Abstract
Background
Seldom performance evaluation and diagnosis comparison studies were reported for different chemiluminescent immunoassay (CLIA) kits approved under an emergency approval program for SARS‐CoV‐2 infection.
Methods
A total of 100 and 105 serum separately from non‐infected populations and COVID‐19 patients were detected with SARS‐CoV‐2 IgM and IgG kits. The characteristics including precision, functional sensitivity, linearity, and accuracy were evaluated for Axceed, iFlash, and Maglumi CLIA kits.
Results
Maglumi and iFlash had the best analytical sensitivity for IgM and IgG, respectively. Axceed kits had a linearity response in the detected concentration. The clinical sensitivity of Axceed, iFlash, and Maglumi was 68.0%, 64.9%, and 63.9% with a specificity of 99.0%, 96.0%, and 100% for IgM, 85.6%, 97.9%, and 94.8% with a specificity of 97.0% for IgG. ROC analysis indicated all kits had a diagnostic power greater than 0.9. Notably, either IgM or IgG kits obtained a poor agreement (Kappa value from 0.397 to 0.713) with others. Among 38 recovered patients, 94.7% had an effective immune response, and both seropositive IgM and IgG accounted for the biggest proportion (medium, 42 days onset), then followed by the single seropositive IgG (medium, 50 days onset) in Ab profile.
Conclusion
The performance of CLIA kits satisfied the diagnosis of SARS‐CoV‐2 infection. Both positive of IgG and IgM contributes to improve the specificity, and a positive one will enhance the sensitivity.
Keywords: chemiluminescent immunoassay, clinical diagnosis, performance comparison, SARS‐CoV‐2 infection, serological tests
Performance of three CLIA kits for SARS‐Cov‐2 antibodies was compared. Diagnosis features of three CLIA kits for SARS‐Cov‐2 infection were reviewed. Dynamic profile of IgM and IgG Ab in recovered COVID‐19 patients was followed up.

1. INTRODUCTION
A spread of SARS‐CoV‐2 virus infection occurred in the whole world. The novel coronavirus (COVID‐19) pandemic brings a severe threat to human health. As of August 27, there are now more than 24 million reported cases of COVID‐19 and 820,000 deaths. 1 Symptoms such as fever, cough, sore throat, body aches, headaches, and other acute respiratory syndromes commonly appeared in most of COVID‐19 patients. However, it is worth noting that a grow number of patients were found to be asymptomatic or false‐negative results in nucleic acid tests, thus increasing the difficulty of patient diagnosis and the risk of virus transmission. 2
Laboratory tests play a pivotal role in the diagnosis and management of COVID‐19. To response the serious COVID‐19 pandemic in United States and improve diagnostic efficiency for SARS‐CoV‐2 infection, a study on protocol to validate the sufficiency of home‐collected samples to detection of SARS‐CoV‐2 RNA and antibodies was reported. Although the nucleic acid testing is the gold standard for COVID‐19 diagnosis, the accuracy is generally restricted by specimen collection, transportation, and RNA extracted operation. Besides, the method was proved to be time‐consuming, labor‐intensive, and at high risk of infection. 3 , 4 The SARS‐CoV‐2 infected patients were confirmed by a positive nucleic acid result, but a negative result cannot absolutely exclude the infection possibility because of 50% positive rate found in nucleic acid testing. 5
Less expensive and easy implementable serological tests are urgently needed for an accurate diagnosis of SARS‐CoV‐2 infection, contact tracing, and epidemiological studies. Excitedly, the immune system produces virus‐specific antibody, including immunoglobulin M (IgM) and immunoglobulin G (IgG) in response to COVID‐19. Serological antibody detection emerging as an auxiliary diagnostic method for COVID‐19 has attracted extensive concerns. 6 The assay of SARS‐CoV‐2 IgM and IgG should be applied as an additional non‐invasive method for the infection. It was reported that the positive virus‐specific IgG was 100% within three weeks and IgM reached 94.1% within 20‐22 days after symptom onset. 7 The profile of IgM and IgG can not only make an assisted diagnosis, but also characterize the disease course, investigate epidemiologic features, as well as accelerate vaccine development. 8 , 9 , 10 Except the diagnostic efficacy of IgM and IgG has been proved in SARS‐CoV‐2 infection, the performance of commercial immunoassays was compared aiming at a better choice for a detection tool. 11 , 12 The diagnostic sensitivity, specificity, and concordance of SARS‐CoV‐2 assay between Abbott and Euroimmun was compared, the former had fewer false‐positive and false‐negative results than the later. 13 Most of above previous researches focused on the clinical significance of different antibody kits for SARS‐CoV‐2 infection, the diagnostic characteristics just included sensitivity, specificity, and receiver operator curve (ROC) among various immunoassays. Analytical performance involving testing process of SARS‐CoV‐2 antibody kits was closely related to the diagnostic accuracy. There were only several studies reported the analytical performance evaluation in serological antibody kits. Besides of the sensitivity and specificity, the performance containing imprecision, linearity of dilution, and accuracy were evaluated in the MAGLUMI™ 2000 Plus 2019‐nCov IgM and IgG assays (Snibe, Shenzhen, China). 14 To date, more than 10 antibody kits for SARS‐CoV‐2 infection have been approved by the National Medical Products Administration (NMPA). 15 The antibody was mainly detected by chemiluminescence, colloidal gold immunochromatographic, and enzyme‐linked immunosorbent assay. A report showed variable performance values were acquired among six SARS‐CoV‐2 immunoassays containing Abbott Architec, Diasorin Liasison, Euroimmun, Acro, Xiamen Biotime, etc, in comparison with microneutralization. 16 Similarly, a poor qualitative concordance of IgM and IgG results was obtained in our laboratory tests using different SARS‐CoV‐2 immunoassays for the same patient serum. To improve the serodiagnosis of SARS‐CoV‐2 infection, laboratory should carefully investigate the analytical and diagnostic performance of different immunoassays. Various chemiluminescent immunoassay (CLIA) kits were approved under an emergency approval program due to the automation operation and high efficiency. Hence, the data on performance evaluation and diagnosis comparison studies for different CLIA kits in SARS‐CoV‐2 detection were demanded. This study aimed to evaluate the analytic performance, diagnostic characteristics of three antibody test kits, as well as to investigate the kinetics of antibodies titers in COVID‐19 patients.
2. MATERIALS AND METHODS
2.1. Sample collection and handling
The patients were enrolled from hospital clinics, wards and quarantine stations (from March 13 to April 10, 2020) and performed with a SARS‐CoV‐2 nucleic acid testing according to a standard laboratory protocol. The residual serum used for routine laboratory tests was collected for serological IgM and IgG detection. The serum samples included 46 with or without acute viral respiratory tract infections in fever clinics, 54 inpatients, 60 previously, and 45 currently infected with SARS‐CoV‐2. The serum was centrifuged at 3000g for 10 minutes (Allegra X‐15R, Beckman Coulter, USA), aliquoted and stored at −20°C prior to IgG and IgM determination. To avoid repeated thawing and refreezing, the serum was analyzed as soon as possible or kept within three days at 2 ~ 8°C. This study was reviewed and approved by the Ethical Committee of Shenzhen Luohu People's Hospital (Approval No. 2020‐SZLH‐LW‐011).
2.2. Nucleic acid testing
Upper respiratory nasopharyngeal swabs were collected. Specimens acquisition, preservation, transportation, and detection were strictly in accordance with standard operating procedure. The system NP968‐C (TianLong medtl., China) was used to extract RNA from PCR cell media and then detected with registered kits from BioGerm (No. 20200127F, Shanghai BioGerm Medical Biotechnology Co., Ltd, China) and Liferiver (No. P20200208, Shanghai ZJ Bio‐Tech CO., Ltd), respectively.
2.3. SARS‐CoV‐2 IgM and IgG detection
Serological tests were conducted on three fully automatic CLIA platforms. Those CLIA instruments include Axceed 260 (Bioscience Diagnostic Technology Co.Ltd., Tianjin, China), iFlash3000 (YHLO Biotech Co., Ltd., Shenzhen, China), and Maglumi 800 (New Industries Biomedical Engineering Co., Ltd., Shenzhen, China). The SARS‐CoV‐2 IgM and IgG kits were provided by their manufacturers. For Axceed IgM (Lot No. G202004305) and IgG (Lot No. G202004307) kits, 20 µL of serum samples were added into 980 µL sample diluent, followed by vertexing 5s (Vortex‐Genie2, Scientific Industries, USA). After standing for 15mins, 75 µL of diluted samples were introduced into the Axceed 260 system. As for iFlash (Lot No. 20200304) and Maglumi (Lot No. 2722000201, 2722000202) kits, 20 µL of the serum was directly drawn into the automatic platform and detected. The response signal is expressed in a relative light unit (RLU). The concentration of IgM or IgG antibody is positively correlated with RLU and calculated by the RLU value and a built‐in calibration curve. The cutoff value exceeding 1S/CO, 10.0AU/mL, and 1.0AU/mL was determined as positive IgM or IgG providing by the Axceed, iFlash, and Maglumi kits.
The serum pools for analytical characteristics and clinical samples for diagnosis were analyzed on all of three CLIA platforms according to the instructions of CLIA kits.
2.4. Analytical and clinical performance evaluation
The analytical performance consisting of precision, functional sensitivity, and linearity was evaluated following CLSI protocol EP15‐A2 and EP17‐A 17 , 18 and NCCLS protocol EP12‐A. 19 Considering repeatability and between‐day variability for IgG and IgM detection, the precision was evaluated by using three serum pools at low (negative), medium (around the cutoff value) and high levels. Each level sample was measured four repetitions per day within a five‐day period. The repeatability and within‐laboratory precision were calculated and expressed as coefficient of variation (CV%).
The functional sensitivity was defined as the lowest IgM/IgG concentration. Because lack of the available standard blood serum plate for comparison of the detection limits of SARS‐CoV‐2 IgM and IgG CLIA kits, the same serum pool was serially diluted with negative serum pool used as the alternative one. A total of 8 ~ 10 stepwise serum pools were analyzed for repetitive 20 measurements within five consecutive days at each concentration. The positive probabilistic curve was plotted with dilution times and positive results ratio (%).
Linearity was evaluated by mixing patient serum pool with negative sample pool at a ratio of 1:0, 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, 1:128, 1:256, and 1:512. The linear regression analysis was performed by using the levels at expected and measured concentrations. The regression equation and the correlated coefficient were calculated.
2.5. Data Statistics
Statistical analysis was conducted with SPSS 19.0 for Mann‐Whitney U tests, χ2 test, Kappa and McNemar evaluation between grouped comparison. p value below 0.05 was considered statistically significant. The Kappa value below 0.75 was considered poor consistency among different kits’ results. Clinical diagnosis as the gold standard, the sensitivity, specificity, and the coincident rate were calculated using a fourfold table. Receiver operating characteristic (ROC) and the area under curve (AUC) were performed by GraphPad Prism 5 for diagnosis of patients infected with SARS‐CoV‐2 using the three kits.
3. RESULTS
3.1. Repeatability and within‐laboratory precision
The imprecision of repeatability and within‐laboratory precision tests for three IgG and IgM kits was summarized in Table S1. The CV% was between 0.62% to 5.54% for IgG kits at three levels. The impression ranged from 0.70% to 5.58% for IgM kits, except Axceed kits obtained a high CV% of 12.97% and 16.82% at the low level (0.02 ± 0.00 S/CO). Most of the measured CV% was less than 6.0% or a manufacturer declaration.
3.2. Functional sensitivity
The positive probabilistic curves for SARS‐CoV‐2 IgM and IgG kits were shown in Figure 1. The highest dilution times produced 95% positive results (C95) reflecting the excellent detection limit. The same serum pool diluted by 93.44 times, 54.6 times, and 6.70 times produced 95% positive results (C95) for Maglumi, Axceed, and iFlash IgM kits, indicating the former IgM kit had the best analytical sensitivity than others. For IgG kits, the functional sensitivity in descending order was as follows: iFlash, Maglumi, and Axceed with the serum diluted by 43.8 times, 10.1 times, and 6.02 times. Besides, the gray zone ranging from 5% positive results (C5) to C95 contributes to compare the precision of qualitative results around the cutoff value by different kits. The narrower range of gray zone demonstrated the kits had a greater precision around the cutoff value. The samples approximate to the cutoff level could acquire good consistency diagnosed results (positive or negative). The gray zone ranging from C5 to C95 in Figure 1 was 6.7, 1.48, and 14.06 dilution times for Axceed, iFlash, Maglumi IgM kits, while 1.57, 4.0, 1.7 dilution times for IgG kits. iFlash IgM and Maglumi IgG kits had the best precision for diagnosis the samples around the cutoff level. In summary, Axceed IgM and IgG kits had the best sensitivity and qualitative precision.
Figure 1.
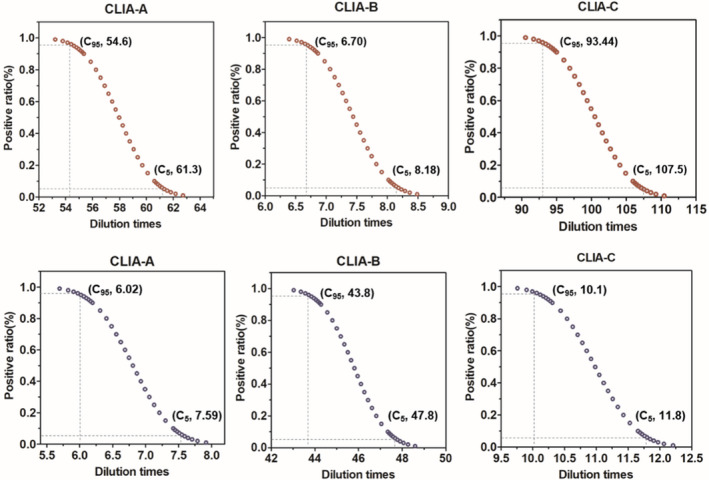
Positive probability curves of the SARS‐CoV‐2 IgM (red line) and IgG (blue line). The same serum pool was diluted approximate to the cutoff value and 20 aliquots per concentration were analyzed within 5 days by CLIA‐A (Axceed), CLIA‐B (iFlash) and CLIA‐C (Maglumi) kits. The x‐axis shows the dilution times and the y‐axis shows positive rates determined by the cutoff value of the kits. The dotted line is produced by GraphPad Prism 5. C95 and C5 shows the 95% and 5% positive rate, respectively
3.3. Linearity assessment
In Figure S1, the good linearity was obtained with Axceed kits in the concentration range of 0.06‐25.9 S/CO (R 2 = 0.9991) and 0.07‐11.37 S/CO (R 2 = 0.9854) for IgM and IgG antibody, respectively. The iFlash IgM kit was obtained a linear concentration between 4.87 AU/mL and 258.72 AU/mL, while a non‐linearity was observed for iFlash IgG kit ranging from 6.75 AU/mL to 134.2 AU/mL. Both of Maglumi IgG and IgM kits exhibited a non‐linear trend in the investigated concentration.
3.4. Diagnosis accuracy comparison
The diagnosis standard was based on RT‐PCR and clinical diagnosis such as image examination. A total of 100 serum from non‐infected groups and 105 serum from COVID‐19 patients were detected with SARS‐CoV‐2 IgM and IgG kits. Among 105 patient serum, five samples were collected within 24‐48 hours of onset of symptoms, most were from individuals appearing fever, cough, sluggish, headache, and chest tightness. Besides, three samples were from asymptomatic patients, and two were positive in IgG (S/CO 1.184, cutoff 1.0) or IgM (S/CO 7.8, cutoff 1.0) detected by CLIA kits. The diagnosis feature in Table S2 showed Axceed IgM kits had a better sensitivity reaching 68.0% with a specificity of 99.0% in comparison with other kits. Although IgG kits had a slightly higher sensitivity (85.6%), specificity for all the kits was 97%. Either IgM or IgG CLIA kits yielded an overall concordance of 80.7% (95% CI, 74.6%‐85.6%). CLIA IgG kits acquired a higher Yuden index (≥0.82) than IgM (≥0.61). The AUC of ROC analysis (Figure 2) provides a good indicator to comparing the diagnostic power among different kits. The IgG antibody yielded a diagnostic power for SARS‐CoV‐2 at 0.9812 (95%CI: 0.9625‐0.9999), 0.9831 (95%CI: 0.9599‐1.006), and 0.9737 (95%CI: 0.9486‐0.9988), with the IgM diagnostic power at 0.9789 (95%CI: 0.9590‐0.9987), 0.9198 (95%CI: 0.8805‐0.9591), 0.8973 (95%CI: 0.8503‐0.9442) by Axceed, iFlash, and Maglumi kits, respectively. The coincidence evaluation of diagnostic results was performed. Either IgM or IgG kits obtained a poor agreement with others (IgM: Axceed vs iFlash, kappa 0.499; iFlash vs Maglumi, kappa 0.585; Axceed vs Maglumi, kappa 0.556; IgG: Axceed vs iFlash, kappa 0.444; Axceed vs Maglumi, kappa 0.397) except for iFlash vs Maglumi IgG kits (kappa 0.713).
Figure 2.
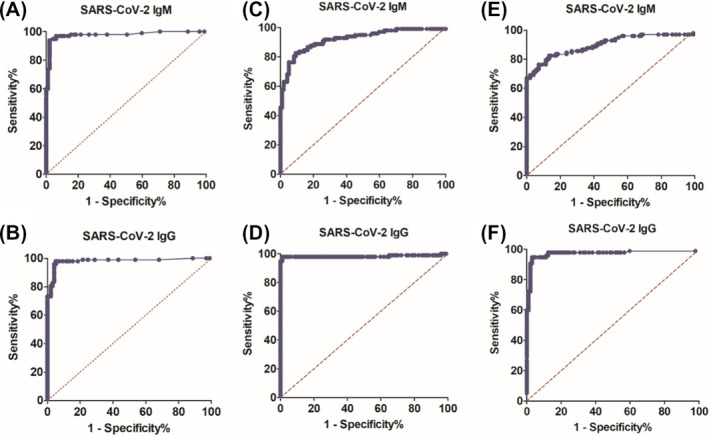
Comparison of diagnostic effect of Axceed (A and B), iFlash (C and D) and Maglumi (E and F). Receiver operating characteristic (ROC) curve and area under the ROC curve (AUR) were analyzed by SARS‐CoV‐2 infected group (n = 105) and control group (n = 100)
3.5. Serological IgM and IgG dynamics in COVID‐19 patients
The serological antibodies profile in confirmed COVID‐19 patients detected with three CLIA kits and RNA testing was listed in Table 1. The χ 2 test attested that there were no remarkable difference (p > .05) on the diagnostic mode (RNA+/IgM+/IgG + and RNA+/IgM‐/IgG‐) vs (RNA‐/IgM+/IgG + and RNA‐/IgM‐/IgG‐) using CLIA combined RNA kits. Moreover, the concentration detected in different modes was compared and no significant difference was found (p > .05) by U test. The application of antibody profile will facilitate defining the clinical infection phase. In general concepts for immune response after virus infection, both of positive IgM and IgG or only positive IgM suggested the patient suffering from an acute infection. In fact, the recovered COVID‐19 patients had cleared the SARS‐CoV‐2 but also were detected with IgM antibody. Among 38 recovered patients, 36 patients (94.7%) had an effective immune response and produced the SARS‐CoV‐2 antibody.
Table 1.
The SARS‐CoV‐2 RNA diagnostic result and synchronous serological antibodies diagnostic result, level and positive ration in 105 COVID‐19 patients
| Profile mode | Axceed (S/CO) | iFlash (AU/mL) | Maglumi (AU/mL) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | RNA | IgM | IgG | IgM | IgG | Po. Ratio | IgM | IgG | Po. Ratio | IgM | IgG | Po. Ratio |
| 1 | + | ‐ | ‐ | 0.05‐0.410 | 0.10‐0.23 | 5.71% | 0.25‐2.82 | 0.35‐3.05 | 4.76% | 0.06‐0.799 | 0.05‐0.73 | 8.57% |
| 2 | + | + | ‐ | 7.80‐16.97 | 0.24‐0.94 | 5.71% | N.A. | N.A. | 0% | 3.66 | 0.05 | 0.95% |
| 3 | + | + | + | 1.75‐22.05 | 1.22‐38.2 | 15.24% | 15.3‐213.5 | 51.3‐539.6 | 22.86% | 1.234‐8.911 | 4.53‐200 | 23.81% |
| 4 | + | ‐ | + | 0.26‐0.93 | 1.16‐19.4 | 16.19% | 0.86‐9.93 | 19.2‐84.0 | 14.29% | 0.363‐0.968 | 1.29‐6.02 | 9.52% |
| 5 | ‐ | ‐ | ‐ | 0.06‐0.98 | 0.10‐0.96 | 5.71% | 0.14‐0.98 | 0.54‐1.57 | 1.90% | 0‐0.26 | 0‐0.03 | 1.90% |
| 6 | ‐ | + | ‐ | 2.66‐3.69 | 0.49‐0.82 | 2.86% | N.A. | N.A. | 0% | N.A. | N.A. | 0 |
| 7 | ‐ | + | + | 1.28‐30.61 | 1.0‐42.85 | 40.0% | 10.3‐485.5 | 42.5‐468.3 | 35.24% | 1.097‐20.48 | 2.17‐98.72 | 35.24% |
| 8 | ‐ | ‐ | + | 0.33‐0.89 | 1.1‐36.79 | 8.57% | 1.03‐9.82 | 48.6‐122.9 | 20.0% | 0‐0.852 | 1.24‐45.81 | 20.0% |
Po. Ratio represents positive ratio; ‐ and + represents negative and positive result, respectively; N.A. represents not available.
The concentration of serological antibodies in all included serum was dramatically scattered in two groups (Figure 3A,B). The U test indicated the mean concentration in patients was significantly different from that in non‐infected group. Those tested serum consisted of two time periods after symptom onset, eight samples within two days and 97 serum from 20 days to 70 days. In Figure 3C,D, the antibodies concentration was observed by regular variation along with increasing of onset time. The IgM antibody gradually decreased after the symptom onset from 20 days to >60 days, while IgG antibody increased between four weeks and eight weeks, followed by a declined trend.
Figure 3.
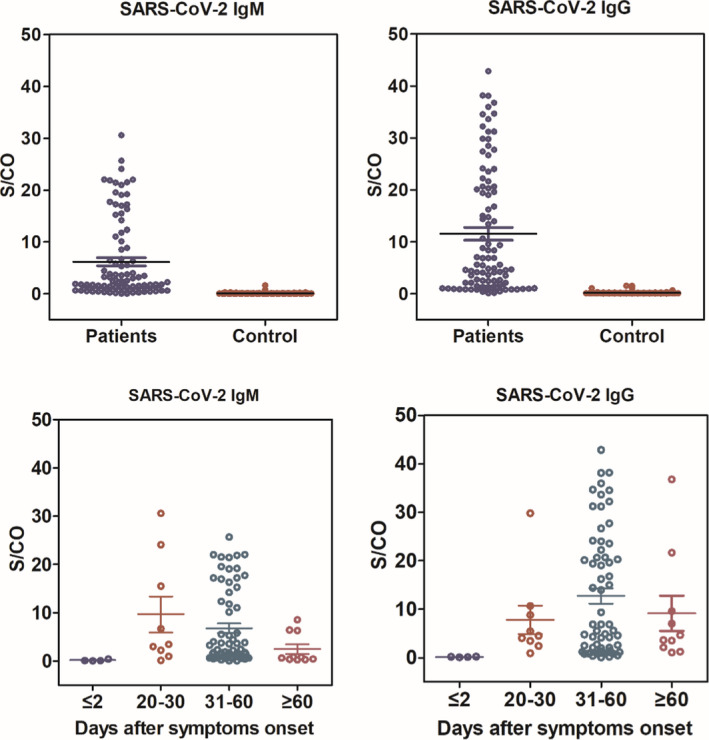
IgM and IgG antibodies response against SARS‐CoV‐2 from patients in comparison with that from control population (A and B), a Mann‐Whitney U test was used and P value below 0.05 was considered statistically significant. Kinetics of antibody responses against SARS‐CoV‐2 following infection from patients (C and D), the x‐axis represents the days of ill onset and synchronous date of IgM and IgG detection, the y‐axis shows the response value detected by Axceed
Six COVID‐19 patients were followed up, the dynamic antibody titers were observed over the follow‐up period (Figure 4). In four ordinary patients, a gradual decrease in IgM and IgG levels was observed after 20 days since illness onset. During the observation period, the individual kept seropositive IgG and IgM, except for patient 9 (after 62 days). With symptom onset after 62 days, patient 9 was observed a seroconversion for IgM. A senior (72 years old) and a prepartal woman (31 years old) were diagnosed as severe COVID‐19 patients because of acute respiratory distress syndrome (ARDS) on admission. The senior was traced to family clustering infection and suffered from coronary heart disease. After multiple transfusions of plasm (300 mL) of convalescent patients, the senior was found in presence of a high titer of IgG (31.22 S/CO). Thirty‐eight days after symptom onset later, IgM was found seronegative and the major clinical symptom improved. As for the prepartal woman, she underwent cesarean, followed by therapy with hemofiltration, anti‐infective drugs, extracorporeal membrane oxygenation (ECMO), and once plasm (300 mL) of convalescent patients. However, IgM persisted high within two months; afterward, the titer remarkably decreases and IgG responds to be positive in a month and a half later. The measurement of IgM and IgG titers facilitate physicians to evaluate the patients recovered situation and guide symptomatic therapy.
Figure 4.
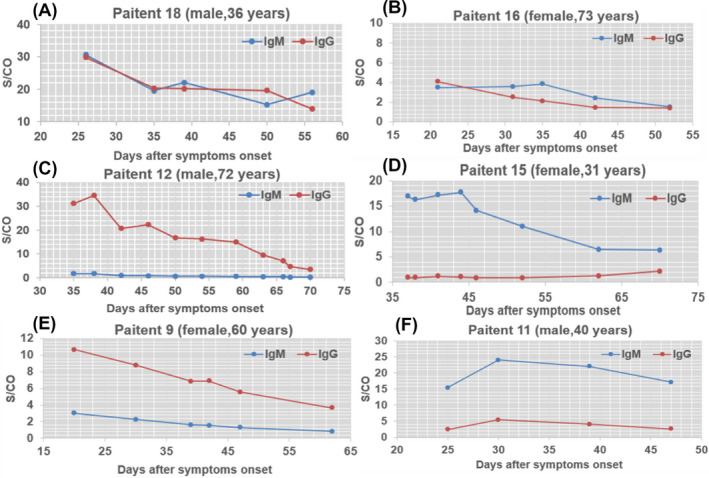
Dynamic profile of IgM and IgG antibodies in representative COVID‐19 patients after symptoms onset. The changes of the levels of antibodies in serum of six patients was presented, the x‐axis represents the days of ill onset, the y‐axis shows the response value of IgM and IgG antibodies in patient serum detected by Axceed. IgG is depicted in blue, and IgM is depicted in orange
4. DISCUSSION
This study presents an analytical performance evaluation and diagnosis comparison for three COVID‐19 chemiluminescent immunoassays for monitoring dynamic profile of SARS‐CoV‐2 IgG and IgM on a panel of 105 samples from individuals with confirmed SARS‐CoV‐2. Two asymptomatic SARS‐CoV‐2‐infected cases were detected by immunoassay and confirmed with RNA. In the early infection period (≤two days), viral RNA testing maintained a higher positive rate than SARS‐CoV‐2 IgG and IgM detection (25%~38%). In agreement with previous publications, 9 , 20 as the infection period extension and medical treatment, the positive rate by viral RNA testing declined remarkably (65.57%, 26.22%, and 6.56% from the first week to the third week, data had not been published). Our results demonstrated serological tests as an effective complementary way for detecting COVID‐19 patients, especially in tracing the asymptomatic cases. There are a number of studies published on the performance comparison of diagnostic sensitivity and specificity of various commercial immunoassays aiming at a better choice for a detection kit. However, less research was conducted on the analytical characteristics of testing process which laid a foundation for diagnosis. Herein, the parameters consisting of repeatability and within‐laboratory precision, functional sensitivity, and linearity of dilute serum pool were verified and compared among three CLIA kits prior to the clinical diagnosis. The functional sensitivity and linearity showed a difference among three automatic CLIA platforms.
As for diagnosis, the good agreement was proved in the overall clinical sensitivity (63.9%~68.0% for IgM, 85.6%~97.9% for IgG) and specificity (96.0%~100.0% for IgM, 97.0% for IgG) among these CLIA kits. In another study, 21 the diagnostic sensitivity of IgM vs IgG (60% vs 86.67%) by an iFlash CLIA kit was similar with our results. However, the diagnosis results for individual patients were not well consistent (the kappa value from 0.397 to 0.713) based on the chemiluminescent method. The obvious difference was observed in diagnostic sensitivity between Axceed and the other two kits. The phenomenon probably derived from reagent composition (referred to Table S3), especially recombinant antigen. Malgumi and iFlash IgG kit achieved a higher sensitivity in comparison with Axceed, it was probably due to the former two identified IgG against the dual target (Nucleocapside and Spiked proteins). 22 Axceed CLIA kit tested IgM and IgG antibodies against the receptor binding domain (RBD) of SARS‐CoV‐2 S1 which was one of subunits of S protein and were expected to be more specific. The capture patter for IgM probably likely contributes to high functional sensitivity (93.44 dilution times) and specificity (100%) than indirect way in comparison with other kits. As shown in Figure 5, Both IgG and IgM antibodies were detected by indirect format in the design of Axceed and iFlash kits, while IgM was recognized by a capture pattern in Malgumi kit. The false‐positive cases were detected with 4/100, 3/100 and 7/100 by Axceed, Malgumi, and iFlash, respectively. Except for antigen components and detection patter, the cross‐reactivity might derive from non‐specific binding of plasma proteins, non‐specific antibodies (such as heterophile antibodies), rheumatoid factors, complements, or drugs. Serum was diluted before introducing it into the Axceed 260 system, while serum was directly draw into the iFlash and Maglumi automatic platform. This might benefit the performance evaluation, decreasing the false‐positive results, it might as well underestimate the seroprevalence of SARS‐CoV‐2 antibodies. Safety operation with the infected serum also was important, and previous report showed the heat inactivation at 56°C significantly decreased the level of SARS‐CoV‐2 antibodies. 23 In the study, automatic CLIA system effectively reduced the professional exposing risk in compared with immunochromatographic assay when the infected serums are analyzed. In the study, the sensitivity of IgM is lower than IgG, the patients enrolled mostly were in the medium or recovered term of infection, leading to a lower sensitivity in IgM.
Figure 5.
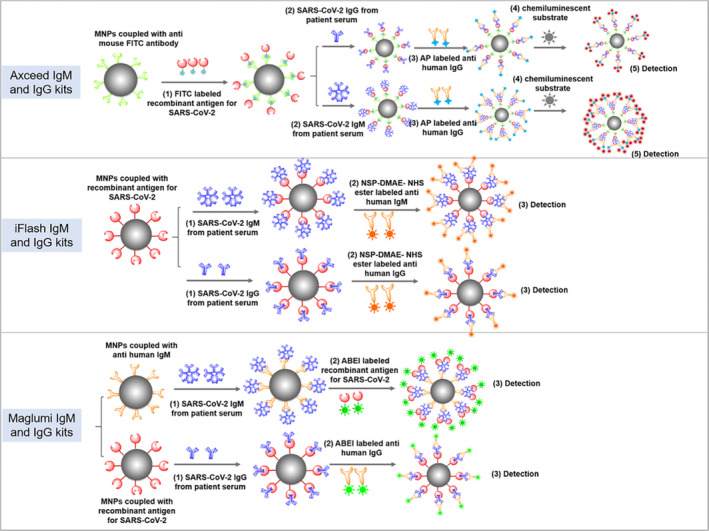
The detection principle and flow diagram of Axceed, iFlash and Maglumi chemiluminescent immunoassays for SARS‐CoV‐2 IgM and IgG. Axceed and iFlash kits applied an indirect immunofluorescence for the detection of IgG and IgM antibodies, Malgumi IgM kit used an antigen capture pattern
From the antibodies profile, both seropositive IgM and IgG accounted for the biggest proportion (onset days: medium, 42 days), then followed by the single seropositive IgG (onset days: medium, 50 days). The positive of IgG and IgM contributes to improve specificity of IgG, and a positive one is valuable to enhance the sensitivity in the early‐infected phase. Even though CLIA kit belongs to a qualitative diagnosis tool, the light intensity given in the system provided a way for antibodies determination in the patients. With a linearity investigation, Axceed kit was perfectly suitable for monitoring the dynamic levels of IgM and IgG in the patients in comparison with other kits. The results may derive from pre‐dilution by 50 times with buffer prior to injection and the matrix effect from human serum was greatly reduced. The dynamic measurement of IgM and IgG titers facilitate to judge the infected or recovered situation, further guide symptomatic therapy. Serological screening also was proved to be an excellent approach for asymptomatic patient diagnosis. A patient had contact with patients from infected area in epidemiology survey and was detected with positive IgM by Axceed kit and positive IgG by iFlash and Maglumi kits with high titers. However, the patient was found only a slight elevation of the amplification curve in repetitive RNA tests prior to confirmation. Six recovered patients maintained SARS‐CoV‐2 IgM and IgG negative status after discharge. The phenomenon was also reported in previous study. 24 It is worth noting that the IgM clearance was not always accompanied by the rising of IgG. 7 The study also had limitations. Due to lack of standard serological serum plates, the performance evaluation replied on a serial of home‐made diluted serum, the real concentration remained unknown. The serum collected focused on early‐infected and recovered patients, a whole seroconversion phase was restricted to be followed up.
In summary, thanks to automatic operation, the CLIA kits were valuable to rapidly and effectively screen the infected patients in controlling of SARS‐CoV‐2 outbreak. Under an emergency approval, different CLIA remained unsatisfied consistency for individual diagnostic results. Further verification was demanded in more populations from different infected area. The follow‐up measurement of Ab titers enhanced the knowledge of Ab elimination during a recovered period and contributed to guide symptomatic therapy.
CONFLICT OF INTERESTS
The authors state that there are no financial, personal, or professional conflicts of interests that may hinder this work.
Supporting information
Figure S1 and Tables S1‐S3
ACKNOWLEDGMENT
This study was supported by the National Natural Science Foundation of China (No. 81703699), Science and Technology Planning Project of Shenzhen City of China (No. JCYJ20180306172209668), the discipline construction ability promotion project of Shenzhen health and population family planning commission (No. SZXJ2017018) and Sanming Project of Medicine in Shenzhen (No. SZSM201601062). We greatly appreciate the kits support from Bioscience Diagnostic Technology, YHLO Biotech, and New Industries Biomedical Engineering.
Dou X, Wang E, Hu J, et al. Comparison of three automatic chemiluminescent immunoassays for monitoring dynamic profile of SARS‐CoV‐2 IgG and IgM. J Clin Lab Anal.2021;35:e23681 10.1002/jcla.23681
Xiaowen Dou and Enyun Wang the author works equally.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCE
- 1. World Health Organization . WHO Coronavirus Disease (COVID‐19) Dashboard. https://covid19.who.int/. Accessed August 27, 2020.
- 2. Lauer SA, Grantz KH, Bi Q, et al. The Incubation Period of Coronavirus Disease 2019 (COVID‐19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172(9):577‐582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zou L, Ruan F, Huang M, et al. SARS‐CoV‐2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382(12):1177‐1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xie X, Zhong Z, Zhao W, et al. Chest CT for Typical 2019‐nCoV Pneumonia: relationship to Negative RT‐PCR Testing. Radiology. 2020;296:E41‐E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Younes N, Al‐Sadeq DW, Al‐Jighefee H, et al. Challenges in Laboratory Diagnosis of the Novel Coronavirus SARS‐CoV‐2. Viruses. 2020;12(6):582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Long QX, Liu BZ, Deng HJ, et al. Antibody responses to SARS‐CoV‐2 in patients with COVID‐19. Nat Med. 2020;26:845‐848. [DOI] [PubMed] [Google Scholar]
- 8. Guo L, Ren L, Yang S, et al. Profiling Early Humoral Response to Diagnose Novel Coronavirus Disease (COVID‐19). Clin Infect Dis. 2020;71(15):778‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020;71(16):2027‐2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie J, Ding C, Li J, et al. Characteristics of patients with coronavirus disease (COVID‐19) confirmed using an IgM‐IgG antibody test. J Med Virol. 2020;92(10):2004‐2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. National SARS‐CoV‐2 Serology Assay Evaluation Group . Performance characteristics of five immunoassays for SARS‐CoV‐2: a head‐to‐head benchmark comparison. Lancet Infect Dis. 2020;20(12):1390‐1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kontou PI, Braliou GG, Dimou NL, et al. Antibody tests in detecting SARS‐CoV‐2 infection: a meta‐analysis. Diagnostics (Basel). 2020;10(5):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tang MS, Hock KG, Logsdon NM, et al. Clinical Performance of Two SARS‐CoV‐2 Serologic Assays. Clin Chem. 2020;66(8):1055‐1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Padoan A, Cosma C, Sciacovelli L, et al. Analytical performances of a chemiluminescence immunoassay for SARS‐CoV‐2 IgM/IgG and antibody kinetics. Clin Chem Lab Med. 2020;58(7):1081‐1088. [DOI] [PubMed] [Google Scholar]
- 15. National Medical Products Administration . Medical devices. http://app1.nmpa.gov.cn/data_nmpa/face3/base.jsp?tableId=26&tableName=TABLE26&title=%E5%9B%BD%E4%BA%A7%E5%99%A8%E6%A2%B0&bcId=118103058617027083838706701567&CbSlDlH0=qAksqaqSWslSWslSWw2U.yyWmpyLmvMytJyfeXC2GaEqqkl. Accessed August 21, 2020.
- 16. Jääskeläinen AJ, Kuivanen S, Kekäläinen E, et al. Performance of six SARS‐CoV‐2 immunoassays in comparison with microneutralisation. J Clin Virol. 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. CLSI . User verification of precision and estimation of bias: Approved guideline‐Third edition. In. CLSI document EP15‐A2. Clinical and Laboratory Standards Institute: Wayne. PA; 2014.
- 18. CLSI . Protocols for determination of limits of detection and limits of quantita‐tion. approved guideline‐second edition. In. CLSI document EP17‐A2. Clinical and Laboratory Standards Institute: Wayne PA; 2014.
- 19. NCCLS . User Protocol for Evaluation of Quantitate Test of Performance; Approved guideline. In. NCCLS document EP12‐A [ISBN 1‐56238‐468‐6]. National Committee for Clinical Laboratory Standards: Wayne, Pennsylvania, 19087‐1898, USA 2002; 2002. [Google Scholar]
- 20. Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465‐469. [DOI] [PubMed] [Google Scholar]
- 21. Lijia S, Lihong S, Huabin W, et al. Serological chemiluminescence immunoassay for the diagnosis of SARS‐CoV‐2 infection. J Clin Lab Anal. 2020;35:e23466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DomBourian MG, Annen K, Huey L, et al. Analysis of COVID‐19 convalescent plasma for SARS‐CoV‐2 IgG using two commercial immunoassays. J Immunol Methods. 2020;486:112837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hu X, An T, Situ B, et al. Heat inactivation of serum interferes with the immunoanalysis of antibodies to SARS‐CoV‐2. J Clin Lab Anal. 2020;35:e23411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brandstetter S, Roth S, Harner S, et al. Symptoms and immunoglobulin development in hospital staff exposed to a SARS‐CoV‐2 outbreak. Pediatr Allergy Immunol. 2020;31(7):841‐847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 and Tables S1‐S3
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


