Abstract
Background & Aims
Inflammatory bowel disease (IBD) is a polygenic disorder characterized principally by dysregulated inflammation impacting the gastrointestinal tract. However, there also is increasing evidence for a clinical association with stress and depression. Given the role of the hypothalamus in stress responses and in the pathogenesis of depression, useful insights could be gleaned from understanding its genetic role in IBD.
Methods
We conducted genetic correlation analyses on publicly available genome-wide association study summary statistics for depression and IBD traits to identify genetic commonalities. We used partitioned linkage disequilibrium score regression, leveraging our ATAC sequencing and promoter-focused Capture C data, to measure enrichment of IBD single-nucleotide polymorphisms within promoter-interacting open chromatin regions of human embryonic stem cell-derived hypothalamic-like neurons (HNs). Using the same data sets, we performed variant-to-gene mapping to implicate putative IBD effector genes in HNs. To contrast these results, we similarly analyzed 3-dimensional genomic data generated in epithelium-derived colonoids from rectal biopsy specimens from donors without pathologic disease noted at the time of colonoscopy. Finally, we conducted enrichment pathway analyses on the implicated genes to identify putative IBD dysfunctional pathways.
Results
We found significant genetic correlations (rg) of 0.122 with an adjusted P (Padj) = 1.4 × 10-4 for IBD: rg = 0.122; Padj = 2.5 × 10-3 for ulcerative colitis and genetic correlation (rg) = 0.094; Padj = 2.5 × 10-3 for Crohn’s disease, and significant approximately 4-fold (P = .005) and approximately 7-fold (P = .03) enrichment of IBD single-nucleotide polymorphisms in HNs and colonoids, respectively. We implicated 25 associated genes in HNs, among which CREM, CNTF, and RHOA encode key regulators of stress. Seven genes also additionally were implicated in the colonoids. We observed an overall enrichment for immune and hormonal signaling pathways, and a colonoid-specific enrichment for microbiota-relevant terms.
Conclusions
Our results suggest that the hypothalamus warrants further study in the context of IBD pathogenesis.
Keywords: HPA, Stress, Colonoids, hESC
Abbreviations used in this paper: ACTH, adrenocorticotrophic hormone; ATAC, Assay for Transposase-Accessible Chromatin; CD, Crohn’s disease; CNTF, –; cRE, cis-regulatory element; CRH, corticotrophin-releasing hormone; GWAS, genome-wide association study; hESC, human Embryonic Stem Cell; HN, hypothalamic-like neuron; HPA, hypothalamus–pituitary–adrenal; IBD, inflammatory bowel disease; LD, linkage disequilibrium; LDSC, linkage disequilibrium score regression; MS, multiple sclerosis; NE, norepinephrine; Padj, adjusted P value; PSC, primary sclerosing cholangitis; rg, genetic correlation; RHOA, ras homolog gene family, member A; SNP, single-nucleotide polymorphism; UC, ulcerative colitis
Graphical abstract
Summary.
Inflammatory bowel disease is associated with stress and depression. These 2 comorbidities are influenced by the hypothalamus. Integrating our 3-dimensional genomic data with publicly available genome-wide association study data, our results implicate a subset of inflammatory bowel disease loci conferring their effect via the hypothalamus. Our findings warrant further investigation.
Inflammatory bowel disease (IBD) is an immune-mediated trait, consisting principally of Crohn’s disease (CD) and ulcerative colitis (UC), caused by inflammation of the gastrointestinal tract with the disease course ranging from chronically active to intermittent/rare flares. Multiple genetic and environmental factors are known to contribute to the pathogenesis of IBD. More than 230 independent genetic loci for IBD have been reported from genome-wide association studies (GWAS) to date, with many implicated in host-microbiome immunologic-mediated interactions.1, 2, 3, 4, 5
The environmental factors responsible for triggering the initial presentation and subsequent relapses, as well as their related mechanisms, are not clearly understood. Psychosocial stress is considered one such factor and the majority of IBD patients report a link between their illness and their stress levels6; indeed, clinicians have observed an exacerbation of symptoms caused by emotional conflicts.7 In addition, multiple case studies point to adverse life events as causal factors in IBD relapses,8 and psychosocial stress can induce and reactivate gut inflammation in animal models of colitis.9 Finally, as many as a third of IBD patients also are affected by depression,10 a mood disorder characterized by dysregulated stress responses.11
The hypothalamus orchestrates an intricate network of physiological and behavioral responses via processes mediated by the hypothalamus–pituitary–adrenal (HPA) axis and the sympathetic adrenomedullary network.12 The HPA axis modulates cortisol release via coordinated feedback interactions between the hypothalamus and the pituitary and adrenal glands.13
The HPA axis also has been implicated in the pathogenesis of depression.14,15 Furthermore, IBD patients are more susceptible to depression,10 with the incidence increasing in association with the active phase of inflammation.16 However, the possible physiological links between depression and IBD remain unclear.17 Stress as a comorbid factor in both IBD and depression render the hypothalamus a possible mediator of IBD.
We performed genetic correlation analyses between IBD and depression to assess the degree of genetic commonality and followed this analysis by: (1) partitioned linkage disequilibrium (LD) score regression to measure the enrichment of IBD-associated risk variants in human Embryonic Stem Cell (hESC)-derived, hypothalamic-like neuron (HN), promoter-interacting, open chromatin regions (by intersecting publicly available IBD summary statistics with our Assay for Transposase-Accessible Chromatin (ATAC) sequencing and promoter-focused Capture C data); (2) variant-to-gene mapping to implicate putative causal IBD genes expressed in human embryonic stem cell–derived HNs and in primary hypothalamic tissue; (3) generating ATAC sequencing and promoter-focused Capture C libraries in epithelial-derived colonoids (whose barrier function can be impaired in IBD) to conduct the same analyses as in HNs; and (4) assessing the newly implicated genes, both in the HNs and colonoids, for pathway enrichment to implicate shared, as well as tissue-specific, pathways possibly influenced by IBD-associated genetic variants.
Results
Genetic Correlations
To explore genetic commonalities between IBD and depression, we conducted linkage disequilibrium score regression (LDSR) genetic correlation analyses,18 leveraging the Psychiatric Genomics Consortium of UK Biobank depression summary statistics19 and the summary statistics of 11 autoimmune diseases available in the LD Hub Test Center (http://ldsc.broadinstitute.org),20 namely IBD, UC, CD, asthma, multiple sclerosis (MS), primary sclerosing cholangitis (PSC), rheumatoid arthritis, celiac disease, lupus, and eczema.
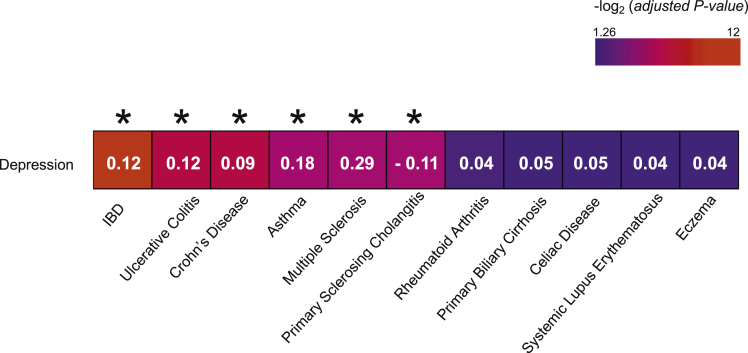
Among the 11 autoimmune diseases, IBD was the most statistically significant trait positively correlated with depression (Figure 1). IBD yielded a genetic correlation coefficient (rg) of 0.122 with an adjusted P value of 1.4 × 10-4, followed by UC (rg = 0.122; adjusted P value [Padj] = 2.5 × 10-3) and CD (rg = 0.094 Padj = 2.5 × 10-3). It also was noted that asthma and multiple sclerosis were highly correlated with depression. Although primary sclerosing cholangitis also was correlated significantly with depression, we did not observe such a relationship with celiac disease and rheumatoid arthritis, despite these traits being pathologically related to IBD.21,22
Figure 1.
Genetic correlation estimates for 11 autoimmune diseases with depression. Numbers indicate the genetic correlation coefficients (rg) estimated with the LD-Hub test center, using precalculated LD scores. Colors indicate the statistical significance (-log2 [adjusted P value]) of the correlations. ∗Adjusted P value < .05.
Genome-Wide ATAC Sequencing and Promoter-Focused Capture C
We compared 2 cell models: hESC-derived HNs and colonoids by leveraging ATAC sequencing–defined open chromatin maps and high-resolution, genome-scale, promoter-focused Capture C atlases.23,24 The HNs data sets were previously derived in our laboratory.23 Colonoids were derived from rectal biopsy specimens of 3 donors without pathologic disease noted at the time of colonoscopy.25, 26, 27 From colonoids, we generated 3 ATAC sequencing libraries that were sequenced and subsequently analyzed with the ENCODE pipeline (https://github.com/kundajelab/atac_dnase_pipelines), yielding 72,440 open chromatin conservative peaks. The Capture C libraries yielded high coverage (an average of ∼1.7 billion reads per each colonoid library), with an average of 54% valid reads pairs and 77% capture efficiency. Finally, similarly to what we did with the HNs,23 we leveraged the colonoid data sets to call significant interactions using the CHiCAGO pipeline,28 and performed analyses at 1-fragment resolution to identify short-distance interactions, and at 4-fragment resolution analyses to identify long-distance interactions, and then merged the results.
Partitioned LD Score Regression
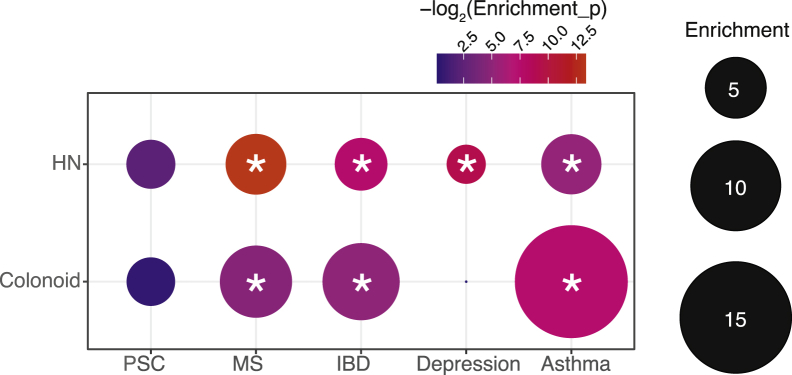
We investigated if IBD-associated variants were enriched within promoter-interacting open chromatin regions of HNs. We intersected the ATAC sequencing and promoter-focused Capture C data from HNs with the International IBD Genetics Consortium IBD summary statistics and performed partitioned LD score regression (LDSR). We observed an approximately 4-fold significant enrichment (P = .005) of IBD single-nucleotide polymorphisms (SNPs) within hypothalamic promoter-interacting, open-chromatin regions (Figure 2).
Figure 2.
Heritability enrichment of IBD, depression, asthma, MS, and PSC loci within HNs and colonoid. The circle size represents the fold enrichment of IBD SNPs within hypothalamic or colonoid promoter-interacting open chromatin regions. Colors indicate the statistical significance (-log2 [P value]) of the enrichments. ∗P < .05.
We extended these analyses to depression and the other autoimmune traits significantly correlated with depression described earlier (Figure 1): namely, asthma, MS, and PSC. Depression yielded an approximately 2.5-fold significant enrichment (P = .001), while both asthma and MS had a significant approximate 5-fold enrichment (P = .028; P = 9.89 × 10-5 ); in contrast, PSC correlated negatively with depression and there was no evidence of hypothalamic enrichment for the associated SNPs (Figure 2).
As a comparison, we also ran partitioned LDSR intersecting the same set of IBD SNPs with ATAC sequencing and promoter-focused Capture C data from colonoids, yielding an approximately 7-fold significant enrichment (P = .03). We extended these analyses to asthma, MS, PSC, and depression. Surprisingly, asthma showed a significant approximately 15-fold enrichment (P = .006) and MS showed an approximately 6-fold enrichment (P = .04); while there was no evidence for PSC or depression enrichment (Figure 2).
Partitioned LDSR therefore showed that IBD loci were enriched significantly both in HNs and colonoid promoter-interacting, open-chromatin regions. The enrichment in colonoids was higher with respect to the hypothalamic data, suggesting, not surprisingly, that IBD-associated loci operate more strongly in the gastrointestinal tract, but, importantly, our data also implicate the hypothalamus to a high degree.
Variant to Gene Mapping
To identify putative IBD functional effector genes, we leveraged published GWAS data, along with the ATAC sequencing and promoter-focused Capture C data described earlier.
We retrieved 332 statistically significant independent sentinel SNPs reported in published IBD GWAS and identified 10,035 proxy SNPs with an r2 > 0.8.1, 2, 3,5,29,30 To focus on just those variants residing within open-chromatin regions in either HNs or colonoids, and therefore putatively functional, we overlapped these proxies with ATAC sequencing peaks. This step identified a total of 471 open proxies for IBD (273 in colonoids, 198 in HNs).
To identify the IBD open proxies that contact the promoters of putative effector genes in HNs and colonoids, we constrained the Capture C data with the ATAC sequencing data for each cell type. We focused on open proxies with r2 > 0.8 that contacted open gene promoter regions, given that this r2 value represents strong linkage disequilibrium with each reported sentinel SNP. Indeed, approximately 70% of relevant proxies corresponded to members of previously described credible sets.5
Implicated Genes
We identified 25 gene promoters contacted by IBD open proxies in the HNs (Table 1, Supplementary Table 1). Gene expression was verified using both our RNA sequencing data generated in HNs23 (Table 1) and data derived from hypothalamic tissue in the GTEx database (Supplementary Table 2).
Table 1.
Variant-to-Gene Mapping Implicated IBD Putative Effector Genes in HNs
| Implicated genes | PubMed hypothalamus/brain | TPM |
|---|---|---|
| CREM | Inhibition of CRH31 | 8.9 |
| Regulation of PENK expression84 | ||
| Circadian rhythm85 | ||
| Regulation of stress-induced corticosterone release in mice86 | ||
| CNTF | Cortical stress responses32 | 2.5 |
| Neurogenesis in feeding centers of hypothalamus87 | ||
| Secretion of pituitary hormones (PRL, GH)38 | ||
| Neuroprotection and axonal regeneration in retina88 | ||
| FAM111A | — | 4 |
| SLC9B2 (or NHA2) | — | 54.9 |
| SNAI2 (or SLUG) | — | 3.5 |
| LMAN2 | — | 52 |
| PRR7 | Specific removal of excitatory synapses and Wnt inhibition41 | 31.8 |
| PRR7-AS1 | — | 8.4 |
| FOXD1 | Differentiation of anterior hypothalamic neurons36 | 0.1 |
| Pituitary luteinizing hormone expression in mice37 | ||
| PRDM1 (or BLIMP-1) | — | 1.7 |
| UBAC2 | — | 13.8 |
| GPR183 (or EBI2) | Central nervous system autoimmunity and T-cell migration | 2 |
| Neuroinflammation89 | ||
| SKAP2 | — | 16.2 |
| TPD52L2 | Invasiveness of glioblastomas90 | 54.2 |
| ATG16L1 | Neuroinflammation of central nervous system55 | 19.2 |
| Epilepsy91 | ||
| NFKB2 | Deficient anterior pituitary with variable immune-deficiency syndrome39 | 3.2 |
| Neuroprotection of dopaminergic neurons92 | ||
| ACO2 | Neurodegenerative disorders93 | 90.8 |
| PHF5A | — | 27.5 |
| DENND1B | — | 7.1 |
| TCTA | — | 64.7 |
| RHOA | Ghrelin/leptin sensitivity in a subset of hypothalamic neurons35 | 261.5 |
| Social stress responses in dopaminergic neurons33 | ||
| Depression-like behaviors via dendritic remodeling of dopaminergic neurons34 | ||
| STARD3 | — | 26.6 |
| CBX3 | Neural differentiation94 | 43.7 |
| Glioma proliferation95 | ||
| HNRNPA2B1 | — | 82.1 |
| GPR22 | — | 43.7 |
NOTE. Genes implicated leveraging the publicly available IBD summary statistics and ATAC sequencing and promoter-focused Capture C libraries generated in HNs; their reported known functions and their mean relative expression in HNs in TPM were according to RNA sequencing data.
GH, growth hormone; PRL, prolactin; TPM, transcript per million.
Eleven of the implicated genes have known functions in the brain, although there is no current evidence to support a role for the other 14 genes. Among genes with known functions in the hypothalamus/brain, CREM, CNTF, and RHOA are directly involved in stress response regulation: CREM encodes a transcriptional repressor that regulates the secretion of corticotrophin-releasing hormone (CRH) and, in turn, corticosterone31; Ciliary neurotrophic factor (CNTF) is a polypeptide hormone that modulates cortical stress responses promoting norepinephrine release32; ras homolog gene family, member A is a monomeric guanosine triphosphatase that in mice mediates social stress responses and depressive-like behavior in dopaminergic neurons33,34 and regulates food intake altering hypothalamic tyrosine hydroxylase neuron sensitivity to ghrelin and leptin and the expression of orexigenic hypothalamic neuropeptide agouti-related peptide and neuropeptide Y.35
Forkhead Box D1 is required for the development of hypothalamic cell populations responsible for the secretion of hormones such as CRH.36 Both Forkhead Box D1 and CNTF regulate the secretion of pituitary hormones,37,38 while mutations in NFKB2 are causative for variable immunodeficiency and deficient anterior pituitary with variable immune deficiency syndrome, a condition characterized by immunodeficiency and anterior pituitary hormone deficiency, including adrenocorticotrophic hormone (ACTH).39,40
In the brain, PRR7 encodes for a synapse-to-nucleus messenger, which regulates excitatory synaptogenesis by inhibiting Wnt.41 Hypothalamic Wnt signaling participates in systemic glucose and energy homeostasis.42
The ACO2, GPR183, and ATG16L1 gene products are associated with neuroinflammation and autoimmunity in the central nervous system, and GPR183 also enhances the migration/infiltration of autoreactive T cells in the brain.43, 44, 45 ATG16L1 encodes a key autophagy protein critical in handling bacteria in IBD pathogenesis and is associated strongly with CD.46 The DENND1B locus is associated strongly with asthma,47 a trait in which dysfunction of the HPA axis has been implicated.48,49
In colonoids, associated IBD loci contacted 43 genes (Table 2, Supplementary Table 3) that are expressed in the sigmoid and transverse colon, and the small intestine (GTEx database) (Table 2). Seven genes (ACO2, FOXD1, LMAN2, PHF5A, PRR7, PRR7-AS1, and SKAP2) were implicated in both HNs and colonoids; FOXD1, PRR7, and SKAP2 encode proteins with known roles in immune regulation,50, 51, 52 and PHF5A mediates stress resistance in a model of colorectal cancer.53
Table 2.
Variant-to-Gene Mapping Implicated IBD Putative Effector Genes in Colonoids
| Genes | TPM sigmoid colon | TPM transverse colon | TPM small intestine-terminal ileum |
|---|---|---|---|
| ACO2 | 70.53 | 82.89 | 82.42 |
| ASCC2 | 27.81 | 29.43 | 32.96 |
| C1orf106 | 0.52 | 14.09 | 18.08 |
| C5orf56 | 5.881 | 4.29 | 6.56 |
| CBLL1 | 16.63 | 10.98 | 11.46 |
| CD6 | 0.47 | 2.54 | 14.41 |
| CDH3 | 0.24 | 0.19 | 17.05 |
| CEBPG | 23.55 | 29.01 | 30.61 |
| COG5 | 11.78 | 9.55 | 10.4 |
| CUL2 | 19.02 | 12.54 | 12.61 |
| DAG1 | 60.8 | 39.55 | 29.11 |
| DLD | 57.23 | 51.36 | 36.65 |
| DOCK7 | 7.67 | 4.89 | 3.83 |
| DUS4L | 3.9 | 3.33 | 3.83 |
| FAM213A | 27.37 | 25.59 | 19.45 |
| FNDC3A | 26.5 | 20.54 | 23.5 |
| FOXD1 | 0.12 | 0.07 | 0.58 |
| GDF9 | 0.54 | 0.4 | 0.48 |
| GNPDA1 | 25.58 | 17.39 | 21.6 |
| HOXA-AS2 | 4.79 | 4.21 | 3.92 |
| HOXA-AS3 | 0.65 | 1.54 | 1.29 |
| HOXA3 | 9.73 | 11.47 | 9.45 |
| HOXA5 | 23.7 | 29.19 | 22.48 |
| HOXA9 | 25.13 | 27.34 | 19.39 |
| LMAN2 | 77.46 | 96.25 | 94.96 |
| MAML2 | 7.28 | 6.98 | 8.37 |
| PHF5A | 37.89 | 32.55 | 34.48 |
| PRR7 | 1.13 | 1.42 | 2 |
| PRR7-AS1 | 0.22 | 0.28 | 0.5 |
| SATB1 | 26.37 | 14.94 | 7.84 |
| SBNO2 | 33.46 | 30.04 | 41.32 |
| SKAP2 | 14.97 | 15.56 | 19.82 |
| SLC2A13 | 5.06 | 4.88 | 4.11 |
| SPATA7 | 6.36 | 3.71 | 2.84 |
| SPRED2 | 13.19 | 13.68 | 12.4 |
| TEF | 34.89 | 19.16 | 16.04 |
| TMED10 | 87.99 | 80.77 | 75.48 |
| TNFSF15 | 3.17 | 1.38 | 1.58 |
| TRIM25 | 17.32 | 25.54 | 33.6 |
| TSPAN14 | 8.97 | 10.93 | 13.33 |
| UQCRQ | 163.4 | 184.7 | 155.5 |
| ZFP90 | 12.06 | 7.14 | 9.29 |
| TBC1D5 | 12.42 | 8.78 | 10.91 |
NOTE. Genes implicated leveraging the publicly available IBD summary statistic and ATAC sequencing and promoter-focused Capture C libraries generated in colonoids and their relative expression in the sigmoid and transverse colon, and the small intestine in TPM according to GTEx.
TPM, transcript per million.
Pathway Analyses
The implicated IBD effector genes in both the HNs and the colonoids were assessed for pathway enrichment through ConsensusPathDB (http://consensuspathdb.org).54 We initially ran enrichment analyses pooling the implicated genes from both cellular settings. We only considered pathways with an adjusted P value less than .05.
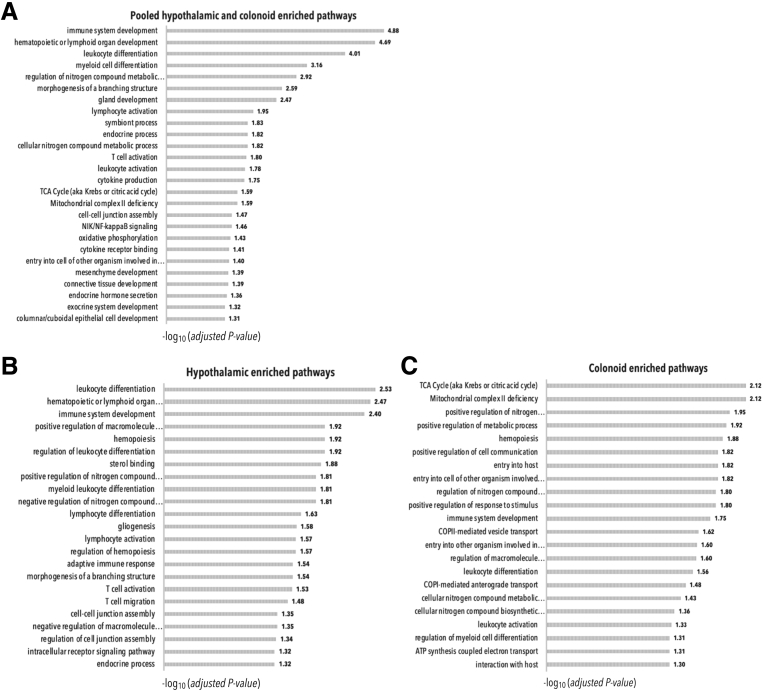
We observed an enrichment for immune-related pathways (eg, immune system development, hematopoietic/lymphoid organ development, leukocyte/myeloid cell differentiation, lymphocyte /T-cell/leukocyte activation, cytokine production); respiratory chain-related pathways (eg, tricarboxylic acid cycle, mitochondrial complex II deficiency, oxidative phosphorylation, nitrogen compound metabolic process); hormone-related pathways (eg, endocrine hormone secretion, endocrine process, exocrine system development); tissue development pathways (mesenchyme, connective tissue, columnar/cuboidal epithelial cells, gland); and symbiosis-related pathways (eg, symbiont entry into cells of other organism involved in symbiotic interaction) (Figure 3A, Supplementary Table 4).
Figure 3.
Pathway enrichment in HNs and/or colonoids. Enriched pathways for (A) hypothalamic- and colonoid-, (B) only hypothalamic-, or (C) only colonoid-implicated genes identified using ConsensusPathDB. The selection of significant enriched pathways (adjusted P value < .05) is reported. Bars indicate the statistical significance (-log10 [adjusted P value]) of the enrichments. ATP, adenosine triphosphate; COPI, coat protein; TCA, tricarboxylic acid cycle.
We then ran the same analyses using the hypothalamic and colonoid implicated gene lists separately. We again considered only pathways with Padj < .05. We observed similar enrichments (Figure 3B and C, Supplementary Tables 5 and 6). The 25 hypothalamic genes showed enrichment for various immune pathways, for the endocrine process, nitrogen compound regulation, gliogenesis, cell junction assembly, and branching structure morphogenesis, as also seen in the pooled analysis. The hypothalamic genes were enriched specifically for sterol binding (Figure 3B, Supplementary Table 5). The 43 colonoid genes were enriched for various pathways associated with oxidative stress/respiratory chain reactions and symbiosis. They also were enriched for endocrine hormone secretion and immune-related pathways (Figure 3C, Supplementary Table 6).
Taken together, the overall enrichment pathway results suggested a role for immunity, nitrogen compound metabolism, and hormones in IBD, with a specific role for symbiosis and host interaction in the colonoids and sterol binding in the HNs.
Discussion
Our results implicate a role for the hypothalamus in the genetic susceptibility to IBD. IBD is a complex polygenetic and multifactorial disease that principally affects the immune system and the gastrointestinal tract. However, the phenotype is also highly impacted by stress6: psychosocial stress is a known environmental trigger of IBD onset and flares7,8 and IBD patients manifest high comorbidity with depression,16,55 a stress-related disorder involving the hypothalamus and characterized by a deregulated HPA axis.11,15
We investigated how the genetics of IBD potentially might influence both the hypothalamus and the HPA axis by leveraging both public GWAS reports and our own genomic data. We investigated potential genetic relationships between IBD and stress using depression as a key proxy. We leveraged depression GWAS results, owing to the paucity of GWAS efforts focused on stress, with other stress-related GWAS also being relatively limited and largely focused on male military cases suffering from post-traumatic stress disorder,56,57 a condition triggered by a life-changing traumatic event rather than everyday psychosocial stress. Smaller studies on genetic determinants of stressful life events showed a significant and robust genetic association between stressful life events and depression.58 Despite these correlations, the genetic studies of response to stress-related events are not uniform, with various different triggers being investigated. Each individual trigger study is much smaller than the large, relatively uniform studies of depression, thus supporting our use of depression as the best proxy for stress to run the most powered genetic correlation analyses possible. Using linkage disequilibrium score regression (LDSR) analyses based on selected GWAS, among 11 autoimmune diseases, IBD, CD, and UC were correlated the most significantly with depression (Figure 1). The genetic correlation scores were modest but robust (rg = 0.12, 0.12, and 0.09 for IBD, UC, and CD, respectively), given that in contrast the rg value between CD and UC themselves is 0.54 (http://ldsc.broadinstitute.org/lookup).
We conducted partitioned LDSR to assess the enrichment of IBD loci in promoter-interacting, open-chromatin regions of hESC-derived HNs and colonoids (Figure 2). Colonoids in this study were used principally to contrast with findings in the hypothalamic context. Not surprisingly, we observed a greater enrichment in colonoids (∼7-fold enrichment), thus validating our approach, but also giving a sense of the relative degree of the hypothalamic contribution to the pathogenesis of IBD. The significant approximately 4-fold enrichment supports the hypothesis that HNs are indeed a relevant cell type for IBD genetics. We also observed significant enrichment for MS and asthma SNPs in HNs and colonoid putative regulatory elements. The HPA axis is dysregulated in some MS patients, and hypothalamic lesions are correlated with the disease severity and outcomes.59, 60, 61 MS is associated with neurogenic bowel dysfunction,62 and recent studies have implicated a role for microbial breeches in intestinal barrier integrity in its pathogenesis.63,64 Asthma also is characterized by a dysregulation of the HPA axis49 and by breeches in respiratory epithelium barrier integrity leading to exacerbation of inflammatory responses.65 We speculate that similar abnormalities might occur in the gut, predisposing to a deregulation in the gut microbiome66 and to characteristic inflammatory responses to food allergens.67
A variant-to-gene mapping–based search for IBD SNPs variants in HNs was used to investigate the potential role of IBD genetics within the hypothalamus. We implicated CREM (Table 1), which in the hypothalamus encodes an inducible cAMP early repressor,68 which binds to the CRH promoter to terminate CRH expression in response to stressors.31 However, in the pituitary gland, CRH promotes the expression of adrenocorticotrophic hormone, which in turn stimulates cortisol release from the adrenal gland. Thus, by repressing CRH release, inducible cAMP early repressor represses cortisol secretion and inhibits the HPA axis.69
We speculate that in IBD, hypothalamic expression of CREM is dysregulated in some genetically predisposed individuals, leading to suboptimal activation of the HPA axis with consequent overactivation of the immune response in the presence of a stressor. Supporting this hypothesis, in 1 small study, IBD steroid-naïve pediatric subjects showed a hyporeactive HPA axis and increased proinflammatory cytokine release in response to psychosocial stressors.70
Moreover, LEW/N rats, which have lower levels of hypothalamic CRH and circulating corticosterone, that is, murine cortisol, are more susceptible to both inflammatory disorders and induced colitis.71 In experiments that aimed to assess a potential role for the neuroimmunoendocrine process mediated by adrenal glands in IBD, adrenalectomized experimental dextran sodium sulfate colitis mice show more severe inflammation and a higher clinical score. Administration of corticosterone to the adrenalectomized dextran sodium sulfate mice eliminates this phenotype.72
Another implicated hypothalamic gene is CNTF. CNTF encodes ciliary neurotrophic factor, which is released in response to acute stress and is necessary for the synthesis of cortical norepinephrine (NE) in norepinephrinergic neurons of the locus coeruleus. CNTF recruits secretagogin and extracellular signal-regulated kinase 1 to increase NE production. NE in turn activates and maintains neurons in the long-lasting excitatory cortical vigilance necessary to promote more favorable physiological and behavioral responses to stressors.32 Therefore, dysregulation of CNTF expression might lead to an abnormal release of NE, affecting responses to stress. NE, epinephrine, and dopamine are stress-related catecholamines that affect nutrient and ion absorption rates in the gastrointestinal tract in addition to modulating the innate immune system and microbiome interactions.73
In mice, the guanosine triphosphatase RHOA mediates social stress responses and depressive-like behavior via dendritic remodeling of neurons responsible for motivation and reward33,34; specific knockout of RHOA in tyrosine hydroxylase hypothalamic neurons of the arcuate nucleus in mice leads to a higher sensitivity to peripherally administered ghrelin and to abolished response to leptin, promoting food intake and obesity-induced ghrelin resistance.35 Similar modifications of paraventricular hypothalamic nucleus neurons participating in the HPA axis are possible.
NFKB2 mutations are associated with deficient anterior pituitary with variable immune deficiency syndrome, a rare condition characterized by immune and anterior pituitary deficiencies including ACTH.39
ATG16L1 is a known risk factor for CD46 and encodes for a protein required for autophagy,45 while DENND1B has been implicated in asthma.47,48 Our results showed that asthma and IBD also both are correlated with depression (Figure 1B), and their associated SNPs are enriched in promoter-interacting open chromatin regions of HNs and colonoids (Figure 2), suggesting genetic overlap in functions beyond autoimmunity.
We also conducted a variant-to-gene mapping analysis in colonoids, identifying only 7 shared putative effector genes between the 2 cell models (Table 2). Among them, FOXD1, PRR7, and SKAP2 gene products participate in regulating the immune system,50, 51, 52 while PHF5A encodes a regulator of cellular stress resistance in colorectal cancer.53 Although we compared 2 completely different cell models, a number of shared functional genes between HNs and colonoids might have been expected. However, our results reflect the cell-type specificity of chromatin organization and gene regulation, and reflect a possible key aspect of IBD: the same set of genetic variants likely interacting with distinct genes in different cell types. For example, the IBD SNP rs34779708 in the HNs implicated CREM, while in the colonoids the same SNP implicated CUL2 (Supplementary Tables 1 and 3).
We performed enrichment pathway analyses based on the variant-to-gene mapping results. Despite the relatively large number of pathways observed as a consequence of ConsensuPathDB annotations, such that repeated enriched genes showed pathways that are nested and not necessarily independent of each other, we observed a predictably broad enrichment for immune pathways, and enrichment for pathways involving hormonal and endocrine/exocrine systems and for symbiotic pathways and oxidative stress. These associations are consistent with a deregulation of the HPA axis and are in line with current knowledge supporting a role for the microbiome74 and reactive oxygen/nitrogen species75 in the development of IBD (Figure 3, Supplementary Tables 4–6).
Limitations of this study included the absence of a valid GWAS focused on stress/psychosocial stress–related disorders resulting from the complexity of defining and measuring the trait and the lack of in-depth characterization of HPA reactivity in an IBD population. Such data, in the context of currently available IBD, CD, and UC GWAS, could provide more direct insights about the relevance of IBD genetics in the hypothalamus and in hypothalamus-mediated stress responses. Our results warrant functional follow-up evaluation to validate these putative hypothalamic–IBD implicated genes, in particular, CREM, CNTF, and RHOA. Future studies are warranted, in which genes could be knocked out specifically in the PVH neurons of model mice for colitis and then the mice exposed to stress, to investigate how inflammation severity compares with model wild-type mice.
Our results are consistent with a role of the hypothalamus in IBD. We propose that IBD-associated SNPs alter the HPA axis and stress responses predisposing to and/or exacerbating this disease. Our data suggest that IBD genetic risk variants influence both general (eg, immunologic and endocrine) and specific (eg, symbiotic process) pathways in different cell types. These studies provide approaches to extending mechanistic analyses of GWAS data for complex phenotypic traits.
Materials and Methods
Genetic Correlation Analyses
We performed LDSR analyses18 to calculate genetic correlations between pairs of traits using the LD-Hub web interface (http://ldsc.broadinstitute.org). We uploaded the Psychiatric Genomics Consortium of UK Biobank_depression summary statistics19 and we used the GWAS summary statistic available in the LD-Hub test center20 for IBD,2 UC,2 CD,2 asthma,76 MS (http://www.imsgenetics.org), PSC,77 rheumatoid arthritis,78 primary biliary cirrhosis,79 celiac disease,21 lupus,80 and eczema.81 We ran the analyses using default parameters. Quality control of the inputted summary statistics and heritability analysis were performed automatically. The SNPs were filtered automatically and selected on the basis of minor allele frequency >1%, and sample size (X2 > 80). Only summary statistics with more than 450,000 SNPs and more than 5000 individuals were considered. Insertion or deletion of basans, strand-ambiguous SNPs, and SNPs whose alleles did not match those in the 1000 Genomic data were removed. All summary statistics excluded the major histocompatibility complex.20 We adjusted the P values for multiple comparisons using the Benjamini–Hochberg false-discovery rate method and considered 2 traits to be correlated significantly with an adjusted P value < .05.
Partitioned LDSR
Partitioned LDSR estimates the heritability from GWAS summary statistics within a subset of regions of the genome after accounting for LD.18 Partitioned heritability was measured using LDSR v1.0.0 (https://github.com/bulik/ldsc) to identify enrichment of GWAS signals among cis-regulatory elements (cREs) in HNs and colonoids as previously performed.23 Briefly, the annotation and heritability estimates for HNs and colonoids were generated using bed files containing the position of the cRE (open chromatin regions located proximal to a promoter (-1500/+500 bp of transcription start site) + open chromatin regions located within overlapping regions with ±500 bp buffers. We considered a cRE as open if the normalized ATAC sequencing signal exceeded a cut-off value of fragments per kilobase >1. The HNs and the colonoids were compared with the baseline model for the European ancestry, downloaded from https://github.com/bulik/ldsc/wiki/Partitioned-Heritability. The results were visualized as bubble plots using ggplot2 (https://cran.r-project.org/web/packages/ggplot2/index.html), with the circle size representing the fold enrichment of cREs compared with the base annotation, and the color indicating the statistical significance (-log [P value]). We selected a subset of autoimmune GWAS data for partitioned LDSR based on genetic correlation results; we chose the autoimmune diseases that correlated significantly with depression. We tested asthma,76 IBD (https://www.ibdgenetics.org), MS (http://www.imsgenetics.org), and PSC,77 and also depression 19 We selected GWAS summary statistics available on LD-Hub.20 Because the asthma GWAS was a meta-analysis, partitioned LDSR was conducted using the summary statistics from the fixed-effects model.
Cell Models
HNs were generated and validated previously.23,82 Colonoids were derived following established protocols.25,27 Briefly, at the time of endoscopy, 1–2 biopsy specimens from the rectum were placed in culture media with antibiotics and then incubated in chelation buffer to isolate the epithelial crypts. The biopsy specimen was incubated in chelation buffer to isolate the epithelial crypts. Purified crypts were plated in Matrigel (Corning, Corning, NY) droplets and then overlaid with media. The overlaid media, partially produced by conditioned media, contained growth factors (epidermal growth factor, R-spondin, Noggin, and Wnt3a) for stem cell expansion. Approximately 100–200 colonoids were grown per well (∼100,000 live cells/well). The colonoids were passaged weekly at a ratio of 1:2 or 1:3 based on density. At each passage, an aliquot of 100,000 cells was processed for ATAC sequencing and to control for passage effects. To obtain at least 2.5 million cells for Capture-C, the colonoids were passaged for expansion 4–6 times. Three replicates were processed from subjects with no pathologic disease.
ATAC Sequencing
ATAC sequencing libraries were generated and analyzed as previously described.23,24 Briefly, 100,000 colonoids were collected and pelleted at 550 relative centrifugal force for 5 minutes at 4°C. The pellet was resuspended in 50 μL chilled lysis buffer and then centrifuged again at 550 rcf for 10 minutes at 4°C. The pelleted DNA then was tagged using Tn5 Transposase (cat. FC-121–1030; Illumina, San Diego, CA) and incubated for 45 minutes at 37°C. Next, the DNA was purified with the MinElute Kit (cat. 28004; Qiagen, Germantown, MD) and eluted in elution buffer. Purified tagged DNA fragments were polymerase chain reaction–amplified using Nextera primers and NEB-Next High-Fidelity Polymerase Chain Reaction Master Mix (cat. M0541; New England Labs, Woburn, MD) to generate libraries, which then were cleaned with Agencourt AMPureXP beads (cat. A63880; BeckmanCoulter, Brea, CA) and bioanalyzed. Finally, the libraries were paired-end sequenced using the Illumina NovaSeq platform. Open chromatin regions were called using the ENCODE ATAC sequencing pipeline (https://www.encodeproject.org/atac-seq), selecting the resulting irreproducibility discovery rate conservative peaks (with all coordinates referring to Homo sapiens genome assembly GRCh37). We defined a genomic region as open if it had a 1-bp overlap with an ATAC sequencing peak.
ATAC sequencing libraries for HNs were generated and analyzed previously in our laboratory.23
Promoter-Focused Capture C
We generated colonoid Capture C libraries following an established protocol.23,24 Each library then was sonicated using a QSonica (QSonica, Newtown, CT) Q800R to obtain DNA fragments with an average size of 350 bp. DNA fragments were purified using AMPureXP beads (Agencourt) and measured via Qubit (Invitrogen, Carlsbad, CA) fluorometer. Fragment quality and sizes were assessed on a Bioanalyzer (Agilent Technologies, Carlsbad, CA) 2100 using a 1000 DNA Chip. DNA ends were repaired and adaptors were ligated using the SureSelectXT Library Prep Kit (Agilent). After a clean-up step, the sizes and concentrations of DNA fragments were checked again. To generate high-complexity libraries, adaptor ligated DNA fragments were hybridized with a custom-designed capture panel (Agilent)24 using the SureSelectXT capture kit (Agilent). Each captured library was first paired-end sequenced on 1-lane HiSeq 4000 sequencing (100-bp read length) for quality control and then sequenced on an S2 flow cells on an Illumina NovaSeq (50-bp read length). Data were analyzed as previously described.24
Promoter-focused Capture-C libraries from HNs were generated and analyzed previously in our laboratory.23
Genetic Loci Included in Variant-to-Gene Mapping
We used 332 loci from published IBD GWAS studies.1, 2, 3,5,29,30 To derive proxy SNPs, we used SNiPA83 with Homo sapiens genome assembly GRCh37 as human reference assembly, 1000 Genomes phase 1v3 as the variant set, a European population; Ensembl 87 for genome annotation, and an LD threshold of r2 > 0.8.
Enrichment Pathway Analyses
Enrichment pathway analyses were performed using the ConsensusPathDB platform over-representation analyses.54 Our selected genes were searched in predefined Gene Ontology–based gene sets using a cut-off P value of ≤.05 using the default background gene set. ConsensuPathDB calculated P values using the hypergeometric test, which then were corrected by the false-discovery rate method. We considered pathways with at least 2 overlapping genes and with an adjusted P value less than .05 as significant.
Acknowledgments
The authors would like to acknowledge the Daniel B. Burke Endowed Chair for Diabetes Research.
CRediT Authorship Contributions
Chiara Lasconi, PhD (Conceptualization: Lead; Formal analysis: Lead; Investigation: Lead; Writing – original draft: Lead; Writing – review & editing: Lead);
Matthew C Pahl, PhD (Data curation: Lead; Formal analysis: Lead; Visualization: Lead; Writing – review & editing: Lead);
Diana L Cousminer, PhD (Formal analysis: Supporting; Investigation: Supporting; Supervision: Supporting; Writing – review & editing: Supporting);
Claudia A Doege, MD (Methodology: Supporting; Resources: Lead; Writing – review & editing: Supporting);
Alessandra Chesi, PhD (Data curation: Lead; Supervision: Supporting; Writing – review & editing: Supporting);
Kenyaita M Hodge, Graduate Student (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting);
Michelle E Leonard, BA (Data curation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting);
Sumei Lu, BS (Data curation: Supporting; Project administration: Supporting; Writing – review & editing: Supporting);
Matthew E Johnson, PhD (Data curation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting);
Chun Su, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Project administration: Supporting; Supervision: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting);
Reza K Hammond, PhD (Data curation: Supporting; Formal analysis: Supporting; Writing – review & editing: Supporting);
James A Pippin, BA (Data curation: Supporting; Investigation: Supporting; Methodology: Lead; Project administration: Lead; Supervision: Equal; Writing – review & editing: Supporting);
Natalie A Terry, MD, PhD (Data curation: Equal; Investigation: Equal; Resources: Supporting; Supervision: Supporting; Writing – review & editing: Supporting);
Louis R Ghanem, MD, PhD (Data curation: Lead; Investigation: Equal; Resources: Supporting; Writing – review & editing: Equal);
Rudolph L Leibel, MD (Data curation: Supporting; Investigation: Supporting; Methodology: Supporting; Supervision: Equal; Writing – review & editing: Lead);
Andrew D Wells, PhD (Conceptualization: Supporting; Data curation: Lead; Formal analysis: Supporting; Funding acquisition: Lead; Methodology: Supporting; Project administration: Equal; Resources: Equal; Writing – original draft: Supporting; Writing – review & editing: Lead);
Struan Grant, Ph.D. (Supervision: Lead).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by National Institutes of Health R01 HL143790 and R01 HG010067.
Supplementary Material
References
- 1.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.-P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Büning C., Cohain A., Cichon S., D’Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., International IBD Genetics Consortium (IIBDGC) Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J.Z., van Sommeren S., Huang H., Ng S.C., Alberts R., Takahashi A., Ripke S., Lee J.C., Jostins L., Shah T., Abedian S., Cheon J.H., Cho J., Daryani N.E., Franke L., Fuyuno Y., Hart A., Juyal R.C., Juyal G., Kim W.H., Morris A.P., Poustchi H., Newman W.G., Midha V., Orchard T.R., Vahedi H., Sood A., Sung J.J.Y., Malekzadeh R., Westra H.-J., Yamazaki K., Yang S.-K., Barrett J.C., Franke A., Alizadeh B.Z., Parkes M., K T.B., Daly M.J., Kubo M., Anderson C.A., Weersma R.K. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellinghaus D., Jostins L., Spain S.L., Cortes A., Bethune J., Han B., Park Y.R., Raychaudhuri S., Pouget J.G., Hübenthal M., Folseraas T., Wang Y., Esko T., Metspalu A., Westra H.-J., Franke L., Pers T.H., Weersma R.K., Collij V., D’Amato M., Halfvarson J., Jensen A.B., Lieb W., Degenhardt F., Forstner A.J., Hofmann A., International IBD Genetics Consortium (IIBDGC) International Genetics of Ankylosing Spondylitis Consortium (IGAS) International PSC Study Group (IPSCSG) Genetic Analysis of Psoriasis Consortium (GAPC) Psoriasis Association Genetics Extension (PAGE) Schreiber S., Mrowietz U., Juran B.D., Lazaridis K.N., Brunak S., Dale A.M., Trembath R.C., Weidinger S., Weichenthal M., Ellinghaus E., Elder J.T., Barker J.N.W.N., Andreassen O.A., McGovern D.P., Karlsen T.H., Barrett J.C., Parkes M., Brown M.A., Franke A. Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–518. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turpin W., Goethel A., Bedrani L., Croitoru Mdcm K. Determinants of IBD heritability: genes, bugs, and more. Inflamm Bowel Dis. 2018;24:1133–1148. doi: 10.1093/ibd/izy085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang H., Fang M., Jostins L., Mirkov M.U., Boucher G., Anderson C.A., Andersen V., Cleynen I., Cortes A., Crins F., D’Amato M., Deffontaine V., Dmitrieva J., Docampo E., Elansary M., Farh K.K.-H., Franke A., Gori A.-S., Goyette P., Halfvarson J., Haritunians T., Knight J., Lawrance I.C., Lees C.W., Louis E., Mariman R., Meuwissen T., Mni M., Momozawa Y., Parkes M., Spain S.L., Théâtre E., Trynka G., Satsangi J., Sommeren S van, Vermeire S., Xavier R.J., Weersma R.K., Duerr R.H., Mathew C.G., Rioux J.D., McGovern D.P.B., Cho J.H., Georges M., Daly M.J., Barrett J.C. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh S., Graff L.A., Bernstein C.N. Do NSAIDs, antibiotics, infections, or stress trigger flares in IBD? Am J Gastroenterol. 2009;104:1298–1313. doi: 10.1038/ajg.2009.15. quiz 1314. [DOI] [PubMed] [Google Scholar]
- 7.Bhatia V., Tandon R.K. Stress and the gastrointestinal tract. J Gastroenterol Hepatol. 2005;20:332–339. doi: 10.1111/j.1440-1746.2004.03508.x. [DOI] [PubMed] [Google Scholar]
- 8.Mawdsley J.E., Rampton D.S. Psychological stress in IBD: new insights into pathogenic and therapeutic implications. Gut. 2005;54:1481–1491. doi: 10.1136/gut.2005.064261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reber S.O. Stress and animal models of inflammatory bowel disease--an update on the role of the hypothalamo-pituitary-adrenal axis. Psychoneuroendocrinology. 2012;37:1–19. doi: 10.1016/j.psyneuen.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Goodhand J.R., Wahed M., Mawdsley J.E., Farmer A.D., Aziz Q., Rampton D.S. Mood disorders in inflammatory bowel disease: relation to diagnosis, disease activity, perceived stress, and other factors. Inflamm Bowel Dis. 2012;18:2301–2309. doi: 10.1002/ibd.22916. [DOI] [PubMed] [Google Scholar]
- 11.Juruena M.F. Early-life stress and HPA axis trigger recurrent adulthood depression. Epilepsy Behav. 2014;38:148–159. doi: 10.1016/j.yebeh.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 12.Sun Y., Li L., Xie R., Wang B., Jiang K., Cao H. Stress triggers flare of inflammatory bowel disease in children and adults. Front Pediatr. 2019;7:432. doi: 10.3389/fped.2019.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herman J.P., McKlveen J.M., Ghosal S., Kopp B., Wulsin A., Makinson R., Scheimann J., Myers B. Regulation of the hypothalamic-pituitary-adrenocortical stress response. Compr Physiol. 2016;6:603–621. doi: 10.1002/cphy.c150015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dedovic K., Ngiam J. The cortisol awakening response and major depression: examining the evidence. Neuropsychiatr Dis Treat. 2015;11:1181–1189. doi: 10.2147/NDT.S62289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bao A.-M., Swaab D.F. The human hypothalamus in mood disorders: the HPA axis in the center. IBRO Rep. 2019;6:45–53. doi: 10.1016/j.ibror.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abautret-Daly Á., Dempsey E., Parra-Blanco A., Medina C., Harkin A. Gut-brain actions underlying comorbid anxiety and depression associated with inflammatory bowel disease. Acta Neuropsychiatr. 2018;30:275–296. doi: 10.1017/neu.2017.3. [DOI] [PubMed] [Google Scholar]
- 17.Do J., Woo J. From gut to brain: alteration in inflammation markers in the brain of dextran sodium sulfate-induced colitis model mice. Clin Psychopharmacol Neurosci. 2018;16:422–433. doi: 10.9758/cpn.2018.16.4.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., Ripke S., Day F.R., Consortium ReproGen, Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium. Purcell S., Stahl E., Lindstrom S., Perry J.R.B., Okada Y., Raychaudhuri S., Daly M.J., Patterson N., Neale B.M., Price A.L. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard D.M., Adams M.J., Clarke T.-K., Hafferty J.D., Gibson J., Shirali M., Coleman J.R.I., Hagenaars S.P., Ward J., Wigmore E.M., Alloza C., Shen X., Barbu M.C., Xu E.Y., Whalley H.C., Marioni R.E., Porteous D.J., Davies G., Deary I.J., Hemani G., Berger K., Teismann H., Rawal R., Arolt V., Baune B.T., Dannlowski U., Domschke K., Tian C., Hinds D.A., Trzaskowski M., Byrne E.M., Ripke S., Smith D.J., Sullivan P.F., Wray N.R., Breen G., Lewis C.M., McIntosh A.M. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22:343–352. doi: 10.1038/s41593-018-0326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J., Erzurumluoglu A.M., Elsworth B.L., Kemp J.P., Howe L., Haycock P.C., Hemani G., Tansey K., Laurin C., Pourcain B.S., Warrington N.M., Finucane H.K., Price A.L., Bulik-Sullivan B.K., Anttila V., Paternoster L., Gaunt T.R., Evans D.M., Neale B.M. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33:272–279. doi: 10.1093/bioinformatics/btw613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubois P.C.A., Trynka G., Franke L., Hunt K.A., Romanos J., Curtotti A., Zhernakova A., Heap G.A.R., Adány R., Aromaa A., Bardella M.T., van den Berg L.H., Bockett N.A., de la Concha E.G., Dema B., Fehrmann R.S.N., Fernández-Arquero M., Fiatal S., Grandone E., Green P.M., Groen H.J.M., Gwilliam R., Houwen R.H.J., Hunt S.E., Kaukinen K., Kelleher D., Korponay-Szabo I., Kurppa K., MacMathuna P., Mäki M., Mazzilli M.C., McCann O.T., Mearin M.L., Mein C.A., Mirza M.M., Mistry V., Mora B., Morley K.I., Mulder C.J., Murray J.A., Núñez C., Oosterom E., Ophoff R.A., Polanco I., Peltonen L., Platteel M., Rybak A., Salomaa V., Schweizer J.J., Sperandeo M.P., Tack G.J., Turner G., Veldink J.H., Verbeek W.H.M., Weersma R.K., Wolters V.M., Urcelay E., Cukrowska B., Greco L., Neuhausen S.L., McManus R., Barisani D., Deloukas P., Barrett J.C., Saavalainen P., Wijmenga C., van Heel D.A. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attalla M.G., Singh S.B., Khalid R., Umair M., Epenge E. Relationship between ulcerative colitis and rheumatoid arthritis: a review. Cureus. 2019;11:e5695. doi: 10.7759/cureus.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pahl M.C., Doege C.A., Hodge K.M., Littleton S.H., Leonard M.E., Lu S., Rausch R., Pippin J.A., Bradfield J.P., Hammond R.K., Boehm K., Berkowitz R.I., Lasconi C., Su C., Chesi A., Johnson M., Wells A.D., Voight B.F., Leibel R.L., Cousminer D.L., Grant S.F.A. Cis-regulatory architecture of human ESC-derived hypothalamic neuron differentiation aids in variant-to-gene mapping of relevant common complex traits. bioRxiv. 2020;2020:146951. doi: 10.1038/s41467-021-27001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chesi A., Wagley Y., Johnson M.E., Manduchi E., Su C., Lu S., Leonard M.E., Hodge K.M., Pippin J.A., Hankenson K.D., Wells A.D., Grant S.F.A. Genome-scale Capture C promoter interactions implicate effector genes at GWAS loci for bone mineral density. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahe M.M., Sundaram N., Watson C.L., Shroyer N.F., Helmrath M.A. Establishment of human epithelial enteroids and colonoids from whole tissue and biopsy. J Vis Exp. 2015;97:52483. doi: 10.3791/52483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato T., Stange D.E., Ferrante M., Vries R.G.J., Van Es J.H., Van den Brink S., Van Houdt W.J., Pronk A., Van Gorp J., Siersema P.D., Clevers H. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 27.VanDussen K.L., Marinshaw J.M., Shaikh N., Miyoshi H., Moon C., Tarr P.I., Ciorba M.A., Stappenbeck T.S. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–920. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cairns J., Freire-Pritchett P., Wingett S.W., Várnai C., Dimond A., Plagnol V., Zerbino D., Schoenfelder S., Javierre B.-M., Osborne C., Fraser P., Spivakov M. CHiCAGO: robust detection of DNA looping interactions in Capture Hi-C data. Genome Biol. 2016;17:127. doi: 10.1186/s13059-016-0992-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J.C., Biasci D., Roberts R., Gearry R.B., Mansfield J.C., Ahmad T., Prescott N.J., Satsangi J., Wilson D.C., Jostins L., Anderson C.A., UK IBD Genetics Consortium. Traherne J.A., Lyons P.A., Parkes M., Smith K.G.C. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet. 2017;49:262–268. doi: 10.1038/ng.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.-G., Heap G., Nimmo E.R., Edwards C., Henderson P., Mowat C., Sanderson J., Satsangi J., Simmons A., Wilson D.C., Tremelling M., Hart A., Mathew C.G., Newman W.G., Parkes M., Lees C.W., Uhlig H., Hawkey C., Prescott N.J., Ahmad T., Mansfield J.C., Anderson C.A., Barrett J.C. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Aguilera G. Cyclic AMP inducible early repressor mediates the termination of corticotropin releasing hormone transcription in hypothalamic neurons. Cell Mol Neurobiol. 2009;29:1275–1281. doi: 10.1007/s10571-009-9423-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alpár A., Zahola P., Hanics J., Hevesi Z., Korchynska S., Benevento M., Pifl C., Zachar G., Perugini J., Severi I., Leitgeb P., Bakker J., Miklosi A.G., Tretiakov E., Keimpema E., Arque G., Tasan R.O., Sperk G., Malenczyk K., Máté Z., Erdélyi F., Szabó G., Lubec G., Palkovits M., Giordano A., Hökfelt T.G., Romanov R.A., Horvath T.L., Harkany T. Hypothalamic CNTF volume transmission shapes cortical noradrenergic excitability upon acute stress. EMBO J. 2018;37 doi: 10.15252/embj.2018100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis T.C., Gaynor A., Chandra R., Fox M.E., Lobo M.K. The selective RhoA inhibitor rhosin promotes stress resiliency through enhancing D1-medium spiny neuron plasticity and reducing hyperexcitability. Biol Psychiatry. 2019;85:1001–1010. doi: 10.1016/j.biopsych.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fox M.E., Chandra R., Menken M.S., Larkin E.J., Nam H., Engeln M., Francis T.C., Lobo M.K. Dendritic remodeling of D1 neurons by RhoA/Rho-kinase mediates depression-like behavior. Mol Psychiatry. 2020;25:1022–1034. doi: 10.1038/s41380-018-0211-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skov L.J., Ratner C., Hansen N.W., Thompson J.J., Egerod K.L., Burm H., Dalbøge L.S., Hedegaard M.A., Brakebusch C., Pers T.H., Perrier J.-F., Holst B. RhoA in tyrosine hydroxylase neurones regulates food intake and body weight via altered sensitivity to peripheral hormones. J Neuroendocrinol. 2019;31 doi: 10.1111/jne.12761. [DOI] [PubMed] [Google Scholar]
- 36.Newman E.A., Kim D.W., Wan J., Wang J., Qian J., Blackshaw S. Foxd1 is required for terminal differentiation of anterior hypothalamic neuronal subtypes. Dev Biol. 2018;439:102–111. doi: 10.1016/j.ydbio.2018.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gumbel J.H., Patterson E.M., Owusu S.A., Kabat B.E., Jung D.O., Simmons J., Hopkins T., Ellsworth B.S. The forkhead transcription factor, Foxd1, is necessary for pituitary luteinizing hormone expression in mice. PLoS One. 2012;7 doi: 10.1371/journal.pone.0052156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez Castro C., Carbia Nagashima A., Páez Pereda M., Goldberg V., Chervin A., Carrizo G., Molina H., Renner U., Stalla G.K., Arzt E. Effects of the gp130 cytokines ciliary neurotropic factor (CNTF) and interleukin-11 on pituitary cells: CNTF receptors on human pituitary adenomas and stimulation of prolactin and GH secretion in normal rat anterior pituitary aggregate cultures. J Endocrinol. 2001;169:539–547. doi: 10.1677/joe.0.1690539. [DOI] [PubMed] [Google Scholar]
- 39.Brue T., Quentien M.-H., Khetchoumian K., Bensa M., Capo-Chichi J.-M., Delemer B., Balsalobre A., Nassif C., Papadimitriou D.T., Pagnier A., Hasselmann C., Patry L., Schwartzentruber J., Souchon P.-F., Takayasu S., Enjalbert A., Van Vliet G., Majewski J., Drouin J., Samuels M.E. Mutations in NFKB2 and potential genetic heterogeneity in patients with DAVID syndrome, having variable endocrine and immune deficiencies. BMC Med Genet. 2014;15:139. doi: 10.1186/s12881-014-0139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klemann C., Camacho-Ordonez N., Yang L., Eskandarian Z., Rojas-Restrepo J.L., Frede N., Bulashevska A., Heeg M., Al-Ddafari M.S., Premm J., Seidl M., Ammann S., Sherkat R., Radhakrishnan N., Warnatz K., Unger S., Kobbe R., Hüfner A., Leahy T.R., Ip W., Burns S.O., Fliegauf M., Grimbacher B. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. 2019;10:297. doi: 10.3389/fimmu.2019.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee S.H., Shin S.M., Zhong P., Kim H.-T., Kim D.-I., Kim J.M., Heo W.D., Kim D.-W., Yeo C.-Y., Kim C.-H., Liu Q. Reciprocal control of excitatory synapse numbers by Wnt and Wnt inhibitor PRR7 secreted on exosomes. Nat Commun. 2018;9:3434. doi: 10.1038/s41467-018-05858-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Helfer G., Tups A. Hypothalamic Wnt signalling and its role in energy balance regulation. J Neuroendocrinol. 2016;28:12368. doi: 10.1111/jne.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khodagholi F., Shaerzadeh F., Montazeri F. Mitochondrial aconitase in neurodegenerative disorders: role of a metabolism-related molecule in neurodegeneration. Curr Drug Targets. 2018;19:973–985. doi: 10.2174/1389450118666170816124203. [DOI] [PubMed] [Google Scholar]
- 44.Mutemberezi V., Buisseret B., Masquelier J., Guillemot-Legris O., Alhouayek M., Muccioli G.G. Oxysterol levels and metabolism in the course of neuroinflammation: insights from in vitro and in vivo models. J Neuroinflammation. 2018;15:74. doi: 10.1186/s12974-018-1114-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y., Zhou D., Ren Y., Zhang Z., Guo X., Ma M., Xue Z., Lv J., Liu H., Xi Q., Jia L., Zhang L., Liu Y., Zhang Q., Yan J., Da Y., Gao F., Yue J., Yao Z., Zhang R. Mir223 restrains autophagy and promotes CNS inflammation by targeting ATG16L1. Autophagy. 2019;15:478–492. doi: 10.1080/15548627.2018.1522467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brain O., Cooney R., Simmons A., Jewell D. Functional consequences of mutations in the autophagy genes in the pathogenesis of Crohn’s disease. Inflamm Bowel Dis. 2012;18:778–781. doi: 10.1002/ibd.21832. [DOI] [PubMed] [Google Scholar]
- 47.Sleiman P.M.A., Flory J., Imielinski M., Bradfield J.P., Annaiah K., Willis-Owen S.A.G., Wang K., Rafaels N.M., Michel S., Bonnelykke K., Zhang H., Kim C.E., Frackelton E.C., Glessner J.T., Hou C., Otieno F.G., Santa E., Thomas K., Smith R.M., Glaberson W.R., Garris M., Chiavacci R.M., Beaty T.H., Ruczinski I., Orange J.S., Orange J.M., Allen J., Spergel J.M., Grundmeier R., Mathias R.A., Christie J.D., von Mutius E., Cookson W.O.C., Kabesch M., Moffatt M.F., Grunstein M.M., Barnes K.C., Devoto M., Magnusson M., Li H., Grant S.F.A., Bisgaard H., Hakonarson H. Variants of DENND1B associated with asthma in children. N Engl J Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 48.Priftis K.N., Papadimitriou A., Nicolaidou P., Chrousos G.P. Dysregulation of the stress response in asthmatic children. Allergy. 2009;64:18–31. doi: 10.1111/j.1398-9995.2008.01948.x. [DOI] [PubMed] [Google Scholar]
- 49.Akcan N., Bahceciler N.N. Headliner in physiology and management of childhood asthma: hypothalamic-pituitary-adrenal axis. Curr Pediatr Rev. 2020;16:43–52. doi: 10.2174/1573396315666191026100643. [DOI] [PubMed] [Google Scholar]
- 50.Lin L., Peng S.L. Coordination of NF-kappaB and NFAT antagonism by the forkhead transcription factor Foxd1. J Immunol 1950. 2006;176:4793–4803. doi: 10.4049/jimmunol.176.8.4793. [DOI] [PubMed] [Google Scholar]
- 51.Hrdinka M., Dráber P., Stepánek O., Ormsby T., Otáhal P., Angelisová P., Brdicka T., Paces J., Horejsí V., Drbal K. PRR7 is a transmembrane adaptor protein expressed in activated T cells involved in regulation of T cell receptor signaling and apoptosis. J Biol Chem. 2011;286:19617–19629. doi: 10.1074/jbc.M110.175117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Togni M., Swanson K.D., Reimann S., Kliche S., Pearce A.C., Simeoni L., Reinhold D., Wienands J., Neel B.G., Schraven B., Gerber A. Regulation of in vitro and in vivo immune functions by the cytosolic adaptor protein SKAP-HOM. Mol Cell Biol. 2005;25:8052–8063. doi: 10.1128/MCB.25.18.8052-8063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z., Yang X., Liu C., Li X., Zhang B., Wang B., Zhang Y., Song C., Zhang T., Liu M., Liu B., Ren M., Jiang H., Zou J., Liu X., Zhang H., Zhu W.-G., Yin Y., Zhang Z., Gu W., Luo J. Acetylation of PHF5A modulates stress responses and colorectal carcinogenesis through alternative splicing-mediated upregulation of KDM3A. Mol Cell. 2019;74:1250–1263.e6. doi: 10.1016/j.molcel.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 54.Kamburov A., Stelzl U., Lehrach H., Herwig R. The ConsensusPathDB interaction database: 2013 update. Nucleic Acids Res. 2013;41:D793–D800. doi: 10.1093/nar/gks1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gracie D.J., Williams C.J.M., Sood R., Mumtaz S., Bholah M.H., Hamlin P.J., Ford A.C. Poor correlation between clinical disease activity and mucosal inflammation, and the role of psychological comorbidity, in inflammatory bowel disease. Am J Gastroenterol. 2016;111:541–551. doi: 10.1038/ajg.2016.59. [DOI] [PubMed] [Google Scholar]
- 56.Schijven D., Geuze E., Vinkers C.H., Pulit S.L., Schür R.R., Malgaz M., Bekema E., Medic J., van der Kust K.E., Veldink J.H., Boks M.P., Vermetten E., Luykx J.J. Multivariate genome-wide analysis of stress-related quantitative phenotypes. Eur Neuropsychopharmacol. 2019;29:1354–1364. doi: 10.1016/j.euroneuro.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 57.Stein M.B., Chen C.-Y., Ursano R.J., Cai T., Gelernter J., Heeringa S.G., Jain S., Jensen K.P., Maihofer A.X., Mitchell C., Nievergelt C.M., Nock M.K., Neale B.M., Polimanti R., Ripke S., Sun X., Thomas M.L., Wang Q., Ware E.B., Borja S., Kessler R.C., Smoller J.W. Army Study to Assess Risk and Resilience in Servicemembers (STARRS) Collaborators. Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of US Army Soldiers. JAMA Psychiatry. 2016;73:695–704. doi: 10.1001/jamapsychiatry.2016.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke T.-K., Zeng Y., Navrady L., Xia C., Haley C., Campbell A., Navarro P., Amador C., Adams M.J., Howard D.M., Soler A., Hayward C., Thomson P.A., Smith B.H., Padmanabhan S., Hocking L.J., Hall L.S., Porteous D.J., Deary I.J., McIntosh A.M. Genetic and environmental determinants of stressful life events and their overlap with depression and neuroticism. Wellcome Open Res. 2019;3:11. doi: 10.12688/wellcomeopenres.13893.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burfeind K.G., Yadav V., Marks D.L. Hypothalamic dysfunction and multiple sclerosis: implications for fatigue and weight dysregulation. Curr Neurol Neurosci Rep. 2016;16:98. doi: 10.1007/s11910-016-0700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huitinga I., De Groot C.J., Van der Valk P., Kamphorst W., Tilders F.J., Swaab D.F. Hypothalamic lesions in multiple sclerosis. J Neuropathol Exp Neurol. 2001;60:1208–1218. doi: 10.1093/jnen/60.12.1208. [DOI] [PubMed] [Google Scholar]
- 61.Kantorová E., Poláček H., Bittšanský M., Baranovičová E., Hnilicová P., Čierny D., Sivák Š., Nosáľ V., Zeleňák K., Kurča E. Hypothalamic damage in multiple sclerosis correlates with disease activity, disability, depression, and fatigue. Neurol Res. 2017;39:323–330. doi: 10.1080/01616412.2016.1275460. [DOI] [PubMed] [Google Scholar]
- 62.Preziosi G., Gordon-Dixon A., Emmanuel A. Neurogenic bowel dysfunction in patients with multiple sclerosis: prevalence, impact, and management strategies. Degener Neurol Neuromuscul Dis. 2018;8:79–90. doi: 10.2147/DNND.S138835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camara-Lemarroy C.R., Metz L., Meddings J.B., Sharkey K.A., Wee Yong V. The intestinal barrier in multiple sclerosis: implications for pathophysiology and therapeutics. Brain. 2018;141:1900–1916. doi: 10.1093/brain/awy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirby T.O., Ochoa-Repáraz J. The gut microbiome in multiple sclerosis: a potential therapeutic avenue. Med Sci (Basel) 2018;6:69. doi: 10.3390/medsci6030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swindle E.J., Collins J.E., Davies D.E. Breakdown in epithelial barrier function in patients with asthma: identification of novel therapeutic approaches. J Allergy Clin Immunol. 2009;124:23–34. doi: 10.1016/j.jaci.2009.05.037. [DOI] [PubMed] [Google Scholar]
- 66.Frati F., Salvatori C., Incorvaia C., Bellucci A., Di Cara G., Marcucci F., Esposito S. The role of the microbiome in asthma: the gut–lung axis. Int J Mol Sci. 2018;20:123. doi: 10.3390/ijms20010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Samadi N., Klems M., Untersmayr E. The role of gastrointestinal permeability in food allergy. Ann Allergy Asthma Immunol. 2018;121:168–173. doi: 10.1016/j.anai.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 68.Molina C.A., Foulkes N.S., Lalli E., Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 69.Aguilera G., Kiss A., Liu Y., Kamitakahara A. Negative regulation of corticotropin releasing factor expression and limitation of stress response. Stress. 2007;10:153–161. doi: 10.1080/10253890701391192. [DOI] [PubMed] [Google Scholar]
- 70.Mackner L., Pajer K., Glaser R., Crandall W. S2022 relationships between depression, cytokines, and cortisol in pediatric inflammatory bowel disease. Gastroenterology. 2010;138:S2022. [Google Scholar]
- 71.Million M., Taché Y., Anton P. Susceptibility of Lewis and Fischer rats to stress-induced worsening of TNB-colitis: protective role of brain CRF. Am J Physiol. 1999;276:G1027–G1036. doi: 10.1152/ajpgi.1999.276.4.G1027. [DOI] [PubMed] [Google Scholar]
- 72.Souza PR de, Sales-Campos H., Basso P.J., Nardini V., Silva A., Banquieri F., Alves V.B.F., Chica J.E.L., Nomizo A., Cardoso CR. de B. Adrenal-derived hormones differentially modulate intestinal immunity in experimental colitis. Mediat Inflamm. 2016;2016 doi: 10.1155/2016/4936370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mittal R., Debs L.H., Patel A.P., Nguyen D., Patel K., O’Connor G., Grati M., Mittal J., Yan D., Eshraghi A.A., Deo S.K., Daunert S., Liu X.Z. Neurotransmitters: the critical modulators regulating gut-brain axis. J Cell Physiol. 2017;232:2359–2372. doi: 10.1002/jcp.25518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nishida A., Inoue R., Inatomi O., Bamba S., Naito Y., Andoh A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin J Gastroenterol. 2018;11:1–10. doi: 10.1007/s12328-017-0813-5. [DOI] [PubMed] [Google Scholar]
- 75.Yuksel M., Ates I., Kaplan M., Arikan M.F., Ozin Y.O., Kilic Z.M.Y., Topcuoglu C., Kayacetin E. Is oxidative stress associated with activation and pathogenesis of inflammatory bowel disease? J Med Biochem. 2017;36:341–348. doi: 10.1515/jomb-2017-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., Heinzmann A., Simma B., Frischer T., Willis-Owen S.A.G., Wong K.C.C., Illig T., Vogelberg C., Weiland S.K., von Mutius E., Abecasis G.R., Farrall M., Gut I.G., Lathrop G.M., Cookson W.O.C. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 77.Ji S.-G., Juran B.D., Mucha S., Folseraas T., Jostins L., Melum E., Kumasaka N., Atkinson E.J., Schlicht E.M., Liu J.Z., Shah T., Gutierrez-Achury J., Boberg K.M., Bergquist A., Vermeire S., Eksteen B., Durie P.R., Farkkila M., Müller T., Schramm C., Sterneck M., Weismüller T.J., Gotthardt D.N., Ellinghaus D., Braun F., Teufel A., Laudes M., Lieb W., Jacobs G., Beuers U., Weersma R.K., Wijmenga C., Marschall H.-U., Milkiewicz P., Pares A., Kontula K., Chazouillères O., Invernizzi P., Goode E., Spiess K., Moore C., Sambrook J., Ouwehand W.H., Roberts D.J., Danesh J., Floreani A., Gulamhusein A.F., Eaton J.E., Schreiber S., Coltescu C., Bowlus C.L., Luketic V.A., Odin J.A., Chopra K.B., Kowdley K.V., Chalasani N., Manns M.P., Srivastava B., Mells G., Sandford R.N., Alexander G., Gaffney D.J., Chapman R.W., Hirschfield G.M., de Andrade M., Rushbrook S.M., Franke A., Karlsen T.H., Lazaridis K.N., Anderson C.A. Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. 2017;49:269–273. doi: 10.1038/ng.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., Kochi Y., Ohmura K., Suzuki A., Yoshida S., Graham R.R., Manoharan A., Ortmann W., Bhangale T., Denny J.C., Carroll R.J., Eyler A.E., Greenberg J.D., Kremer J.M., Pappas D.A., Jiang L., Yin J., Ye L., Su D.-F., Yang J., Xie G., Keystone E., Westra H.-J., Esko T., Metspalu A., Zhou X., Gupta N., Mirel D., Stahl E.A., Diogo D., Cui J., Liao K., Guo M.H., Myouzen K., Kawaguchi T., Coenen M.J.H., van Riel P.L.C.M., van de Laar M.A.F.J., Guchelaar H.-J., Huizinga T.W.J., Dieudé P., Mariette X., Bridges S.L., Zhernakova A., Toes R.E.M., Tak P.P., Miceli-Richard C., Bang S.-Y., Lee H.-S., Martin J., Gonzalez-Gay M.A., Rodriguez-Rodriguez L., Rantapää-Dahlqvist S., Arlestig L., Choi H.K., Kamatani Y., Galan P., Lathrop M., RACI consortium. GARNET consortium. Eyre S., Bowes J., Barton A., de Vries N., Moreland L.W., Criswell L.A., Karlson E.W., Taniguchi A., Yamada R., Kubo M., Liu J.S., Bae S.-C., Worthington J., Padyukov L., Klareskog L., Gregersen P.K., Raychaudhuri S., Stranger B.E., De Jager P.L., Franke L., Visscher P.M., Brown M.A., Yamanaka H., Mimori T., Takahashi A., Xu H., Behrens T.W., Siminovitch K.A., Momohara S., Matsuda F., Yamamoto K., Plenge R.M. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506:376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cordell H.J., Han Y., Mells G.F., Li Y., Hirschfield G.M., Greene C.S., Xie G., Juran B.D., Zhu D., Qian D.C., Floyd J.A.B., Morley K.I., Prati D., Lleo A., Cusi D., Canadian-US PBC Consortium. Italian PBC Genetics Study Group, UK-PBC Consortium. Gershwin M.E., Anderson C.A., Lazaridis K.N., Invernizzi P., Seldin M.F., Sandford R.N., Amos C.I., Siminovitch K.A. International genome-wide meta-analysis identifies new primary biliary cirrhosis risk loci and targetable pathogenic pathways. Nat Commun. 2015;6:8019. doi: 10.1038/ncomms9019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bentham J., Morris D.L., Graham D.S.C., Pinder C.L., Tombleson P., Behrens T.W., Martín J., Fairfax B.P., Knight J.C., Chen L., Replogle J., Syvänen A.-C., Rönnblom L., Graham R.R., Wither J.E., Rioux J.D., Alarcón-Riquelme M.E., Vyse T.J. Genetic association analyses implicate aberrant regulation of innate and adaptive immunity genes in the pathogenesis of systemic lupus erythematosus. Nat Genet. 2015;47:1457–1464. doi: 10.1038/ng.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paternoster L., Standl M., Waage J., Baurecht H., Hotze M., Strachan D.P., Curtin J.A., Bønnelykke K., Tian C., Takahashi A., Esparza-Gordillo J., Alves A.C., Thyssen J.P., den Dekker H.T., Ferreira M.A., Altmaier E., Sleiman P.M., Xiao F.L., Gonzalez J.R., Marenholz I., Kalb B., Yanes M.P., Xu C.-J., Carstensen L., Groen-Blokhuis M.M., Venturini C., Pennell C.E., Barton S.J., Levin A.M., Curjuric I., Bustamante M., Kreiner-Møller E., Lockett G.A., Bacelis J., Bunyavanich S., Myers R.A., Matanovic A., Kumar A., Tung J.Y., Hirota T., Kubo M., McArdle W.L., Henderson A.J., Kemp J.P., Zheng J., Smith G.D., Rüschendorf F., Bauerfeind A., Lee-Kirsch M.A., Arnold A., Homuth G., Schmidt C.O., Mangold E., Cichon S., Keil T., Rodríguez E., Peters A., Franke A., Lieb W., Novak N., Fölster-Holst R., Horikoshi M., Pekkanen J., Sebert S., Husemoen L.L., Grarup N., de Jongste J.C., Rivadeneira F., Hofman A., Jaddoe V.W., Pasmans S.G., Elbert N.J., Uitterlinden A.G., Marks G.B., Thompson P.J., Matheson M.C., Robertson C.F., Australian Asthma Genetics Consortium (AAGC) Ried J.S., Li J., Zuo X.B., Zheng X.D., Yin X.Y., Sun L.D., McAleer M.A., O’Regan G.M., Fahy C.M., Campbell L.E., Macek M., Kurek M., Hu D., Eng C., Postma D.S., Feenstra B., Geller F., Hottenga J.J., Middeldorp C.M., Hysi P., Bataille V., Spector T., Tiesler C.M., Thiering E., Pahukasahasram B., Yang J.J., Imboden M., Huntsman S., Vilor-Tejedor N., Relton C.L., Myhre R., Nystad W., Custovic A., Weiss S.T., Meyers D.A., Söderhäll C., Melén E., Ober C., Raby B.A., Simpson A., Jacobsson B., Holloway J.W., Bisgaard H., Sunyer J., Hensch N.M.P., Williams L.K., Godfrey K.M., Wang C.A., Boomsma D.I., Melbye M., Koppelman G.H., Jarvis D., McLean W.I., Irvine A.D., Zhang X.J., Hakonarson H., Gieger C., Burchard E.G., Martin N.G., Duijts L., Linneberg A., Jarvelin M.-R., Noethen M.M., Lau S., Hübner N., Lee Y.-A., Tamari M., Hinds D.A., Glass D., Brown S.J., Heinrich J., Evans D.M., Weidinger S. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L., Meece K., Williams D.J., Lo K.A., Zimmer M., Heinrich G., Martin Carli J., Leduc C.A., Sun L., Zeltser L.M., Freeby M., Goland R., Tsang S.H., Wardlaw S.L., Egli D., Leibel R.L. Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J Clin Invest. 2015;125:796–808. doi: 10.1172/JCI79220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Arnold M., Raffler J., Pfeufer A., Suhre K., Kastenmüller G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics. 2015;31:1334–1336. doi: 10.1093/bioinformatics/btu779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Borsook D., Smirnova O., Behar O., Lewis S., Kobierski L.A. PhosphoCREB and CREM/ICER: positive and negative regulation of proenkephalin gene expression in the paraventricular nucleus of the hypothalamus. J Mol Neurosci. 1999;12:35–51. doi: 10.1385/JMN:12:1:35. [DOI] [PubMed] [Google Scholar]
- 85.Schwartz W.J., Aronin N., Sassone-Corsi P. Photoinducible and rhythmic ICER–CREM immunoreactivity in the rat suprachiasmatic nucleus. Neurosci Lett. 2005;385:87–91. doi: 10.1016/j.neulet.2005.05.018. [DOI] [PubMed] [Google Scholar]
- 86.Conti A.C., Kuo Y.-C., Valentino R.J., Blendy J.A. Inducible cAMP early repressor regulates corticosterone suppression after tricyclic antidepressant treatment. J Neurosci. 2004;24:1967–1975. doi: 10.1523/JNEUROSCI.4804-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kokoeva M.V., Yin H., Flier J.S. Neurogenesis in the hypothalamus of adult mice: potential role in energy balance. Science. 2005;310:679–683. doi: 10.1126/science.1115360. [DOI] [PubMed] [Google Scholar]
- 88.Wen R., Tao W., Li Y., Sieving P.A. CNTF and retina. Prog Retin Eye Res. 2012;31:136–151. doi: 10.1016/j.preteyeres.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wanke F., Moos S., Croxford A.L., Heinen A.P., Gräf S., Kalt B., Tischner D., Zhang J., Christen I., Bruttger J., Yogev N., Tang Y., Zayoud M., Israel N., Karram K., Reißig S., Lacher S.M., Reichhold C., Mufazalov I.A., Ben-Nun A., Kuhlmann T., Wettschureck N., Sailer A.W., Rajewsky K., Casola S., Waisman A., Kurschus F.C. EBI2 is highly expressed in multiple sclerosis lesions and promotes early CNS migration of encephalitogenic CD4 T cells. Cell Rep. 2017;18:1270–1284. doi: 10.1016/j.celrep.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 90.Qiang Z., Jun-Jie L., Hai W., Hong L., Bing-Xi L., Lei C., Wei X., Ya-Wei L., Huang A., Song-Tao Q., Yun-Tao L. TPD52L2 impacts proliferation, invasiveness and apoptosis of glioblastoma cells via modulation of Wnt/β-catenin/snail signaling. Carcinogenesis. 2018;39:214–224. doi: 10.1093/carcin/bgx125. [DOI] [PubMed] [Google Scholar]
- 91.Gan J., Cai Q., Qu Y., Zhao F., Wan C., Luo R., Mu D. miR-96 attenuates status epilepticus-induced brain injury by directly targeting Atg7 and Atg16L1. Sci Rep. 2017;7:10270. doi: 10.1038/s41598-017-10619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cao J.P., Wang H.J., Yu J.K., Liu H.M., Gao D.S. The involvement of NF-kappaB p65/p52 in the effects of GDNF on DA neurons in early PD rats. Brain Res Bull. 2008;76:505–511. doi: 10.1016/j.brainresbull.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 93.Marelli C., Hamel C., Quiles M., Carlander B., Larrieu L., Delettre C., Sarzi E., Chretien D., Rustin P., Koenig M., Guissart C. ACO2 mutations: a novel phenotype associating severe optic atrophy and spastic paraplegia. Neurol Genet. 2018;4:e225. doi: 10.1212/NXG.0000000000000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Huang C., Su T., Xue Y., Cheng C., Lay F.D., McKee R.A., Li M., Vashisht A., Wohlschlegel J., Novitch B.G., Plath K., Kurdistani S.K., Carey M. Cbx3 maintains lineage specificity during neural differentiation. Genes Dev. 2017;31:241–246. doi: 10.1101/gad.292169.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao S.-P., Wang F., Yang M., Wang X.-Y., Jin C.-L., Ji Q.-K., Li S., Zhao X.-L. CBX3 promotes glioma U87 cell proliferation and predicts an unfavorable prognosis. J Neurooncol. 2019;145:35–48. doi: 10.1007/s11060-019-03286-w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.