Abstract
Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) is an essential insect pollinator in oil palm plantations. Recently, researches have been undertaken to improve pollination efficiency using this species. A fundamental understanding of the genes related to this pollinator behavior is necessary to achieve this goal. Here, we present the draft genome sequence, annotation, and simple sequence repeat (SSR) marker data for this pollinator. In total, 34.97 Gb of sequence data from one male individual (monoisolate) were obtained using Illumina short-read platform NextSeq 500. The draft genome assembly was found to be 269.79 Mb and about 59.9% of completeness based on Benchmarking Universal Single-Copy Orthologs (BUSCO) assessment. Functional gene annotation predicted about 26.566 genes. Also, a total of 281.668 putative SSR markers were identified. This draft genome sequence is a valuable resource for understanding the population genetics, phylogenetics, dispersal patterns, and behavior of this species.
Keywords: Whole-genome sequencing, NGS, Simple Sequence Repeat, Weevil, Curculionidae, Oil Palm, Pollinator, Genomics
Specifications Table
| Subject | Omics: Genomics |
| Specific subject area | Insects, Coleoptera, Oil Palm, Weevil, Whole-genome sequencing (WGS) |
| Type of data | Table |
| Figure | |
| Raw DNA sequencing reads | |
| Draft genome assembly | |
| Repeat elements file | |
| Simple sequence repeat file | |
| Genome annotation file | |
| How data were acquired | Paired-end sequencing on Illumina Nextseq 500 platform. |
| Data format | Raw – Fastq |
| Analyzed – Fasta, gff | |
| Parameters for data collection | DNA from one male adult individual (monoisolate) was used. |
| Description of data collection | DNA from the whole-body was extracted. DNA purity and concentration were measured before sequencing. DNA sequences obtained by Illumina Nextseq 500 platform followed by de novo assembly using SPAdes. |
| Data source location | Institution: Research and Development, PT. Astra Agro Lestari Tbk |
| City/Town/Region: Pangkalan Lada, Kalimantan Tengah | |
| Country: Indonesia | |
| Latitude and longitude for collected samples/data: | |
| (2°25′28.6″S, 111°47′26.8″E). | |
| Data accessibility | All data in this article are available at NCBI, BioProject number PRJNA637822. |
| Direct URL to data: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA637822 | |
| Whole-genome sequence data are accessible at NCBI under GenBank accession number JACGEL000000000 | |
| Direct URL to data: https://www.ncbi.nlm.nih.gov/nuccore/JACGEL000000000.1 | |
| The raw sequence data with this article are accessible under SRA accession number SRR12726955-SRR12726958. | |
| Direct URL to data: | |
| https://www.ncbi.nlm.nih.gov/sra/?term=SRR12726955 | |
| https://www.ncbi.nlm.nih.gov/sra/?term=SRR12726956 | |
| https://www.ncbi.nlm.nih.gov/sra/?term=SRR12726957 | |
| https://www.ncbi.nlm.nih.gov/sra/?term=SRR12726958 | |
| Related research article | A. Apriyanto, V.B. Tambunan, The complete mitochondrial genome of oil palm pollinating weevil, Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae), Mitochondrial DNA Part B. 5(3) (2020) 3450–3452. https://doi.org/10.1080/23802359.2020.1823899 |
Value of the Data
-
•
This article provides the draft genome sequence data of Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) and thus addresses a knowledge gap of genome sequence within the order Coleoptera.
-
•
The draft genome sequence of this species will be useful for entomologists interested in functional genomics, population genetics, phylogenetics, and selection by breeding.
-
•
This dataset can be used as a reference for future complete genome assembly of this species.
-
•
The newly developed SSR markers dataset in this report should be useful tools for assessing the genetic diversity, conservation, and bio management of this species.
1. Data Description
Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) is an essential insect pollinator in oil palm plantations. This species is native to the tropical Africa region but introduced into Asia, including Indonesia [1]. The introduction of this weevil species into oil palm plantations successfully improved fruit set, increased the yield of oil palm, and reducing the need for assisted pollination [2]. Recent studies of this species have only focused on analyzing genetic diversity and species identification [1], [3], [4]. Interestingly, several divergent mitochondrial lineages in this species have been discovered based on the information of cytochrome c oxidase subunit I (COI) and cytochrome c oxidase subunit II (COII) gene sequences [1], [3]. Our recent study successfully obtained the complete mitochondrial genome of E.kamerunicus from the partial dataset from this report, representing the first complete mitogenome for this species [5]. Nevertheless, the genomic resources of E.kamerunicus remain underdeveloped compared with many other agricultural insect species.
This article presents the first draft genome assembly, annotation, and SSR marker data of the oil palm pollinating weevil, Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae). All raw sequencing reads data (34.97 Gb) used for genome assembly were deposited in the NCBI Short Read Archive (SRA) database. All of these SRA data are retrievable under the accession number SRR12726955-SRR12726958.
The assembled draft genome was constructed using the filtered reads, which is about 73.71% of the total raw sequence reads. The final draft genome assembly was 269.79 Mb containing 364.527 scaffolds with 31.71% GC (Table 1). The genome project information has been deposited in the NCBI GenBank under the Bioproject ID: PRJNA637822. The whole-genome sequencing (WGS) data can be retrieved from the NCBI GenBank under accession JACGEL000000000.
Table 1.
Draft genome assembly statistic of E.kamerunicus.
| Statistics | JACGEL000000000 |
|---|---|
| Number of scaffolds | 364,527 |
| Number of scaffolds (>= 0 bp) | 364,527 |
| Number of scaffolds (>= 1000 bp) | 82,506 |
| Largest scaffolds (bp) | 16,904 |
| Total length (bp) | 269,798,182 |
| Total length (>= 0 bp) | 269,798,182 |
| Total length (>= 1000 bp) | 145,163,070 |
| N50 | 1084 |
| N75 | 568 |
| L50 | 72,645 |
| L75 | 157,429 |
| GC (%) | 31.71 |
The assembled E.kamerunicus genome analyzed with BUSCO tools showed 59.9% completeness, indicating the genome to be of good quality. We found about 638 complete orthologs genes (C: 59.9%), 632 orthologs complete genes and single-copy (S: 59.3%), 6 orthologs complete genes and duplicated (D: 0.6%), 323 orthologs fragmented genes (F: 30.3%) and 105 missing genes (M: 9.8%).
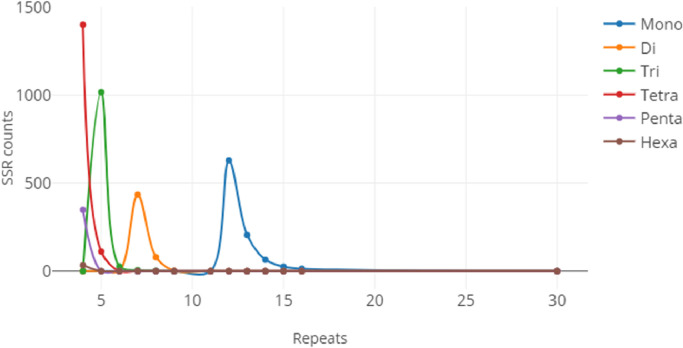
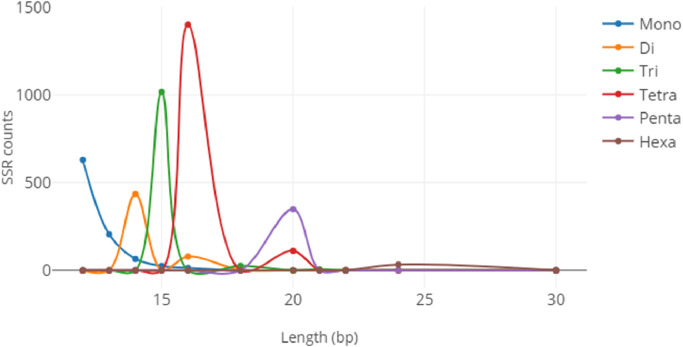
The assembled draft genome of E.kamerunicus was used to identify simple sequence repeat (SSR) or microsatellite markers. In this dataset, we reported about 4.396 perfect SSRs (pSSRs), 3 compound SSRs (cSSRs), 251.377 imperfect SSRs (iSSRs), and 25.892 variable number tandem repeats (VNTRs) inside the E.kamerunicus genome. The annotation files of pSSRs, cSSRs, iSSRs, and VNTRs are provided in Supplementary file S1-S4, respectively. The distribution of perfect SSRs (pSSRs) based on their motif and sequence length can be seen in Fig. 1, Fig. 2, respectively.
Fig. 1.
Perfect SSR distribution for each SSR type based on the number of repeats.
Fig. 2.
Perfect SSR distribution for each SSR type based on sequence length (bp).
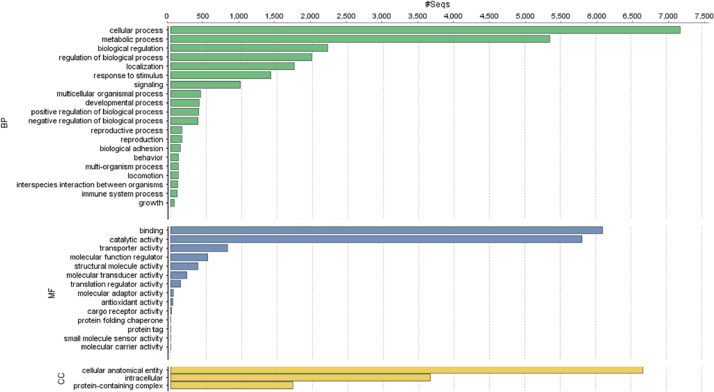
Table 2 provides the detailed information of repetitive elements detected in this assembled genome. The annotation data of repetitive elements can be found in Supplementary file S5. Functional gene annotation pipeline predicted about 26.566 genes, 14.145 were found to have GO term assigned to them. The GO term classification and distribution can be seen in Fig. 3. The genome annotation data can be found in Supplementary file S6.
Table 2.
Summary of repetitive elements in the assembled genome of E.kamerunicus. Most repeats fragmented by insertions or deletions have been counted as one element.
| Repeat class/family | Number of elements | Length occupied |
|---|---|---|
| SINEs | 9 | 630 |
| Penelope | 15 | 1110 |
| LINEs | 402 | 25,989 |
| L2/CR1/Rex | 78 | 4842 |
| R1/LOA/Jockey | 60 | 3771 |
| R2/R4/NeSL | 27 | 1748 |
| RTE/Bov-B | 29 | 2891 |
| LTR elements | 143 | 10,416 |
| BEL/Pao | 43 | 3194 |
| Ty1/Copia | 4 | 346 |
| Gypsy/DIRS1 | 96 | 6876 |
| DNA transposons | 376 | 25,824 |
| hobo-Activator | 76 | 6178 |
| Tc1-IS630-Pogo | 230 | 14,626 |
| Other (Mirage, P-element, Transib) | 4 | 349 |
| Rolling-circles | 36 | 2939 |
| Unclassified | 440 | 30,196 |
| Small RNA | 35 | 1999 |
| Satellites | 2 | 93 |
| Low complexity | 193 | 9749 |
Fig. 3.
Histogram representing the gene ontology distribution of the annotated E.kamerunicus genes. The functionally annotated genes were assigned to three main GO categories: Biological Process (BP), Molecular Function (MF), and Cellular Component (CC).
2. Experimental Design, Materials and Methods
2.1. Sample collection and sequencing
Elaeidobius kamerunicus samples were captured from oil palm female inflorescence during its anthesis. All of the samples were originally collected from an oil palm plantation of PT. Gunung Sejahtera Ibu Pertiwi, Kalimantan Tengah, Indonesia, with geospatial coordinate (2°25′28.6″S 111°47′26.8″E). The samples were then identified based on their morphological characteristic. One male of E.kamerunicus (monoisolate) was then selected for the determination of the genome sequence.
Total genomic DNA was extracted using the gSYNC™ DNA Extraction Kit (Geneaid) following the manufacturer's instructions. The quantity and quality of genomic DNA were measured using NanoDrop spectrophotometer (Thermo Fisher Scientific) and Qubit fluorometer (Invitrogen), followed by visualization on 0.8% agarose gel.
The library for NGS was prepared using NexteraXT library prep kit, and their quality and quantity were determined using Agilent Tapestation 4200 (Agilent), Qubit fluorometer (Invitrogen), and ABI 7500 Fast System qPCR (Applied Biosystems). The library sizes of about 350–600 bp were used for sequencing. Four paired-end libraries were generated using the Illumina NextSeq 500 sequencing platform.
2.2. Genome assembly and evaluation
The quality of the reads was assessed with the FastQC v. 0.11.2 software [6]. Genome assembly requires the sequencing quality of each base of the read at the level of Q30 (Phred scale). The raw reads were trimmed using the Trimmomatic v. 0.17 software [7]. K-mer length estimation for genome assembly was conducted using Kmergenie software [8]. Paired and unpaired high quality reads were taken as an input to the SPAdes v. 3.10.1 genomic assembler [9] with the following options: -careful -k 17, 19, 21, 23, 25, 31, 33, 35, 37, 41, 43, 45, 47, 51, 53, 55, 57, 61. Scaffolds that were <200 bp in length were removed manually. The contaminants of foreign DNA, such as remaining adapters/vectors, organellar DNA, or contamination, were removed during submission to the NCBI GenBank database. The genome assembly statistics were obtained using QUAST software [10]. The completeness of E.kamerunicus genome assembly data was evaluated using BUSCO v. 3 analysis [11] against the Arthropoda database (odb9), consisting of 1066 orthologs constructed from 60 species.
2.3. Identification of putative simple sequence repeat (SSR)
The SSR mining data in the E.kamerunicus genome was performed using Krait v. 1.3.3 software [12]. Four types of genetic variation, such as perfect SSRs (pSSRs), compound SSRs (cSSRs), imperfect SSRs (iSSRs), and variable number tandem repeats (VNTRs), were analyzed.
2.4. Repeat identification, masking, and genome annotation
The repetitive contents detection, such as transposable elements, retroelements, and total interspersed repeats, were detected using RepeatMasker v. 4.1 [13] with default parameters and insect Dfam repeat database [14]. The repeat-masked scaffold sequences were subjected to functional gene annotation. Functional annotation and gene ontology (GO) mapping of the final set of predicted protein sequences was carried out by OmicsBox v. 1.3.11 [15], [16].
Ethics Statement
Not applicable. No ethics protocols are required for Coleoptera in Indonesia.
CRediT Author Statement
Ardha Apriyanto: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data curation, Visualization, Writing, Reviewing, and Editing; Van Basten Tambunan: Conceptualization, Methodology, Investigation, Resources, and Writing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgments
This research was fully funded by PT. Astra Agro Lestari Tbk. We thank all the Research and Development team of PT. Astra Agro Lestari Tbk, especially Mr Adi Pancoro, Mr Satyoso Harjotedjo, and Mr Cahyo Sri Wibowo. The author wishes to convey special thanks to Mr Santosa and Mr M. Hadi Sugeng, CEO and R&D Director of PT. Astra Agro Lestari Tbk, respectively.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.106745.
Appendix. Supplementary Materials
References
- 1.Haran J., Ndzana Abanda R.F.X., Benoit L., Bakoume C., Beaudoin-Ollivier L. Multilocus phylogeography of the world populations of Elaeidobius kamerunicus (Coleoptera, Curculionidae), pollinator of the palm Elaeis guineensis. Bull. Entomol. Res. 2020;110:654–659. doi: 10.1017/S0007485320000218. [DOI] [PubMed] [Google Scholar]
- 2.Li K., Tscharntke T., Saintes B., Buchori D., Grass I. Critical factors limiting pollination success in oil palm: a systematic review. Agric. Ecosyst. Environ. 2019;280:152–160. doi: 10.1016/j.agee.2019.05.001. [DOI] [Google Scholar]
- 3.Tambunan V.B., Apriyanto A., Ajambang W., Etta C.E., Sahari B., Buchori D., Hidayat P. Molecular identification and population genetic study of Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) from Indonesia, Malaysia and Cameroon based on mitochondrial gene. Biodiversitas. 2020;21(7):3263–3270. doi: 10.13057/biodiv/d210749. [DOI] [Google Scholar]
- 4.Haran J.M., Beaudoin-Ollivier L., Benoit L., Kuschel G. Revision of the palm-pollinating weevil genus Elaeidobius Kuschel, 1952 (Curculionidae, Curculioninae, Derelomini) with descriptions of two new species. Eur. J. Taxon. 2020;684:1–32. doi: 10.5852/ejt.2020.684. [DOI] [Google Scholar]
- 5.Apriyanto A., Tambunan V.B. The complete mitochondrial genome of oil palm pollinating weevil, Elaeidobius kamerunicus Faust. (Coleoptera: Curculionidae) Mitochondrial DNA Part B. 2020;5(3):3450–3452. doi: 10.1080/23802359.2020.1823899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.FASTQC software. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, 2019 (Accessed 12 Jan 2019).
- 7.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chikhi R., Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30(1):31–37. doi: 10.1093/bioinformatics/btt310. [DOI] [PubMed] [Google Scholar]
- 9.Bankevich A. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seppey M., M Manni, Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 12.Du L., Zhang C., Liu Q., Zhang X., Yue B. Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics. 2018;34(4):681–683. doi: 10.1093/bioinformatics/btx665. [DOI] [PubMed] [Google Scholar]
- 13.RepeatMasker software. https://repeatmasker.org, 2019 (Accessed 12 January 2019).
- 14.Hubley R., Finn R.D., Clements J., Eddy S.R., Jones T.A., W Bao, Smit A.F., Wheeler T.J. The Dfam database of repetitive DNA families. Nucleic Acids Res. 2016;44(D1):D81–D89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.OmicsBox software. https://www.biobam.com/omicsbox, 2019 (Accessed 12 April 2019).
- 16.Götz S., Garcia-Gomez J.M., Terol J., Williams T.D., Nagaraj S.H., Nueda M.J., Robles M., Talon M., Dopazo J., Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36(10):3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.