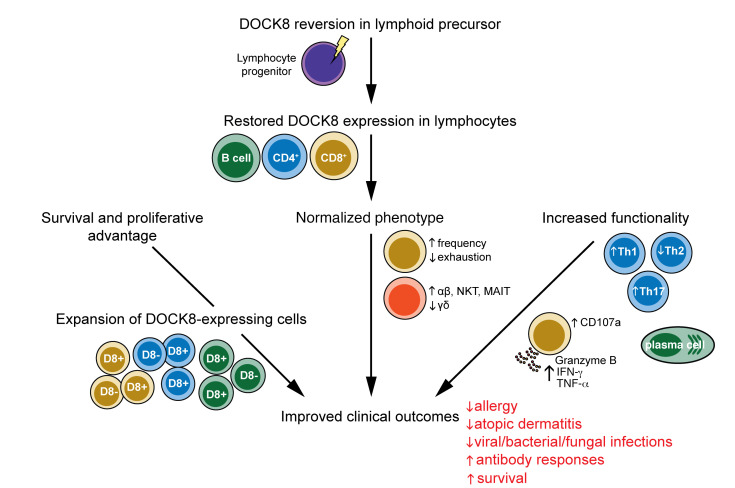
Abstract
Inborn errors of immunity cause monogenic immune dysregulatory conditions such as severe and recurrent pathogen infection, inflammation, allergy, and malignancy. Somatic reversion refers to the spontaneous repair of a pathogenic germline genetic variant and has been reported to occur in a number of inborn errors of immunity, with a range of impacts on clinical outcomes of these conditions. DOCK8 deficiency due to biallelic inactivating mutations in DOCK8 causes a combined immunodeficiency characterized by severe bacterial, viral, and fungal infections, as well as allergic disease and some cancers. Here, we describe the clinical, genetic, and cellular features of 3 patients with biallelic DOCK8 variants who, following somatic reversion in multiple lymphocyte subsets, exhibited improved clinical features, including complete resolution of infection and allergic disease, and cure over time. Acquisition of DOCK8 expression restored defective lymphocyte signalling, survival and proliferation, as well as CD8+ T cell cytotoxicity, CD4+ T cell cytokine production, and memory B cell generation compared with typical DOCK8-deficient patients. Our temporal analysis of DOCK8-revertant and DOCK8-deficient cells within the same individual established mechanisms of clinical improvement in these patients following somatic reversion and revealed further nonredundant functions of DOCK8 in human lymphocyte biology. Last, our findings have significant implications for future therapeutic options for the treatment of DOCK8 deficiency.
Keywords: Immunology, Infectious disease
Keywords: β cells, Adaptive immunity, T cells
Introduction
Primary immunodeficiencies (PIDs) or inborn errors of immunity (IEI) are caused by mutations in single genes that compromise the function of the immune system (1). Somatic reversion is the spontaneous repair of a pathogenic germline genetic variant, resulting in improvement of the defective molecular phenotype in progeny cells. This repair may result in restoration of either expression or function of a hypomorphic or WT allele and protein. As the immune system is a regenerating system, genomic reversion provides the opportunity for somatically corrected cells to replace the preexisting immune repertoire if the revertant cells have a selective advantage over mutant cells. Somatic reversion has been associated with milder clinical phenotypes in patients with numerous PIDs (1, 2). The first example of somatic reversion in a PID was in a patient with adenosine deaminase (ADA) deficiency who was born with no ADA activity and developed severe combined immunodeficiency (SCID). However, a B cell line generated when he was 14 years old exhibited 50% ADA activity, which resulted from correction of a pathogenic splice site mutation (3). Additional ADA-deficient patients with SCID have since been identified, with evidence of somatic reversion in their B cells, T cells, or both lymphocyte lineages (4–6). While the majority of these reversion events corrected the original germline pathogenic variant, one patient acquired a second deletion mutation which suppressed aberrant splicing due to a cryptic splice site created by the initial mutation (6).
These studies of ADA-SCID have been extended to additional PIDs, which established that somatic reversion is not uncommon in some IEI caused by loss-of-function mutations, where correction of the pathogenic variant is sufficient to provide a selective survival or growth advantage over the majority of immune cells that continue to bear the pathogenic genetic lesion in their germline (1, 2). Thus, X-linked SCID patients were also reported with evidence of somatic reversion in IL2RG in their T cells, but not B cells, NK cells, or monocytes, with the reversion event likely occurring in early T cell precursors (7, 8). Similarly, reversion has been detected in T cells in patients with SCID due to mutations in CD247 (CD3ζ) (9–11). Beyond SCID, reversion has been found to occur in approximately 10% of patients with Wiskott-Aldrich syndrome (WAS), with functional WASP being predominantly expressed by T cells, and to a lesser extent in NK and B cells, but not monocytes in any of the patients tested (12, 13). Somatic reversion has also been found predominantly in memory CD8+ T cells of patients with leukocyte adhesion deficiency (LAD) type 1 due to mutations in ITGB2 (CD18) (14, 15), ARPC1B deficiency (16), or X-linked lymphoproliferative disease (XLP1) due to mutations in SH2D1A (SAP) (17, 18), and primarily in T cells in patients with pathogenic variants in RAG1 (19), JAK3 (20), or CARD11 (21). Reversion was detected in CD4+ T cells, CD8+ T cells, NK cells, and a small proportion of B cells, but not myeloid cells, in one patient with X-linked ectodermal dysplasia and immunodeficiency (XL-EDA-ID) due to IKBKG (NEMO) mutations (22). Somatic correction has also been reported in patients with pathogenic gain-of-function variants in SAMD9 or CXCR4 in which the mechanism can commonly involve loss of the defective allele through deletion of the genetic locus (23–25). Last, a high proportion of patients with autosomal recessive hyper-IgE syndrome due to biallelic mutations in DOCK8 were found to exhibit reversion in multiple lymphocyte lineages, but predominantly T cells, through a variety of repair mechanisms, including original site and secondary site mutations as well as gene conversion (26).
The impact of somatic reversion, and subsequent reexpression of a functional protein in immune cells, on the clinical outcomes of PID patients has been assessed. Somatic reversion in IL2RG is associated with a milder form of immunodeficiency compared with typical X-SCID patients, in which the improved clinical condition has been successfully managed with prophylactic antibiotics and immunoglobulin replacement infusions in one patient, while another patient still required hematopoietic stem cell transplantation (HSCT) (7, 8). Similarly, outcomes for revertant XLP1 patients were generally improved compared with classic XLP1 patients who were SAP-deficient (17, 18). However, somatic reversion in ADA or WAS has been reported to have variable effects on clinical outcomes. This includes ADA-deficient patients with extended survival without the need for ongoing therapy (3, 4, 6) or patients still requiring HSCT (27), and WAS patients with reduced incidence and severity of infections and eczema (28, 29), to a reported lack of correlation between reversion and disease severity (13). In contrast to these examples, somatic reversion in CD247 in SCID (9–11), ITGB2 in several LAD1 patients (14, 15), or in the single reported IKBKG revertant patient (22) had no influence on clinical phenotype of these conditions, whereas reversion in CARD11 actually resulted in severe disease in the form of Omenn syndrome (21). With the exception of less atopic burden and possible longer survival, overall outcomes for somatically reverted DOCK8-deficient patients did not differ significantly from typical DOCK8-deficient patients, with several patients still requiring curative HSCT (26). As cases of reversion have generally only been reported over the past 2 decades, the long-term implications of reversion on patients with PID have not yet been extensively reported. However, the study of reversion in XLP-1 patients included 2 patients who remained disease free over a period of 15 years (18) as well as a patient aged 37 years with only mild clinical features (17). Additionally, a recent report detailed an extensive follow-up of an X-SCID patient who was found to have undergone somatic reversion of IL2RG by the age of 6 years, and has remained free of significant infection over the last 12 years, requiring only prophylactic immunoglobulin substitution due to the lack of reversion in the B lineage (30). These observations from albeit limited cases of XLP-1 and X-SCID suggest that somatic reversion can provide long-term protective immunity in individuals who would otherwise require life-saving interventions such as HSCT.
During the course of our studies into DOCK8 deficiency (31–33), we identified 3 patients with pathogenic biallelic compound heterozygous germline variants in DOCK8 who nonetheless demonstrated expression of DOCK8 protein in the majority of their CD8+ T cells, memory CD4+ T and B cells, and some NK cells. Detailed genetic, molecular, cellular, and functional analyses revealed somatic reversion in these lymphocyte subsets, with reexpression of DOCK8 improving survival, proliferation, and function of DOCK8+ lymphocytes in vivo and in vitro. Furthermore, the clinical phenotype of these patients dramatically improved over time, with amelioration of EBV viremia, severe allergic disease, allergen-specific IgE levels, and eosinophilia, as well as cessation of severe recurrent infections. These findings establish that somatic reversion in DOCK8 can greatly improve the clinical condition of some patients. Our results provide insight into strategies for novel treatments for DOCK8 deficiency, such as gene therapy and adoptive cellular therapies.
Results
Case details.
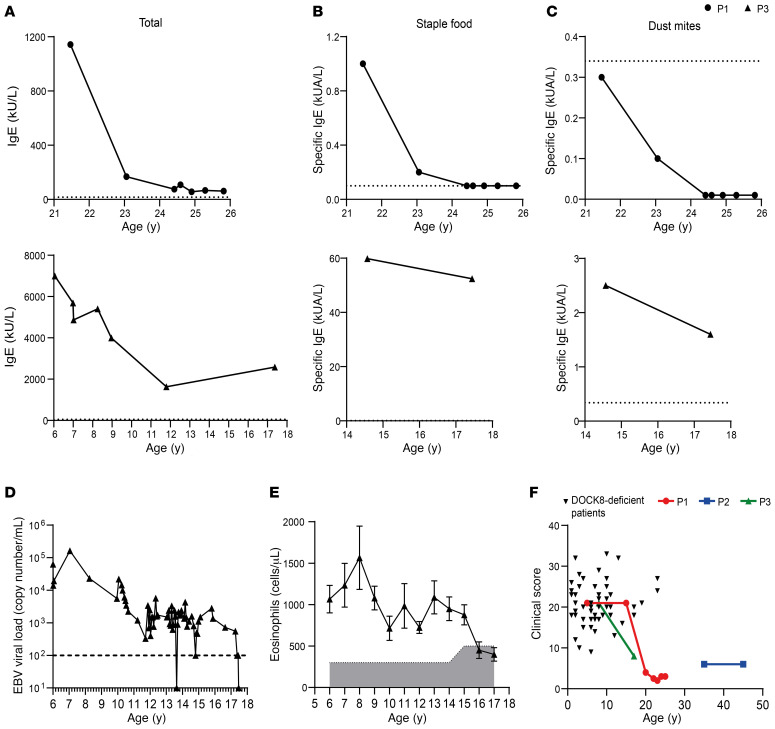
Patient 1 (P1) is a 25-year-old female born to nonconsanguineous parents of Argentinian origin. The patient had a history of infections, including 6 episodes of pneumonia, on one occasion complicating varicella infection, recurrent otitis media, onychomycosis at age 4–6 years, and oral and vaginal thrush. In early life she suffered severe atopic dermatitis as well as milk allergy during infancy. She had normal levels of serum IgG, IgA, and IgM in childhood, but increased IgE (peak 1835 IU/mL). Vaccine antibody responses to measles were negative despite vaccination, and immunity against rubella vaccine waned over time, yet varicella serology remained positive after infection. Whole genome sequencing identified a maternally inherited 2 bp deletion in exon 8 of DOCK8, causing a frameshift and introduction of a premature stop codon at amino acid residue 294 (chr9:g.325693_325694del, NM_203447.3:c.850_851del, p.(Leu284Valfs*10). This patient was also found to have a novel missense variant in the canonical splice acceptor site of intron 20 of DOCK8 (Chr9:g. 376975A > G, c.2206-2A > G), which is predicted to prevent splicing. The variant was not present in either parent but was present on Sanger sequencing of patient buccal epithelia, confirming this to be a germline de novo variant. The use of long-range sequencing enabled phasing of the 2 variants and determined that the de novo mutation was present on the paternal allele, rendering the patient compound heterozygous for DOCK8 deficiency. Despite this molecular diagnosis and her clinical history, the patient experienced a general improvement in health from mid-adolescence, has had no major infections in recent years, and has been consistently negative for EBV despite a significant proportion of DOCK8-deficient patients presenting with EBV viremia (34). Additionally, serum levels of total and allergen-specific IgE have impressively decreased over time (Figure 1, A–C). P1 has protective antibody levels against tetanus toxoid but showed no response to pneumococcal polysaccharide vaccine.
Figure 1. Improved clinical outcomes in atypical DOCK8-deficient patients over time.
(A–C) Serum was collected at different time points from 2 atypical DOCK8-deficient patients (P1: circles, top; P3: triangles, bottom) and was tested by ImmunoCAP assay for concentrations of (A) total, (B) staple food–specific, and (C) dust mite–specific IgE. Dotted line indicates average value of healthy donors (n = 3–5). (D) EBV viral load in peripheral blood of P3 was assessed at numerous time points. The dashed line indicates the limit of detection. (E) Eosinophil numbers in peripheral blood of P3 were assessed at numerous ages. Error bars indicate SEM and the shaded area indicates references ranges for healthy age-matched donors. (F) Clinical score based on the NIH scoring system (26) was calculated for DOCK8-deficient patients (n = 53) and for patients P1, P2, and P3 at various ages.
Patient 2 (P2) is a 45-year-old male born to nonconsanguineous White parents who presented at 37 years of age with bilateral proximal bronchiectasis. He had a history of skin rash from 6 weeks of age, and from age 5 years experienced recurrent otitis media, sinusitis, and multiple pneumonias, and moderate to severe psoriasis from 16 years of age. He was found to carry a maternally inherited missense mutation in exon 41 of DOCK8, resulting in the introduction of a premature stop site at amino acid R1763 (chr9:g.441349C>T, NM_203447.3:c.5287C>T p.(Arg1763*), and a paternally inherited large 40 kb duplication spanning exons 15–26 of DOCK8 (chr9:g.364057_404510dup). Thus, this patient was also compound heterozygous for DOCK8 deficiency. Recently, serum Ig levels have been found to be normal and P2 has protective antibody levels against tetanus toxoid, but antibody levels to pneumococcal polysaccharide waned to undetectable levels 4 years after vaccination. No pneumonias have been experienced since the patient commenced prophylactic cotrimoxazole in 2015. EBV viral load has been measured as low to moderate but the patient shows no clinical signs of EBV-related disease. Notably, the patient has reached an age that is unusual among nontransplanted DOCK8-deficient patients, for whom event-free survival at 30 years of age is only 4% (35). Furthermore, although malignancy occurs in 15%–20% of DOCK8-deficient patients, with a median age of detection at 12 years and a cumulative increase in incidence with each decade of life (e.g., ~50% at 30 years)(35), P2 remains malignancy free at age 45 years.
Patient 3 (P3) is an 18-year-old male born to nonconsanguineous parents of African descent (Mali). The patient presented with severe atopic dermatitis from early childhood with recurrent episodes of subcutaneous abscesses. From the age of 3 years, he suffered from recurrent and extended molluscum contagiosum and flat warts related to human papilloma virus (HPV) types 3 and 5. The lesions were resistant to IFN-α treatment and topical and systemic antiviral treatment (cidofivir). Severe HPV-related oral lesions occurred from the age of 10 years and recurred after each resection between 10–14 years. The patient also experienced recurrent herpes simplex virus (HSV) perineal skin infections during childhood. T-dependent antibody responses were abnormal and so IVIg was administered. The patient was found to be compound heterozygous for 2 inherited variants in DOCK8: a maternally inherited 2 bp deletion in exon 41 causing a truncating frameshift chr9:g.441371_441372del, NM_203447.3:c.5309_5310del, p.(Leu1770Hisfs*14) as well as a paternally inherited large 268 kb deletion spanning exons 1–29 (chr9:g. 139481_407477del). From his mid-teens, HPV-related lesions became less problematic and his atopic dermatitis improved. The elevated levels of total and allergen-specific serum IgE (Figure 1, A–C), as well as chronic EBV viremia (Figure 1D) which is observed in 20%–30% of DOCK8-deficient patients (34), also declined over time. Additionally, eosinophilia, which occurs in more than 96% of DOCK8-deficient patients (35), decreased in P3 from 8 years of age, normalizing by age 17 years (Figure 1E). P3’s affected younger sister had a much more severe disease course and consequently underwent HSCT. Following the success of this treatment for one child, and the availability of a 10/10 matched-unrelated donor, the family chose to also treat P3 with transplantation, despite his improved clinical state.
Quantitation of clinical improvement in DOCK8-deficient patients.
To quantify clinical status and improvements in these DOCK8-deficient patients, we used the NIH scoring system for DOCK8 deficiency, reported by Jing et al. (26), that enumerates viral, bacterial, and fungal infections, hyper-IgE and atopic diseases (allergy, eczema, asthma), vascular disease, malignancy, and mortality. First, we collected clinical information from 53 DOCK8-deficient patients, aged 1–23 years (mean ± SEM: 7.6 ± 0.76 years). The average clinical score for these patients was 20.7 ± 5.2 (mean ± SEM; range: 9–33; Figure 1F). We then applied this scoring matrix to P1, P2, and P3. The initial disease score for P1 was 21 at age 5 years. This declined dramatically to a score of 4 at age 20 years, and has remained at this level for the past 5 years (Figure 1F). A clinical score for P2 during early childhood and adolescence was not available; however, a score of 6 in their fourth and fifth decades of life is well below that of the DOCK8-deficient patients examined here, all of whom were much younger than P2 (Figure 1F). Similar to P1, the clinical score of P3 at age 10 years was 21, and this decreased to 8 at 17 years of age (Figure 1F). Thus, the clinical score of the 3 novel DOCK8-deficient patients described here was drastically lower than that of typical DOCK8-deficient patients, with clear evidence of dramatic improvements in measurable clinical outcomes for P1 and P2 over time.
DOCK8-expressing cells are present in numerous lymphocyte populations and are enriched in the memory compartments.
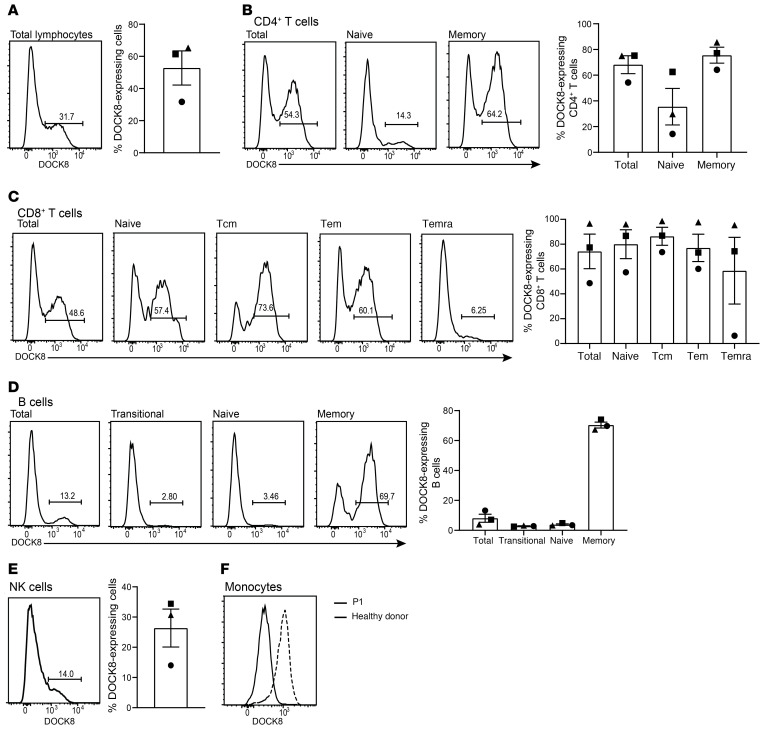
As P1, P2, and P3 had been identified as having biallelic variants in DOCK8 but were experiencing milder clinical outcomes than most DOCK8-deficient patients (Figure 1F), PBMCs from the 3 patients were assessed for DOCK8 expression by flow cytometry. Unlike all DOCK8-deficient patients that we assessed previously (n = 20) (31–33), peripheral blood from each of these 3 patients contained a subset of lymphocytes that clearly expressed DOCK8 protein (32%–65% of total lymphocytes, Figure 2A). When defined lymphocyte populations were investigated, we found that all subsets had significant populations of DOCK8+ cells. Thus, approximately 65% of CD4+ T cells (Figure 2B), approximately 75% of CD8+ T cells (Figure 2C), approximately 10% of B cells (Figure 2D), and approximately 25% of NK cells (Figure 2E) expressed detectable levels of DOCK8. Interestingly, the memory compartment of both CD4+ T cells and B cells displayed a higher proportion of DOCK8-expressing cells than the corresponding naive compartment (Figure 2, B and D). Memory CD4+ T cells on average had twice the number of DOCK8-expressing cells than naive CD4+ T cells (Figure 2B), while the disparity in B cell subsets was striking with almost 9 times as many memory B cells being DOCK8+ compared with naive B cells (8% vs 70%, Figure 2D). Curiously, the difference in proportions of DOCK8-expressing cells among CD8+ T cell subsets was not as marked as in CD4+ T cells and B cells, with more than 50% of naive, central memory (Tcm), and effector memory (Tem) CD8+ T cells being DOCK8+ (Figure 2C). There was no DOCK8 expression detected in monocytes from any of these 3 patients, compared with typical unimodal DOCK8 expression in monocytes from healthy donors (Figure 2F). It is also of note that the level of DOCK8 expression seen in the DOCK8-expressing populations of the 3 patients was comparable to the level expressed by monocytes from the healthy donors (Figure 2).
Figure 2. DOCK8-expressing cells are present in numerous lymphocyte populations and enriched in the memory compartments.
Patient PBMCs were stained for surface expression of CD4, CD8, CD20, CD56, CCR7, CD45RA, CD10, and CD27, fixed, permeabilized, and then stained intracellularly to detect DOCK8. Percentage of cells positive for DOCK8 expression in (A) total lymphocytes; (B) total, naive (CCR7+CD45RA+), and memory (CCR7±CD45RA–) CD4+ T cells; (C) total, naive, Tcm (CCR7+CD45RA–), Tem (CCR7–CD45RA–), and Temra (CCR7–CD45RA+) CD8+ T cells; (D) total, transitional (CD10+CD27–), naive (CD10–CD27–), and memory (CD10–CD27+) B cells; (E) CD3–CD56+ NK cells; and (F) monocytes were determined by flow cytometry. Circles, P1; squares, P2; triangles, P3. Solid line histograms depict P1 and dashed line histograms depict a healthy donor. Error bars represent SEM
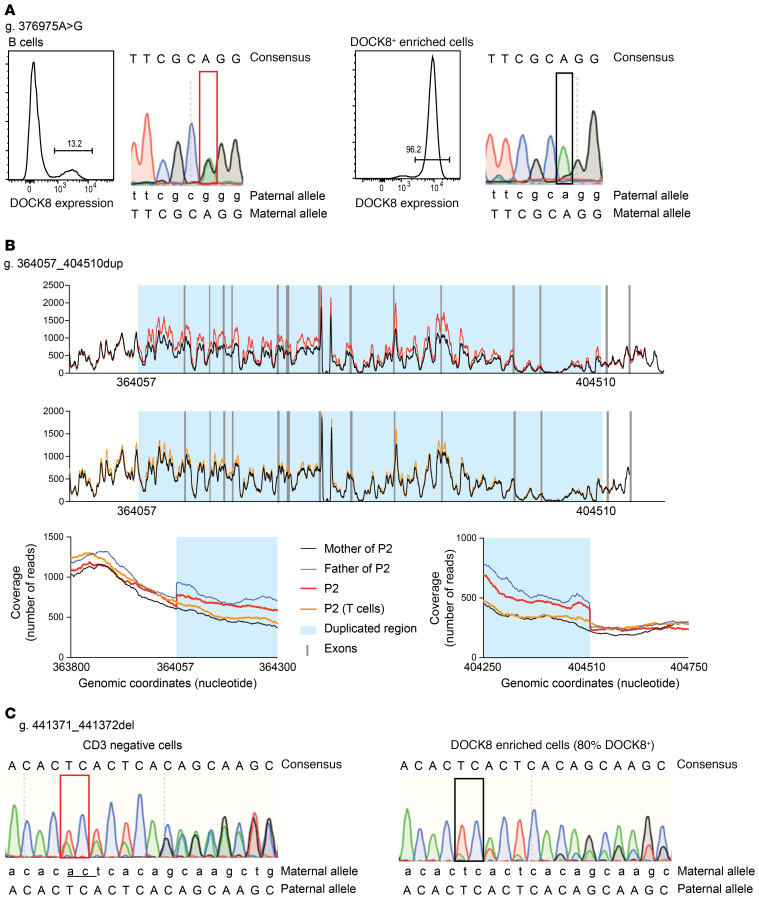
Our results indicated that these patients had likely undergone somatic reversion and repair of at least one of the mutant DOCK8 alleles. To confirm this, we examined somatic reversion at the genetic level by isolating lymphocytes enriched for high proportions of DOCK8+ cells and sequencing across the mutation sites at each allele in the patients. For P1, CD8+ T cells were expanded in vitro; under these conditions 96% expressed DOCK8 (Figure 3A). Sequencing revealed that the missense mutation at the exon 20 splice acceptor site had been repaired (Figure 3A) but not the 2 bp deletion in exon 8 (data not shown). T cell blasts generated from P2 (95% DOCK8+) displayed a reversion of the large duplication (Figure 3B) but not the missense mutation in exon 41 (data not shown). Last, sorted T cells from P3 (80% DOCK8+) exhibited reversion of the 2 bp deletion in exon 41 (Figure 3C) but not the large deletion affecting exons 1–29 (data not shown). Thus, all patients achieved DOCK8 expression in their lymphocytes via the genetic repair of one of the original pathogenic mutations. The genetic repair was most surprising in P2 as exact deletion of a large duplicated region occurred. However, similar reversion of a large duplication on the X chromosome has been previously reported in a XL-EDA-ID (NEMO) patient (22) as well as patients with Fanconi anemia (36, 37). The presence of revertant cells in multiple lymphocyte lineages, but not monocytes, in all patients suggests genetic repair occurred in a common lymphoid progenitor, which then expanded as DOCK8 expression conferred a selective advantage. It is also possible, however, that genetic repair took place in a more immature cell (e.g., hematopoietic stem cell) but conferred a selective advantage only to lymphocytes, resulting in no detectable expansion of DOCK8-expressing cells in the myeloid lineage. Overall, we identified reversion in 13% (3/23) of patients with biallelic mutations in DOCK8.
Figure 3. Genetic repair in DOCK8-revertant patients.
(A) P1: sequencing across the mutation site was undertaken with DNA extracted from B cells and CD8+ T cells isolated by magnetic separation from PBMCs following expansion with TAE beads and IL-2 for 8 days. Flow cytometry was used to determine proportions of DOCK8+ cells. (B) P2: sequencing of the DOCK8 region was undertaken with DNA extracted from whole blood from P2 (top, in red), PHA-blast T cells from P2 (middle, in orange) and compared with his parents. Mother was used as a nonduplicated control (in black). The father carries the duplication (in blue). Coverage/number of reads is represented as a function of genomic position/coordinates. Duplicated region, visualized by an increased coverage in P2 and his father, is highlighted by light blue color. The bottom panels are zoomed on the 2 breakpoints of duplication. (C) P3: sequencing across the mutation site was undertaken using DNA extracted from CD3– (left) or CD3+ (right) T cells after magnetic separation from PBMCs. Flow cytometry established that 80% of the CD3+ T cells were DOCK8+. The 2 bp deletion is underlined.
Lymphocytes from DOCK8-revertant patients exhibit an intermediate-to-complete cellular phenotype of DOCK8 deficiency, similar to healthy donors.
Having definitively established that these 3 patients had undergone somatic reversion at one mutant DOCK8 allele, we next analyzed the composition and phenotype of their lymphocyte subsets and compared these with typical DOCK8-deficient patients. Furthermore, as DOCK8 protein expression in these patients’ lymphocytes was bimodal (i.e., negative and positive), it was possible to compare pathogenic DOCK8-negative (DOCK8–) and repaired DOCK8-expressing (DOCK8+) populations within the same individual.
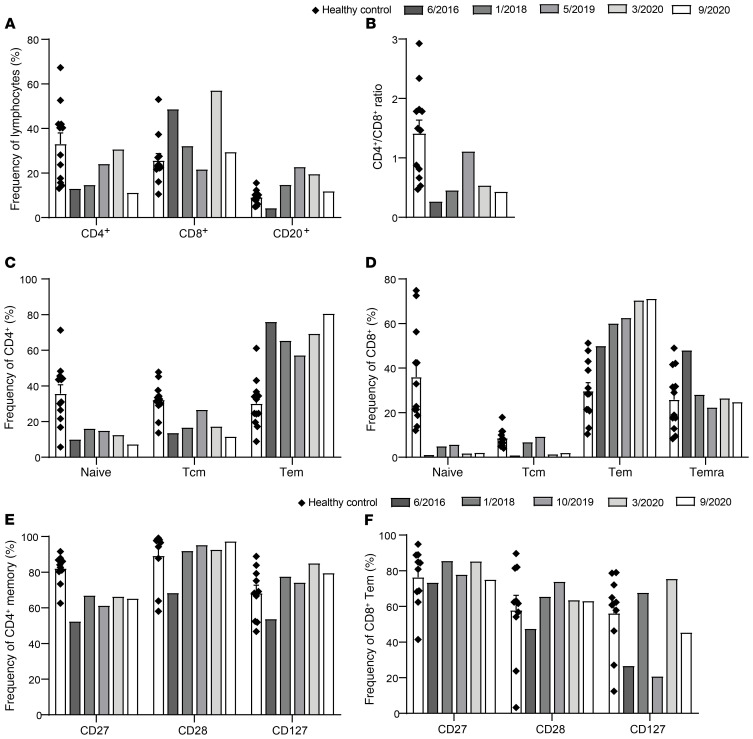
Similar to DOCK8-deficient patients, the proportion of total CD4+ T cells in DOCK8-revertant patients was reduced compared with healthy donors, whereas NK cells were intact. In contrast, DOCK8 reversion overcame the CD8+ T cell deficit and restrained the apparent increase in proportion of B cells that is characteristic of DOCK8-deficient patients (Figure 4A) (31). DOCK8 reversion also improved the skewed proportions of αβ and γδ T cells, which are typically reduced and increased respectively (31), in 2 of 3 DOCK8-deficient patients (Figure 4B), and increased proportions of MAIT and NKT cells such that the frequencies of these T cell subsets were restored to levels observed in healthy donors (Figure 4, C and D) (31, 38).
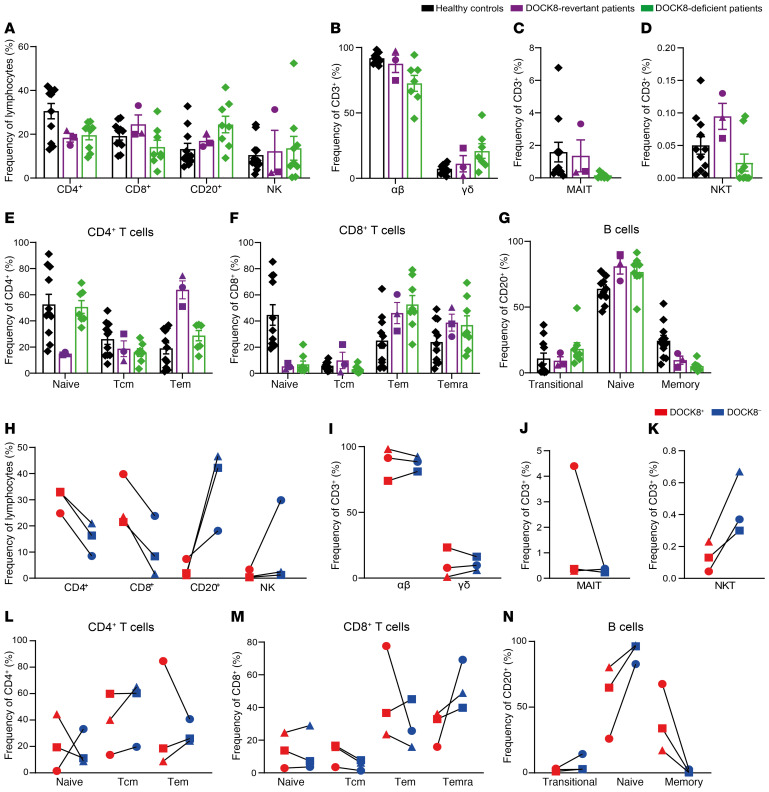
Figure 4. Patient phenotype displays some aspects of DOCK8 deficiency due to DOCK8– cells.
PBMCs from healthy donors (n = 11), DOCK8-revertant patients (n = 3), and DOCK8-deficient patients (n = 7–9) were surface stained for CD3, CD4, CD8, CD20, CD56, CCR7, CD45RA, CD10, CD27, CD161, Vαβ, Vγδ, Vα7.2, Vα24, and Vβ11, fixed, permeabilized, and then intracellularly stained to detect DOCK8. Frequencies of (A) CD4+ T cells, CD8+ T cells, B cells (CD20+), and NK cells (CD3–CD56+); (B) αβ (CD3+ Vαβ+) and γδ (CD3+Vγδ+) T cells; (C) MAIT cells (CD3+CD161+Vα7.2+); (D) NKT cells (CD3+Vβ11+ Vα24+); (E) naive, Tcm and Tem CD4+ T cells; (F) naive, Tcm, Tem, and Temra CD8+ T cells; and (G) transitional, naive, and memory B cells were determined by flow cytometry. (H–N) The frequencies of the populations in A–G within the DOCK8– and DOCK8+ populations of the DOCK8-revertant patients. Circles, P1; squares, P2; triangles, P3. Error bars represent SEM.
The in vivo differentiation of lymphocyte subsets was next interrogated. This revealed that total CD4+ T cells in DOCK8-revertant patients were skewed toward Tem cells at the expense of naive cells, a phenotype that contrasts with DOCK8-deficient patients (Figure 4E) (32). However, this may have been influenced by the younger age of the DOCK8-deficient patients. CD8+ T cells in DOCK8-revertant patients were also skewed away from naive cells with an accumulation of Tem and Temra (CCR7–CD45RA+) cells which is also characteristic of DOCK8-deficient patients (Figure 4F) (31, 39). The distribution of B cell subsets in DOCK8-revertant patients was also similar to DOCK8-deficient patients (Figure 4G) (15, 32).
While the phenotype of total cells in DOCK8-revertant patients was intermediate to DOCK8-deficient patients and healthy donors, DOCK8-negative cells detected in the revertant patients phenocopied typical DOCK8 deficiency, while the corresponding revertant DOCK8+ cells in the patients resembled those from healthy donors. This was seen most clearly in the pattern of T, B, and NK cell frequency in total lymphocytes (Figure 4H), as well as αβ and γδ T cells (Figure 4I) and MAIT cells (Figure 4J), but not NKT cells (Figure 4K). Despite not being present in the DOCK8-deficient patients, Tem skewing of CD4+ T cells was observed in DOCK8– cells from 2 revertant patients (Figure 4L) whereas an increase in Temra cells in the CD8+ T cell compartment, seen in DOCK8-deficient patients, was present in DOCK8– cells from all revertant patients (Figure 4M). The pattern of B cell differentiation of DOCK8– cells in DOCK8-revertant patients replicated DOCK8-deficient patients with increased naive and substantially decreased memory cells (Figure 4N) (15, 31). However, acquisition of DOCK8 expression in the B lineage enabled differentiation of naive cells into memory B cells, evidenced by the significant enrichment of memory cells within the DOCK8+ subset (Figure 4N). Thus, the DOCK8+ B cell compartment in the DOCK8-revertant patients resembled that of healthy donors (Figure 4N).
DOCK8-revertant T cells exhibit fewer features of exhaustion than DOCK8-deficient cells.
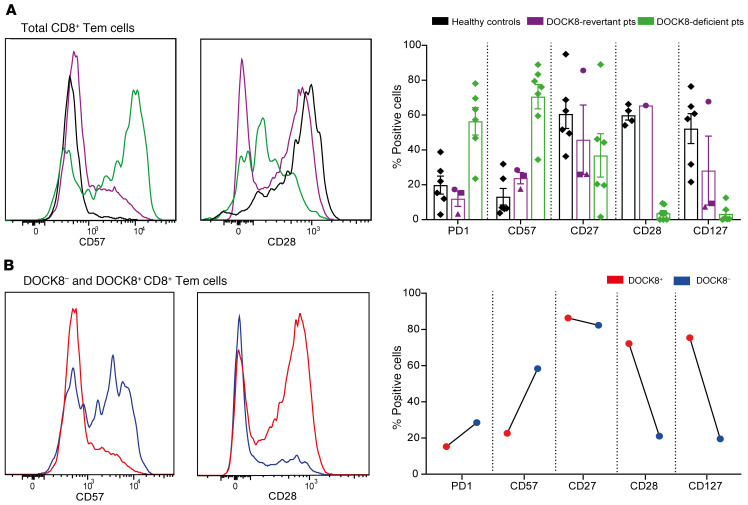
We previously reported that T cells from DOCK8-deficient patients acquire a phenotypic signature characteristic of chronic activation or exhaustion, especially for CD8+ Tem cells (31, 32, 39). Therefore, we examined the phenotype of total CD8+ Tem cells in DOCK8-revertant patients, as well as cells that could be identified as DOCK8– or DOCK8+. CD8+ Tem cells from DOCK8-revertant patients showed milder signs of chronic activation compared with the prominent exhaustion signature of DOCK8-deficient patients, evidenced by less upregulation of PD1 and CD57 in all patients and reduced downregulation of CD27, CD28, and CD127, particularly in P1 (Figure 5A). This altered phenotype of CD8+ Tem cells was due to restored DOCK8 expression modulating exhaustion, with fewer DOCK8+ cells expressing PD1 and CD57 and more expressing CD27, CD28, and CD127 than DOCK8– cells (Figure 5B). It has been previously hypothesized that the exhausted phenotype of DOCK8-deficient T cells resulted from frequent and chronic encounters with viral pathogens (39). However, since both DOCK8– and DOCK8+ cells were exposed to the same environment, our findings suggest DOCK8 deficiency intrinsically biases T cells to prematurely enter an exhausted state of differentiation.
Figure 5. DOCK8-revertant cells exhibit fewer signs of exhaustion than DOCK8-deficient cells.
PBMCs from healthy donors (n = 4–6), DOCK8-revertant patients (n = 3), and DOCK8-deficient patients (n = 5–7) were stained for surface expression of CD8, CCR7, CD45RA, PD1, CD57, CD27, CD28, and CD127, fixed, and intracellularly stained for DOCK8. The expression of exhaustion markers on (A) total CD8+ CD45RA–CCR7– Tem cells and (B) DOCK8+ and DOCK8– cells was then determined by flow cytometry. Circles, P1; squares, P2; triangles, P3. Histograms depict P1. Error bars represent SEM.
TCR repertoire diversity in DOCK8-revertant patients.
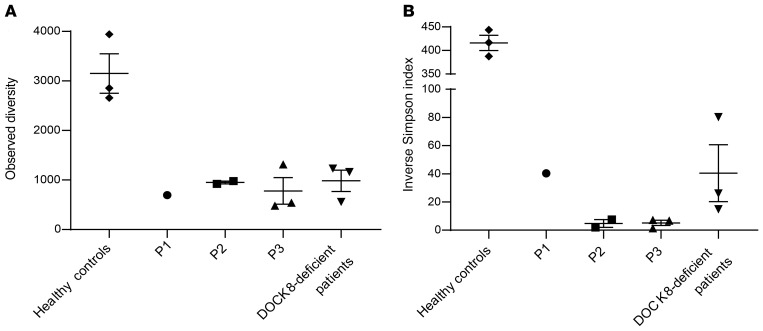
It was previously found that DOCK8-expressing T cells in DOCK8-revertant patients have a limited TCR repertoire, dominated by only a few TCR Vβ clonotypes, suggesting that the accumulation of DOCK8-expressing cells results from the selective outgrowth of a limited number of clones (26). For this reason, we performed TCR Vβ analysis with high-throughput sequencing to gain insights into the diversity of circulating T cells in the DOCK8-revertant patients. For P3, we were able to analyze gDNA extracted from whole blood collected at 3 different time points (6, 12, and 17 years of age). TCR Vβ gene repertoire at age 6 years (i.e., time of clinical diagnosis and the beginning of clinical improvement) showed minimal diversity with one dominant expanded clone (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI142434DS1). Progressive contraction of this clone can be seen at the ages of 12 and 17 years (Supplemental Figure 1). To compare the number of different clones (observed diversity) and the evenness of distribution across the different clones (inverse Simpson index), repertoires from P1, P2 at 2 different time points, 3 healthy controls, and 3 DOCK8-deficient patients were also determined (Supplemental Figure 1). As expected, we found a reduced observed diversity in DOCK8-deficient patients and DOCK8-revertant patients compared with controls (Figure 6A). Surprisingly, the inverse Simpson index of DOCK8-revertant P2 and P3 was even lower than that of DOCK8-deficient patients (Figure 6B). Thus, TCR repertoire diversity of DOCK8-revertant patients was limited. We next sought to analyze the repertoire specifically in DOCK8-expressing cells, using flow cytometry to evaluate global TCR Vβ usage in DOCK8– and DOCK8+ cells within the CD4+ memory and CD8+ Tem compartments in each patient. While there were differences in TCR Vβ usage across the individual patients, we did not observe a skewed repertoire in DOCK8+ cells in CD4+ memory T or CD8+ Tem cells in the revertant patients (Supplemental Figure 2).
Figure 6. Diversity of the TCR repertoire is not improved in DOCK8-revertant patients.
TCR beta genes were sequenced from genomic DNA from healthy donors (n = 3), DOCK8-revertant patients (P1, P2 at 2 time points, P3 at 3 time points), and DOCK8-deficient patients (n = 3). TCR repertoire diversity was estimated in all samples after down-sampling transformation (23,838 sequences) by (A) plotting the observed diversity (number of different clones in the down-sampled file) and (B) calculating the inverse Simpson index, which computes repertoire richness and evenness. Error bars represent SEM.
Restored expression of DOCK8 by somatic reversion improves TCR signalling, lymphocyte function, proliferation, and survival.
To establish the functional consequences of DOCK8 reversion, we next studied lymphocyte responses from DOCK8-revertant patients in vitro. Global tyrosine phosphorylation in response to TCR engagement was examined in T cell blasts from P2 and P3 and compared with DOCK8-deficient and healthy control T cell blasts. Phosphorylation of intracellular substrates was markedly decreased in T cell blasts from DOCK8-deficient patients compared with healthy donors (Supplemental Figure 3). However, this was restored to an almost normal level in P2 and P3 T cell blasts (Supplemental Figure 3). Additionally, tyrosine phosphorylation was higher in DOCK8+ T cells compared with DOCK8– T cells in P1 in response to TCR cross-linking (Supplemental Figure 3). T cells from Dock8-deficient mice have lower baseline levels of F-actin and a reduced increase in F-actin upon induction compared with WT mice (40). Thus, to further investigate the impact of DOCK8 on the cellular response to TCR signaling, we assessed cytoskeletal function by measuring actin polymerization in T cells from P1. Similar to findings in mice (40), we observed that F-actin content was higher in DOCK8+ cells than in DOCK8– cells in CD4+ and CD8+ T cells from P1 before and after activation (Supplemental Figure 3).
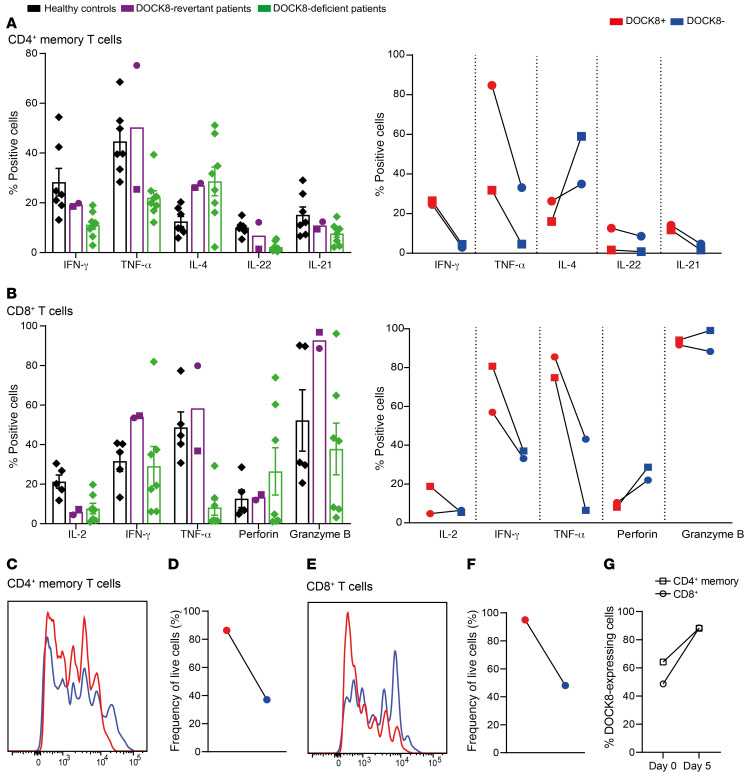
Next, the functionality of CD4+ memory T cells was assessed in P1 and P2 by analyzing cytokine production after 5 days of in vitro culture. The pattern of cytokine production by CD4+ memory T cells from DOCK8-revertant patients resembled that of DOCK8-deficient patients, evidenced by the previously reported characteristic skewing to Th2 cytokines (IL-4), and decreased production of Th1 (IFN-γ), Th17 (IL-22), and Tfh (IL-21) cytokines (Figure 7A) (31, 32). However, analysis of DOCK8-deficient (DOCK8–) and DOCK8-revertant (DOCK8+) memory CD4+ T cells in the revertant patients established that reexpression of DOCK8 corrected the cytokine profile. Thus, the proportions of CD4+ memory T cells producing IFN-γ, TNF-α, IL-22, and IL-21 were greater, and those producing IL-4 were lower, in DOCK8-revertant versus DOCK8-deficient cells in the same patients (Figure 7A). Overall, the cytokine profile of DOCK8-revertant memory CD4+ T cells essentially matched that of memory CD4+ T cells from healthy donors.
Figure 7. T cell function, proliferation, and survival is improved in DOCK8-revertant cells.
Sorted CD4+ memory T cells and CD8+ T cell from healthy donors (n = 5–9), DOCK8-revertant patients (circles, P1; squares, P2) and DOCK8-deficient patients (n = 7) were cultured for 5 days with TAE beads (anti-CD2, -CD3, -CD28). After restimulation with PMA/ionomycin, (A) CD4+ memory T cells were stained for intracellular expression of DOCK8 and the cytokines IFN-γ, TNF-α, IL-4, IL-22, and IL-21 in total cells and DOCK8+ and DOCK8– cells. (B) CD8+ T cells were stained for intracellular expression of DOCK8 and the cytokines IL-2, IFN-γ, and TNF-α, as well as perforin and granzyme B. (C–F) Proliferation (C, E) and survival (D, F) of DOCK8+ and DOCK8– CD4+ memory T cells and CD8+ T cells from P1 were determined by dilution of CFSE and staining with a live/dead marker. (G) The frequency of DOCK8-expressing cells in CD4+ memory T cells and CD8+ T cells before and after 5-day culture was determined in P1. Error bars are SEM.
Cytokine production by total CD8+ Tem cells from DOCK8-revertant patients was much less impacted than that by CD8+ Tem cells from DOCK8-deficient patients following in vitro stimulation, with the exception of IL-2 (Figure 7B). Despite this, the capacity of DOCK8– Tem cells to produce IFN-γ and TNF-α was greatly diminished compared with DOCK8+ revertant Tem cells (Figure 7B), similar to the findings for CD4+ memory T cells. Similarly, while expression of the degranulation marker CD107a was markedly reduced in DOCK8-deficient patient CD8+ T cells compared with CD8+ T cells from healthy donors, the defect in DOCK8-revertant patient CD8+ T cells was modest (Supplemental Figure 3).
Labeling of cells with CFSE and a live/dead marker allowed proliferation and survival, respectively, to be measured. Proliferation and survival of CD4+ memory and CD8+ T cells in DOCK8-revertant patients were only mildly reduced compared with healthy controls, whereas that of T cells from DOCK8-deficient patients was substantially reduced (data not shown). Further analysis established that DOCK8-expressing CD4+ memory (Figure 7, C and D) and CD8+ (Figure 7, E and F) T cells isolated from blood of a DOCK8-revertant patient had substantial proliferative and survival advantages over DOCK8– T cells, evidenced by increased rounds of cell division and greater viability of DOCK8+ T cells over the 5-day culture period than matched DOCK8– cells. The combined effect of this improvement in proliferation and survival was that after 5 days of in vitro culture the proportion of DOCK8-expressing cells among live cells increased by 38% in CD4+ memory T cells and 82% in CD8+ T cells (Figure 7G).
Last, we examined memory B cells. While total memory B cells from P1 exhibited comparable survival to those from healthy donors in vitro, analyzing DOCK8 expression also revealed a dramatic survival advantage of DOCK8+ over DOCK8– memory B cells. Consistent with this, there was a subsequent increase in proportions of DOCK8+ cells among live cells after 5 days of in vitro culture compared with prior to culture (Supplemental Figure 3).
DOCK8-revertant lymphocyte populations accumulate over time.
The observation that DOCK8-expressing revertant T and B cells preferentially expanded during a 5-day in vitro culture period led us to investigate whether this also occurred in vivo. We collected serial blood samples from P1 and assessed DOCK8 expression over a 51-month time frame. This longitudinal analysis revealed that proportions of all lymphocytes expressing DOCK8 increased steadily over time, from 18% at initial analysis to more than 50% 4 years later (Figure 8A). Total B cells did not show this same sharp increasing trajectory of progressively accumulating DOCK8+ cells. Rather, the proportions of total B cells expressing DOCK8 slowly increased over time and this was due mostly to an expanding population of DOCK8+ memory B cells (Figure 8B).
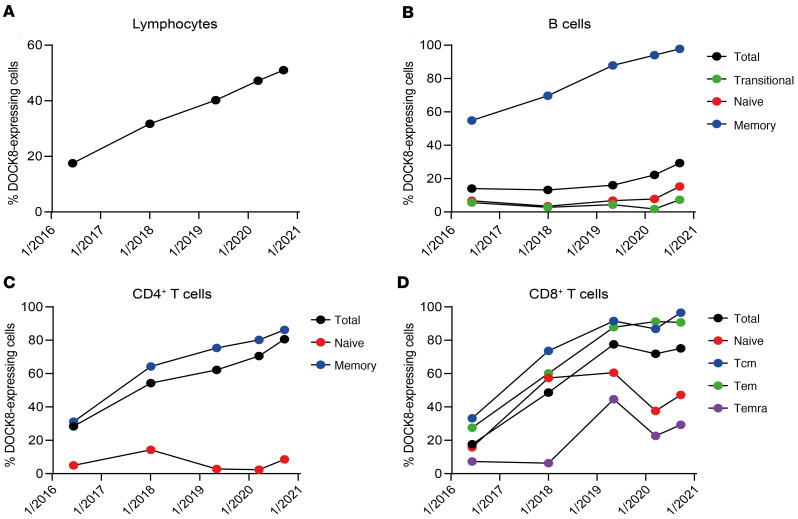
Figure 8. DOCK8-expressing populations expand over time.
PBMCs from P1 taken at various time points were stained for surface expression of CD4, CD8, CD20, CCR7, CD45RA, CD10, and CD27, fixed, permeabilized, and intracellularly stained to detect DOCK8. Percentages of cells expressing DOCK8 in (A) total lymphocytes; (B) total, transitional, naive, and memory B cells; (C) total, naive, and memory CD4+ T cells; and (D) total, naive, Tcm, Tem, and Temra CD8+ T cells were determined by flow cytometry.
DOCK8-expressing total CD4+ T cells also increased considerably over time (28%–80%) and again this was due to predominant expansion within the memory compartment (Figure 8C). DOCK8 expression in total as well as all subsets of CD8+ T cells increased to a similar extent at most time points (Figure 8D). This increase in revertant cells in numerous lymphocytes over time may be due to a reservoir of revertant stem cells, which continues to populate the circulating lymphocytes. Additionally, the presence of reversion in numerous lymphocyte subsets and the preferential expansion of these revertant B and T cells over time due to an intrinsic growth and proliferative advantage unequivocally demonstrates that DOCK8 plays a key role in numerous fundamental aspects of lymphocyte biology.
The increase in DOCK8-revertant cells correlates with improvements in immune cell phenotype.
As the proportion of DOCK8-expressing CD4+ and CD8+ T cells increased over time, we also investigated whether this impacted the overall phenotype of these cells in a temporal manner. Over time the frequencies of B cells and CD4+ T cells increased, while those of CD8+ T cells decreased, such that frequencies of B cells and CD8+ T cells were equivalent to the average of healthy donors at the most recent time point of analysis (Figure 9A). Alterations to the frequencies of T cell subsets also improved the CD4+/CD8+ T cell ratio (Figure 9B). While in vivo differentiation of CD4+ T cells fluctuated over time (Figure 9C), CD8+ T cell differentiation improved, with the frequency of the most differentiated subset decreasing over time (Temra cells, Figure 9D), coupled within an increase in the penultimately differentiated subset (Tem cells, Figure 9D). The accelerated loss of costimulatory receptors, indicative of chronic activation/exhaustion, also improved over time as expression of CD27, CD28, and CD127 increased on both CD4+ memory (Figure 9E) and CD8+ Tem (Figure 9F) T cells. Thus, somatic repair of a mutant DOCK8 allele was sufficient to restore protein expression and subsequent proliferation, survival, differentiation, and function of CD4+ T, and CD8+ T and B cells, thereby explaining the improved clinical phenotype of these patients.
Figure 9. Increases in DOCK8-expressing cells correlate with improvements in phenotype.
PBMCs from healthy donors (n = 10–12) and P1 taken at various time points were stained for surface expression of CD4, CD8, CD20, CCR7, CD45RA, CD10, CD27, CD28, and CD127. Frequencies of (A) CD4+ T cells, CD8+ T cells, and B cells (CD20+); (B) CD4+/CD8+ ratio; (C) naive, Tcm, and Tem CD4+ T cells; (D) naive, Tcm, Tem, and Temra CD8+ T cells; exhaustion marker–expressing (E) CD4+ CD45RA–CCR7+/– memory T cells; and (F) CD8+ CD45RA–CCR7– effector memory T cells were determined by flow cytometry. Error bars represent SEM.
Discussion
IEI compromise the development, differentiation, and function of cells of the immune system, resulting in recurrent, severe, and often fatal clinical outcomes. However, there is considerable phenotypic diversity among individuals with mutations in the same gene, and even among family members who share the same genotype. While this heterogeneity can result from the qualitative impact of the genetic variant on the function of the encoded proteins (e.g., hypomorphic vs null mutations) or the nature of inheritance (e.g., monoallelic vs biallelic) additional factors such as genetic polymorphisms, epigenetic regulation, and environmental exposures can also influence clinical severity. A further, albeit rare, genetic modulator of clinical outcome of IEI is somatic reversion (1, 2). Somatic reversion has been reported in numerous PIDs but remarkably, despite the detection of immune cell subsets expressing functional protein and exhibiting restored functionality, somatic reversion has only been found to result in improved clinical outcomes for some of these conditions (1). This has been proposed to be due to a number of factors including reversion being limited to certain lymphocyte subsets (e.g., T cells but not B cells), a restricted TCR repertoire in revertant T cells and persisting functional defects of nonrevertant myeloid cells.
Here, we identified 3 patients with compound heterozygous inactivating mutations in DOCK8. While each patient exhibited many of the typical features of DOCK8 deficiency, each experienced remarkable clinical improvement of their disease over time. Detailed molecular studies demonstrated that each patient underwent somatic reversion at one of the pathogenic DOCK8 alleles, resulting in not only restored expression of DOCK8 protein in lymphocyte populations, but the accumulation of DOCK8+ cells over time. The ability to simultaneously detect DOCK8-deficient and DOCK8-expressing immune cells in these patients allowed us to ascertain that rescue of DOCK8 expression by somatic reversion significantly improved survival, differentiation, and function of revertant lymphocytes in vivo, ex vivo and in vitro. Thus, we conclude that somatic reversion in DOCK8-deficient lymphocytes can be sufficient to reduce the incidence and burden of infectious diseases, control EBV viremia, correct eosinophilia, and repress the exaggerated levels of total and allergen-specific IgE and eosinophils thereby resolving allergic disease. The lack of malignancy in P1 and P2 also suggests that long-term effects of somatic reversion in one allele of DOCK8 may protect against malignancy, which is common in DOCK8-deficient patients. These improvements are clearly evidenced by quantifiable reductions in clinical score compared with DOCK8-null patients, as well as over time for P1 and P2. Mechanistically, these clinical improvements are mediated by restored functionality of (a) cytotoxic CD8+ T cells, (b) CD4+ T cells producing antiviral/fungal/bacterial cytokines (IFN-γ/TNF-α/ Th17/IL-21), (c) memory B cells, and (d) contraction of Th2-type proallergenic cells. Consistent with these findings, a previous report of a patient with partial DOCK8 deficiency demonstrated normal proliferation, exhaustion, and differentiation of T cell subsets as well as normal IFN-γ and TNF-α production (41), while a patient with a truncated DOCK8 protein with hypomorphic function experienced milder clinical symptoms including a lack of severe viral infections, atopy, or elevated serum IgE (42).
The discrepancy in cytokine production between DOCK8– and DOCK8+ cells despite differentiating in the same cytokine milieu indicates that these defects are T cell intrinsic and result from DOCK8-deficiency, rather than from chronic antigen exposure. These findings provide support for our previous hypothesis that Th2 skewing may be due to weakened TCR signaling in DOCK8-deficient T cells (32). This is also suggested by improved TCR-induced tyrosine phosphorylation in DOCK8+ T cells in P1, P2, and P3 compared with corresponding DOCK8-deficient T cells in P1. Increased F-actin levels in DOCK8+ T cells compared with DOCK8– T cells in P1 also suggest that stronger TCR signaling may be due to increased stability of the immune synapse.
Somatic reversion has previously been reported in a cohort of 16 patients with recessive DOCK8 mutations (26). However, this study did not determine the extent of reversion among all defined lymphocyte subsets, nor assess the consequences of restored DOCK8 expression on lymphocyte function (26). In doing so, we have now revealed that reversion was predominantly restricted to memory B cells, memory CD4+ T cells, and most CD8+ T cells. Naive CD4+ T cells and NK cells had lower but appreciable levels of reversion. It was suggested that the higher rates of turnover in T cells and memory cells contributes to their higher levels of reversion in DOCK8 deficiency (26). The importance of delineating DOCK8 expression among lymphocyte subsets following reversion is evidenced by our study of B cells. Thus, the frequency of reversion in total B cells was low, but by interrogating B cell subsets it became clear that reversion was highly enriched in memory B cells. This established a nonredundant role for DOCK8 in the generation and/or maintenance of memory B cells and subsequent long-lived humoral immunity, consistent with a deficit in memory B cells and impaired Ag-specific Ab responses in typical DOCK8-deficient patients (11, 31), and restored humoral immunity in DOCK8-revertant patients. Similarly, the observation that reversion varied between naive and memory CD4+ T cells but was consistent among the CD8+ T cell compartment, suggests a possible differential role for DOCK8 in CD4+ and CD8+ T cell differentiation.
Intriguingly, Jing et al. (26) reported that DOCK8 reversion had minimal impact on clinical outcome and was thus insufficient for disease improvement. Indeed, several patients still required curative HSCT (26). There are several possible explanations for these divergent findings between our study and this earlier study. First, the patients in our study displayed reversion in NK cells while the majority of previously reported patients had less than 5% repaired NK cells. Second, the patients in our study exhibited functional T cells whereas functionality was not tested in the previous study. Given the importance of NK and T cells in effective antiviral and antibacterial immunity (43, 44), and the key roles of DOCK8 in regulating function of these cytotoxic lymphocytes (33, 39, 45), these cellular differences may account for the discrepancy in clinical improvement between the 2 patient groups. Jing et al. also suggested that the restricted TCR repertoire of DOCK8-expressing cells compromises the ability of these cells to combat the wide range of infections experienced by DOCK8-deficient patients (26). While TCR Vβ staining of DOCK8-expressing T cells in the revertant patients studied here showed a broader TCR repertoire than TCR Vβ staining of previous DOCK8-revertant patients (26), sequencing of TCR Vβ genes in total T cells of P1, P2, and P3 revealed low diversity, even more so than in typical DOCK8-deficient patients. This could result from preferential expansion of a limited set of revertant T cell clones associated with recurrent infections experienced by the patients. Additionally, the number of revertant precursors present in the thymus could also restrict the diversity of the progeny they yield.
TCR diversity varies greatly between CD4+ and CD8+ T cells as well as naive and memory T cells. Specifically, CD4+ T cells have greater diversity than CD8+ T cells, while naive cells having greater diversity than memory cells within both CD4+ and CD8+ T cell populations (46, 47). Our findings established that DOCK8-revertant patients have not only decreased CD4+ and increased CD8+ T cells but also skewing of both CD4+ and CD8+ T cells toward memory cells. Thus, the DOCK8-revertant patients have a much higher frequency of T cells that typically display lower diversity, which would impact assessment of diversity. We observed that, over time, CD4+ T cells increased and CD8+ T cells decreased and both CD4+ and CD8+ T cells moved away from memory skewing in P1; therefore, TCR diversity would be expected to increase in a temporal manner. Further investigation of TCR diversity in revertant patients will require TCR Vβ gene sequencing of purified T cell populations (e.g., memory CD4+ T cells) as well as DOCK8-expressing cells (e.g., DOCK8+ memory CD4+ T cells). It is also likely that the age of the patient at time of reversion influences disease resolution. The majority of the previously reported cohort of DOCK8-revertant patients were adults when reversion was discovered, whereas P1 and P3 were adolescents when clinical improvement began. Additionally, given the advanced age of P2 when he presented with symptoms following a long period without incident, it is likely that he was also of a young age when reversion occurred. The degree of clinical severity prior to reversion may also impact the degree of clinical improvement due to somatic reversion in DOCK8.
Our results provide valuable insight into novel treatment strategies for patients with DOCK8 deficiency. While HSCT is standard of care for DOCK8 deficiency, donors are not always available. Furthermore, HSCT involves significant risks such as graft vs host disease, viral reactivation, and decreased donor chimerism over time (48). Our findings from these revertant patients provide evidence that a high level of DOCK8 expression in all lymphocytes is not required to achieve clinical improvement. This suggests that gene therapy, even with modest transfection of autologous hematopoietic stem cells, would be a promising prospect for treating DOCK8 deficiency. Additional findings from our study provide support for this proposal. First, in X-SCID patients who exhibit reversion in IL2RG, or have undergone gene therapy, the few cells present that express functional γc acquire a survival advantage and thus dominate the lymphocyte compartment over time (7, 49). The fact we observed rapid expansion of DOCK8-expressing revertant cells during in vitro culture, as well as the accumulation of these cells over time in vivo, also reveals that DOCK8 endows a survival advantage to lymphocytes. Second, we observed striking parallels between improvements in the clinical, phenotypic, and functional features of DOCK8-revertant patients, and individual lymphocyte subsets within these patients, and those of DOCK8-deficient patients who have undergone HSCT (31). As neither we nor Jing et al. detected reversion in monocytes of DOCK8-deficient patients, it is likely that clinical improvement can be achieved through DOCK8 expression exclusively in the lymphoid lineage without the need for viral transduction of myeloid cells. Together, these conclusions further suggest the suitability of gene therapy as a treatment for DOCK8 deficiency. Our findings also raise the possibility of a cell-based therapy either to treat DOCK8 deficiency or accelerate clinical improvement of patients with evidence of DOCK8 reversion. In this setting, lymphocytes from histocompatible donors, or revertant lymphocytes isolated from patients, could be expanded in vitro by appropriate stimulation to enrich for pathogen-specific and/or DOCK8-expressing cells before allogeneic or autologous adoptive transfer into DOCK8-deficient patients. In conclusion, we have shown that somatic genetic reversion of pathogenic DOCK8 mutations can lead to clinical improvements and long-term benefits on immunity, host defense, and tumorigenesis in DOCK8 deficiency. Thus, patients with milder clinical symptoms should be investigated for reversion in all lymphocyte subsets.
Methods
Human samples.
PBMCs and plasma were collected from healthy volunteers (Australian Red Cross LifeBlood, Etablissement Français du Sang), DOCK8-deficient patients reported previously (31), and DOCK8-revertant patients (P1, P2, P3) either at a single or multiple time points. Plasma was analyzed for total and allergen-specific IgE (food or mite mix) (32). EBV load was quantified by qPCR. Clinical score was calculated using criteria from Jing et al. (26).
Flow cytometry.
Immunofluorescence staining was performed according to standard flow cytometry methods (31, 50) with slight modifications (DOCK8 Alexa Fluor 647 replacing KLRG1APC). DOCK8 expression was determined on fixed cells, permeabilized with saponin, and stained with rabbit anti-DOCK8 followed by Alexa Fluor 647-anti-rabbit IgG heavy and light chain. TCR Vβ repertoire was determined using a TCR Vβ repertoire kit. Antibodies used are detailed in Supplemental Table 1.
Lymphocyte isolation and functional analysis.
Memory CD4+ T cells were isolated after excluding Tregs and sorting CD4+CD45RA– cells. CD8+ T cells were isolated as CD8+CD4– lymphocytes. Memory CD4+ and CD8+ T cells were cultured with T cell activation and expansion (TAE) beads (anti-CD2, -CD3, -CD28 mAbs). Cells were labeled with CFSE and proliferation determined by assessing CFSE dilution. Cells were harvested and stained with Zombie live/dead marker to determine viability. Intracellular cytokine staining was conducted on fixed and permeabilized cells.
Targeted resequencing by NGS (capture by hybridization approach).
A custom capture by hybridization approach was used as previously described (51, 52). Biotinylated single-strand DNA probes targeting all the regions of interest were designed to cover a 190 kb chromosomal region including the DOCK8 gene on chromosome 9 (chr9:246,837-437,749 according to the GRCh38.p12 assembly of the human reference genome). The capture by hybridization libraries were sequenced using a paired-end mode 100+100 bases on an Illumina NovaSeq6000, to produce approximately 64 million clusters per sample. After demultiplexing, sequences were aligned to the reference human genome hg19 using the Burrows-Wheeler Aligner (53). The mean depth of coverage per sample was approximately 500×. Downstream processing was carried out with the Genome Analysis Toolkit (GATK), SAMtools, and Picard, following documented best practices (http://www.broadinstitute.org/gatk/guide/topic?name=best-practices). Base pair coverage was plotted with the genomic position and analyzed to find the breakpoints of duplication.
Long read sequencing.
Synthetic long read sequencing was performed to phase genomic variants in P1. To prepare the libraries, 10x Genomics Chromium Genome Library Kit (https://www.10xgenomics.com/solutions/genome/). Libraries were created as per the manufacturer’s instructions, loaded onto Illumina HiSeq X sequencer, and paired-end sequencing was performed. The raw data from the sequencers were converted to FastQ file format using Illumina’s bcl2fastq 2.16.0. The FastQ files were used as input into 10x Genomics Long Ranger Analysis Pipelines (version 2.1.6) for further analysis (https://support.10xgenomics.com/genome-exome/software/pipelines/latest/what-is-long-ranger). The pipeline used a reference genome available from 10x Genomics (version b37-2.1.0). Results were visualized using 10x Genomics software Loupe.
TCR repertoire sequencing.
To perform TCR repertoire analysis, genomic DNA was extracted from blood samples or PBMCs. The TRB repertoire was assessed by 1-step Next-Generation Sequencing using 100 ng DNA, EuroClonality-NGS amplicon primers (54). Sequencing was performed on Illumina MiSeq sequencers, using 2 × 250 bp version 2 chemistry. Primary analysis of TCR repertoire was carried out with IMGT/HighV-QUEST (55). VDJtools software was used to evaluate repertoire diversity and composition (56). Nonfunctional clones were filtered out and downsampling transformation was performed for better intersample comparison. These data have been deposited in the NCBI database (accession number: PRJNA680367).
Study approval.
This study was approved by the Sydney Local Health District RPAH Zone Human Research Ethics Committee and Research Governance Office, Royal Prince Alfred Hospital, Camperdown, New South Wales, Australia (protocol X16-0210/LNR/16/RPAH/257); the South East Sydney Local Health District Human Research Ethics Committee, Prince of Wales/Sydney Children’s Hospital, Randwick, New South Wales, Australia (protocol HREC/11/POWH/152); the Comité de Protection des Personnes de l’Ile de France II and the French advisory committee, Paris, France (protocol NGS-PID IMIS2015-02). Written informed consent was obtained from participants or their guardians.
Author contributions
BAP designed the research, performed experiments, analyzed and interpreted results, and wrote the manuscript. MF designed the research, performed experiments, and analyzed and interpreted results. PEG, ALS, L Burnett, SC, CM, YS, and CB performed and analyzed genetic data. CA, EM, and JPDV performed TCR repertoire analysis. SW, RL, MM, CP, BN, AF, PEG, AK, WT, L Bezrodnik, JMS, JP, JBZ, AA, LH, and HA provided patient samples, clinical details and genetics, and managed patients. SL analyzed and interpreted results and provided funding for the project. SGT and CSM designed the research, analyzed and interpreted results, wrote the manuscript, and provided funding for the project. All authors commented on the manuscript. BAP, together with CSM and SGT, wrote the majority of the original manuscript, as well as the revised versions. For this reason, BAP is listed in print as the first co–first author of this study.
Supplementary Material
Acknowledgments
This study was supported by the National Health and Medical Research Council (NHMRC) of Australia (1060303, 1088215, 1127157, 1139865), the Office of Health and Medical Research of the New South Wales government, the Jeffrey Modell Foundation, and the John Brown Cook Foundation, the Centre de Référence Déficits Immunitaires Héréditaires (CEREDIH), and the ERA-Net for Research Programmes on Rare Diseases (E-Rare: EuroCID, ANR RANR187; no. 061-2016-050). BAP was supported by a Research Training Program Scholarship awarded by the Australian government. SGT was a Principal Research Fellow (1042925) of the NHMRC, and is currently a recipient of an NHMRC Leadership 3 Investigator grant. CSM is supported by an Early-Mid Career Research Fellowship from the New South Wales government.
Version 1. 12/08/2020
In-Press Preview
Version 2. 02/01/2021
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(3):e142434.https://doi.org/10.1172/JCI142434.
Contributor Information
Bethany A. Pillay, Email: b.pillay@garvan.org.au.
Mathieu Fusaro, Email: mathieu.fusaro@gmail.com.
Paul E. Gray, Email: Paul.Gray1@health.nsw.gov.au.
Leslie Burnett, Email: l.burnett@garvan.org.au.
Liliana Bezrodnik, Email: lbezrodnik@yahoo.com.ar.
Alisa Kane, Email: a.kane@garvan.org.au.
Chrystelle Abdo, Email: chrystelle.abdo@gmail.com.
Sarah Winter, Email: sarahwinter_36@hotmail.com.
Samuel Chevalier, Email: samuel.chevalier@aphp.fr.
Romain Levy, Email: rmn.levy@gmail.com.
Cécile Masson, Email: cecile.masson@inserm.fr.
Yohann Schmitt, Email: yohann.schmitt@institutimagine.org.
Marion Malphettes, Email: marion.malphettes@aphp.fr.
Elizabeth Macintyre, Email: elizabeth.macintyre@aphp.fr.
Jean-Pierre De Villartay, Email: jean-pierre.de-villartay@inserm.fr.
John B. Ziegler, Email: j.ziegler@unsw.edu.au.
Joanne M. Smart, Email: Joanne.smart@rch.org.au.
Jane Peake, Email: j.peake@uq.edu.au.
Asghar Aghamohammadi, Email: aghamohammadi@tums.ac.ir.
Lennart Hammarström, Email: lennart.hammarstrom@ki.se.
Hassan Abolhassani, Email: hassan.abolhassani@ki.se.
Capucine Picard, Email: capucine.picard@inserm.fr.
Alain Fischer, Email: alain.fischer@aphp.fr.
Sylvain Latour, Email: sylvain.latour@inserm.fr.
Benedicte Neven, Email: benedicte.neven@aphp.fr.
Cindy S. Ma, Email: c.ma@garvan.org.au.
References
- 1.Revy P, et al. Somatic genetic rescue in Mendelian haematopoietic diseases. Nat Rev Genet. 2019;20(10):582–598. doi: 10.1038/s41576-019-0139-x. [DOI] [PubMed] [Google Scholar]
- 2.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40(10):721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirschhorn R, et al. Somatic mosaicism for a newly identified splice-site mutation in a patient with adenosine deaminase-deficient immunodeficiency and spontaneous clinical recovery. Am J Hum Genet. 1994;55(1):59–68. [PMC free article] [PubMed] [Google Scholar]
- 4.Hirschhorn R, et al. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat Genet. 1996;13(3):290–295. doi: 10.1038/ng0796-290. [DOI] [PubMed] [Google Scholar]
- 5.Ariga T, et al. T-cell lines from 2 patients with adenosine deaminase (ADA) deficiency showed the restoration of ADA activity resulted from the reversion of an inherited mutation. Blood. 2001;97(9):2896–2899. doi: 10.1182/blood.V97.9.2896. [DOI] [PubMed] [Google Scholar]
- 6.Arredondo-Vega FX, et al. Adenosine deaminase deficiency with mosaicism for a “second-site supplementalressor” of a splicing mutation: decline in revertant T lymphocytes during enzyme replacement therapy. Blood. 2002;99(3):1005–1013. doi: 10.1182/blood.V99.3.1005. [DOI] [PubMed] [Google Scholar]
- 7.Speckmann C, et al. Clinical and immunologic consequences of a somatic reversion in a patient with X-linked severe combined immunodeficiency. Blood. 2008;112(10):4090–4097. doi: 10.1182/blood-2008-04-153361. [DOI] [PubMed] [Google Scholar]
- 8.Stephan V, et al. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N Engl J Med. 1996;335(21):1563–1567. doi: 10.1056/NEJM199611213352104. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser FMP, et al. Protein functionality as a potential bottleneck for somatic revertant variants. J Allergy Clin Immunol. 2021;147(1):391–398. doi: 10.1016/j.jaci.2020.04.045. [DOI] [PubMed] [Google Scholar]
- 10.Marin AV, et al. Primary T-cell immunodeficiency with functional revertant somatic mosaicism in CD247. J Allergy Clin Immunol. 2017;139(1):347–349.e8. doi: 10.1016/j.jaci.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 11.Rieux-Laucat F, et al. Inherited and somatic CD3ζ mutations in a patient with T-cell deficiency. N Engl J Med. 2006;354(18):1913–1921. doi: 10.1056/NEJMoa053750. [DOI] [PubMed] [Google Scholar]
- 12.Ariga T, et al. Spontaneous in vivo reversion of an inherited mutation in the Wiskott-Aldrich syndrome. J Immunol. 2001;166(8):5245–5249. doi: 10.4049/jimmunol.166.8.5245. [DOI] [PubMed] [Google Scholar]
- 13.Stewart DM, et al. The phenomenon of spontaneous genetic reversions in the Wiskott-Aldrich syndrome: a report of the workshop of the ESID Genetics Working Party at the XIIth Meeting of the European Society for Immunodeficiencies (ESID). Budapest, Hungary October 4–7, 2006. J Clin Immunol. 2007;27(6):634–639. doi: 10.1007/s10875-007-9121-z. [DOI] [PubMed] [Google Scholar]
- 14.Tone Y, et al. Somatic revertant mosaicism in a patient with leukocyte adhesion deficiency type 1. Blood. 2007;109(3):1182–1184. doi: 10.1182/blood-2006-08-039057. [DOI] [PubMed] [Google Scholar]
- 15.Uzel G, et al. Reversion mutations in patients with leukocyte adhesion deficiency type-1 (LAD-1) Blood. 2008;111(1):209–218. doi: 10.1182/blood-2007-04-082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brigida I, et al. T-cell defects in patients with ARPC1B germline mutations account for combined immunodeficiency. Blood. 2018;132(22):2362–2374. doi: 10.1182/blood-2018-07-863431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoshino A, et al. Modification of cellular and humoral immunity by somatically reverted T cells in X-linked lymphoproliferative syndrome type 1. J Allergy Clin Immunol. 2019;143(1):421–424.e11. doi: 10.1016/j.jaci.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 18.Palendira U, et al. Expansion of somatically reverted memory CD8+ T cells in patients with X-linked lymphoproliferative disease caused by selective pressure from Epstein-Barr virus. J Exp Med. 2012;209(5):913–924. doi: 10.1084/jem.20112391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crestani E, et al. RAG1 reversion mosaicism in a patient with Omenn syndrome. J Clin Immunol. 2014;34(5):551–554. doi: 10.1007/s10875-014-0051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ban SA, et al. Combined immunodeficiency evolving into predominant CD4+ lymphopenia caused by somatic chimerism in JAK3. J Clin Immunol. 2014;34(8):941–953. doi: 10.1007/s10875-014-0088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs S, et al. Omenn syndrome associated with a functional reversion due to a somatic second-site mutation in CARD11 deficiency. Blood. 2015;126(14):1658–1669. doi: 10.1182/blood-2015-03-631374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishikomori R, et al. X-linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T-cell development and/or survival. Blood. 2004;103(12):4565–4572. doi: 10.1182/blood-2003-10-3655. [DOI] [PubMed] [Google Scholar]
- 23.Narumi S, et al. SAMD9 mutations cause a novel multisystem disorder, MIRAGE syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792–797. doi: 10.1038/ng.3569. [DOI] [PubMed] [Google Scholar]
- 24.Buonocore F, et al. Somatic mutations and progressive monosomy modify SAMD9-related phenotypes in humans. J Clin Invest. 2017;127(5):1700–1713. doi: 10.1172/JCI91913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McDermott DH, et al. Chromothriptic cure of WHIM syndrome. Cell. 2015;160(4):686–699. doi: 10.1016/j.cell.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jing H, et al. Somatic reversion in dedicator of cytokinesis 8 immunodeficiency modulates disease phenotype. J Allergy Clin Immunol. 2014;133(6):1667–1675. doi: 10.1016/j.jaci.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moncada-Velez M, et al. Somatic mosaicism caused by monoallelic reversion of a mutation in T cells of a patient with ADA-SCID and the effects of enzyme replacement therapy on the revertant phenotype. Scand J Immunol. 2011;74(5):471–481. doi: 10.1111/j.1365-3083.2011.02593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wada T, et al. Somatic mosaicism in Wiskott-Aldrich syndrome suggests in vivo reversion by a DNA slippage mechanism. Proc Natl Acad Sci U S A. 2001;98(15):8697–8702. doi: 10.1073/pnas.151260498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie JW, et al. In vivo reversion of an inherited mutation in a Chinese patient with Wiskott-Aldrich syndrome. Hum Immunol. 2015;76(6):406–413. doi: 10.1016/j.humimm.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Kury P, et al. Long-term robustness of a T-cell system emerging from somatic rescue of a genetic block in T-cell development. EBioMedicine. 2020;59:102961. doi: 10.1016/j.ebiom.2020.102961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay BA, et al. Hematopoietic stem cell transplant effectively rescues lymphocyte differentiation and function in DOCK8-deficient patients. JCI Insight. 2019;5:e127527. doi: 10.1172/jci.insight.127527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tangye SG, et al. Dedicator of cytokinesis 8-deficient CD4+ T cells are biased to a TH2 effector fate at the expense of TH1 and TH17 cells. J Allergy Clin Immunol. 2017;139(3):933–949. doi: 10.1016/j.jaci.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kearney CJ, et al. DOCK8 drives Src-dependent NK cell effector function. J Immunol. 2017:ji1700751. doi: 10.4049/jimmunol.1700751. [DOI] [PubMed] [Google Scholar]
- 34.Tangye SG, et al. Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol. 2020;40(1):24–64. doi: 10.1007/s10875-019-00737-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aydin SE, et al. DOCK8 deficiency: clinical and immunological phenotype and treatment options - a review of 136 patients. J Clin Immunol. 2015;35(2):189–198. doi: 10.1007/s10875-014-0126-0. [DOI] [PubMed] [Google Scholar]
- 36.Asur RS, et al. Somatic mosaicism of an intragenic FANCB duplication in both fibroblast and peripheral blood cells observed in a Fanconi anemia patient leads to milder phenotype. Mol Genet Genomic Med. 2018;6(1):77–91. doi: 10.1002/mgg3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Virts EL, et al. AluY-mediated germline deletion, duplication and somatic stem cell reversion in UBE2T defines a new subtype of Fanconi anemia. Hum Mol Genet. 2015;24(18):5093–5108. doi: 10.1093/hmg/ddv227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crawford G, et al. DOCK8 is critical for the survival and function of NKT cells. Blood. 2013;122(12):2052–2061. doi: 10.1182/blood-2013-02-482331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Randall KL, et al. DOCK8 deficiency impairs CD8 T cell survival and function in humans and mice. J Exp Med. 2011;208(11):2305–2320. doi: 10.1084/jem.20110345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janssen E, et al. A DOCK8-WIP-WASp complex links T cell receptors to the actin cytoskeleton. J Clin Invest. 2016;126(10):3837–3851. doi: 10.1172/JCI85774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz-García R, et al. A case of partial dedicator of cytokinesis 8 deficiency with altered effector phenotype and impaired CD8+ and natural killer cell cytotoxicity. J Allergy Clin Immunol. 2014;134(1):218–221. doi: 10.1016/j.jaci.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 42.Kienzler A-K, et al. Hypomorphic function and somatic reversion of DOCK8 cause combined immunodeficiency without hyper-IgE. Clin Immunol. 2016;163:17–21. doi: 10.1016/j.clim.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jouanguy E, et al. Human inborn errors of immunity to herpes viruses. Curr Opin Immunol. 2020;62:106–122. doi: 10.1016/j.coi.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sallusto F. Heterogeneity of human CD4+ T cells against microbes. Annu Rev Immunol. 2016;34(1):317–334. doi: 10.1146/annurev-immunol-032414-112056. [DOI] [PubMed] [Google Scholar]
- 45.Mizesko MC, et al. Defective actin accumulation impairs human natural killer cell function in patients with dedicator of cytokinesis 8 deficiency. J Allergy Clin Immunol. 2013;131(3):840–848. doi: 10.1016/j.jaci.2012.12.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qi Q, et al. Diversity and clonal selection in the human T-cell repertoire. Proc Natl Acad Sci U S A. 2014;111(36):13139–13144. doi: 10.1073/pnas.1409155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robins HS, et al. Overlap and effective size of the human CD8+; T cell receptor repertoire. Science Translational Medicine. 2010;2(47):47ra64. doi: 10.1126/scitranslmed.3001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aydin SE, et al. Hematopoietic stem cell transplantation as treatment for patients with DOCK8 deficiency. J Allergy Clin Immunol Pract. 2019;7(3):848–855. doi: 10.1016/j.jaip.2018.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hacein-Bey-Abina S, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346(16):1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 50.Payne K, et al. OMIP–063: 28–color flow cytometry panel for broad human immunophenotyping. Cytometry A. 2020;97(8):777–781. doi: 10.1002/cyto.a.24018. [DOI] [PubMed] [Google Scholar]
- 51.Grandin V, et al. A RAB27A duplication in several cases of Griscelli syndrome type 2: An explanation for cases lacking a genetic diagnosis. Hum Mutat. 2017;38(10):1355–1359. doi: 10.1002/humu.23274. [DOI] [PubMed] [Google Scholar]
- 52.Rosain J, et al. A variety of alu-mediated copy number variations can underlie IL-12Rβ1 deficiency. J Clin Immunol. 2018;38(5):617–627. doi: 10.1007/s10875-018-0527-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brüggemann M, et al. Standardized next-generation sequencing of immunoglobulin and T-cell receptor gene recombinations for MRD marker identification in acute lymphoblastic leukaemia; a EuroClonality-NGS validation study. Leukemia. 2019;33(9):2241–2253. doi: 10.1038/s41375-019-0496-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alamyar E, et al. IMGT(®) tools for the nucleotide analysis of immunoglobulin (IG) and T cell receptor (TR) V-(D)-J repertoires, polymorphisms, and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol Biol. 2012;882:569–604. doi: 10.1007/978-1-61779-842-9_32. [DOI] [PubMed] [Google Scholar]
- 56.Shugay M, et al. VDJtools: Unifying post-analysis of T cell receptor repertoires. PLoS Comput Biol. 2015;11(11):e1004503. doi: 10.1371/journal.pcbi.1004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.