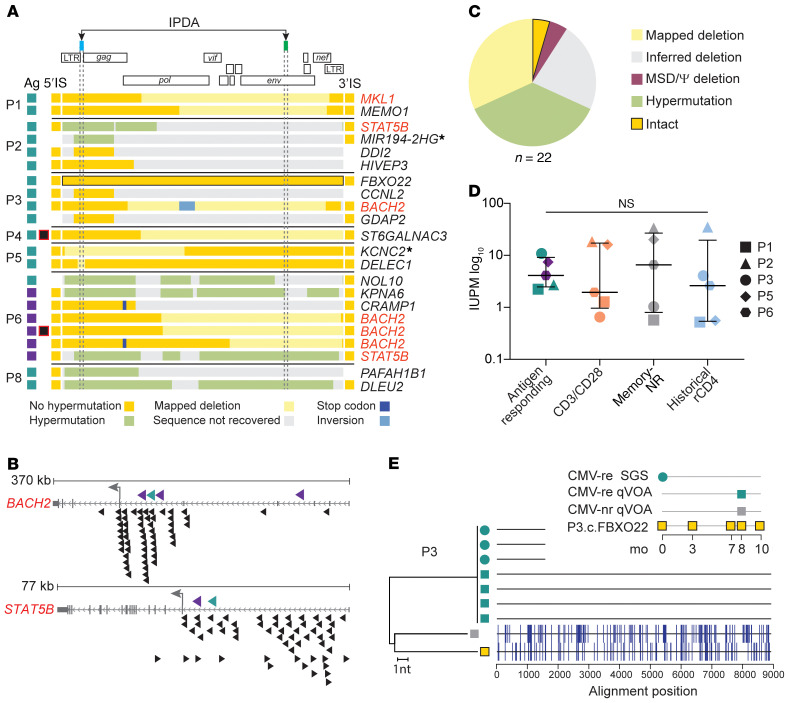
To confirm that the identical proviral sequences result from clonal expansion, we recovered integration sites using linker-mediated PCR (LM-PCR) on whole-genome–amplified DNA from pools of antigen-responding cells that contained identical proviruses (Figure 1C). We retrieved integration sites of 22 expanded clones, 16 from CMV-responding cells and 6 from Gag-responding cells (Figure 3A and Supplemental Table 3). Most were within introns (19 of 22), consistent with previous studies (45, 46). There was no bias in orientation relative to the host gene (9 in the same and 13 in the opposite orientation). Ten integration sites were in cancer-related genes; among these, we found 1 provirus in MKL1, 4 in BACH2, and 2 in STAT5B. HIV-1 integration has been previously described in these 3 genes (Supplemental Table 3). Integration in BACH2 and STAT5B has been linked to HIV-1 persistence due to gene activation by promoter insertion (23). The proviruses identified here shared the same features of those previously found in individuals on ART (Figure 3B). Strikingly, all 4 proviruses in BACH2 were in the same orientation relative to host gene transcription and upstream of the BACH2 translation start site, similar to the 55 BACH2 integrants identified previously (see Methods). Moreover, despite defects including deletions and/or inversions (Figure 3A), these proviruses retained the 5′ long terminal repeat (LTR) and splicing donor sequences required to generate LTR-driven chimeric transcripts (47, 48). Similarly, the 2 proviruses in STAT5B were in the same orientation as STAT5B in intron 1, upstream of the translation start site, as with 42 of the 57 proviruses previously described. These unique features, likely the result of postintegration positive selection, suggest that some HIV-1–infected clones not only proliferate in response to antigen, but also gain a survival advantage from the effects of HIV-1 integration. However, 15 of the 22 proviruses from antigen-responding expanded clones showed integration sites in loci not associated with proliferation of HIV-1–infected cells (Supplemental Table 3). Moreover, some of these were near or within genes with only trace to low mRNA levels in lymphocytes, as their expression is restricted to other tissues (see Methods). CD4+ T cell clones carrying these proviruses have probably undergone extensive proliferation, with negligible contribution of HIV-1 integration–related effects.
Figure 3. Characterization of defective and infectious proviruses from antigen-responding CD4+ T cell clones.
(A) Genome sequences and integration sites (IS) recovered from proviruses in antigen-responding clones obtained from each participant. Each horizontal bar represents 1 provirus found in CMV- or Gag-responding cells (indicated by teal and purple boxes, respectively). Sequence features are color coded (see legend). The intact provirus from participant P3 is highlighted in black. Captured host-proviral junctions are depicted as squares flanking the horizontal bars, and the gene symbols listed on the right show the gene containing or closest to (indicated by an asterisk) the integration site. Genes previously linked to the persistence of HIV-1–infected cells are highlighted in red. Proviruses with asymmetrical aberrant integration are marked with black and red squares. (B) Integration sites in BACH2 and STAT5B found in CMV- and Gag-responding clones (teal and purple, respectively) compared with those previously reported in individuals on ART (black). Arrowhead direction represents proviral orientation relative to host gene transcription. The small gray arrows show host gene transcriptional orientation, and the large gray arrows show the translation start site. (C) Summary of the proviral sequences in A. (D) Frequency of infected cells carrying inducible replication-competent proviruses from 5 participants. Horizontal bars show the median and interquartile values. Statistical significance was determined using a 1-way ANOVA. (E) NJ tree including u5-gag sequences from p24-positive qVOA wells, gDNA SGS, and provirus P3.c.FBXO22, sampled according to the inset legend. The highlighter plot shows mismatches from the top sequences in the tree.
Antigen-responding clones carry both defective and infectious proviruses.
To investigate whether clones of HIV-1–infected, antigen-responding cells carried intact or defective proviruses, we recovered the partial or full-length sequences of the proviruses for which we identified integration sites (Figure 3, A and C). As expected, most genomes (13 of 22) were defective because of mapped or inferred deletions or G-to-A hypermutation (see Supplemental Results). However, we detected 1 intact genome. Despite the limited sampling, this proportion of intact and defective proviruses was consistent with a previous analysis of the proviral landscape in CD4+ T cells (49).
We also carried out quantitative viral outgrowth assays (qVOAs) (50) (Figure 3, D and E, and Supplemental Figure 7) on the sorted cell populations and observed outgrowth from all cell populations from all participants. Infected cell frequencies in infectious units per million (IUPM) were not significantly different for CMV- or Gag-responding cells than for cells responding to anti-CD3/anti-CD28 stimulation or for nonresponding memory cells. In addition, we observed no difference relative to historical qVOAs using resting CD4+ T cells from the same individuals (Figure 3D), suggesting that, at least in these participants, the HIV-1 reservoir was not enriched in cells responding to the antigens tested.
To determine whether the induced infectious proviruses were from clonally expanded cells, we sequenced the HIV-1 RNA from the supernatants of p24-positive wells (Figure 3E and Supplemental Figure 7). In participant P3, we detected viral outgrowth in 4 independent wells seeded with CMV-responding cells (corresponding to 10.8 IUPM). These replication-competent viruses had identical near-full-length genome sequences that were genetically distinct from 1 outgrowth virus recovered from CMV-nonresponding cells (1.03 IUPM). In addition, these 4 identical isolates matched three HIV-1 DNA SGSs found in CMV-responding cells 8 months previously. Interestingly, we did not detect outgrowth of the intact provirus integrated into the FXBO22 gene, which was the most abundant sequence among HIV-1–infected, CMV-responding cells from participant P3 (frequency of 0.2) (Supplemental Figure 4) and persisted across multiple time points (Figure 3E), suggesting a lack of inducibility or the presence of missense mutations that would affect replicative fitness.
These results provide strong evidence that CD4+ T cell clones carrying an inducible replication-competent provirus can be selected over time in response to CMV. However, the size of infectious proviral clones within antigen-specific populations varied, as in the study from Mendoza et al. (37). In participant P2, cells responding to anti-CD3/anti-CD28 and CMV-nonresponding cells had a markedly higher frequency of cells carrying inducible, replication-competent proviruses (18.3 and 33.5 IUPM, respectively, versus 2.7 IUPM in CMV-responding cells). This was likely due to a single large clone specific for an antigen other than CMV (Supplemental Figure 4). Proviral sequences identical to this outgrowth virus were found in PBMCs from 8 months previously, when they represented the most abundant viral variant in both CD3/CD28 and CMV- or Gag-nonresponding memory cells (frequency of 0.15, 0.49, and 0.32, respectively) (Figure 2A and Supplemental Figure 4). Thus, other factors, including antigens not explored here, can lead to massive expansion of clones carrying infectious proviruses.
TCRβ repertoire mirrors the clonality of proviral populations.
VDJ recombination generates a TCR repertoire with a theoretical diversity exceeding the total body number of T cells (up to 1015 cells) (51). Therefore, we used VDJ rearrangements as T cell barcodes to compare clonality in CMV- and Gag-responding cells with that in nonresponding memory T cells and cells activated by anti-CD3/anti-CD28. Despite comparable sequencing depth (Supplemental Figure 8), we found that species richness for TCRβ sequences in CMV- and Gag-responding cells was significantly lower than that for CD3/CD28-responding cells (Figure 4A), and the oligoclonality indices for CMV-responding cells were not only higher than those for CD3/CD28-stimulated cells but also than those for Gag-responding cells (Figure 4B). Thus, patterns of clonality for the whole responding CD4+ T cell population mirrored those of the HIV-1–infected cells within that cell population. Oligoclonality of TCRβ sequences correlated with oligoclonality of proviruses within the relevant cell populations (Spearman’s coefficient r = 0.52, P = 0.01), and samples from CMV-responding cells clustered away from anti-CD3/anti-CD28 and Gag-responding cells (Figure 4C), suggesting that adaptive immune responses globally affected the expansion of HIV-1–infected clones, supporting previous studies describing inflation of memory cell populations driven by CMV (30).
Figure 4. Antigen-responding cells show higher clonality and evidence of convergent selection.
(A) TCR diversity, estimated by the Chao index (93), was lower in antigen-responding cells. Gray circles represent memory cells nonresponsive to either CMV or Gag stimulation (teal and purple borders, respectively). (B) Gini coefficients based on TCRs from CMV-responding cells showed the highest clonality. (C) Correlation of Gini coefficients based on TCR and proviral populations. Spearman’s r value and linear correlation with 95% CIs are shown. (D) log10 abundance of productive TCRs from CMV-responding cells from participant P1 collected at month 6 and month 9 of the study. (E) VDJ sequences from the 10 most abundant clonotypes encoding CASRGSTEAFF. (F) Summed abundance of all degenerate and expanded CDR3β sequences. (G) Frequency of TCR clusters normalized by CDR3β input. Clusters were filtered on the basis of CDR3β of 2 or greater and a Fisher’s exact test P < 0.0001. (H) TCR clusters are plotted on the basis of the number of total CDR3β and Vβ gene scores. Lower scores indicate more homogeneous Vβ. Circle size is scaled to the total sum of TCR templates in each group, indicating clonal expansion of cells within the cluster. (I) Representative TCR clusters involving more than 1 participant, displayed as networks and CDR3β sequence logos. Nodes represent each CDR3β sequence, with circle colors based on participant and circle size representing clonal expansion. Edge colors highlight antigen stimulation (teal for CMV and purple for Gag). CDR3β logos display amino acid representation at each position. The core motif shared by the convergent cluster is colored. The table shows cluster characteristics and shared HLA alleles (see Supplemental Table 4 for additional details). Exclamation marks indicate enrichment of the participants’ HLA allele contributing to the cluster. Horizontal bars show the median and interquartile values. Statistical significance was determined by 1-way ANOVA.
To estimate the distribution and the relative sizes of CMV- and Gag-responding T cell clones within the entire CD4+ T cell repertoire, we identified TCRβ sequences with a significantly (P < 0.01) higher abundance in CMV- and Gag-responding cells than in cells activated by anti-CD3/anti-CD28 stimulation (see Methods). Across 7 participants, we identified an average of 210 (SD ± 103) and 137 (SD ± 82) T cell clones enriched in CMV- and Gag-specific cells, respectively (Supplemental Figure 8). Of note, significantly fewer clones were enriched upon either stimulation (average of 16, SD ± 10, P = 0.0004), likely reflecting cross-reactivity or nonspecific activation. Interestingly, although these CMV- and Gag-reactive T cell clones represented only a small percentage of all CD4+ T cells (average of 2.5% and 1%, respectively; Supplemental Figure 8, C and D), they were among the top expanded T cell clones in the anti-CD3/anti-CD28–responding cell population. CMV-responding T cell clones were particularly dominant (Supplemental Figure 8), as was the case in the analysis of clonal proviral populations among HIV-1–infected cells (Figure 2D and Supplemental Figure 4).
To prove that the striking clonality of CMV-responding CD4+ T cells resulted from antigen-dependent selection and not homeostatic proliferation, we examined the most expanded clonotypes (defined here as cell clones sharing an identical VDJ β sequence) and found that many shared the same amino acid sequence in the CDR3β region, despite different VDJ rearrangements at the nucleotide level. These so-called “degenerate” TCRs are signatures of convergent immune responses, selected over time for binding to specific peptide-loaded MHC molecules (52, 53). Figure 4, D and E, shows one such degenerate TCR. TCRβ sequencing on CMV-responding CD4+ T cells collected 3 months apart from participant P1 showed overlapping distributions of clonal frequencies (suggesting stability over time), including 19 different clonotypes with an identical CDR3β amino acidic sequence (CASRGSTEAFF) (Figure 4E). These rearrangements represented the most abundant CDR3β sequences among CMV-responding cells (7% and 10%, for the 2 time points). Degenerate expanded clones of this type were significantly more abundant in antigen-responding cells (median of 2.8% for CMV and 2% for Gag, respectively) compared with nonresponding and anti-CD3/anti-CD28–stimulated cells (median of 0.5% and 0.003%, respectively) (Figure 4F and Supplemental Figure 8), supporting the hypothesis that CD4+ T cell responses against these 2 chronic infections undergo a process of convergent selection.
To further explore clonal selection based on TCR specificity, we performed a TCR cluster analysis of shared structural features using the GLIPH2 algorithm, which groups TCRs into clusters predicted to bind the same peptide-MHC complex (54). TCRs from CMV-responding cells had a significantly higher frequency of clusters than did those from anti-CD3/anti-CD28–stimulated cells (median of 4.6 vs. 0.8 clusters every 1000 TCR input sequences, P = 0.005; Figure 4G). In addition, a higher proportion of clusters from CMV-responding cells were larger in size (and included a higher number of clonotypes), included expanded clones, and showed a restricted V gene use (P < 0.05; Figure 4H), supporting an overall higher degree of selection toward convergent immune responses. Finally, when we used GLIPH2 on TCRs sampled from all participants (n = 8 for CMV and n = 7 for Gag), we found convergent clusters including CDR3s from multiple participants, likely representing public responses against shared immunodominant epitopes. Four exemplary clusters are described in Figure 4I and Supplemental Table 4. The CDR3 sequences in these clusters showed a restricted use of V and J genes, shared significant motif residues, and were often degenerate. Moreover, the individuals contributing to these clusters shared 1 or more class II HLA alleles. Although it is not technically feasible to extend these TCR analyses to the rare subset of CMV-responding cells that harbor latent proviruses, our results strongly support the conclusion that the clonality of CMV-responding cells is the result of antigen-driven proliferation and not a homeostatic process that occurs independently of TCR specificity.
Coupled quantification of provirus and VDJ rearrangements.
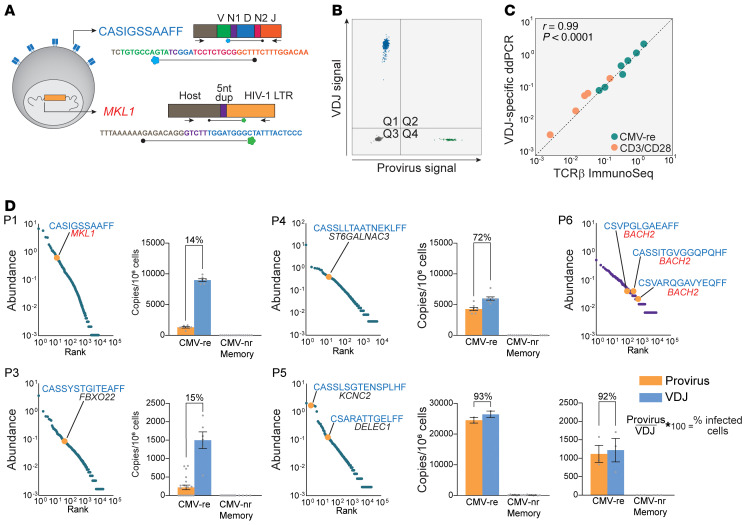
To better understand the nature of antigen-specific CD4+ clones carrying proviruses, we developed a method to identify their cognate TCRβ sequences. We sequenced the TCRβ repertoire from multiple whole-genome–amplified cell pools containing the same provirus and searched for the unique VDJ rearrangement that recurred across all pools carrying a provirus with a specific integration site. To confirm the assignment to TCR/provirus pairs, we leveraged the extraordinarily high diversity of the TCR repertoire and adopted combinatorial statistics previously used to confidently pair α and β chains (see Methods and Supplemental Figure 9A) (55). The patterns of co-occurrence observed for assigned pairs of proviruses and TCRβ sequences indicated that they belonged to the same T cell clones and did not occur together simply by chance (P value between 10–3 and 10–13) (Supplemental Figure 9B). We identified 8 unique pairs across 6 participants, 5 pairs from CMV-responding cells and 3 from Gag-responding cells (Figure 5). T cell clones carrying specific proviruses ranked among the most abundant CMV-responding TCR sequences (Figure 5D). As expected, the TCRs of the 3 Gag-specific clones paired with proviruses were less abundant, in agreement with our analyses of clonality of Gag-responding cells.
Figure 5. Analysis of VDJ and provirus pairs belonging to the same antigen-responding clone.
(A) ddPCR design for duplex quantification of VDJ and proviral copies from gDNA of sorted cells. (B) Representative ddPCR 2D plot of duplex amplification of CASIGSSAAFF and cognate provirus integrated into the MKL1 gene. (C) Quantification of clonotypes by VDJ-specific ddPCR strongly correlated with TCRβ ImmunoSeq. Five CMV-responding clonotypes (described in D) were quantified in sorted cells responding to CMV or anti-CD3/anti-CD28 stimulation; axes represent log10 abundance. (D) log10-ranked abundance plots of CMV- of Gag-responding cells showing HIV-1–infected clonotypes for which both the VDJ rearrangement and the integration site were identified (highlighted in orange). For each pair, bar graphs show the frequency of provirus (orange) and VDJ (blue) copies in CMV-responding and nonresponding memory cells. Provirus-to-VDJ ratios were used to calculate the percentage of a given clone that was HIV-1 infected.
To estimate the fraction of a given CMV-specific CD4+ clone that carried the associated provirus, we designed duplex digital droplet PCR (ddPCR) assays to directly count, within the same sample of sorted cells, both the rearranged VDJ sequence and the host-provirus junction belonging to 5 CMV-specific clones (Figure 5, A–C). These corresponding VDJ and proviral sequences were highly enriched in CMV-responding cells but absent or rare in memory cells not responding to CMV. We calculated the ratio between proviral copies (infected cells) and VDJ copies (total cells in the antigen-specific clone) (Figure 5D). We observed 2 patterns: in 3 clones from 3 participants, only a fraction of all the cells comprising the clone carried the proviruses (range, 14%–72%), suggesting that these clones were expanded before a member of the clone became infected and subsequently proliferated. Conversely, in 2 clones from participant P5, almost 100% of the cells comprising the clone carried the relevant provirus, suggesting that extensive antigen-driven clonal expansion happened after the infection event in one of the progenitor cells of that clone (Figure 6, A and B).
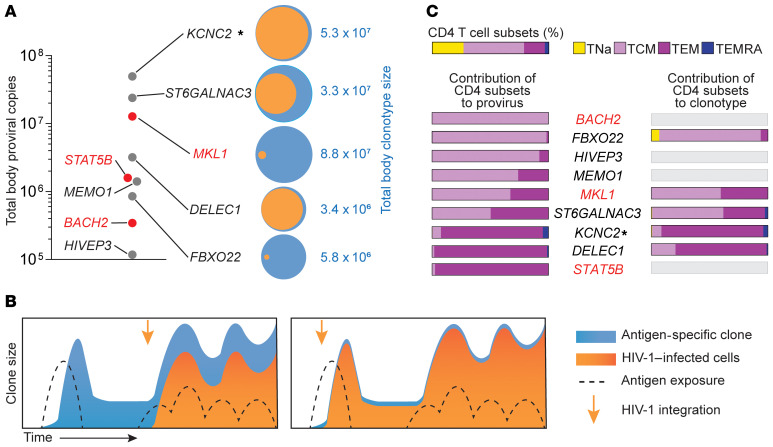
Figure 6. Total body clone sizes and contribution of CD4+ T cell subsets.
(A) Proviral and VDJ frequencies were used to back-calculate total body clone sizes. Clones with integration sites in genes previously linked to the persistence of infected cells are highlighted in red. Venn diagrams show the fraction of a clonotype (orange) carrying its cognate provirus (blue). (B) Two scenarios of infection-expansion dynamics of antigen-driven clonal selection. The left panel shows infection of a clone already expanded in response to antigen, whereas the right panel shows selection occurring after an early infection event. (C) Contribution of CD4+ T cell memory subsets to clones based on provirus and VDJ measurements. Memory subsets are defined as shown in Supplemental Figure 10. Horizontal bars show the relative contribution of memory subsets for each clone. Gene symbols show the genes containing or closest to (indicated by an asterisk) the integration site. Genes previously linked to the persistence of HIV-1–infected cells are highlighted in red.
To investigate the contribution of T cell differentiation to the persistence of these antigen-specific clones, we applied the ddPCR assay to CD4+ T cells subsets sorted on the basis of CCR7 and CD45RA expression levels (Supplemental Figure 10). Although we observed the expected proportions of naive (TNa), central memory (Tcm), effector memory (Tem), and effector memory CD45RA+ (TEMRA) T cells within total CD4+ T cells, most proviral copies were found in Tcm and Tem subsets (Figure 6C and Supplemental Figure 10), in accordance with previous studies based on HIV-1 DNA (19, 25, 26), HIV-RNA (56), and on specific expanded clones carrying proviruses (57, 58). However, we observed a wide variation in the relative contribution of Tcm or Tem cells for different proviruses (from 0% to 100%), suggesting that adaptive immune responses to individual epitopes are heterogeneous and likely influenced by other factors, such as the frequency of peptide-MHC stimulation, TCR affinity (59) and T cell activation. More important, despite this variation, we observed a similar contribution of Tcm and Tem subsets between proviral and VDJ copies of the same clone (Figure 6C), supporting the hypothesis that infected and uninfected cells within an antigen-responding T cell clone are under the same differentiation-proliferation program. This finding further supports the idea that expansion of HIV-1–infected T cell clones is mostly regulated by T cell physiology rather than HIV-1–mediated effects.
To understand the extent of expansion of these HIV-1–infected, CMV-specific clones, we estimated their total body size. We observed striking clone sizes ranging between 105 and 108 cells (Figure 6A). The number of divisions needed to reach such sizes (mean of 21, SD ± 2.9) is achievable, considering the short doubling time of activated T cells (60), but it does not account for cell death (during clonal contraction) and the fact that antigen-driven expansion occurs only for rare (specific) clones and not for the whole T cell population. To investigate whether clones of this size could result from homeostatic proliferation, we calculated the likelihood that a clone could reach a given size by chance if the entire population of CD4+ T cells was maintained by a constant, balanced process of division and death (see Supplemental Methods and Supplemental Table 5). For the sizes of the HIV-1–infected CMV-responsive clones described here, the probabilities approached zero even for high turnover rates, supporting a scenario in which nonrandom events drove the proliferation of rare cells (antigen-specific clonotypes) rather than a homogeneous process of “cohort homeostasis.” Thus, the major driver of infected cell proliferation, and hence HIV-1 persistence, is the response to antigen and not to specificity-independent homeostatic or integration site–related proliferation.
Discussion
The requirement for lifelong treatment because of the HIV-1 reservoir further hinders the scalability and maintenance of HIV-1 care worldwide (61). Thus, a better understanding of the mechanisms of HIV-1 persistence is needed. Two recent studies investigated the immunological factors contributing to the proliferation of the HIV-1 reservoir: Mendoza et al. characterized proviral sequences from cells reactive to viral antigens (37), whereas Gantner et al. profiled TCRβ sequences from cells with inducible p24 expression (58). Here, by combining provirus, integration site, and TCRβ analyses from persistent clones with known specificities, we provide insights into the importance of antigen-driven proliferation, relative to other drivers of clonal expansion, such as integration site effects and homeostatic proliferation. Through the analysis of antigen-responding cells, we identified proviral populations resulting from clonal expansion in all participants. Some clonal proviruses were present at multiple time points for up to 25 months. In CMV-responding cells, proviral populations showed high clonality. Analyses of TCRβ repertoires confirmed the overall higher clonality in CMV-responding cells. TCRs from these cells were characterized by degenerate and often convergent CDR3β sequences, further supporting the conclusion that proliferation of infected cells is driven by adaptive immune responses affecting all cells, not just infected ones. To untangle the role of antigenic pressure relative to HIV-1 insertional mutagenesis, we determined the integration sites of infected antigen-responding clones. We identified CMV- and Gag-specific clones with proviruses in genes previously linked to the proliferation of HIV-1–infected cells (STAT5B, BACH2, MKL1). This is the first evidence to our knowledge that some HIV-1–infected cells persist with more than 1 selective advantage, as recently described in CAR-T cell clones (62, 63). Nevertheless, many CMV-specific clones carried proviruses in loci unlikely to affect T cell phenotype and survival. In addition, the quantification of proviral and VDJ sequences belonging to the same clonotype indicated that in some cases only a fraction of the T cell clone is infected, suggesting antigen-driven clonal expansion before the HIV-1 integration event and subsequent proliferation of infected cells. Together, our work suggests that, although proviruses can disrupt the expression of host genes and promote survival (48), the effect of HIV-1 integration is not indispensable for the expansion of antigen-specific infected cells, in accordance with a recent study on large clones carrying infectious proviruses responsible for low-level viremia (64).
T cell activation reverses latency, which is how the reservoir was discovered. But the extensive proliferation observed here is hard to reconcile with latency reversal, which should render cells susceptible to cytopathic effects or immune clearance. Our results further support the hypothesis that infected cells undergo extensive proliferation without necessarily inducing viral expression (18, 65). The identification of common antigen-specific responses driving the proliferation of cells in the HIV-1 reservoir could have implications for novel therapeutics strategies (36). Such antigens could reverse latency or drive the differentiation of infected cells toward terminal effector subsets with shorter half-lives (66, 67). Sources of antigens driving clonal expansion, like CMV, could be targeted to both reduce chronic inflammation and accelerate reservoir decay (68, 69). CD4+ T cells can be differentially susceptible to HIV-1 according to their specificity (70). For example, although the autocrine production of β-chemokines can protect some CMV-specific cells from HIV-1 infection (71), HIV-1–specific cells are preferentially infected during acute infection and viral recrudescence (35). Since we used a different experimental approach (activation-induced markers CD40L and CD69 instead of intracellular staining of effector molecules) and studied individuals who started ART during chronic infection and were on suppressive ART for years, we were not able to determine whether CMV- and Gag-specific CD4+ T cells had a significantly different frequency of infected cells or confirm the observations from previous studies. However, we observed a striking heterogeneity across participants in the distribution and relative size of antigen-specific clones carrying HIV-1 genomes. Similarly, the recovery of replication-competent virus from antigen-specific cells varied for each participant and showed no reservoir enrichment, overall, in either CMV- or Gag-responding cells. Despite the small number of individuals and the limited range of antigens tested in this study, our results suggest that the specificity of HIV-1–infected cells differs extensively from person to person. In addition, our findings further support the dynamic nature of the reservoir (29). Fluctuations of clone sizes due to episodic exposure to cognate antigen can further complicate efforts to quantify the reservoir and its decay on the basis of measurements of total or intact proviral DNA (72, 73). Future research should investigate other ubiquitous pathogens that typically coinfect individuals living with HIV-1 and commensal microbials that play a role in chronic antigenic stimulation. Moreover, antigen-independent mechanisms that contribute to the reservoir structure need to be explored, such as the proliferation induced by T cell depletion that occurs upon ART introduction (74) or post-therapeutic ablation (31, 75). Similarly, steady-state homeostatic proliferation, which allows the persistence of long-term memory in the absence of antigen, likely plays a role in the maintenance of the HIV-1 reservoir.
In conclusion, our work provides a better understanding of the complex forces driving clonal expansion, showing that antigens can shape the fate of infected cells independently from the effects of HIV-1 integration. Antigen-driven clonal expansion and selection cause HIV-1–infected clones to persist indefinitely, proliferate, and reach sizes that make eradication daunting, except with strategies such as bone marrow transplantation that completely eliminate the adaptive immune repertoire (76–78).
Methods
Study participants.
Characteristics of the study participants are provided in Supplemental Table 1. Participants were HIV-1–infected adults on suppressive ART initiated during chronic infection, with undetectable plasma HIV-1 RNA levels (<50 copies/mL) for 4 or more years. Additional inclusion criteria were CD4+ T cell counts above 400 cells/μL and positive CMV serology. Peripheral blood samples (up to 180 mL) were collected at multiple time points (Supplemental Figure 2). For participants P9 and P10, leukapheresis was performed at a single time point.
Isolation of CD8-depleted PBMCs.
PBMCs were isolated and depleted of CD8+ T cells in a single step by density centrifugation using Ficoll Paque PLUS (GE Healthcare Life Sciences), SepMate tubes (STEMCELL Technologies), and the RosetteSep Human CD8 Depletion Cocktail (STEMCELL Technologies) following the manufacturers’ instructions. For leukapheresis samples, total PBMCs were isolated by density centrifugation and viably frozen. PBMCs were thawed, rested overnight in RPMI with 10% FBS, and depleted of CD8+ T cells using a negative selection step (CD8 MicroBead Kit, LD columns, and QuadMACS Separator; all from Miltenyi Biotec) according to the manufacturer’s instructions.
Isolation of antigen-responding CD4+ T cells.
To identify antigen-responding CD4+ T cells, CD8-depleted PBMCs were stimulated with antigen and costimulatory molecules (CD28+CD49d; BD Biosciences; 0.5 μg/mL) in the presence of CD40-blocking antibody to reduce CD40L internalization (Miltenyi Biotec; 1 μg/mL) and 10 μM enfuvirtide (T20) to prevent new infection events. We stimulated 10–100 million PBMCs with either lysates of CMV-infected fibroblasts (Virusys, 10 μg/mL) or overlapping Gag 15 mer peptides (HIV-1 Gag peptide pool, JPT peptides, 1 μg/peptide/mL). For each stimulation, aliquots of PBMCs (5–15 million cells) were cultured with no antigens (costimulation-only control) or with anti-CD3/anti-CD28 antibodies bound to magnetics beads as a positive, nonspecific control(25 μL/106 cells, Thermo Fisher Scientific). Cells were stimulated for 18 hours and then washed, incubated with FcgR Block (BD Pharmingen) at 25°C for 10 minutes, and then stained for 30 minutes on ice with an APC-labeled antibody against CD3 (BioLegend; clone UCHT1), phycoerythrin-Cy7–labeled (PE-Cy7–labeled) antibody against CD4 (BioLegend; clone RPA-T4), BV421-labeled antibody against CD45RO (BioLegend; clone UCHL1), PE-labeled antibody against CD154 (BioLegend; clone 24-31), FITC-labeled antibody against CD69 (BioLegend; clone FN50), and PE-Cy5–labeled antibodies against CD14 (Thermo Fisher Scientific; clone 61D3), CD16 (BioLegend; clone 3G8), and CD20 (BioLegend; clone 2H7). Dead cells were excluded using propidium iodide. Cells stained with single fluorophore-labeled antibodies, costimulation-only controls, and positive controls were used to set sorting gates. Cells were sorted using either the Beckman Coulter MoFlo Legacy or the XDP cell sorter. A representative gating strategy and sorting logic is provided in Figure 1 and Supplemental Figure 1A. To test nonspecific activation of cells upon stimulation with CMV and Gag antigens, PBMCs from 3 healthy donors negative for HIV-1 and CMV were stimulated (Supplemental Figure 1C).
Extraction of genomic DNA.
Sorted cells were pelleted and immediately lysed for genomic DNA (gDNA) extraction. If samples had more than 106 cells, the QIAamp DNA Mini Kit (QIAGEN) was used, otherwise the Quick-DNA Microprep Kit (Zymo Research) was used according to the manufacturer’s instructions. NanoDrop 2000 and Qubit 3 Broad Range (Thermo Fisher Scientific) were used to quantify gDNA concentrations.
HIV-1 DNA single-genome sequencing.
gDNA was subjected to limiting dilution nested PCR using Platinum Taq High Fidelity DNA Polymerase (Thermo Fisher Scientific) as previously described (79). To maximize the chances of identifying identical proviral sequences across participants, we used a database of 431 near-full-length proviral sequences (80) to identify a 1.5 kb amplicon covering a region that was the least affected by deletions. In addition, degenerate positions were introduced into the primer sequences to allow amplification of sequences containing mismatches caused by viral diversification or APOBECG/F-induced G-to-A mutations. Primers were designed to amplify the U5-gag region (HXB2 positions 584-1841). A second set of primers was used to amplify a region of the envelope (env) gene (HXB2 positions 6980–8036). Outer PCRs were performed with the following cycles: 94°C for 2 minutes, 44 cycles of 94°C for 30 seconds, 50°C for 30 seconds, 72°C for 2 minutes, followed by 72°C for 3 minutes and a hold at 4°C. Inner PCRs were performed with the following cycles: 94°C for 2 minutes, 41 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes, and then 72°C for 3 minutes and a hold at 4°C. PCR product from positive reactions was purified and sent for direct Sanger sequencing (GENEWIZ). The oligonucleotide sequences used for PCR and Sanger sequencing are listed in Supplemental Table 6.
WGA.
Antigen-responding cells from follow-up samples were sorted into 96-well plates in small pools (range, 15–500 cells) to achieve 30% or fewer HIV-1 DNA–positive wells. Sorted pools were then subjected to WGA by multiple displacement amplification (MDA) using the Advance Single Cell Repli-G Kit (QIAGEN) following the manufacturer’s protocol. Whole-genome–amplified samples were diluted 1:20 in 10 mM Tris HCl, pH 8.0, quantified by Qubit 3 Broad Range (Thermo Fisher Scientific), and used as templates for HIV-1 bulk PCR (same methods used for single-genome sequencing). Sanger sequencing from positive PCR reactions allowed the selection of HIV-1–positive wells containing individual proviruses of interest.
Integration site analysis.
Integration site analysis was performed using ligation-mediated PCR. In brief, whole-genome–amplified DNA from pools of cells positive for proviruses of interest were fragmented by sonication. DNA adapters were ligated onto the DNA ends. PCR was carried out between the adaptor and LTR ends, and then sequences were determined from both ends of the PCR products using the Illumina method as previously described (81, 82). Abundance of individual clones was measured by quantifying the numbers of positions of DNA linkers associated with each unique integration site sequence, which scores the numbers of DNA chains at the start of the procedure associated with each cell clone (the sonic abundance method, ref. 83). Some samples were analyzed with the Lenti-X Integration Site Analysis Kit (Takara Bio) using primers designed to amplify both LTR ends. Libraries were prepared to capture the integration sites at both 5′ and 3′ host-provirus junctions. When necessary, the primers were modified to reflect sequence variation in the HIV LTRs within the participant studied. The oligonucleotide sequences used for integration site analysis are provided in Supplemental Table 6. The code base for the bioinformatic pipeline used to process high-throughput sequencing data is available at Zenodo: https://zenodo.org/record/3885688#.X_M-iKpKiMI Cancer-related genes were determined as previously described (81, 82). Expression of genes in CD4+ T cells and in other tissues was assessed through the Human Cell Atlas (http://immunecellatlas.net/) and the GTEx Project portal (https://www.gtexportal.org/home/). Only integration sites recovered from more than 1 sample and/or confirmed with a second method were used in the final analysis (see Supplemental Table 3). To recover integration sites in BACH2 and STAT5B genes from previous publications, we used the NCI Retrovirus Integration Database (84). Only data from studies in vivo were used for the analysis in Figure 3B.
Near-full-length proviral sequencing.
To recover proviral sequences from whole-genome –amplified DNA, 3 different methods were used on the basis of the type of provirus. Given the average size of DNA fragments generated by WGA (~3000 nt), 5 overlapping nested PCR amplicons (~2000 nt each) were used as previously described (43). Proviruses with potential large deletions (inferred by the failure of 1 or more of the overlapping PCRs and intact proviral DNA assay [IPDA]) were amplified with outer and nested primers as described by Hiener et al. (26). Given the large deletions encompassing one of the LTR regions, some proviruses could not be amplified with this method. To bypass this problem, we designed primers in the human genome flanking the integration site. These were paired with HIV-specific primers annealing to proviral regions outside of inferred deletions and used for nested PCR amplification using Platinum Taq High Fidelity DNA Polymerase (Thermo Fisher Scientific) with modifications of the single-genome sequencing protocol described above. Outer PCRs were performed with the following parameters: 94°C for 2 minutes, 44 cycles of 94°C for 30 seconds, 50°C for 30 seconds, 72°C for X minutes, and then 72°C for 3 minutes and a hold at 4°C. Inner PCRs were performed with the following parameters: 94°C for 2 minutes, 41 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for X minutes, and then 72°C for 3 minutes and a hold at 4°C. The duration of the elongation step (X minutes) was adjusted to the estimated maximum size of the amplicon (1 minute every 1000 nt). Primer details are provided in Supplemental Table 6.
IPDA.
The IPDA was performed as previously described (80). Because of the sequence variation, a custom ψ-probe was designed for participant P2 (FAM-TGGCGTACTCACCAGG-MGBNFQ; Applied Biosystems), and a custom ψ-forward primer was designed for participant P3 (CAGGACTCGGCTTGCTGAGC).
qVOA.
We performed qVOAs as previously described (50). MOLT-4/CCR5 cells were added on day 2, and culture supernatants were examined for the p24 viral capsid protein by ELISA (PerkinElmer) after 14, 21, and 28 days. The results are expressed as IUPM CD4+ T cells, calculated using the maximal likelihood as previously described (85). Supernatants positive for p24 were collected, spun at 400g for 10 minutes to remove cells and debris, and stored at –80°C. Cell-free HIV-RNA was extracted as previously described (86) and used for cDNA synthesis with SuperScript IV Reverse Transcriptase (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. Reverse transcription was conducted at 55°C for 50 minutes followed by inactivation at 85°C for 10 minutes and a hold at 4°C, and cDNA was treated with RNAse H (Thermo Fisher Scientific) (1 unit) at 37°C for 20 minutes. For cDNA synthesis, 2 separate reactions were used for the 5′ and 3′ halves of the HIV-1 genome, with the following gene-specific primers: SCO5R (AGCTCTTCGTCGCTGTCTCCGCTT) and PARO (TTTTTGAAGCACTCAAGGCAAG). Five overlapping PCR amplicons were used to reconstruct partial or near-full-genome sequences as previously described (43, 87). The primers used for direct Sanger sequencing have been previously published (18).
Bioinformatics analysis of HIV-1 sequences.
Raw data from Sanger sequencing were analyzed in Geneious to resolve base call conflicts and eliminate sequences with poor quality or double peaks (more than one HIV-1 variant per PCR reaction). Sequence contigs were aligned with ClustalW (88), G-to-A hypermutants were identified with Hypermut 2.0 (https://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html), and codon alignments were analyzed with GeneCutter (https://www.hiv.lanl.gov/content/sequence/GENE_CUTTER/cutter.html) to identify premature stop codons and frameshifts. To confirm the lack of genetic defects affecting infectivity, we used the Proviral Sequence Annotation & Intactness Test (Proseq-IT) (89). ElimDupes was used to identify and collapse identical sequences (https://www.hiv.lanl.gov/content/sequence/elimdupesv2/elimdupes.html). Neighbor-joining (NJ) trees were constructed in MEGA 7.0 (90) with a subtype-specific HIV-1 consensus as the outgroup. The phylogenetic structure was tested by bootstrap analysis (1000 replicates). To estimate the oligoclonality of proviral populations, we used the Gini coefficient of inequality (91), calculated in RStudio with the ineq R package (https://cran.rproject.org/web/packages/ineq/index.html) and corrected for small samples (92). This measurement of sample dispersion provides an estimate of whether the proviral sequences within a sample are evenly distributed (values approaching 0) or dominated by groups of identical sequences (values approaching 1).
TCRβ immunosequencing.
gDNA was isolated from cell samples, quantified as described above, and diluted in Tris-acetate-EDTA to a concentration of 10 ng/μL (for a total of up to 1 μg per sample). TCRβ sequencing data were generated using the ImmunoSEQ hsTCRB assay, version 4, in survey mode (Adaptive Biotechnologies).
TCRβ analysis.
TCRβ sequencing data were analyzed with ImmunoSeq Analyzer 3.0 (Adaptive Biotechnologies). Diversity analyses were conducted on nucleotide sequences from productive VDJ rearrangements. The lower bound estimate of diversity was calculated with the improved Chao index (93). TCR clonality estimates were based on the Gini coefficient (as described above) calculated in R with the Immunarch package (https://cloud.r-project.org/package=immunarch). Differential abundance analysis of VDJ rearrangements between populations of sorted cells was conducted in ImmunoSeq Analyzer 3.0 as previously described (94). Rearrangements with different nucleotide sequences encoding for the same CDR3β amino acid sequences were considered distinct clonotypes. Identification and tallying of degenerate CDR3β sequences were performed in Windows Excel. GLIPH2 (54) was used to cluster TCRs on the basis of predicted binding to peptide-loaded MHC. The GLIPH2 pipeline was applied to TCR data sets obtained from individual participants and merged sets from all participants. For clustering based on global similarity, member CDR3s need to be of the same length and differ at the same position. Amino acids were interchangeable only if they had a positive score in BLOSUM-62 matrix (54). Analysis and visualization of significant CDR3β convergence groups were performed in RStudio. Additional details on the 4 TCR clusters shown in Figure 4I are provided in Supplemental Table 4. Visualization of CDR3β networks and sequence logos was conducted with Gephi (https://gephi.org/) and Weblogo (95), respectively. The HLA haplotypes of the participants are shown in Supplemental Table 7.
T cell subset analysis.
A fraction of CD8-depleted PBMCs, isolated as described above, were dedicated to sort CD4+ T cell subsets. Cells were incubated with FcgR Block (BD Pharmingen) at room temperature for 10 minutes. Cells were stained, followed by a 30-minute incubation on ice, with an APC-labeled antibody against CD3 (BioLegend; clone UCHT1), PE-Cy7–labeled antibody against CD4 (BioLegend; clone RPA-T4), BV421-labeled antibody against CD45RA (BD Biosciences; clone HI100), PE-labeled antibody against CCR7 (BioLegend; clone G043H7), and PE-Cy5–labeled antibodies against CD14 (Thermo Fisher Scientific; clone 61D3), CD16 (BioLegend; clone 3G8), and CD20 (BioLegend; clone 2H7). Dead cells were excluded using propidium iodide. Cells stained with single fluorophore-labeled antibodies were used to set sorting gates. A representative gating strategy and sorting logic is provided in Supplemental Figure 10A. CD45RA expression was used to distinguish naive-like (Na) and terminally differentiated cells from memory cells. Tcm cells were distinguished from Tem cells by the expression of CCR7. TNa, Tcm, Tem, and effector memory CD45RA-positive (EMRA) cell subsets were sorted using either the Beckman Coulter MoFlo Legacy or the XDP cell sorters.
Quantification of specific proviruses and VDJ rearrangements.
See Supplemental Methods.
Assignment of TCR-provirus pairs from antigen-reactive clonotypes.
See Supplemental Methods.
Total body size estimates and likelihood of homeostatic proliferation.
See Supplemental Methods.
Data and software availability.
The HIV-1 sequences are available in the NCBI’s GenBank (accession numbers MW255985-MW256411, MW262767-MW262790 and MW309883-MW309909). Integration site sequencing data can be found in the NCBI’s Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/PRJNA637643). The code base for the bioinformatic pipeline used to process high-throughput integration site sequencing data is available at the following site: https://zenodo.org/record/3885688#.X_M0VKpKiMI Integration site data have also been submitted to the NCI’s Retrovirus Integration Database (RID) (https://rid.ncifcrf.gov). TCRβ sequencing data can be accessed through the ImmuneAccess database (https://clients.adaptivebiotech.com/pub/simonetti-2020-jci).
Statistics.
Descriptive statistics, tests for normality, Spearman’s correlation, 2-tailed Student’s t test, and 1-way ANOVA were used to determine statistical significance with GraphPad Prism 8.0 (GraphPad Software). A P value of less than 0.05 was considered significant, unless otherwise stated.
Study approval.
The Johns Hopkins IRB and the UCSF Committee on Human Research approved this study. All participants provided written informed consent before enrollment in this study.
Author contributions
FRS, JDS, and RFS conceptualized the study. FRS, GPS, JD, KR, SAB, KJK, JAW, and JL performed experiments. HZ and JBM performed cell sorting. KM, HER, CLN, JKE, FDB, and FRS conducted integration site analysis. ALH provided the mathematical model of homeostatic proliferation. FRS analyzed the data and generated the figures. SAB, RH, and SGD provided participant samples. FRS and RFS wrote the manuscript with input from all authors.
Supplementary Material
Acknowledgments
We thank the participants who volunteered to take part in this study and their families. We thank Guido Massaccesi and Andrea L. Cox for providing PBMCs from donors with known cell-mediated responses to CMV. We thank Srona Sengupta, Annie A. Antar, Janelle Montagne, and H. Benjamin Larman for discussions leading to this work, and Monica Sullivan for the administrative support. This work was supported by the NIH Martin Delaney, Beat-HIV (UM1 AI126620), and Delaney AIDS Research Enterprise (DARE) (UM1 AI12661) Collaboratories; the Howard Hughes Medical Institute; and the Bill and Melinda Gates Foundation (OPP1115715). This work was also supported by NIH grants DP5OD019851, U19-AI117950, R01AI129661, and R01CA241762; the Penn Center for AIDS Research (P30AI045008); and the PennCHOP Microbiome Program.
Version 1. 12/10/2020
In-Press Preview
Version 2. 02/01/2021
Electronic publication
Footnotes
Conflict of interest: Aspects of the intact proviral DNA assay (IPDA) are the subject of a patent application (“Compositions and methods Related to characterizing proviral reservoirs,” PCT/US16/28822) filed by Johns Hopkins University, and RFS is one of the inventors on this application. RFS is a consultant on cure-related HIV research for Merck and AbbVie.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(3):e145254.https://doi.org/10.1172/JCI145254.
Contributor Information
Francesco R. Simonetti, Email: fsimonetti@jhmi.edu.
Hao Zhang, Email: hzhang5@jh.edu.
Garshasb P. Soroosh, Email: gsoroosh@jhmi.edu.
Jiayi Duan, Email: jduan7@jhu.edu.
Kyle Rhodehouse, Email: krhodeh1@jhmi.edu.
Alison L. Hill, Email: alhill@fas.harvard.edu.
Subul A. Beg, Email: subul.beg@gmail.com.
Kevin McCormick, Email: kevinmcc@pennmedicine.upenn.edu.
Hayley E. Raymond, Email: hayley.raymond@pennmedicine.upenn.edu.
Christopher L. Nobles, Email: chris.l.nobles@gmail.com.
John K. Everett, Email: everj@pennmedicine.upenn.edu.
Kyungyoon J. Kwon, Email: kjkwon29@gmail.com.
Jennifer A. White, Email: jauslan2@jhmi.edu.
Jun Lai, Email: lai@jhmi.edu.
Joseph B. Margolick, Email: jmargol1@jhu.edu.
Rebecca Hoh, Email: rhoh@php.ucsf.edu.
Steven G. Deeks, Email: sdeeks@php.ucsf.edu.
Frederic D. Bushman, Email: bushman@mail.med.upenn.edu.
Janet D. Siliciano, Email: Jsilicia@jhmi.edu.
Robert F. Siliciano, Email: rsiliciano@jhmi.edu.
References
- 1.Finzi D, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 2.Wong JK, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siliciano JD, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 5.Crooks AM, et al. Precise quantitation of the latent HIV-1 reservoir: implications for eradication strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner TA, et al. An increasing proportion of monotypic HIV-1 DNA sequences during antiretroviral treatment suggests proliferation of HIV-infected cells. J Virol. 2013;87(3):1770–1778. doi: 10.1128/JVI.01985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reeves DB, et al. A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat Commun. 2018;9(1):1–16. doi: 10.1038/s41467-017-02088-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McManus WR, et al. HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J Clin Invest. 2019;129(11):4629–4642. doi: 10.1172/JCI126714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coffin JM, et al. Clones of infected cells arise early in HIV-infected individuals. JCI Insight. 2019;4(12):e128432. doi: 10.1172/jci.insight.128432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamichi H, et al. Lifespan of effector memory CD4+ T cells determined by replication-incompetent integrated HIV-1 provirus. AIDS. 2014;28(8):1091–1099. doi: 10.1097/QAD.0000000000000223. [DOI] [PubMed] [Google Scholar]
- 14.Cohn LB, et al. HIV-1 integration landscape during latent and active infection. Cell. 2015;160(3):420–432. doi: 10.1016/j.cell.2015.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorenzi JC, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci U S A. 2016;113(49):E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med. 2017;214(4):959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bui JK, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 2017;13(3):e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29(6):848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Bosque A, et al. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7(10):e1002288. doi: 10.1371/journal.ppat.1002288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandergeeten C, et al. Interleukin-7 promotes HIV persistence during antiretroviral therapy. Blood. 2013;121(21):4321–4329. doi: 10.1182/blood-2012-11-465625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushman FD. Retroviral insertional mutagenesis in humans: evidence for four genetic mechanisms promoting expansion of cell clones. Mol Ther. 2020;28(2):352–356. doi: 10.1016/j.ymthe.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda T, et al. Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J Infect Dis. 2007;195(5):716–725. doi: 10.1086/510915. [DOI] [PubMed] [Google Scholar]
- 25.Jaafoura S, et al. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T cells. Nat Commun. 2014;5:5407. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hiener B, et al. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep. 2017;21(3):813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Restifo NP, Gattinoni L. Lineage relationship of effector and memory T cells. Curr Opin Immunol. 2013;25(5):556–563. doi: 10.1016/j.coi.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shan L, et al. Transcriptional reprogramming during effector-to-memory transition renders CD4+ T cells permissive for latent HIV-1 infection. Immunity. 2017;47(4):766–775.e3. doi: 10.1016/j.immuni.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Z, et al. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci U S A. 2018;115(11):E2575–E2584. doi: 10.1073/pnas.1720665115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abana CO, et al. Cytomegalovirus (CMV) epitope-specific CD4+ T cells are inflated in HIV+ CMV+ subjects. J Immunol. 2017;199(9):3187–3201. doi: 10.4049/jimmunol.1700851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henrich TJ, et al. Human immunodeficiency virus type 1 persistence following systemic chemotherapy for malignancy. J Infect Dis. 2017;216(2):254–262. doi: 10.1093/infdis/jix265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu F, et al. Sequential dysfunction and progressive depletion of candida albicans-specific CD4 T cell response in HIV-1 infection. PLoS Pathog. 2016;12(6):e1005663. doi: 10.1371/journal.ppat.1005663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones RB, et al. Short communication: HIV type 1 accumulates in influenza-specific T cells in subjects receiving seasonal vaccination in the context of effective antiretroviral therapy. AIDS Res Hum Retroviruses. 2012;28(12):1687–1692. doi: 10.1089/aid.2012.0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hey-Nguyen WJ, et al. HIV-1 DNA is maintained in antigen-specific CD4+ T cell subsets in patients on long-term antiretroviral therapy regardless of recurrent antigen exposure. AIDS Res Hum Retroviruses. 2019;35(1):112–120. doi: 10.1089/aid.2018.0235. [DOI] [PubMed] [Google Scholar]
- 35.Douek DC, et al. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417(6884):95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 36.Cohn LB, et al. The biology of the HIV-1 latent reservoir and implications for cure strategies. Cell Host Microbe. 2020;27(4):519–530. doi: 10.1016/j.chom.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mendoza P, et al. Antigen-responsive CD4+ T cell clones contribute to the HIV-1 latent reservoir. J Exp Med. 2020;217(7):e20200051. doi: 10.1084/jem.20200051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sylwester AW, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202(5):673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. Heterogeneity of CD4+ and CD8+ T-cell responses to cytomegalovirus in HIV-infected and HIV-uninfected men who have sex with men. J Infect Dis. 2014;210(3):400–404. doi: 10.1093/infdis/jiu093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann DE, et al. Comprehensive analysis of human immunodeficiency virus type 1-specific CD4 responses reveals marked immunodominance of gag and nef and the presence of broadly recognized peptides. J Virol. 2004;78(9):4463–4477. doi: 10.1128/JVI.78.9.4463-4477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morou A, et al. Altered differentiation is central to HIV-specific CD4+ T cell dysfunction in progressive disease. Nat Immunol. 2019;20(8):1059–1070. doi: 10.1038/s41590-019-0418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patro SC, et al. Combined HIV-1 sequence and integration site analysis informs viral dynamics and allows reconstruction of replicating viral ancestors. Proc Natl Acad Sci U S A. 2019;116(51):25891–25899. doi: 10.1073/pnas.1910334116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Einkauf KB, et al. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest. 2019;129(3):988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turpin J, et al. Impact of hepatitis B Virus coinfection on human T-lymphotropic virus type 1 clonality in an indigenous population of central Australia. J Infect Dis. 2019;219(4):562–567. doi: 10.1093/infdis/jiy546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schroder AR, et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110(4):521–529. doi: 10.1016/S0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 46.Han Y, et al. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol. 2004;78(12):6122–6133. doi: 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cesana D, et al. HIV-1-mediated insertional activation of STAT5B and BACH2 trigger viral reservoir in T regulatory cells. Nat Commun. 2017;8(1):498. doi: 10.1038/s41467-017-00609-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu R, et al. Single-cell transcriptional landscapes reveal HIV-1-driven aberrant host gene transcription as a potential therapeutic target. Sci Transl Med. 2020;12(543):eaaz0802. doi: 10.1126/scitranslmed.aaz0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Laird GM, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradley P, Thomas PG. Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu Rev Immunol. 2019;37:547–570. doi: 10.1146/annurev-immunol-042718-041757. [DOI] [PubMed] [Google Scholar]
- 52.Wendel BS, et al. The receptor repertoire and functional profile of follicular T cells in HIV-infected lymph nodes. Sci Immunol. 2018;3(22):eaan8884. doi: 10.1126/sciimmunol.aan8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia Q, et al. Diversity index of mucosal resident T lymphocyte repertoire predicts clinical prognosis in gastric cancer. Oncoimmunology. 2015;4(4):e1001230. doi: 10.1080/2162402X.2014.1001230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang H, et al. Analyzing the Mycobacterium tuberculosis immune response by T-cell receptor clustering with GLIPH2 and genome-wide antigen screening. Nat Biotechnol. 2020;38(10):1194–1202. doi: 10.1038/s41587-020-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howie B, et al. High-throughput pairing of T cell receptor α and β sequences. Sci Transl Med. 2015;7(301):301ra131. doi: 10.1126/scitranslmed.aac5624. [DOI] [PubMed] [Google Scholar]
- 56.Grau-Exposito J, et al. A novel single-cell FISH-flow assay identifies effector memory CD4+ T cells as a major niche for HIV-1 transcription in HIV-infected patients. mBio. 2017;8(4):e00876–17. doi: 10.1128/mBio.00876-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson EM, et al. Dynamic shifts in the HIV proviral landscape during long term combination antiretroviral therapy: implications for persistence and control of HIV infections. Viruses. 2020;12(2):136. doi: 10.3390/v12020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gantner P, et al. Single-cell TCR sequencing reveals phenotypically diverse clonally expanded cells harboring inducible HIV proviruses during ART. Nat Commun. 2020;11(1):4089. doi: 10.1038/s41467-020-17898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schober K, et al. Reverse TCR repertoire evolution toward dominant low-affinity clones during chronic CMV infection. Nat Immunol. 2020;21(4):434–441. doi: 10.1038/s41590-020-0628-2. [DOI] [PubMed] [Google Scholar]
- 60.De Boer RJ, et al. Different dynamics of CD4+ and CD8+ T cell responses during and after acute lymphocytic choriomeningitis virus infection. J Immunol. 2003;171(8):3928–3935. doi: 10.4049/jimmunol.171.8.3928. [DOI] [PubMed] [Google Scholar]
- 61.Ndung’u T, et al. Why and where an HIV cure is needed and how it might be achieved. Nature. 2019;576(7787):397–405. doi: 10.1038/s41586-019-1841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fraietta JA, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. 2018;558(7709):307–312. doi: 10.1038/s41586-018-0178-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nobles CL, et al. CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J Clin Invest. 2020;130(2):673–685. doi: 10.1172/JCI130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Halvas EK, et al. HIV-1 viremia not suppressible by antiretroviral therapy can originate from large T cell clones producing infectious virus. J Clin Invest. 2020;130(11):5847–5857. doi: 10.1172/JCI138099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Musick A, et al. HIV infected T cells can proliferate in vivo without inducing expression of the integrated provirus. Front Microbiol. 2019;10:2204. doi: 10.3389/fmicb.2019.02204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Macallan DC, et al. Rapid turnover of effector-memory CD4+ T cells in healthy humans. J Exp Med. 2004;200(2):255–260. doi: 10.1084/jem.20040341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grossman Z, et al. ‘Rinse and replace’: boosting T cell turnover to reduce HIV-1 reservoirs. Trends Immunol. 2020;41(6):466–480. doi: 10.1016/j.it.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 68.Hunt PW, et al. Valganciclovir reduces T cell activation in HIV-infected individuals with incomplete CD4+ T cell recovery on antiretroviral therapy. J Infect Dis. 2011;203(10):1474–1483. doi: 10.1093/infdis/jir060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gianella S, et al. Replication of human herpesviruses is associated with higher HIV DNA levels during antiretroviral therapy started at early phases of HIV infection. J Virol. 2016;90(8):3944–3952. doi: 10.1128/JVI.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Geldmacher C, Koup RA. Pathogen-specific T cell depletion and reactivation of opportunistic pathogens in HIV infection. Trends Immunol. 2012;33(5):207–214. doi: 10.1016/j.it.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Casazza JP, et al. Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection. PLoS Pathog. 2009;5(10):e1000646. doi: 10.1371/journal.ppat.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peluso MJ, et al. Differential decay of intact and defective proviral DNA in HIV-1-infected individuals on suppressive antiretroviral therapy. JCI Insight. 2020;5(4):e132997. doi: 10.1172/jci.insight.132997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gandhi RT, et al. Selective decay of intact HIV-1 proviral DNA on antiretroviral therapy. J Infect Dis. doi: 10.1093/infdis/jiaa532. [published online August 21, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. doi: 10.1101/2020.04.06.20053231. Tettamanti Boshier FA, et al. Blind uneven proliferation of CD4 + T cells during primary infection generates the majority of the HIV reservoir. [preprint]. Posted on medRxiv April 10, 2020. [DOI]
- 75.Eberhard JM, et al. Vulnerability to reservoir reseeding due to high immune activation after allogeneic hematopoietic stem cell transplantation in individuals with HIV-1. Sci Transl Med. 2020;12(542):eaay9355. doi: 10.1126/scitranslmed.aay9355. [DOI] [PubMed] [Google Scholar]
- 76.Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–698. doi: 10.1056/NEJMoa0802905. [DOI] [PubMed] [Google Scholar]
- 77.Henrich TJ, et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis. 2013;207(11):1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta RK, et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature. 2019;568(7751):244–248. doi: 10.1038/s41586-019-1027-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Zyl GU, et al. No evidence of HIV replication in children on antiretroviral therapy. J Clin Invest. 2017;127(10):3827–3834. doi: 10.1172/JCI94582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bruner KM, et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566(7742):120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berry CC, et al. INSPIIRED: quantification and visualization tools for analyzing integration site distributions. Mol Ther Methods Clin Dev. 2017;4:17–26. doi: 10.1016/j.omtm.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sherman E, et al. INSPIIRED: a pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol Ther Methods Clin Dev. 2017;4:39–49. doi: 10.1016/j.omtm.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Berry CC, et al. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28(6):755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shao W, et al. Retrovirus integration database (RID): a public database for retroviral insertion sites into host genomes. Retrovirology. 2016;13(1):47. doi: 10.1186/s12977-016-0277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rosenbloom DI, et al. Designing and interpreting limiting dilution assays: general principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect Dis. 2015;2(4):ofv123. doi: 10.1093/ofid/ofv123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palmer S, et al. Selection and persistence of non-nucleoside reverse transcriptase inhibitor-resistant HIV-1 in patients starting and stopping non-nucleoside therapy. AIDS. 2006;20(5):701–710. doi: 10.1097/01.aids.0000216370.69066.7f. [DOI] [PubMed] [Google Scholar]
- 87.Lee GQ, et al. Prevalence and clinical impacts of HIV-1 intersubtype recombinants in Uganda revealed by near-full-genome population and deep sequencing approaches. AIDS. 2017;31(17):2345–2354. doi: 10.1097/QAD.0000000000001619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson JD, et al. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shao W, et al. HIV proviral sequence database: a new public database for near full-length HIV proviral sequences and their meta-analyses. AIDS Res Hum Retroviruses. 2020;36(1):1–3. doi: 10.1089/aid.2019.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kumar S, et al. MEGA7: molecular evolutionary genetics analysis version 7.0 For bigger datasets. Mol Biol Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gillet NA, et al. Strongyloidiasis and infective dermatitis alter human T lymphotropic virus-1 clonality in vivo. PLoS Pathog. 2013;9(4):e1003263. doi: 10.1371/journal.ppat.1003263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Einsiedel L, et al. Human T-lymphotropic virus type 1c subtype proviral loads, chronic lung disease and survival in a prospective cohort of Indigenous Australians. PLoS Negl Trop Dis. 2018;12(3):e0006281. doi: 10.1371/journal.pntd.0006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chiu CH, et al. An improved nonparametric lower bound of species richness via a modified good-turing frequency formula. Biometrics. 2014;70(3):671–682. doi: 10.1111/biom.12200. [DOI] [PubMed] [Google Scholar]
- 94.DeWitt WS, et al. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J Virol. 2015;89(8):4517–4526. doi: 10.1128/JVI.03474-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.