Abstract
Background
Identifying point mutations in 23S rRNA closely associated with clarithromycin resistance can increase the eradication rate of Helicobacter pylori (H pylori). In this study, we verified the sensitivity, specificity, and reliability of a newly developed loop‐mediated isothermal amplification (LAMP) assay kit to detect H pylori and 2143G and 2182C mutations in 23S rRNA.
Methods
LAMP assay to detect H pylori and a mutant strain with 2143G and 2182C was conducted with the Isopollo® H pylori & ClaR kit. A prospective, open‐label, observational study was conducted to validate the reliability of the LAMP assay in both a development cohort and a bedside direct LAMP cohort.
Results
The LAMP assay had good sensitivity, as it could detect as few as 10–100 copies of H pylori and mutants with 2143G and 2182C in 23S rRNA, and good specificity, as it did not react with other bacterial species. In the development cohort with 622 participants, the LAMP assay showed good agreement with RUT for detecting H pylori (kappa value 0.923, P < .001) and had exactly the same results as sequencing analysis for 2143G and 2182C point mutations. The direct LAMP cohort including 93 patients had 97.7% (42/43) of concordance in detecting 2143G and 2182C point mutations compared to the PCR‐based sequencing analysis.
Conclusion
The Isopollo® H pylori & ClaR LAMP assay was a valid method for detecting H pylori and for 2143G and 2182C point mutations in 23S rRNA in a clinical setting.
Keywords: Helicobacter pylori, loop‐mediated isothermal amplification, point mutation, validation
We propose a valid loop‐mediated isothermal amplification (LAMP) assay for detecting H. pylori and for 23S rRNA point mutations to improve the eradication rate of H pylori. The mucosal biopsy sample obtained by upper gastroduodenoscopic procedure is subjected to the LAMP assay with one‐step RNA extraction. The LAMP assay had good sensitivity and specificity, as it could detect as few as 10–100 copies of H pylori and mutants with 2143G and 2182C in 23S rRNA, and was reliable compared to the rapid urease test (RUT) and PCR‐based sequencing analysis. This assay is a relatively simple and cost‐effective to detect the point mutations related to the clarithromycin resistance of H pylori in bedside. The choice of a tailored eradication regimen based on its result could be a good way to decrease the eradication failure associated with clarithromycin resistance.
1. INTRODUCTION
Helicobacter pylori (H pylori) is a gram‐negative rod that resides in the deeper portion of the gastric mucosal layer and on the surface of gastric epithelial cells. 1 , 2 More than half of the world's population is infected with H pylori, and it is found in 90% of people with gastric ulcer diseases. 3 H pylori synthesizes urease, which produces ammonia and neutralizes the acidic environment of the stomach, and proteases, causing damage to the inner layer of the stomach and consequently causing gastric ulcers. 1 Chronic mucosal damage by H pylori can lead to gastric cancer over time. 4 , 5 , 6
An increasing disease burden from H pylori infection has increased the need for the eradication of this bacteria, which would theoretically prevent approximately 150 000 deaths from gastric cancer over the next 5 years. 7 The first‐line eradication regimen approved by the health insurance system of Korea and many other countries is clarithromycin‐based triple therapy. 8 Along with a recent increase in antibiotic resistance, including clarithromycin, the eradication rate of H pylori has declined to 60% in some countries. 9 This has led to a need for a tailored eradication strategy according to the results of antibiotic resistance. 10
Currently, the gold‐standard method for identifying resistance to clarithromycin is culturing H pylori and performing sensitivity tests. 11 However, H pylori is difficult to culture in clinical settings because it requires microaerobic and high‐CO2 culture conditions. 11 Several lines of evidence suggest that the 2143A > G mutation in H pylori 23S ribosomal RNA (rRNA) is related to clarithromycin resistance and eradication failure, 12 , 13 , 14 , 15 and screening for this mutation is likely to be an effective alternative method to identify clarithromycin resistance in H pylori. 12 We previously reported that the presence of 2143G in H pylori 23S rRNA is an independent risk for eradication failure, which reached approximately 60% in response to clarithromycin‐based triple therapy, and patients with 2143A‐2182C mutations showed a moderate risk with approximately 10% eradication failure. 15 However, the polymerase chain reaction (PCR)–based method to amplify DNA fragments and identify targeted mutations is also time‐consuming, 16 and it is emerged the need for simple method to detect mutations. 17
Loop‐mediated isothermal amplification (LAMP) is a single‐tube technique that amplifies DNA at a constant temperature of 60‐65°C. 18 LAMP can amplify a target sequence with high selectivity using two sets of primers because it recognizes the target with six distinct sequences. 18 Compared to PCR‐based amplification, LAMP does not require sophisticated equipment, such as a thermal cycler, and efficiently produces a larger amount of DNA that can be easily detected by the naked eye or by measuring the turbidity within an hour. 18 The simplicity and cost‐effectiveness of the LAMP method make it particularly useful for screening or detecting infectious diseases in the field or at the point of care by physicians. 19 In this study, we first optimized the LAMP assay to detect H pylori and 2143A > G and 2182T > C mutations in 23S rRNA, which have been known to be associated with clarithromycin resistance, and compared the results of the LAMP assay to those from a rapid urease test (RUT) and PCR‐based sequencing analysis.
2. PATIENTS AND METHODS
2.1. Sensitivity and specificity tests for the LAMP assay
Control wild‐type H pylori and mutant strain with 2143G and 2182C were obtained from Helicobacter pylori Korean Type Culture Collection (HpKTCC). H pylori DNA was extracted with a QIAamp® DNA Mini Kit (QIAGEN, #51306) according to the manufacturer's protocol. The extracted DNA concentration was measured with NanoDrop™ spectrophotometers (Thermo Scientific). Based on the genome length (bp) and molar mass (g/mol), the copy number of H pylori was calculated: a concentration of 2 ng/μL H pylori DNA corresponded to a copy number of 106. Then, 2 ng/μL H pylori DNA (1 μL of DNA +9 μL of distilled water from an Isopollo® H pylori & ClaR kit) was used to generate a 10‐fold serial dilution up to 2 fg, which corresponds to 1 copy of H pylori, to test the sensitivity of the LAMP assay for detecting H pylori and mutant strains with 2143G and 2182C mutations using the Isopollo® H pylori & ClaR kit (Mmonitor Inc). Details of the LAMP assay are described below in the direct LAMP assay section of the methods. The specificity of the LAMP assay for H pylori was assessed with 10 ng of DNA extracted from H pylori, a mutant strain with 2143G and 2182C mutations, and other bacterial species, such as C jejuni, B subtilis, C perfringens, E aerogenes, C albicans, P aeruginosa, E coli, and H influenza. A total of 10 ng of DNA extracted from H pylori and distilled water were used as positive and negative controls, respectively.
2.2. Patients and upper gastrointestinal endoscopy
This prospective, open‐label, observational study was carried out in Daegu Fatima Hospital from June 2016 to November 2017. The study protocol was approved by the Institutional Review Board of Daegu Fatima Hospital (study number: DFH16DRIS026), and also registered with the Clinical Research Information Service (CRIS; study number: KCT0002668). Patients aged ≥18 years who were willing to undergo an upper gastrointestinal endoscopic examination have been eligible to be enrolled into this study. Patients who had a history of gastrectomy, were pregnant or lactating, had a severe concurrent disease or disability or were unable to understand the consent were excluded. All patients provided a written informed consent before enrollment.
To validate the LAMP assay for H pylori in a clinical setting, we used two groups of patients, the development cohort and the direct LAMP cohort (Table 1). The development cohort was recruited from June 2016 to April 2017; DNA was extracted from mucosal biopsy tissues using a QIAamp® DNA Mini Kit and was subjected to the LAMP assay. The direct LAMP cohort was recruited from May 2017 to November 2017; three pieces of gastric antrum tissue were obtained and subjected to the RUT, PCR/sequencing, and the LAMP assay. For the direct LAMP assay, the biopsy sample was directly emerged in Mmaxpress® DNA kit HS2 (Mmonitor Inc), as described below. For the RUT, one biopsy tissue sample was immediately placed on a Pronto Dry® NEW kit (Medical Instruments Corporation), and the color change was measured at 30 min and 24 hr
TABLE 1.
Demographic data for the 622 participants in the development cohort and the 93 participants in the direct LAMP cohort
| Development cohort | RUT (+) (n = 246) | RUT(−) (n = 376) | P‐value |
|---|---|---|---|
| Male sex | 128 (52.0%) | 188 (50.0%) | .620 |
| Age | 58.4 ± 11.8 | 60.1 ± 13.7 | .098 |
| BMI (kg/m2) | 20.3 ± 9.6 | 19.2 ± 10.3 | .189 |
| Endoscopic findings | |||
| Gastric ulcer | 61 (24.8%) | 79 (21.0%) | .269 |
| Duodenal ulcer | 220 (89.4%) | 354 (94.1%) | .031 |
| Gastric cancer | 15 (6.1%) | 20 (5.3%) | .680 |
| Direct LAMP cohort | RUT (+) (n = 43) | RUT(−) (n = 50) | |
| Male sex | 18 (41.9%) | 25 (50%) | .440 |
| Age | 59.6 ± 12.5 | 56.8 ± 12.9 | .299 |
| BMI (kg/m2) | 23.0 ± 2.7 | 22.9 ± 5.1 | .981 |
| Endoscopic findings | |||
| Gastric ulcer | 8 (18.6%) | 8 (16.0%) | .740 |
| Duodenal ulcer | 39 (90.7%) | 46 (92.0%) | .823 |
| Gastric cancer | 1 (2.3%) | 1 (2.0%) | .914 |
2.3. DNA extraction, PCR, and sequencing analysis
If the RUT was positive, whole DNA was isolated from additional biopsy tissue using a QIAamp® DNA Mini Kit (QIAGEN, #51306) according to the manufacturer's protocol. The concentration of extracted DNA was measured with NanoDrop™ spectrophotometers (Thermo Scientific), and the patient whose extracted DNA concentration was <50 ng/μL was excluded from sequencing analysis. We examined the nucleotide sequence of domain V in the 23S rRNA gene of H pylori by amplifying a segment that was approximately 182 bp using the PCR primers 23S F (5′‐TGA ATG TAA CGA GAT G‐3′, corresponding to H pylori 23S rRNA positions 2052‐2070) and 23S R (5′‐GCC AAA GCC CTT ACT TCA‐3′, positions 2216‐2233). The PCR products were subjected to agarose gel electrophoresis and visualized by staining the gel with ethidium bromide. If a PCR band was found at 182 bp, it was judged to be positive for H pylori. The nucleotide sequencing of amplified DNA was performed by a commercial expert agency (www.solgent.com, Daejeon, South Korea).
2.4. Direct LAMP assay for H pylori and 23S rRNA point mutants
One piece of gastric mucosal biopsy samples was subjected to the direct LAMP assay using the Isopollo® H pylori & ClaR kit and Mmaxpress® DNA kit HS2 (Mmonitor Inc) according to the manufacturer's protocol. To extract DNA, 100 µL of Mmaxpress® DNA kit HS2 was added to the tube containing the gastric mucosal biopsy sample, and it was incubated for 5 min at room temperature (RT). Then, 10 µL of lysate was added to tubes containing a mixture of 1.5 µL of detection primer, 1 µL of Bst DNA polymerase, and 12.5 µL of 2X buffer, resulting in a final volume of 25 µL. Three kinds of detection primers were used, which targeted H pylori and mutant genotypes with 2143G and 2182C mutants in 23S rRNA. The mixed tubes were loaded on an automated incubator (Mmonitor Inc) and amplified at 63°C for 30 min. If target DNA was meaningfully amplified, the hydroxynaphthol blue dye with a violet color changed to a sky‐blue color, which is easily detected by the naked eye. Mmaxpress® DNA kit HS2 was used as a negative control.
2.5. Statistical analysis
To compare the demographic characteristics between the RUT‐positive and RUT‐negative groups, categorical variables were analyzed with a chi‐square test, and continuous variables were analyzed with Student's t test. Differences between the direct LAMP assay and the RUT and PCR test were compared using Cohen's kappa statistics and the McNemar test. A P‐value of <.05 indicated statistical significance. Data were analyzed using SPSS software (version 18.0).
3. RESULTS
3.1. Sensitivity and specificity of the LAMP assay for H pylori and 23S rRNA point mutants
First, the sensitivity of the LAMP assay for H pylori and 23S rRNA point mutants was assessed with commercial heat‐killed H pylori. When 106 copies of stock DNA from H pylori were serially diluted, the LAMP assay could detect as few as 10 copies of H pylori, which was a similar sensitivity to that of qPCR. 20 The LAMP primers targeting the 2143G and 2182C mutations showed a lower degree of sensitivity than the primers for H pylori and could detect as few as 100 copies of mutant H pylori (Figure 1A). In terms of the specificity of the LAMP assay, the primer set for H pylori showed a positive result for both the wild‐type and mutant 2143G and 2182C H pylori strains. In addition, the primer set did not detect other bacterial species, such as C jejuni, B subtilis, C perfringens, E aerogenes, C albicans, P aeruginosa, E coli, and H influenza, at a concentration of 10 ng/μL DNA. The primers for 2143G and 2182C mutations were able to detect H pylori with 2143G and 2182C mutations in 23S rRNA but not wild‐type H pylori or other bacterial species (Figure 1B).
FIGURE 1.
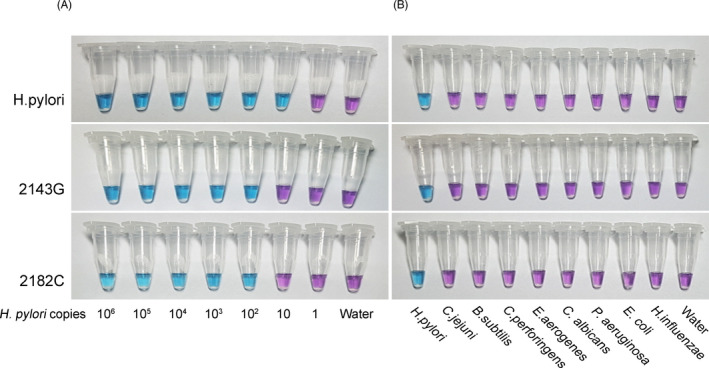
Sensitivity and specificity of the colorimetric LAMP assay. A, The sensitivity of the LAMP assay was assessed using a 10‐fold serial dilution of H pylori DNA. A 2 ng/μL DNA concentration corresponds to 106 copies of H pylori. B, Species specificity using 10 ng of genomic DNAs from various bacteria. The upper row is the result with commercial H pylori, and the second and third rows are the results from 2143G and 2182C mutant strains
3.2. Clinical relevance of the LAMP assay for detecting H pylori and 23S rRNA point mutations with gastric mucosal DNA extracted in a laboratory setting
To test the clinical relevance of this assay, a total of 622 participants were recruited to the development cohort and were subjected to the LAMP assay. Gastric mucosal DNA was extracted with a commercial DNA extraction kit (QIAamp® DNA Mini Kit) in a laboratory setting, and the results were compared to the results of the RUT and PCR assay for H pylori. The positivity rate for H pylori based on the RUT was 39.5% (246/622). When the development cohort was analyzed with the RUT, the frequency of inactive duodenal ulcers was lower in the RUT‐positive group than that in the RUT‐negative group (P = .031), but differences were not observed in other variables, such as sex, age, body mass index (BMI), and the frequency of gastric ulcers and cancer (Table 1).
For the detection of H pylori, the LAMP assay agreed with the RUT for 92% of samples (572/622). The kappa value, a measure of the agreement, was 0.923 (SE 0.016, P < .001), suggesting almost perfect agreement. 21 However, based on the McNemar test, which assessed the consistency of the two tests, the LAMP assay was significantly different from the RUT (P = .017). Among 622 samples, 28 samples (0.05%) failed to yield enough DNA from the extractions (concentrations of less than 50 ng/µL) for LAMP and PCR analysis. PCR analysis, which confirmed that there was a 182 bp band with agarose gel electrophoresis, showed exactly the same result as the LAMP assay for H pylori detection (Table 2). The 246 samples that had a positive band in the PCR analysis were subjected to sequencing analysis to detect 2143A > G and 2182T > C mutations in the 23S rRNA of H pylori, and the results were compared to those of the LAMP assay. Among 246 samples, 2143A and 2143G were detected in 182 and 64 samples, respectively, and 2182T and 2182C were detected in 30 and 216 samples, respectively. LAMP and sequencing analysis showed exactly the same results (Table 2).
TABLE 2.
The results of the LAMP assay compared to those of the RUT and PCR and sequencing analysis for the diagnosis of H pylori and the detection of the A2143G and T2182C point mutations in 23S rRNA
| Development cohort | RUT | PCR | ||||
|---|---|---|---|---|---|---|
| LAMP for H pylori a | Negative | Positive | Statistics b | Negative | Positive | Statistics |
| Negative | 343 | 5 | P = .017 | 348 | 0 | P = 1.0 |
| Positive | 17 | 229 | κ = 0.923 | 0 | 246 | κ = 1.0 |
| LAMP for 2143G or 2182C | Sequencing A2143G | Sequencing T2182C | |||||
|---|---|---|---|---|---|---|---|
| 2143A | 2143G | Statistics | 2182T | 2182C | Statistics | ||
| 2143G | Negative | 182 | 0 | P = 1.0 | |||
| Positive | 0 | 64 | κ = 1.0 | ||||
| 2182C | Negative | 30 | 0 | P = 1.0 | |||
| Positive | 0 | 216 | κ = 1.0 | ||||
DNA used in LAMP analysis was extracted in a laboratory setting with a QIAamp® DNA Mini Kit (QIAGEN, #51306). Twenty‐eight patients with an extracted DNA concentration of <50 ng/µL were excluded from the PCR and LAMP analyses.
P from McNemar test and κ from unweighted pairwise Cohen's Kappa.
3.3. Direct bedside detection of H pylori and 23S rRNA point mutants with the LAMP test
We then tested the validity of the LAMP assay for instant bedside testing. Gastric biopsy samples were directly soaked in Mmaxpress® DNA kit HS2 for 5 min, and 10 μL of each lysate was transferred to tubes containing primers for H pylori and 2143G and 2182C point mutants. After a 30‐min incubation at 63°C, the color change from purple to blue was visually inspected. Representative results from the direct LAMP assay for H pylori and 23S rRNA point mutants are shown in Figure 2, showing that the assay was easy to interpret.
FIGURE 2.

Representative results of the LAMP analysis for the presence of H pylori and genetic mutations in 23S rRNA in gastric biopsy samples. Endoscopic biopsy specimens of gastric mucosa were dissolved in DNA extraction buffer for 10 min. Isolated DNA (2 μL) was added to each tube and analyzed with primers for H pylori and the 2143G and 2182C mutations, and color changes were observed after a 30‐min reaction at 63°C
To evaluate the diagnostic reliability of the direct LAMP assay for the detection of H pylori in a clinical setting, the results were compared to those of the RUT and PCR test. Of the 93 specimens tested, 43 were positive for H pylori based on the RUT, and there was no difference in clinical features according to RUT positivity (Table 1). The direct LAMP assay had 96.7% (90/93) and 98.9% (92/93) concordance rates with the RUT and PCR test, respectively. Cohen's kappa coefficient for the agreement of the direct LAMP assay was 0.935 for the RUT and 0.978 for the PCR test. The McNemar test revealed no statistically significant discordance between the direct LAMP and RUT/PCR tests (P = 1.0; Table 3).
TABLE 3.
A comparison of the direct LAMP assay with the RUT and PCR and sequencing analysis for the diagnosis of H pylori and the detection of the A2143G and T2182C point mutations in 23S rRNA
| Direct LAMP cohort | RUT | PCR | ||||
|---|---|---|---|---|---|---|
| Direct LAMP analysis a | Negative | Positive | Statistics b | Negative | Positive | Statistics |
| Negative | 49 | 2 | P = 1.0 | 50 | 1 | P = 1.0 |
| Positive | 1 | 41 | κ = 0.935 | 0 | 42 | κ = 0.978 |
| LAMP for 2143G or 2182C | Sequencing A2143G | Sequencing T2182C | |||||
|---|---|---|---|---|---|---|---|
| 2143A | 2143G | Statistics | 2182T | 2182C | Statistics | ||
| 2143G | Negative | 29 | 1 | P = 1.0 | |||
| Positive | 0 | 12 | κ = 0.943 | ||||
| 2182C | Negative | 8 | 1 | P = 1.0 | |||
| Positive | 0 | 33 | κ = 0.926 | ||||
Gastric mucosal DNA was directly extracted from a biopsy sample at the bedside with the Mmaxpress® DNA kit HS2 included in the Isopollo® H pylori & ClaR kit (Mmonitor Inc).
P from McNemar test and κ from unweighted pairwise Cohen's Kappa.
The direct LAMP assay results for the detection of the point mutations 2143G and 2182C in 23S rRNA were compared to the sequencing results. Among 43 patients with positive RUT and PCR tests, 2143G was positive in 14 patients in sequencing analysis (32.6%). The direct LAMP assay for 2143G was positive in 13 patients and showed discordance with sequencing for only 1 patient, which represented just a 2.3% (1/43) discordance rate. For the 2182T > C mutation, in sequencing analysis, 35 patients had 2182C, and 8 had 2182T. The direct LAMP assay for 2182C showed a 97.7% (42/43) concordance with sequencing analysis (Table 3).
4. DISCUSSION
In this study, we provide evidence that the LAMP assay has high sensitivity for detecting H pylori and 23S rRNA point mutations and good specificity for distinguishing other bacterial species. The LAMP assay was a reliable method to detect H pylori and 23S rRNA point mutations and showed almost identical results with PCR‐based sequencing analysis. The direct LAMP assay using DNA directly extracted at the bedside also had consistent results with sequencing analysis, demonstrating the potential for this assay to be used to determine a treatment regimen in a clinical setting.
At least 105 copies of bacterial DNA are required for a positive RUT result. 22 On the other hand, quantitative RT‐PCR can detect as few as 10 copies of H pylori. 20 The LAMP assay tested in this study detected 10 copies of H pylori, which was comparable to that of quantitative RT‐PCR. The LAMP assay showed a sensitivity of 100 copies of H pylori for the detection of the point mutations 2143G and 2182C in 23S rRNA, which was a somewhat inferior sensitivity compared to that for the detection of H pylori. However, the LAMP results for the point mutations 2143G and 2182C were fully consistent with the sequencing results in the development and direct LAMP cohorts. These data provide evidence that the sensitivity of the LAMP assay for detecting point mutations in 23S rRNA is comparable to that of PCR‐based sequencing analysis, and this method is appropriate for clinical settings.
Despite the high sensitivity of PCR analysis, its relatively low specificity for H pylori detection has been noted as a drawback. 23 Although it depends on the target primer used, the specificity of PCR analysis for H pylori is usually between 90% and 95%, which is comparable or inferior to that of RUT when culture or histology results were considered the gold standard for detecting H pylori. 23 , 24 , 25 , 26 This finding could have resulted from the low yield of H pylori culture due to the complex microaerobic and high CO2 culture conditions. 11 The LAMP analysis used in this study can overcome the specificity limitations of PCR by testing not only H pylori but also point mutations in 23S rRNA. In particular, the point mutations 2143G and 2182C in 23S rRNA are associated with eradication failure with clarithromycin‐based triple therapy, and individuals with these point mutations need a modified eradication regimen, as discussed below. A higher degree of specificity is essential for this testing which can influence the development of the treatment plan. This LAMP assay, which detects both H pylori and 23S rRNA point mutants, may have advantages over conventional PCR analysis for detecting H pylori only in terms of specificity.
The most important implication of this study is the clinical usefulness of the LAMP assay for detecting the point mutations 2143G and 2182C in 23S rRNA, which are known to be associated with clarithromycin resistance. 13 , 15 Along with increasing antibiotic resistance, the eradication rate of clarithromycin‐based triple therapy has continued to decrease in recent years. 9 A meta‐analysis of 5 studies published from 2006 to 2011 revealed only a 62% eradication rate with clarithromycin‐based triple therapy. 27 For this reason, the recent consensus for H pylori treatment recommended quadruple regimens for 14 days in areas with over 15% clarithromycin resistance. 28 In Korea, the clarithromycin resistance rate of H pylori is up to 20%, and the metronidazole resistance rate is more than 30%. 29 Therefore, the 14‐day quadruple regimens are recommended according to this consensus. 28 However, our previous data showed that the point mutations 2143G and 2182C in 23S rRNA are closely related to the eradication failure rate of clarithromycin‐based triple therapy; patients with 2143G in 23S rRNA had an eradication failure rate of approximately 60%, while the patients without 2143G had an eradication rate of more than 90%, even with 7‐day clarithromycin‐based triple therapy in Korea. 15 This study showed that the LAMP assay can accurately detect 23S rRNA point mutations in a relatively simple manner, and the selection of an H pylori eradication regimen based on the LAMP assay results is able to dramatically increase the success rate of eradication.
The main limitation of this study was that we did not perform a gold‐standard assay, such as culture or histologic staining, to confirm H pylori infection and consequently, we cannot draw conclusions regarding the statistical characteristics of the LAMP assay as a diagnostic test, such as the sensitivity, specificity, and negative and positive predictive value. However, the primary aim of this study was to validate the Isopollo® H pylori & ClaR LAMP assay by comparing it to the RUT and PCR analysis, thus verifying its reliability and clinical usefulness. These data showed that the results from the LAMP assay were almost the same as those from the PCR‐based sequencing analysis; therefore, the statistical characteristics of the LAMP assay are assumed to be similar to those of the PCR‐based sequencing analysis.
In conclusion, the LAMP assay using the Isopollo® H pylori & ClaR kit had good sensitivity and specificity for detecting H pylori and point mutations in 23S rRNA and was reliable compared to the RUT and PCR‐based sequencing analysis. Direct LAMP analysis is a relatively simple and cost‐effective method to detect the point mutations 2143G and 2182C in 23S rRNA of H pylori in bedside, and the choice of a tailored eradication regimen based on its result could be a good way to decrease the eradication failure associated with clarithromycin resistance.
CONFLICT OF INTEREST
Jeon, HS and Kim, SH are employee of Mmonitor Inc, a bioventure company for in vitro diagnostic kits. To ensure the objectivity of this study, the patients’ information was not provided and the whole process was carried out in the blindness. The Isopollo® H pylori & ClaR kit and Mmaxpress® DNA kit HS2 were provided from Mmonitor Inc, company. Park, CG and Han, SW have no conflicts of interest in this study.
AUTHOR CONTRIBUTIONS
CP, HJ, and SH conceptualized and designed the study. CP, SK, and SH analyzed and interpreted the data. SK and CP collected the data and performed experimental procedures. CP, SK, and SH drafted the article. CP, SK, HJ, and SH approved the final article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved by the Institutional Review Board of Daegu Fatima Hospital (study number: DFH16DRIS026), and all participants signed a consent form. This study was registered in the the Clinical Research Information Service (CRIS) (study number: KCT0002668).
Park C‐G, Kim S, Jeon H‐S, Han S. Validation of loop‐mediated isothermal amplification to detect Helicobacter pylori and 23S rRNA mutations: A prospective, observational clinical cohort study. J Clin Lab Anal.2021;35:e23563 10.1002/jcla.23563
Chang‐Geun Park and Seohyeon Kim These authors contributed equally to this article.
Funding information
This research was supported by a grant to SH from the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), which is funded by the Ministry of Health & Welfare, Republic of Korea (Grant Number: HI15C1780).
Contributor Information
Hyo‐Sung Jeon, Email: jeonh@mmonitor.net.
Seungwoo Han, Email: kiefe73@gmail.com, Email: jeonh@mmonitor.net.
REFERENCES
- 1. Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crowe SE. Helicobacter pylori Infection. N Engl J Med. 2019;380:1158‐1165. [DOI] [PubMed] [Google Scholar]
- 3. Hooi JKY, Lai WY, Ng WK, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta‐analysis. Gastroenterology. 2017;153:420‐429. [DOI] [PubMed] [Google Scholar]
- 4. Choi IJ, Kook MC, Kim YI, et al. Helicobacter pylori therapy for the prevention of metachronous gastric cancer. N Engl J Med. 2018;378:1085‐1095. [DOI] [PubMed] [Google Scholar]
- 5. Wroblewski LE, Peek RM Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713‐739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gong X, Zhang H. Diagnostic and prognostic values of anti‐Helicobacter pylori antibody combined with serum CA724, CA19‐9, and CEA for young patients with early gastric cancer. J Clin Lab Anal. 2020;34(7):e23268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Asaka M, Kato M, Graham DY. Strategy for eliminating gastric cancer in Japan. Helicobacter. 2010;15:486‐490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chuah YY, Wu DC, Chuah SK, et al. Real‐world practice and expectation of Asia‐Pacific physicians and patients in Helicobacter pylori eradication (REAP‐HP Survey). Helicobacter. 2017;22(3):e12380. [DOI] [PubMed] [Google Scholar]
- 9. Ramas M, Donday MG, McNicholl AG, Gisbert JP. Efficacy and safety of rifaximin associated with standard triple therapy (omeprazole, clarithromycin and amoxicillin) for H. pylori eradication: a phase IV pilot clinical trial. Gastroenterol Hepatol. 2017;40:658‐662. [DOI] [PubMed] [Google Scholar]
- 10. Suzuki H, Mori H. World trends for H. pylori eradication therapy and gastric cancer prevention strategy by H. pylori test‐and‐treat. J Gastroenterol. 2018;53:354‐361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Megraud F, Lehours P. Helicobacter pylori detection and antimicrobial susceptibility testing. Clin Microbiol Rev. 2007;20:280‐322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ong S, Kim SE, Kim JH, et al. Helicobacter pylori eradication rates with concomitant and tailored therapy based on 23S rRNA point mutation: a multicenter randomized controlled trial. Helicobacter. 2019;24:e12654. [DOI] [PubMed] [Google Scholar]
- 13. Soltermann A, Perren A, Schmid S, et al. Assessment of Helicobacter pylori clarithromycin resistance mutations in archival gastric biopsy samples. Swiss Med Wkly. 2005;135:327‐332. [DOI] [PubMed] [Google Scholar]
- 14. De Francesco V, Margiotta M, Zullo A, et al. Clarithromycin‐resistant genotypes and eradication of Helicobacter pylori . Ann Intern Med. 2006;144:94‐100. [DOI] [PubMed] [Google Scholar]
- 15. Park CG, Kim S, Lee EJ, Jeon HS, Han S. Clinical relevance of point mutations in the 23S rRNA gene in Helicobacter pylori eradication: a prospective, observational study. Medicine (Baltimore). 2018;97:e11835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rimbara E, Sasatsu M, Graham DY. PCR detection of Helicobacter pylori in clinical samples. Methods Mol Biol. 2013;943:279‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farah R, Hamza H, Khamisy‐Farah R. A link between platelet to lymphocyte ratio and Helicobacter pylori infection. J Clin Lab Anal. 2018;32:e22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomita N, Mori Y, Kanda H, Notomi T. Loop‐mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc. 2008;3:877‐882. [DOI] [PubMed] [Google Scholar]
- 19. Gill P, Ghaemi A. Nucleic acid isothermal amplification technologies: a review. Nucleosides Nucleotides Nucleic Acids. 2008;27:224‐243. [DOI] [PubMed] [Google Scholar]
- 20. Morilla A, Melon S, Alvarez‐Arguelles ME, Armesto E, Villar H, de Ona M. Utility of normalized genome quantification of Helicobacter pylori in gastric mucosa using an in‐house real‐time polymerase chain reaction. PLoS ONE. 2017;12:e0178674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McHugh ML. Interrater reliability: the kappa statistic. Biochem Med (Zagreb). 2012;22:276‐282. [PMC free article] [PubMed] [Google Scholar]
- 22. Uotani T, Graham DY. Diagnosis of Helicobacter pylori using the rapid urease test. Ann Transl Med. 2015;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trung TT, Minh TA, Anh NT. Value of CIM, CLO test and multiplex PCR for the diagnosis of Helicobacter pylori infection status in patients with gastritis and gastric ulcer. Asian Pac J Cancer Prev. 2019;20:3497‐3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Burucoa C, Garnier M, Silvain C, Fauchere JL. Quadruplex real‐time PCR assay using allele‐specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J Clin Microbiol. 2008;46:2320‐2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang SW, Yu FJ, Lo YC, et al. The clinical utility of string‐PCR test in diagnosing Helicobacter pylori infection. Hepatogastroenterology. 2003;50:1208‐1213. [PubMed] [Google Scholar]
- 26. Vinette KM, Gibney KM, Proujansky R, Fawcett PT. Comparison of PCR and clinical laboratory tests for diagnosing H. pylori infection in pediatric patients. BMC Microbiol. 2004;4:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Venerito M, Krieger T, Ecker T, Leandro G, Malfertheiner P. Meta‐analysis of bismuth quadruple therapy versus clarithromycin triple therapy for empiric primary treatment of Helicobacter pylori infection. Digestion. 2013;88:33‐45. [DOI] [PubMed] [Google Scholar]
- 28. Fallone CA, Chiba N, van Zanten SV, et al. The toronto consensus for the treatment of Helicobacter pylori infection in adults. Gastroenterology. 2016;151(51–69):e14. [DOI] [PubMed] [Google Scholar]
- 29. Hu Y, Zhu Y, Lu NH. Novel and effective therapeutic regimens for Helicobacter pylori in an era of increasing antibiotic resistance. Front Cell Infect Microbiol. 2017;7:168. [DOI] [PMC free article] [PubMed] [Google Scholar]


