Abstract
Pectobacterium versatile (formerly P. carotovorum) is a recently defined species of soft rot enterobacteria capable of infecting many plant hosts and damaging different tissues. Complex transcriptional regulation of virulence properties can be expected for such a versatile pathogen. However, the relevant information is available only for related species and is rather limited. The PhoPQ two-component system, originally described in pectobacteria as PehRS, was previously shown to regulate a single gene, pehA. Using an insertional phoP mutant of Pectobacterium versatile (earlier—P. carotovorum), we demonstrate that PhoP regulates at least 115 genes with a majority of them specific for pectobacteria. The functions performed by PhoP-controlled genes include degradation, transport and metabolism of plant-derived carbon sources (polygalacturonate, arabinose-containing polysaccharides and citrate), modification of bacterial cell envelope and stress resistance. We also demonstrated PhoP involvement in establishing the order of plant cell wall decomposition and utilisation of the corresponding breakdown products. Based on experimental data and in silico analysis, we defined a PhoP binding site motif and provided proof for its universality in enteric bacteria. Scanning P. versatile genome for the locations of this motif suggested a much larger PhoP regulon enriched with the genes important for a plant pathogen, which makes PhoP a global virulence regulator. Potential PhoP targets include many regulatory genes and PhoP control over one of them, expI, was confirmed experimentally, highlighting the link between the PhoPQ two-component and quorum sensing systems. High concentrations of calcium and magnesium ions were found to abolish the PhoPQ-dependent transcription activation but did not relieve repression. Reduced PhoP expression and minimisation of PhoP dependence of regulon members’ expression in P. versatile cells isolated from potato tuber tissues suggest that PhoPQ system is a key switch of expression levels of multiple virulence-related genes fine-tuned to control the development of P. versatile-host plant pathosystem.
Keywords: Pectobacterium versatile, PhoPQ, two-component system, pectin, arabinose, citrate, virulence
Introduction
Pectobacterium spp. are pectinolytic bacteria that cause soft rot and other diseases in a variety of plants. Until recently, members of this genus were often considered to be broad host range pathogens. However, genomic studies revealed significant diversity within this group, resulting in the ongoing subdivision of the previously described species and elevation of subspecies to species level. That resulted in better separation of the related strains according to their environmental preferences. Candidatus Pectobacterium maceratum was suggested as a new name for a group of strains isolated from potato and cabbage (Shirshikov et al., 2018). Recently, more isolates from various sources, including Chrysanthemum, Iris and water, were united with Ca. P. maceratum strains and renamed as Pectobacterium versatile (Portier et al., 2019). The P. versatile taxon includes two strains that have been well characterised previously, Ecc71 and SCC1. The 3-2 strain used in this work also belongs to this species.
P. versatile (Pve) strains can infect various plant hosts, and the same strain can cause diverse symptoms while infecting distinct tissues of the same plant (e.g., stem blackleg and tuber soft rot in potato) (Portier et al., 2019). Pectobacteria are also known to proliferate stealthily for a while in the vascular tissues before switching to a brute force attack of surrounding tissues via the massive secretion of plant cell wall hydrolases (Toth and Birch, 2005). Our previous work showed that initiation and development of the Pectobacterium-plant pathosystem requires extensive reprogramming of gene expression, both in the pathogen and its host (Gorshkov et al., 2018; Tsers et al., 2020). Such transcriptional reprogramming obviously involves multiple transcription factors. Yet, only a small fraction of about 300 transcription factors have been characterised in pectobacteria. These include pectinolysis and exoenzyme regulators KdgR (Liu et al., 1999), RexZ (Thomson et al., 1999), and GacA (Cui et al., 2001; Hyytiainen et al., 2001), the alternative sigma factor HrpL—the activator of the type III secretion system and its substrate genes (Chatterjee et al., 2002), motility regulators HexA (Harris et al., 1998), and FlhDC (Cui et al., 2008), pectin lyase regulators RdgA and RdgB (Liu et al., 1994) and quorum sensing regulators ExpR and VirR (Cui et al., 2005, 2006; Burr et al., 2006; Sjöblom et al., 2006).
PehR was originally described as a transcriptional activator of the pehA gene encoding endopolygalacturonase, a major virulence factor in soft rot bacteria (Flego et al., 2000). PehR is a response regulator that forms a two-component sensory system (TCS) with the membrane histidine kinase PehS. In the SCC3193 strain, which is currently classified as Pectobacterium parmentieri, inactivation of either pehR or pehS resulted in reduced virulence (Flego et al., 2000). The Ca2+ ion was reported as the PehS sensor ligand (Flego et al., 1997, 2000). It was also suggested that the PehRS system is responsible for the decrease of the polygalacturonase and increase of the pectate lyase activities in response to Ca2+ released from the degraded cell walls (Flego et al., 1997).
The pectobacterial PehRS system received very little attention since its discovery two decades ago. However, information has been accumulated about the orthologous PhoPQ TCS in other bacteria from the order Enterobacterales. In Dickeya dadantii, which belongs to the Pectobacteriaceae family together with Pve, PhoPQ was shown to control (Haque and Tsuyumu, 2005; Manjurul Haque et al., 2012) to cationic antimicrobial peptides (CAMPs) and expression of pectate lyases in response to changes of Mg2+ concentration (Haque and Tsuyumu, 2005; Manjurul Haque et al., 2012). The PhoPQ system has been thoroughly characterised in Salmonella and to a lesser extent in few more Enterobacteriaceae species including Escherichia coli and Yersinia spp. (see Groisman et al., 2013, for a review).
The PhoQ sensor histidine kinase activates PhoP by phosphorylation in response to several stimuli, including low Mg2+ concentration (Véscovi et al., 1996), cationic peptides (Bader et al., 2005), acidity (Prost et al., 2007) and high osmolarity (Yuan et al., 2017). PhoP binds to direct repeats with a consensus gGTTTA which seems to be well conserved in enterobacteria (Perez et al., 2009; Harari et al., 2010). The regulon composition nevertheless varies widely even between closely related bacteria. In Salmonella enterica, over a hundred genes are regulated by PhoP, but only three of them (phoP, phoQ, and slyB) are always under PhoP control in other enterobacteria. Some regulon members are shared by several species, but most PhoP controlled genes are species and even strain-specific (Perez et al., 2009). In summary, PhoP is a global regulator controlling diverse, but always large, regulons in Enterobacterales.
Despite the obvious importance of PhoP in Enterobacterales, its regulon has not been studied so far in Pectobacterium spp. To date, pehA remains the only known PhoP (PehR) target in these bacteria. The global mode of regulation reported for PhoP in other species strongly suggests that in pectobacteria it is likely to control many genes in addition to pehA. Moreover, in the recent work on P. atrosepticum transcriptome profiling in planta, we have noticed downregulation of phoPQ (Gorshkov et al., 2018). Expression level changes in planta combined with a potentially large regulon suggested that PhoP might be an important regulator of virulence properties in pectobacteria.
In this study we demonstrate that PhoP regulon of Pve includes at least 115 genes and incorporates a significant number of Pectobacterium-specific genes. We also try to distinguish the regulon parts directly and indirectly controlled by PhoP, discuss the implications of regulon composition for the regulation of Pectobacterium virulence and show an effect of divalent cations on the PhoP/PhoQ TCS.
Materials and Methods
Bacterial Strains and Growth Conditions
Pve strain JN42 (Myamin et al., 2004) is a spontaneous rifampicin-resistant derivative of the wild type isolate 3-2, originally described as Erwinia carotovora. The Escherichia coli strain XL-1 Blue (Bullock, 1987) was primarily used for plasmid construction and E. coli strain BW 19851 (Metcalf et al., 1994) was used for conjugational transfer of suicide vector pJP5603 (Penfold and Pemberton, 1992) derivatives into Pve. Pve and E. coli were routinely grown in lysogeny broth (LB) or minimal media at 28 and 37°C, respectively. For N-acyl-homoserine lactone bioassay, Chromobacterium violaceum CV026 was grown as described (McClean et al., 1997).
Two minimal media were used throughout this work. Minimal medium A (MMA) was composed of K2HPO4 (10.5 g/l), KH2PO4 (4.5 g/l), (NH4)2SO4 (1 g/l), sodium citrate (0.6 g/l), and 0.2% glycerol. MgSO4 was added to MMA to the final concentration of 0.5 mM. Sodium polypectate (Sigma) or L-arabinose were added to the final concentrations of 0.5 and 0.2% when necessary. To avoid precipitation, the effects of different divalent cation concentrations were studied in minimal medium N (MMN) (Nelson and Kennedy, 1971) containing KCl (5 mM), (NH4)2S04 (7.5 mM), K2S04 (0.5 mM), KH2PO4 (1 mM), Tris-HCl (0.1 M), pH 7.4, and 0.2% glycerol. MgSO4 and CaCl2 were added to MMN to either 10 μM or 10 mM as required.
Antibiotics were used at the following concentrations (μg/ml): ampicillin, 100; gentamycin, 10; rifampicin, 25; kanamycin, 20.
Construction of the phoP Mutant and Complementation Plasmid
To construct a phoP mutant of Pve, the phoP gene sequence was PCR amplified from Pve 3-2 with phoPf and phoPr primers (Supplementary Table 1) and its internal NdeI-PvuII fragment (225 bp) was cloned into the suicide vector pJP5603. The resulting plasmid was mobilised into Pve from E. coli BW 19851 and Pve crossover clones were selected on kanamycin (20 μg/ml) containing plates. Pve disruption was confirmed by PCR with combinations of primers to phoP and suicide vector sequences (phoPf-phoPr, phoPf-pjp2, and phoPr-pjp1, Supplementary Table 1).
The plasmid expressing phoP and phoQ was constructed by cloning the PCR fragment amplified with the Tersus DNA polymerase (Evrogen) and the phoPQf1 and phoPQr primers into the low copy number vector pZH449.
Characteristics of the plasmids used in this work are specified in Supplementary Table 2.
RNA-Seq Analysis
For RNA preparation from Pve 3-2 or phoP-mutant cultures, cells were aerobically grown at 28°C in MMA with sodium polypectate to an optical density at 600 nm (OD600) of 0.4. The details of RNA isolation, cDNA library preparation, sequencing and general characteristics of the RNA-seq data generated have been described (Gogoleva et al., 2020). 84.2 million reads were generated in total with four biological replicates sequenced for each type of the libraries (wild type and phoP mutant). The obtained read sequences corresponding to the Pve genome can be accessed from NCBI’s BioProject under the accession number PRJNA627079.
For quantification of gene expression levels, the coding sequences of Pve 3-2 genome were used as a reference (GenBank accession CP024842). Read pseudo-alignment and transcript quantification was performed using the alignment-free kallisto tool (Bray et al., 2016). The analysis of the differentially expressed genes (DEGs) was carried out with the edgeR package (Robinson et al., 2010). Genes with fold-change ≥ 2 and significant differences in expression levels (FDR < 0.05) were considered as DEGs.
Gene Expression Analysis by qRT-PCR
The total RNA from Pve cells was extracted, treated with DNAse and quantified as described above. One microgram of RNA was used for cDNA synthesis using RevertAid reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer’s instructions. Two microliter of fivefold-diluted cDNA were used as the template for qPCR.
qPCR was performed using a SYBR Green I—containing master mix. Primers for target and reference genes (Supplementary Table 1) were designed using Primer3 software (Untergasser et al., 2012) and checked for specificity with Primer-Blast (Ye et al., 2012). PCR was performed under the following conditions: 95°C for 2 min, followed by 45 cycles at 94°C for 10 s and 60°C for 60 s. After that, melting curve analysis was performed in the temperature range of 60–90°C. The reactions were run and changes in fluorescence emission were detected using a DT-96 quantitative PCR system (DNA Technology, Russia). The amount of fluorescence was plotted as a function of the PCR cycle and converted to Ct values using RealTime_PCR Software (DNA Technology, Russia). The amplification efficiency for all primers was determined using a dilution series of genomic DNA. Additional controls included the omission of reverse transcriptase to measure the extent of residual genomic DNA contamination and template omission. The ffh and gyrA genes, the transcript levels of which were confirmed by the geNorm software (Vandesompele et al., 2002) to be stable under the applied experimental conditions (data not shown), were used for normalisation of the expression of the target genes. At least four biological replicates were performed for each measurement. Relative expression levels and error estimates were calculated using the REST 2009 v. 2.0.13 software (Pfaffl et al., 2002).
Transcription Factor Binding Site Inference and Correction of Genome Annotation
ChIPmunk (Kulakovskiy et al., 2010) was used for the inference of PhoP binding sites within the regulatory regions of the differentially expressed genes. The ChIPhorde version of the algorithm was run, followed by manual filtering of putative operators according to their scores and positions relative to transcription initiation sites. TFBS (Transcription factor binding site) positions in the context of RNA-seq coverage were visualised by SigmoID (Nikolaichik and Damienikan, 2016). Wig files with RNA-seq coverage data were generated by Rockhopper (McClure et al., 2013).
Inference of operator motifs for other TFs was done with the modified algorithm of Sahota and Stormo (2010) as implemented in SigmoID version 2.01. SigmoID was also used for scanning the Pve genome for operator sequences matching known or new motifs. Positions of critical residues required by this algorithm were determined by the interaction service of NPIDB (Zanegina et al., 2016). Alignment of the DNA binding domains was done with hmmalign from the HMMER3 package (Eddy, 2011).
Correction of annotation for PhoP controlled genes was done using search and editing capabilities of SigmoID (Nikolaichik and Damienikan, 2016). The updated annotation of the Pve 3-2 genome was submitted to GenBank under the same accession number.
Virulence Assays
Bacteria were grown overnight on solid LB plates, washed off with 0.85% NaCl solution, centrifuged briefly and resuspended in the same solution, after which cell suspension densities were adjusted to achieve the cell doses 5⋅105 (high inoculum dose) or 1⋅105 (low inoculum dose) per inoculation site. Potato (Solanum tuberosum) cv Krone grade E basic seed tubers were obtained from a local seed potato producer. The tubers were inoculated with 10 μl of cell suspensions using a 200 μl pipette tip close to stolon end of the tuber. The inoculation sites were wrapped in parafilm and tubers were kept in plastic bags for 48 h at 28°C.
Inoculum doses were chosen in preliminary experiments as follows. Minimal cell numbers sufficient for reliable disease development by the wild type strain were taken as low inoculum dose. Higher cell numbers that could stably produce different test results were taken as high inoculum dose.
For Chinese cabbage inoculation, cell suspensions were prepared the same way, but their cell densities were adjusted to the values 1⋅105 (high inoculum dose) or 1⋅103 (low inoculum dose) per inoculation site. Chinese cabbage leaves were soaked in the sodium hypochlorite solution (70–85 g/l of active chlorine) and then flushed with sterile deionised water. Rectangular sections were cut out from the base of each leaf. The obtained sections were soaked in the 96% ethanol and then flamed. The sections were inoculated with 10 μl of cell suspensions using a 200 μl pipette tip. The samples were placed in the sterile Petri dishes and incubated for 48 h at 28°C.
At least 10 tubers or leaf sections were used per each experimental condition. 0.85% NaCl solution was used for control inoculations.
Results
Inactivation of phoP Alters P. versatile Virulence
To examine the role of PhoP in P. versatile interaction with host plants, we compared the properties of the phoP mutant strain with its wild type parent and a complementation strain. Since phoP is the first gene of the phoPQ operon and a polar effect of phoP inactivation on the expression of phoQ was expected, complementation was achieved by a plasmid carrying the whole phoPQ operon together with the full regulatory region.
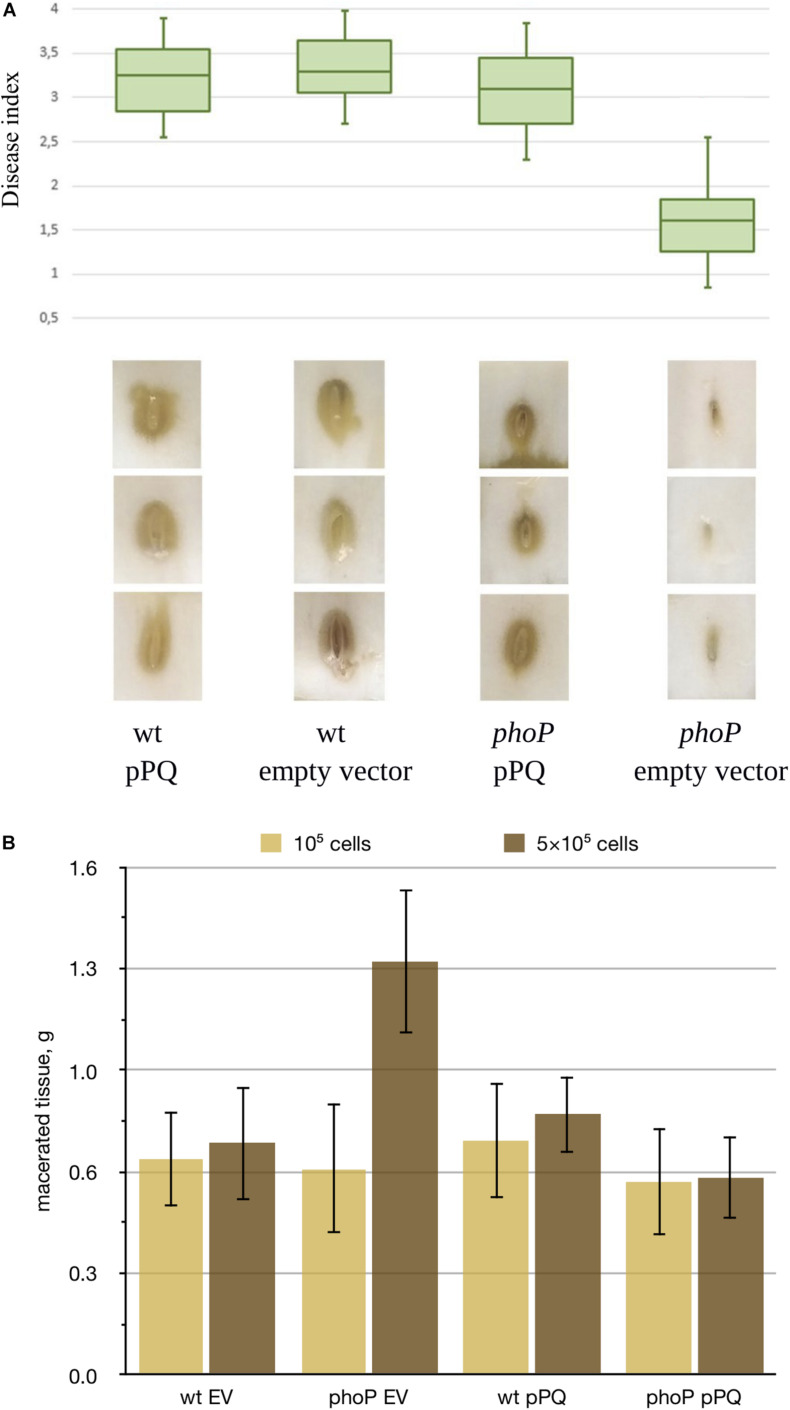
In Chinese cabbage virulence tests carried out at low inoculum doses (103 cells), the mutant strain caused less damage than the wild type strain, and observed virulence defect was fully complemented by the plasmid expressing phoPQ (Figures 1A,B). We could not detect reproducible differences between the strains at higher inoculum doses (105 cells) (data not shown). In potato tubers, we could reliably induce infection only starting with much higher inoculum dose (105 cells) but no difference between the strains was observed. However, the phoP mutant generated 50% more macerated tuber tissue with a dose of 5 × 105 cells (Figure 1B). In this case, the phoP mutant caused more tissue maceration than the wild type strain.
FIGURE 1.
Effect of phoP inactivation on Pve virulence. (A) Boxplot representation of disease symptoms induced in Chinese cabbage leaves by the wild type Pve strain (wt) and its phoP mutant (phoP) carrying either an empty vector (EV) or the same vector with cloned phoPQ operon (pPQ). Middle bar = median; box limit = upper and lower quartile; extremes = minimal and maximal values. Disease index indicates: 0 no symptoms; 1-2 tissue damage mostly localised near inoculation site; 3-4 extensive tissue damage spreading sideways from the inoculation site. A Kruskal-Wallis H-test showed a statistically significant difference in disease indices between the strains (p = 0.002). Three typical leaf sections for each variant are shown below the box plot. (B) Potato tubers inoculated with JN42 wild type (wt) or phoP mutant bacteria carrying either the empty vector pZH449 (EV) or pZH449:phoPQ (pPQ). Two inoculum doses were used: 5⋅105 or 1⋅105 cells per inoculation site. 16 tubers were inoculated for each strain/inoculum dose combination. Macerated tissue was weighed at 48 h post-inoculation. Mean values with 95% confidence intervals are shown.
As the growth rates of the phoP mutant and the wild type strain in MMA were almost the same (Supplementary Figure 1), the differences in disease development show involvement of the PhoPQ two-component system in virulence regulation in Pve. Since low and high inoculum doses somewhat mimic the early and late stages of infection, virulence tests results could indicate a switch in the state of the PhoPQ two-component system during infection.
PhoPQ System Controls Over a Hundred Genes in P. versatile
RNA-seq transcriptional profiling revealed 115 genes with expression level differing between the wild type and the phoP mutant strains. Most of the differentially expressed genes (DEGs) were activated, but 28 were repressed. Tables 1–4 and Supplementary Table 3 depict the genes, whose expression differed significantly (given a false discovery rate of 0.05) by at least twofold. The regulon composition and its control by PhoP are discussed in more detail in the following sections.
TABLE 1.
PhoP-dependent genes of Pve involved in cell envelope modifications, tellurite resistance and anaerobic metabolism.
| Gene name(s) | Locus_tag(s) | Fold changea | Product(s) | Best operator | Scoreb |
| Lipid A remodelling | |||||
| visP | OA04_00090 | 2.73 | Virulence- and stress-related protein | TATTTTTTCTGTTGTTTCA | 10.2 |
| – | OA04_20060 | 3.18 | Acyltransferase 3 | CCGTTTAGCAACCGTTTAG | 10.1 |
| lpxT | OA04_26310 | 4.33 | Lipid A 1-diphosphate synthase | TTTTTTCGTCTTTGTTTGG | 8.4 |
| pgpB | OA04_18890 | 5.59 | Phosphatidylglycerophosphatase B | TTTTTTGAACATGGTTTTT | 10.3 |
| ugd | OA04_31420 | 49.63 | UDP-glucose 6-dehydrogenase | ATGTTTAATTGGCGTTTAA | 10.2 |
| arnBCADTEF | OA04_31410-OA04_31350 | 78.53 | 4-amino-4-deoxy-L-arabinose transferase biosynthesis and lipid A modification | CCGTTTATCTGTAGTTTTA | 8.95 |
| Tellurite resistance | |||||
| terW | OA04_37880 | 6.92 | Putative transcriptional regulator of terZABCDE operon | TGGTTTATCTCTCGTTGAA | 9.8 |
| terZABCDE | OA04_37940-OA04_37990 | 0.16 | Tellurite resistance proteins | ATGTTTCATTAAGGTTTTT | 8.8 |
| – | OA04_10330 | 0.45 | Inner membrane protein, TerC-like | TCGTTTACGTTTCGTTTAA | 10.2 |
| OA04_37930 | OA04_37930 | 0.45 | Hypothetical protein | ATGTTTCATTAAGGTTTTT | 8.8 |
| Anaerobiosis-related genes | |||||
| focA | OA04_25450 | 2.31 | Formate transporter | AGTTTTATGGTTAATTTAT | 7.9 |
| aegA_OA04_45440 | OA04_45430-OA04_45440 | 5.43 | Oxidoreductase Fe-S binding subunit | TGTTTTTTGTCTGGTTTAC | 10.8 |
| hybO | OA04_12810 | 2.03 | Hydrogenase 2 small subunit | TGATTTAACTATAATTTAA | 10 |
| hypAB | OA04_12910-OA04_12900 | 6.85 | Hydrogenase nickel incorporation proteins HypA and HypB | TATTTTTCTACCTCTTTAT | 8.6 |
| hydN | OA04_13040 | 2.71 | Electron transport protein HydN | TTGTTTTTTAATTATTTTA | 11.7 |
| fdhF_2 | OA04_28980 | 2.36 | Formate dehydrogenase H | TGTTTTTGACGATATTAAC | 10.3 |
| nrdD | OA04_04300 | 2.19 | Anaerobic ribonucleoside triphosphate reductase | TATTTTGTCTTTTTTTTAG | 8.8 |
| norV | OA04_09050 | 2.55 | Anaerobic nitric oxide reductase flavorubredoxin | CAGTTTTTAATTTGTTGAT | 10.5 |
| fdnG | OA04_14900 | 5.55 | Formate dehydrogenase, nitrate-inducible, major subunit | ATGTTTTTTATTCGTTATA | 8.1 |
| ccmA | OA04_18220 | 5.88 | Cytochrome c biogenesis protein CcmA | CAATATATTTCCGGTTTAA | 7.7 |
| napF | OA04_18370 | 3.94 | Ferredoxin-type protein | TGTTTTTGTAGGGGTTACA | 5.8 |
| glpA | OA04_42210 | 3.14 | sn-glycerol-3-phosphate dehydrogenase subunit A | TCGTTTAGTGTTCGTTTTT | 8.6 |
| ubiU | OA04_06900 | 5.99 | Ubiquinone biosynthesis protein | TTTTTTATCAGTCGATGAA | 7.3 |
| pepT | OA04_23990 | 4.93 | Tripeptidase T | CAGTTTTTTCCCGATTTAA | 10.7 |
| grcA | OA04_33700 | 6.06 | Stress-induced alternative pyruvate formate-lyase subunit | GGTTTTAAAATTGATTTAA | 10.5 |
aWild type vs. phoP mutant ratio. For operons, the value for the first gene is shown. Supplementary Table 3 contains values for all genes.
bChIPmunk score for the best operator.
TABLE 4.
PhoP-dependent genes of Pve – regulators and unclassified.
| Gene name(s) | Locus_tag(s) | Fold changea | Product(s) | Best operator | Scoreb |
| Regulators | |||||
| glnK | OA04_12080 | 0.49 | Nitrogen regulatory protein P-II | – | – |
| - | OA04_01500 | 0.34 | LysR family transcriptional regulator | – | – |
| sftR | OA04_43750 | 2.34 | LysR family transcriptional regulator SftR | CGGTTTTATTATTTTTTCT | 9.4 |
| cbl | OA04_28870 | 2.71 | Transcriptional regulator CysB-like protein | CCATTTTGCTATGATTTAT | 10.8 |
| Miscellaneous | |||||
| scrK | OA04_04060 | 0.45 | Fructokinase | – | – |
| yjbJ | OA04_05950 | 2.0 | Osmotic stress-induced protein| RpoS regulon | – | – |
| ygdBppdD | OA04_10430-OA04_10440 | 3.58 | Putative pilins | – | – |
| ftp | OA04_17930 | 2.29 | Periplasmic FAD:protein FMN transferase | TTTTTTTCCTTTCATTTGT | 10.3 |
| slyB | OA04_18750 | 2.46 | Outer membrane lipoprotein | CTGTTTATACGCAATTTAA | 9.9 |
| ychH | OA04_21530 | 2.31 | Putative inner membrane stress-induced protein | AATATTTTTTTACGTTTAA | 6 |
| ynfK | OA04_22140 | 5.06 | Putative dithiobiotin synthetase | CATTTTAGCTGTGCTTGTT | 5.7 |
| – | OA04_23860 | 3.76 | hypothetical protein | GGGTATTAATCTTGTTTAA | 7.3 |
| – | OA04_24930 | 2.36 | Protein kinase-like domain protein | ATATTTGATCTGCGTTGAA | 5.4 |
| – | OA04_28020 | 2.03 | 2-dehydropantoate 2-reductase | – | – |
| – | OA04_29260 | 67.65 | Alpha/beta hydrolase superfamily protein | CTATTGAGCCGTGGTTTAA | 8.4 |
| – | OA04_29530 | 2.22 | Putative 2-methylcitrate dehydratase | CGTTTTATTATTTATTTAT | 13.1 |
| – | OA04_29750 | 2.91 | Putative acireductone dioxygenase | TTTTGTTATTTCGATTTAA | 8 |
| – | OA04_32550 | 3.02 | Exported choloylglycine hydrolase | TTGTTTATTTTAGGTTTAT | 13.8 |
| raiA | OA04_34460 | 2.18 | Stationary phase translation inhibitor and ribosome stability factor | CCGTTTTTTTTATGGTTAG | 7.3 |
| gudX | OA04_36650 | 4.2 | Glucarate dehydratase | ACTTTTTACTGAGGTTGGT | 7.5 |
| metF | OA04_43310 | 3.58 | 5,10-methylenetetrahydrofolate reductase | – | – |
aWild type vs. phoP mutant ratio. For operons, the value for the first gene is shown. Supplementary Table 3 contains values for all genes.
bChIPmunk score for the best operator.
TABLE 2.
PhoP-dependent genes of Pve involved in pectin and arabinose degradation and utilisation.
| Gene name(s) | Locus_tag(s) | Fold changea | Product(s) | Best operator | Scoreb |
| Polygalacturonic acid utilisation | |||||
| pelI | OA04_11360 | 0.14 | Pectate lyase PelI | AATTTTATTATTTATTTAT | 12.2 |
| pehA | OA04_11370 | 117.60 | Endo-polygalacturonase | AATTTTATTATTTATTTAT | 12.2 |
| pehN | OA04_12450 | 2.15 | Endo-polygalacturonase | ATTTTTTAGTGAGGTTAAG | 8.1 |
| pelP | OA04_21040 | 9.67 | Endo-pectate lyase | ATCTTTTCATTTTATTTAC | 7.5 |
| pnl | OA04_29050 | 2.63 | Pectin lyase | TTATTTGTTTTTGATTAAA | 9 |
| togT | OA04_08060 | 2.01 | Oligogalacturonide transporter | TATTTGATCTTGCGTTTAT | 8.2 |
| – | OA04_21060-OA04_21050 | 25.51 | KdgM-like porins | ATTTTTTGTAATCATTTCG | 8.9 |
| – | OA04_07070 | 2.00 | Polygalacturonic acid binding protein | – | – |
| – | OA04_12400 | 2.02 | Coagulation factor 5/8 type domain protein | TTATTTTAAAGCGCTTTAC | 7.8 |
| – | OA04_32560 | 2.04 | Coagulation factor 5/8 type domain protein | GTATTTTAAAGCGGTTTAC | 8.7 |
| Arabinose transport and utilisation | |||||
| araD | OA04_19040 | 0.08 | L-ribulose-5-phosphate 4-epimerase | CTGTTTTTTTAGCGTTTCT | 9.8 |
| araC | OA04_22270 | 0.11 | DNA-binding transcriptional regulator AraC | TATTTTATTTGCCATTTTG | 9.1 |
| araFGH | OA04_22300-OA04_22280 | 0.06 | L-arabinose transporter subunits | GGTTTGTGCATACATTTAG | 5.6 |
| araBA | OA04_22310-OA04_22320 | 0.02 | L-ribulokinase and L-arabinose isomerase | GGTTTGTGCATACATTTAG | 5.6 |
| – | OA04_38360-OA04_38370 | 0.19 | Sugar (Glycoside-Pentoside-Hexuronide):cation symporter and exo-α-1,5-L-arabinofuranosidase | ATTTTTGCAACCGATTTCA | 6.2 |
| ytfQRT | OA04_43130-OA04_43110 | 0.14 | Galactofuranose/arabinofuranose ABC transporter subunits | TGATTTTGTGCATATTTAC | 7.3 |
aWild type vs. phoP mutant ratio. For operons, the value for the first gene is shown. Supplementary Table 3 contains values for all genes.
bChIPmunk score for the best operator.
TABLE 3.
PhoP-dependent genes of Pve involved in transmembrane transport.
| Gene name(s) | Locus_tag(s) | Fold changea | Product(s) | Best operator | Scoreb |
| Iron import | |||||
| fetM | OA04_28960 | 2.16 | High affinity Fe2+ permease | GATTTTTACTGTTTTTTAG | 8.2 |
| yiuABC | OA04_33210-OA04_33230 | 8.36 | Ferric-enterobactin ABC-transporter subunits | CTGTATACGCATTGTTTAT | 9 |
| – | OA04_26190 | 9.81 | TonB-dependent receptor | TCGTTTAACTACGGTTTAT | 12.7 |
| entC | OA04_05590c | 4.09 | Enterobactin synthetase and transport proteins | TTTTTTTGTGCATGTTTCA | 7.7 |
| fusB | OA04_08790 | 2.29 | TonB-like protein | CATTTGTATTATTATTTAT | 10.7 |
| – | OA04_13340 | 2.55 | FecI-like RNA polymerase sigma factor | – | – |
| Di- and tricarboxylate sensing and import | |||||
| citW | OA04_25230 | 3.79 | Citrate/acetate antiporter | GAGTTTTAAAGGCGTTTAT | 8.1 |
| citM_OA04_29000 | OA04_28990-OA04_29000 | 0.07 | CitMHS family citrate/H+ symporter and transcriptional regulator | TGGTTTAGACACCGTTTAA | 10.2 |
| – | OA04_29010 | 0.48 | Sodium:dicarboxylate symporter | TAATTTTTTAATTATTTAA | 10.6 |
| tcp | OA04_13410 | 0.39 | Methyl-accepting chemotaxis protein | TGTTTTTATTTGTGTTGAG | 8.9 |
| Exporters | |||||
| – | OA04_04830-OA04_04800 | 5.46 | ABC-2 type exporter | CCGTTTAATTATCGTTTGC | 10.5 |
| – | OA04_05580 | 4.11 | Putative efflux permease | TTTTTTTGTGCATGTTTCA | 7.7 |
| alaE | OA04_38680 | 2.79 | L-alanine exporter | TGTTTTTAAAAAACTTTAA | 7.6 |
| – | OA04_40040 | 2.15 | Putative efflux permease, MFS superfamily | CTATTTATCTCTTTTTATG | 5.3 |
| Other transport | |||||
| mgtA | OA04_05060 | 15.41 | Magnesium transport ATPase | ACGTTGACGTCCGGTTTAG | 6.2 |
| – | OA04_25030 | 3.09 | Glycine betaine/L-proline ABC transporter, ATPase subunit | – | – |
| – | OA04_22250 | 2.10 | Putative inorganic ion transporter | TTTTTTACGCTCGATTTAC | 9.8 |
| mtrB | OA04_14870 | 2.16 | Putative methylthioribose ABC transporter, periplasmic binding protein | CGTTTTATAAATTGATGTC | 5.2 |
| – | OA04_10630 | 2.31 | ABC transporter substrate binding protein | CCGTTTTATTATTTTTTCG | 8.6 |
| gcvB | OA04_10700 | 2.17 | GcvB RNA | – | – |
| – | OA04_29520 | 3.27 | Extracellular solute-binding protein | GAGTATTACCACGATTTAC | 6.3 |
| – | OA04_29540c | 3.39 | ABC transporter permease | CGTTTTATTATTTATTTAT | 13.1 |
| – | OA04_42530 | 2.20 | Extracellular solute-binding protein family 3 | TTGTTTTAACGTGTTGTAA | 6.4 |
| – | OA04_45500 | 4.60 | Extracellular solute-binding protein | TTATATTATAGCTATTTAT | 8.6 |
| – | OA04_05190 | 2.96 | ABC transporter extracellular solute binding protein | – | – |
| – | OA04_02820 | 2.89 | Amino acid ABC transporter substrate binding periplasmic protein | ACGTTGTGTGATGATTTAT | 7.8 |
aWild type vs. phoP mutant ratio. For operons, the value for the first gene is shown. Supplementary Table 3 contains values for all genes.
bChIPmunk score for the best operator.
cOnly the first gene(s) of the larger operon shows differential expression.
The RNA-seq data also confirmed the absence of phoP expression in the mutant cells. Expression of phoQ located immediately downstream in the same operon was barely detectable in the phoP mutant, which is probably due to the polar effect of phoP inactivation. Expression levels of the selected genes from different functional categories were verified by qPCR and found to correlate well with RNA-seq data (Supplementary Figure 2).
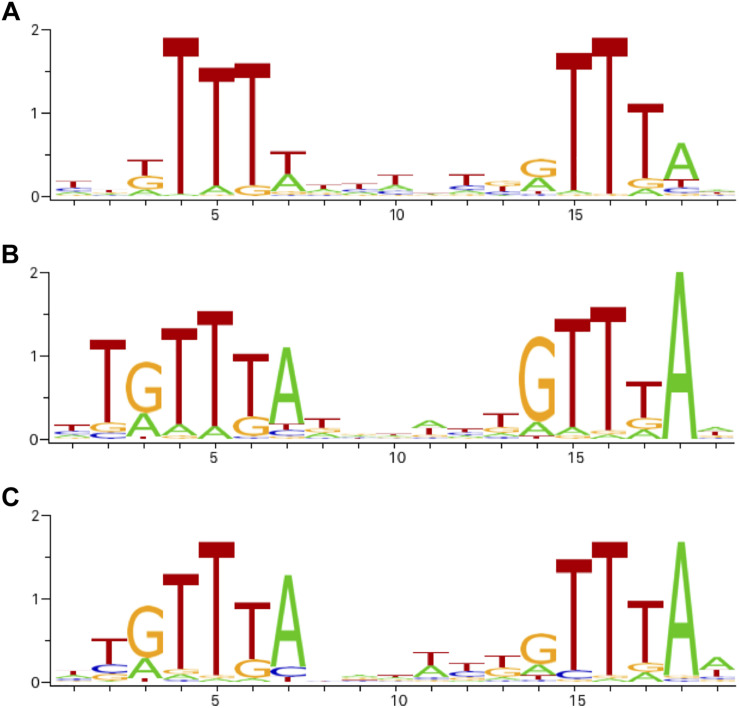
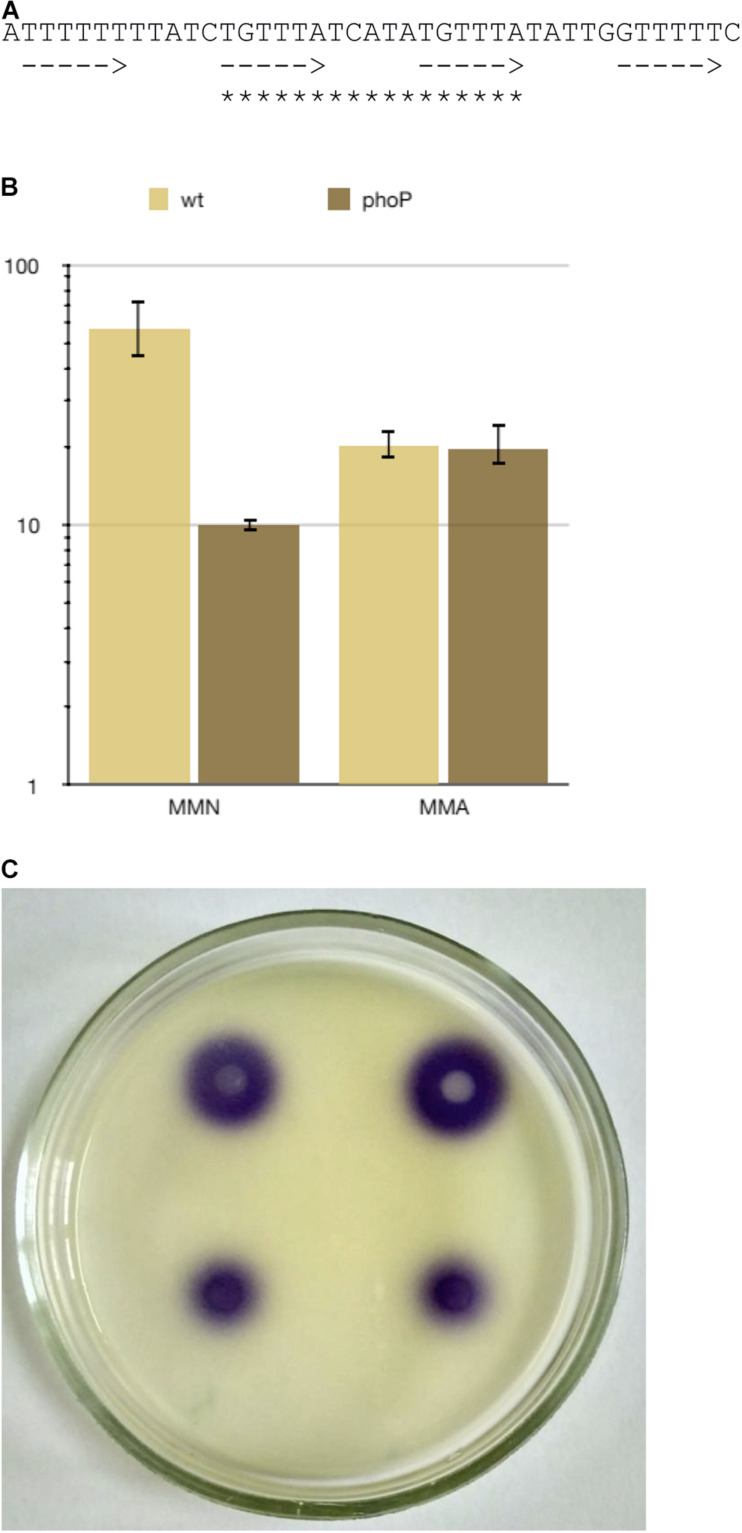
We looked for the presence of PhoP binding sites in the regulatory regions of the differentially expressed genes. ChIPMunk (Kulakovskiy et al., 2010) could locate a relatively weakly conserved 19 bp sequence containing two direct gTTTa repeats (Figure 2A) within most of the 85 regulatory regions of the differentially expressed genes. This sequence resembles the PhoP binding sites reported for other enterobacteria (Figures 2B,C) and was therefore considered the likely PhoP binding site.
FIGURE 2.
Sequence logos of operator motifs for three PhoP orthologues. (A) P. versatile motif made from the sites listed in Table 1. (B) E. coli motif (combined non-redundant RegulonDB and CollecTF data). (C) S. enterica motif (CollecTF data).
The P. versatile PhoP Regulon Part Conserved in Enterobacterales
The universally conserved core of enterobacterial PhoP regulons is known to include only three genes: phoP, phoQ, and slyB (Perez et al., 2009). In Pve, there are high-scoring PhoP-binding sites in front of both the phoPQ operon and the slyB gene. The expression level of slyB is decreased 2.5 times in the phoP mutant (Table 1). Therefore, these three genes are highly likely to be directly controlled by PhoP in Pve.
Additional PhoP regulon members shared by P. versatile with some other enterobacteria are required for cell envelope modifications that improve resistance to antimicrobial peptides and oxidative stress. Most of these modifications alter the structure of the lipid A domain of the outer membrane lipopolysaccharide (LPS). These alterations are known to affect pathogenesis by changing outer-membrane permeability, increasing resistance to antimicrobial peptides and interfering with the ability of the host to recognise LPS as a conserved microbe-associated molecular pattern (see Needham and Trent, 2013, for a review).
The strongest impact (up to 78x decrease of expression levels) phoP inactivation had on the eight genes responsible for the attaching 4-aminoarabinose to the lipid A domain of the LPS. These genes are organised in Pve into two tightly linked operons: monocistronic udg and the arnBCADTEF operon located just after ugd. Multiple high scoring PhoP binding sites are located in front of both ugd and arnB, ensuring direct PhoP control over the whole locus. The addition of 4-aminoarabinose to lipid A reduces the negative charge of the outer membrane and makes the cell more resistant to the CAMPs (Needham and Trent, 2013).
Expression of four more LPS-related genes is activated by PhoP 3–6 fold. The product of OA04_20060 ORF belongs to the acyltransferase 3 family (PF01757) suggesting it might be involved in O-acetylating peptidoglycan (PG) thus increasing resistance to lysozyme (Slauch et al., 1996; Bera et al., 2004). visP (ygiW) homologue from S. typhimurium codes for a virulence- and stress-related protein which binds to peptidoglycan and is required for polymyxin B resistance (Moreira et al., 2013). lpxT codes for the lipid A 1-diphosphate synthase (Touzé et al., 2008b) which increases lipid A negative charge and therefore counteracts the action of arn gene products. pgpBhas undecaprenyl-pyrophosphate phosphatase activity, required for the biosynthesis of the lipid carrier undecaprenyl phosphate (Touzé et al., 2008a). Since LpxT uses undecaprenyl pyrophosphate as a phosphate donor to phosphorylate lipid A (Touzé et al., 2008b), PhoP-dependent PgpB activation might reduce phosphate donor availability for LpxT and hence prevent (or reduce) increase in negative charge of lipid A.
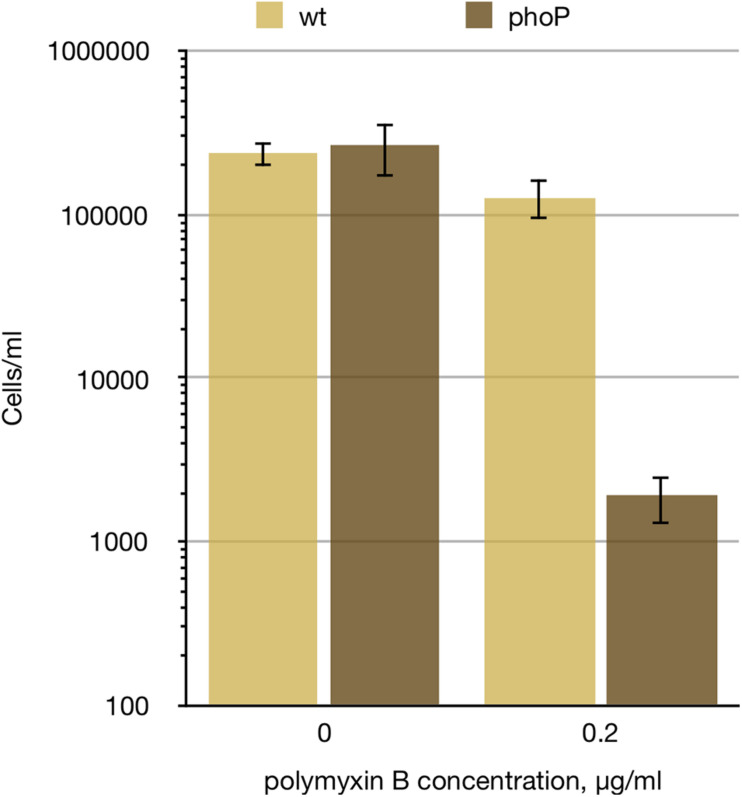
As the products of the ugd-arn locus, VisP, and LpxT have the opposite effects on the lipid A negative charge and the CAMP resistance (and PgpB activity adds to the situation complexity), we have checked the resistance of P. versatile to cationic peptide polymyxin B and found it to be drastically reduced in the phoP mutant (Figure 3). Polymyxin resistance was restored in the phoP mutant to the wild type levels by a plasmid expressing the full phoPQ operon (data not shown). Thus, the net phenotypic result of the PhoP-activated remodelling of the cell envelope in Pve is the activation of the CAMP resistance.
FIGURE 3.
phoP is required for Pve polymyxin resistance. Wild type and phoP mutant Pve cultures were grown aerobically in LB medium to mid-log phase, diluted to 105 cells ml–1 and split in two. 0.2 μg/ml of polymyxin B was added to one of the split cultures and incubation continued for 4 more hours. Surviving cell numbers were determined by plating. Mean values of three biological replicates and 95% confidence intervals are shown.
Pectin Degradation and Utilisation
Degrading pectic compounds of plant cell walls is a characteristic feature of pectinolytic bacteria that is directly responsible for maceration of host plant tissues. PhoP was found to regulate a selection of pectinolysis related genes, activating 11 and repressing one of them.
Just as in P. parmentieri (Flego et al., 2000), PhoP is required in Pve for efficient transcription of the pehA gene encoding the major endo-polygalacturonase. pehA expression demonstrates the strongest response to phoP inactivation among all DEGs (118x), which correlates with the presence of multiple PhoP binding sites in the pehA regulatory region.
Three more PhoP-activated genes code for pectinolytic enzymes: periplasmic endo-pectate lyase PelP (9.7x activation),endo-polygalacturonosidase PehN (2.2x activation) and pectin lyase Pnl (2.6x activation).
Three oligogalacturonate transporter genes are also under positive PhoP control: the oligogalacturonate/cation symporter togT (2x activation) and two neighbour loci OA04_21050 and OA04_21060 (9.1x and 25.5x activation) coding for porins with weak homology to oligogalacturonate-specific porins KdgM and KdgN. The major oligogalacturonate transport and utilisation locus kdgF-kduDpelWtogMNAB-kdgM also has PhoP binding sites in the appropriate regulatory regions, but the observed expression level differences were slightly below the threshold (data not shown), suggesting possible input of additional regulator(s) into transcriptional control of this locus.
Three uncharacterised genes with about twofold activation by PhoP (OA04_07070, OA04_12400, and OA04_32560) encode proteins with family 32 carbohydrate-binding module (CBM32). One well-characterised member of this family, YeCBM32 from Yersinia enterocolitica, binds to polygalacturonic acid (Abbott et al., 2007) and was suggested to bind and retain polygalacturonic acid (PGA) within the periplasm as a substrate for further depolymerisation (Abbott and Boraston, 2008). YeCBM32 has sufficient homology (over 72% identity) to OA04_07070 to assume orthology between the two proteins. Additional evidence for the involvement of OA04_07070 in pectin utilisation is provided by the presence of the signal peptide in the product and a binding site for the major pectinolysis regulator KdgR in front of this gene. Two more proteins with CBM32 are larger, have no signal peptide and their amino acid identity to YeCBM32 is very weak suggesting different function.
Overall, PhoP appears to be a strong activator of extracellular and outer membrane components and a weak activator of periplasmic and cytoplasmic membrane components of polygalacturonate utilisation system. Together, PhoP-activated genes code for a complete set of proteins required for polygalacturonate depolymerisation and transport into the bacterial cytoplasm. However, Pve has more genes encoding PGA depolymerases that showed no PhoP dependence in our experiment. In particular, expression levels of the genes in the major pectate lyase gene cluster (pelA, pelB, pelC, pelZ) were the same in the wild type strain and the phoP mutant. Therefore, we speculate that PhoP activates a subsystem of PGA depolymerisation and transport, finely tuned for a specific condition or a specific variant of PGA modification.
pelI is the only pectinolysis gene repressed (7x) by PhoP. It is located next to pehA, is transcribed in the opposite direction and shares its regulatory region with pehA. PelI is known to be strongly expressed in planta (Jafra et al., 1999). It was also reported to induce a hypersensitive response in plants (Shevchik et al., 1998). Since strong pelI expression can be expected to occur only when the PhoPQ system is inactivated, high pelI expression level in planta may indicate such an inactivation.
Citrate and Dicarboxylate Transport
Two genes of citrate transporters are controlled by PhoP: citM (14x repressed) and citW (3.8x induced).
citW, coding for citrate/acetate antiporter, is located within the citrate fermentation locus. The locus includes citAB operon encoding a citrate responsive two-component system and a divergently transcribed citW, which is followed by the citYCDEFXG operon (coding for the subunits of citrate lyase). All these genes (including citW) were poorly expressed in our study. citW and the whole citrate fermentation locus is expected to be relevant for citrate utilisation under anaerobic conditions while citM, coding for a citrate/proton symporter operates in aerobic environment (Bott, 1997; Kästner et al., 2002).
Besides the transporters and regulators, the tcp gene encoding citrate chemotaxis receptor is activated threefold in the phoP mutant.
One more transporter gene, OA04_29010, is located downstream of the citM operon and codes for a putative sodium:dicarboxylate symporter. Just as the preceding operon, OA04_29010 is negatively controlled by PhoP (2.1x repression).
Citrate and dicarboxylates (e.g., malate) are abundant in plant cells, can be excreted into either apoplast or soil (Meyer et al., 2010) and therefore constitute good nutrients for a plant pathogen. In P. atrosepticum, citrate uptake was attributed to a highly specific transporter Cit1 and shown to be important for virulence (Urbany and Neuhaus, 2008). Cit1 orthologue was expressed poorly underconditions used in this work, so we could not evaluate the involvement of PhoP in its control. However, a good PhoP binding site was located in front of cit1. High scoring PhoP binding sites were also located in the regulatory regions of almost all dicarboxylate transporter genes annotated in the Pve genome: dctPQM, dcuA, dcuC, dcuB, and dctA (more detail available in the “Extended regulon” section). We expect PhoP to affect the expression of at least some of these dicarboxylate transporters in different conditions, e.g., in the presence of dicarboxylates.
Overall, PhoP can be considered to be an important regulator of di- and tricarboxylate transporters. As some of these transporter genes are under negative control while others—under positive control, we speculate that PhoP may be involved in switching between the forms of carboxylate transporters appropriate for different environmental conditions including different stages of infection.
Arabinose Transport and Utilisation
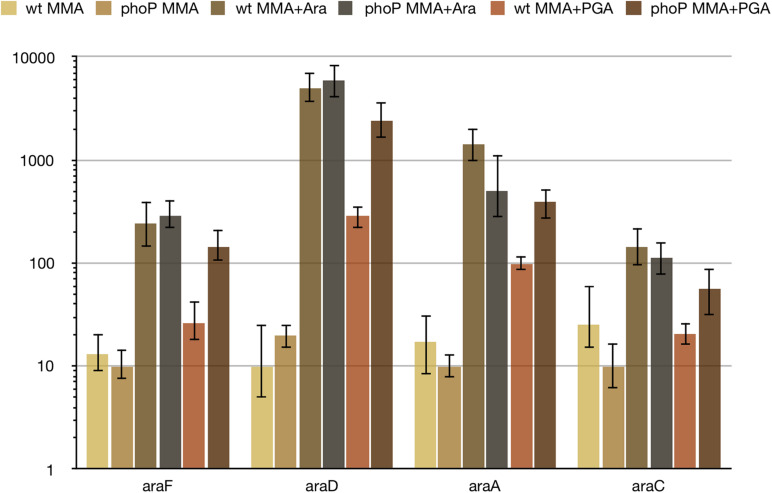
Thirteen genes related to transmembrane arabinose transport and utilisation are organised into five or six operons in Pve and are strongly repressed by PhoP (Table 1). The main arabinose uptake and catabolism locus in Pve includes the divergently transcribed araFGH (transporter) and araBA (catabolism) operons. The araC gene is separated from araH by a 203 bp gap permitting an independent transcription initiation, but the lack of an obvious transcription terminator in this gap can also allow co-transcription with the upstream araFGH genes. The unlinked monocistronic araD codes for the last of the three enzymes required for arabinose catabolism.
The ytfQRTyjfF operon encodes a periplasmic sugar-binding protein YtfQ and three subunits of an ABC transporter. YtfQ was shown to specifically bind the furanose forms of arabinose and galactose (Horler et al., 2009). The OA04_38360-OA04_38370 operon codes for the putative glycoside-pentoside-hexuronide:cation symporter and glycosyl hydrolase which belongs to the subfamily 43.26 according to CAZy classification (Lombard et al., 2014). Well characterised bacterial members of this subfamily with sufficient homology to OA04_38360 product were described as exo-α-1,5-L-arabinofuranosidases specialised at cleaving short arabinooligosaccharides (Matsuo et al., 2000, p. 901; Fujimoto et al., 2010, p. 43; Linares-Pastén et al., 2017). Arabinose is present in plant cell walls mainly in the form of arabinofuranosyl residues of the cell wall polysaccharides and proteoglycans such as pectic arabinan, arabinoxylan, and arabinogalactan-proteins (Kotake et al., 2016). Presumably, the OA04_38360-OA04_38370 and ytfQRTyjfF operon products are involved in transport and utilisation of arabinofuranose containing products of plant cell wall breakdown.
As the OA04_38370 product does not have a signal peptide, it can only degrade cytoplasmic substrates such as α-L-Araf containing oligo- or disaccharides which could be released from plant cell wall by the action of other enzymes. At least two Pve enzymes are relevant: a putative arabinogalactanase encoded by OA04_08560 and an arabinogalactan endo-1,4-beta-galactosidase encoded by ganA of the ganEFGAB operon. Although no differential expression was registered in vitro for any of the two loci encoding secreted arabinogalactanases, both arabinogalactanase genes could be expected to be induced by galactose in planta. Indeed, our recent transcriptome profiling of P. atrosepticum has shown 4- and 24-fold in planta induction of OA04_08560 and ganA orthologues ECA0852 and ECA3128 (Gorshkov et al., 2018).
High scoring PhoP binding sites are located near two out of six arabinose related differentially expressed transcriptional units. A PhoP binding site overlaps the very beginning of the araD reading frame, suggesting direct negative control of araD by PhoP. Another binding site was found in front of the araC gene in a position suitable for repression. Three rather weak PhoP binding sites were found in different ChIPmunk runs within the regulatory region of the OA04_38360-OA04_38370 operon, suggesting the likelihood of direct PhoP control over this operon as well.
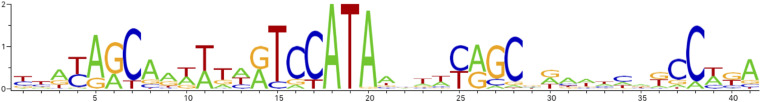
Since araC codes for the arabinose-responsive transcription factor, we searched for AraC binding sites in the Pve genome. First, the AraC operator profile was constructed. For this, we downloaded the AraC_Enterobacterales motif from RegPrecise (Novichkov et al., 2013). However, this short (17 bp) motif corresponds to just a part of the binding site. AraC has two DNA binding domains, both of them can contact DNA and the active AraC form is dimeric (Schleif, 2010), so the real binding site should be larger. We checked the conservation around the sites annotated in RegPrecise and found the conserved region to cover about 40 bp—sufficient to allow binding of four DNA binding domains of the dimeric AraC. The extended part of the motif is less conserved but has the same orientation and an overall resemblance to the conserved part (Figure 4 and Supplementary File 3). This configuration of the operator is suitable for the binding of a tandem of AraC subunits in their activating conformation. The most conserved part of this motif corresponds well to the one recently defined experimentally (Stringer et al., 2014).
FIGURE 4.
AraC binding site sequence logo.
Scanning of the Pve genome with the AraC operator profile has found matches in three locations: in the regulatory region between the araBA and araFGHC operons (two sites), in front of the araD gene and the ytfQRTyjfF operon. Therefore, PhoP dependence of differentially expressed arabinose-related transcriptional units could be explained by direct negative control by PhoP over araD, araC, ytfQRTyjfF and OA04_38360-OA04_38370, combined with positive control via AraC over ytfQRTyjfF, araD and the other two operons that lack PhoP sites in their regulatory regions (araBA, araFGH).
Further investigation of PhoP-dependent control over arabinose utilisation genes has shown, that (i) arabinose is a strong inducer of ara genes and (ii) the observed differential expression depends on the presence of sodium polypectate in the medium (Figure 5). Without polypectate, expression of ara genes does not require PhoP, both with and without arabinose. Sodium polypectate appears to be a weak inducer of arabinose-related operons in the wild type strain, but induction is much stronger in the phoP mutant (Figure 5). PhoPQ two-component system does not seem to respond to polypectate, as most of the other DEGs have the same expression with and without polypectate (data not shown). This suggests indirect regulation by another PhoP-controlled and polypectate responsive transcriptional factor and requires further investigation.
FIGURE 5.
Differential expression of ara genes depends on the presence of polygalacturonate. For qPCR measurement, RNA was isolated from the wild type (wt) or phoP mutant Pve cells grown in MMA, MMA with 0.5% arabinose (MMA + ara) or in MMA with 0.5% sodium polypectate (MMA + PGA). Mean values of relative expression levels with 95% confidence intervals are shown.
Other Transporters
PhoP is known to be involved in the regulation of at least one of the Mg2+ transporters in enteric bacteria (Perez et al., 2009). P. versatile seems to have the only dedicated Mg2+ transporter encoded by mgtA. In the conditions of our RNA-seq experiment with moderate (0.5 mM) Mg2+ concentration mgtA was weakly expressed in the wild type cells and was almost silent in the phoP mutant. At least 10-fold higher expression of the 5′ UTS compared to the coding frame of mgtA (Supplementary Figure 3) hints to a riboswitch-like control of transcription elongation into the coding region of mgtA analogously to the mechanism described for S. enterica (Cromie and Groisman, 2010).
PhoP activates several genes and operons involved in iron acquisition. Since the RNA-seq experiment was performed without iron limitation, these genes had a rather low expression and differential expression in many cases was detected only for the first gene of multigene operons. DEGs related to iron scavenging include fetM encoding high-affinity Fe2+ permease, yiuABC coding for ferric-enterobactin ABC transporter and OA04_26190 encoding TonB-dependent outer membrane siderophore receptor. entC is the first gene of the enterobactin-like siderophore synthesis operon entCEBFA. fusB, the first gene of the fusBACD operon, encodes a TonB-like protein known to be required for import of plant ferredoxin (Wojnowska and Walker, 2019). Differentially expressed FecI-like sigma-factor gene OA04_13340 resides within an apparent TonB-dependent iron acquisition locus of eight genes and is likely to control its transcription.
Three PhoP regulated transporters participate in amino acid import. The OA04_29540-OA04_29570 operon is likely involved in the import and modification of branched-chain amino acids. OA04_42530 codes for the periplasmic binding protein of one more amino acid transporter. OA04_02820 product is annotated as a cystine-binding periplasmic protein.
Three more PhoP-activated loci encode putative efflux transporters. These include L-alanine exporter AlaE (OA04_ 38680), putative multidrug efflux transporter OA04_04830-OA04_04800 and AraJ family efflux permease OA04_40040.
Three additional transporters do not belong to any of the groups mentioned above. mtrB codes for the periplasmic binding subunit of methylthioribose ABC transporter. OA04_25030 encodes the ATPase subunit of glycine betaine/L-proline ABC transporter, OA04_22250—the putative sodium/bile acid cotransporter. The last transporter may act in conjunction with choloylglycine hydrolase encoded by OA04_32550, which is also activated by PhoP.
The potential substrates of four other transporters (OA04_10630, OA04_29520, OA04_45500, and OA04_05190- OA04_05160) could not be inferred even approximately.
All the genes coding for the transporter subunits described above are activated by PhoP. However, the observed effect of phoP inactivation may be indirect in many cases. In particular, we found no PhoP binding sites near mtrB, OA04_25030, OA04_05190 and only weak sites could be located for OA04_38680, OA04_45500, and OA04_02820.
We also note that the GcvB ncRNA, a well-known negative regulator of amino acid transporter genes (Sharma et al., 2011), is 2.2x activated. This effect may partially compensate for the decrease of the transporter gene expression in the phoP mutant. We could not find a PhoP binding site in front of gcvB. Therefore, gcvB expression control by PhoP might be indirect, just as has been reported for E. coli (Raghavan et al., 2011). GcvB has also been reported to control PhoP expression posttranscriptionally by both destabilising its mRNA and decreasing its translation (Coornaert et al., 2013). Such reciprocal repression between PhoP and GcvB, if present in Pve, might result in amplification of any changes in expression of either regulator.
Tellurite Resistance Genes
A group of seven genes (terZABCDE and divergently transcribed OA04_37930) tentatively annotated as coding for tellurite resistance proteins is 2-6x repressed by PhoP. OA04_37930 is the first gene in the operon of five uncharacterised genes (1.7-1.9x repression of the four downstream genes). These genes are a part of a large locus of 18 presumably functionally linked genes (Anantharaman et al., 2012) organised into four or five operons (Supplementary Figure 4). Although the exact function of this locus in vivo is not clear, orthologous genes are required for tellurite resistance in some bacteria (Whelan et al., 1995, 1997; Valková et al., 2007). Tellurite is also thought to induce oxidative stress and tellurite resistance might be the result of decreased sensitivity to the oxidative stress (Chasteen et al., 2009).
At least one PhoP binding site is located in the regulatory region between terZ and OA04_37930, one more is present upstream of the terW gene. terW (6.9x induced) is located a few genes away and was shown to be responsible for transcriptional control of ter genes in E. coli (Valkovicova et al., 2011). The part of the ter operon regulatory region most conserved between E. coli and Pectobacterium sp. sequences contains a 25 bp palindrome (Supplementary Figure 4 and Supplementary File 3). Its position corresponds to the region protected by TerW (Valkovicova et al., 2011) and probably corresponds to the TerW binding site. Three putative TerW binding sites could be located within the ter cluster in Pve: two in the middle of the intergenic region between the divergently transcribed operons (terZABCDE and OA04_37930-OA04_37890) and the third one is in front of terY1. Thus, either the direct repression or a transcriptional cascade, where PhoP activates terW and TerW represses divergent operons, are responsible for the observed PhoP dependence of the genes in the ter locus. Since the terY operon does not respond to phoP inactivation, direct transcriptional control of PhoP over other ter genes is more likely in the conditions studied.
The OA04_10330 gene (2.2x repression) is unlinked to the main ter locus, but codes for a TerC-like membrane protein and may be functionally linked to the ter genes.
Pve cells growing on the medium with low potassium tellurite concentration form characteristic black colonies, indicating the reduction of tellurite to tellurium. We did not notice a significant difference in black colouring between the wild type Pve and the phoP mutant, but the phoP mutant could grow in the MMN medium with potassium tellurite concentration 3 μg ml–1 that is inhibitory to the wild type strain (data not shown).
The actual role of “tellurite resistance” genes of a soft rot pathogen requires separate investigation. Soft rot bacteria can hardly ever encounter significant concentrations of tellurite in their natural environment, so these genes are likely responsible for coping with some other stress. Searching the nr and wgs subdivisions of GenBank shows that “tellurite resistance” genes are present in only three pectobacterial species: P. versatile, P. parmentieri, and P. polaris suggesting such stress is important for the lifestyles of these species.
Anaerobiosis-Related Genes
The transcription profiling experiment described here was performed in aerobic conditions, and most of the anaerobiosis-related genes were expressed poorly. However, the PhoP-dependent positive control over important aspects of anaerobic metabolism was noticeable. The largest group of PhoP-activated genes is required for the detoxification of formate produced by enteric bacteria in anaerobic fermentative conditions. These genes include the formate transporter gene focA and several subunits of formate hydrogenlyase. Expression of the aegA operon, coding for formate metabolism proteins, is also PhoP-dependent. aegA codes for an oxidoreductase that was recently shown to be involved in formate-dependent catabolism of urate (Iwadate and Kato, 2019). A gene downstream aegA, OA04_45440, codes for an uncharacterised [4Fe-4S] ferredoxin subunit of hydrogenase or formate dehydrogenase.
Weakly expressed operons within the hydrogen metabolism gene cluster were PhoP dependent. These include three operons. The first operon is hybOABCDE, coding for subunits of hydrogenase 2. The second one is a hypABCDE operon coding for hydrogenase maturation proteins. The third operon includes genes coding for electron transport protein, which is required for formate dehydrogenase activity (hydN), formate dehydrogenase H (fdhF) and one more hydrogenase maturase (hypF). The Pve 3-2 genome carries a second fdhF paralogue (fdhF_2) which is also PhoP activated. The fdnGHI operon, coding for nitrate inducible formate dehydrogenase N, is PhoP activated too.
Anaerobic nitrate reduction may also be controlled by PhoP. The targets include a PhoP-activated nap operon responsible for periplasmic nitrate reductase production (differential expression was registered for only the first gene napF). Clear differential expression was detected for ccmA, the first gene of the ccmABCD operon coding for the subunits of the heme trafficking system. This system delivers heme to holocytochrome c synthase CcmFGH. And since this cytochrome c biogenesis system is required for maturation of cytochrome c subunits of nitrate and nitrite reductases, PhoP might be required for anaerobic nitrate and nitrite reduction. An additional level of PhoP control over nitrite reduction is provided via regulation of formate dehydrogenase that serves as the electron donor for formate-dependent nitrite reductase coded for by the nrf operon (poorly expressed in this experiment). PhoP also appears to control the norVnorW operon coding for anaerobic nitric oxide reductase.
Anaerobic respiration with fumarate as an electron acceptor may also be PhoP dependent due to control of at least one key gene, glpA. The glpABC operon codes for subunits of anaerobic glycerol-3-phosphate dehydrogenase, which is required for anaerobic respiration with fumarate as a terminal electron acceptor.
Three more PhoP-controlled genes contribute to important aspects of anaerobic metabolism. The nrdD gene codes for anaerobic ribonucleoside triphosphate reductase, which is essential in E. coli for deoxyribonucleotide synthesis during strict anaerobic growth (Garriga et al., 1996). ubiQ is required for anaerobic ubiquinone biosynthesis (Pelosi et al., 2019). GrcA restores pyruvate formate-lyase (PFL) activity after oxidative damage to the main PFL subunit (Wagner et al., 2001; Bowman et al., 2019) and thus may help the pathogen to maintain the high activity of key glycolysis enzyme at microaerobic conditions.
Regulators
Nine regulatory genes showed differential expression in our experiments. Three of the transcription factors (AraC, TerW and OA04_13340) and the regulatory RNA GcvB were already discussed above. glnK codes for the second nitrogen regulatory protein P-II. Since it is functionally equivalent to P-II encoded by glnB, and glnB expression level is much higher (data not shown), the significance of the twofold difference of glnK expression levels should be minor.
Three regulators belong to the LysR family of transcription factors: OA04_01500, SftR and Cbl. OA04_01500 is a member of the RpoN regulon, had very low expression in the nitrogen-rich conditions of this experiment and was therefore unlikely to contribute significantly to the observed expression values.
A LysR family TF annotated as SftR is 2.3x activated by PhoP. We located the only potential operator in Pve genome that might be bound by SftR in the atsR-atsBCA intergenic region (data not shown). Therefore, SftR involvement in the control of the DEGs identified in this experiment is unlikely.
cbl (2.7x activation) codes for a CysB-like protein, whose closest homologue in E. coli controls genes required for aliphatic sulphonate and taurine utilisation and homeostatic response to sulphate starvation, according to RegulonDB. However, the ligand-binding domains of the homologues show low similarity, unlike the highly similar DBDs, which suggests a possibility of binding different ligand(s) by these two TFs. CysB-like proteins were reported to control functions unrelated to sulphur utilisation (Delic-Attree et al., 1997; Imperi et al., 2010; Farrow et al., 2015). A rather high transcription level of cbl in Pve hints at a possibility that this TF controls some DEGs found in this experiment.
One more well-expressed transcription factor belongs to the GntR family. It is coded for by the OA04_29000 gene located downstream of citM, probably in the same operon. The involvement of OA04_29000 and cbl gene products in transcriptional control of the DEGs described here is currently under investigation and will be reported separately.
The Extended PhoP Regulon
Since PhoP is a global transcriptional regulator, PhoP-dependent genes differentially expressed in the single condition studied in this work probably constitute just a subset of the PhoP regulon. To see a broader picture, we analysed potential PhoP operators located near transcriptional units that did not pass differential expression thresholds. Since operator motif based on our experimental dataset (Table 1 and Figure 2A), had relatively low information content, we have looked for the possibility of applying search profiles based on S. enterica and E. coli data which cover a more diverse set of conditions.
First, we have analysed the structures of TF-operator complexes for OmpR family proteins that are present in Protein Data Bank (Berman, 2000). This analysis allowed us to locate the positions of amino acid residues responsible for making specific contacts with DNA bases (Figure 6). Aligning to homologues with known structures shows that PhoP proteins from Pve, E. coli, and S. enterica have identical amino acid residues in the positions making specific contacts with DNA bases (Figure 6). This strongly suggests that indicated proteins must recognise very similar or even identical operator sequence specificities. The differences between their reported operator motifs may reflect the differences between the data sets and algorithms used for their inference. Therefore, the search for additional PhoP binding sites was done by scanning Pve genome sequence with four PhoP operator profiles (Supplementary File 1): the ones based on our experimental data, E. coli data, S. enterica data and profile inferred in silico as described in “Materials and Methods” section.
FIGURE 6.
Alignment of DNA binding domain sequences of the OmpR family TFs. The sequences are aligned with hmmalign according to the Trans_reg_C (PF00486) family model. The amino acid residues of OmpR family proteins that form direct contacts with nucleotide bases were determined by the interaction service of NPIDB (Zanegina et al., 2016). The residues making specific contacts with DNA bases are highlighted in green (hydrogen bonds) and yellow (hydrophobic contacts). Positions with contact observed in at least one sequence are marked with an asterisk. PDB/GenPept/Uniprot IDs followed after a colon by protein names combined with species abbreviations are shown to the right of each sequence. Species names are abbreviated as Eco (Escherichia coli), Mtu (Mycobacterium tuberculosis), Kpn (Klebsiella pneumoniae), Aba (Acinetobacter baumannii), Pve (Pectobacterium versatile), and Sen (Salmonella enterica).
The scan identified a total of 812 genes organised into 254 transcriptional units that could be the targets of transcriptional control by PhoP (Supplementary File 2).
Many of these putative PhoP regulon members are related to plant cell wall degradation and utilisation of carbon sources abundant in plants. Of note are the genes/operons encoding pectate lyases (pelA and pelB), pectin methyl/acetyl esterases (pmeB and paeX), three beta-glucoside transport/utilisation operons (bglHDJ, bglYK arbFBH) and five di/tricarboxylate transporters (dcuA, dcuB, dctA, maeN, and yflS). For most of these genes, the PhoP binding site location suggests an activator role. At least one negative regulatory site (that could block PhoP-dependent transcription activation) could be located between PhoP operator and translation start site (attenuators in front of bglH, bglY, arbF, and possibly paeX, operators for DcuR in front of dcuA, yflS, and dcuB, operators for NarP in front of dcuB and yflS).
A “strong” PhoP binding site is located in front of the expI gene coding for N-acyl-homoserine lactone synthase, a key regulator of quorum sensing. ExpI is known to dramatically affect gene expression, including that of many virulence genes (Liu et al., 2008), therefore PhoP control of expI transcription was studied in more detail. The regulatory region of expI (Figure 7A) has four equally spaced half-sites (two central ones perfectly match the gTTTA consensus) allowing for the binding of two PhoP dimers (Figure 7A). Since PhoP homologues can form oligomers (He et al., 2016), binding of a PhoP tetramer is also possible in this region.
FIGURE 7.
PhoP regulates quorum sensing via transcriptional control of the expI gene. (A) The sequence of the PhoP binding sites in expI promoter region. Arrows indicate possible positions of PhoP monomer binding sites, asterisks—the most likely position for binding of PhoP dimer. (B) expI expression in Pve cultures grown to 108 cells ml–1 in MMN (10 μM Ca2+, 10 μM Mg2+) and MMA (10 μM Ca2+, 0.5 mM Mg2+). (C) N-acyl-homoserine lactone bioassay result. The purple colour indicates N-acyl-homoserine lactone-dependent violacein pigment production by the CV026 C. violaceum indicator strain. Top: the wild type strain, bottom: the phoP mutant.
The RNA-seq data showed a small (22%) difference in expI expression levels between the wild type and the mutant strains grown in MMA with 0.5 mM of magnesium ions (data not shown). However, a fivefold difference was observed if the bacteria were grown in the MMN medium at low concentrations of both calcium and magnesium ions when the PhoPQ system was fully induced (Figure 7B). Higher expI transcription in the wild type strain than in the mutant correlated with the higher production of N-acyl-homoserine lactone (Figure 7C). These results show an intriguing connection between the PhoPQ two-component and quorum-sensing systems in Pve.
Searching the NCBI nr database reveals that the nucleotide sequence very similar to the one shown in Figure 7A is present in many (although not all) Pectobacterium spp. genomes (data not shown), but could not be found outside the genus, making this particular connection between PhoPQ and the quorum sensing system Pectobacterium-specific.
Another regulatory region with multiple potential PhoP binding sites is located in front of the large tss operon coding for the subunits of the type VI secretion system (data not shown). We have not seen a noticeable effect of PhoP inactivation on downstream genes expression in vitro. However, recent work with Pectobacterium brasiliense has reported a strong PhoP-dependent induction of the tss genes in planta (Bellieny-Rabelo et al., 2020) suggesting that additional regulator(s) might block PhoP-dependent expression of the tss genes in vitro while this block is removed in planta.
The expI and tss gene examples confirm that at least some in silico predicted PhoP targets are true regulon members that can be controlled by PhoP in certain conditions. The list of in silico predicted PhoP regulon members (Supplementary File 2) might be helpful in further studies of regulatory networks and different aspects of Pve interaction with its environment, including host plants.
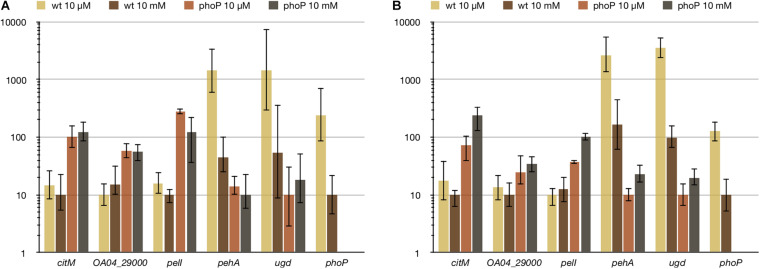
PhoP Regulon Is Activated in Pve by Ca2+ and Mg2+ Limitation
In an attempt to find the conditions responsible for PhoPQ activation in Pve, we checked some environmental stimuli known to be detected by the PhoQ sensor. There was no reaction to cationic peptide polymyxin and low pH (data not shown), but the response to divalent cations was significant. Expression levels of PhoP-activated genes were the highest at low (10 μM) concentrations of both Ca2+ and Mg2+ (Figure 8). Increasing either cation concentration to 10 mM reduced expression levels of PhoP-activated genes. Importantly, expression of phoP was decreased 13 times by 10 mM Mg2+ and 24 times by 10 mM Ca2+. Expression level differences of pehA and ugd caused by divalent cation addition were in the range 16–35 times. Comparison with the phoP mutant strain grown in the same conditions shows that some active PhoP must be present in the wild type cells even after divalent cation addition as phoP inactivation results in larger (91–353 times) drop of pehA and ugd expression levels. We interpret these results by both Mg2+ and Ca2+ being able to bind PhoQ to significantly reduce its kinase activity required for phosphorylation of PhoP resulting in a strong reduction of transcription initiation at PhoP-activated promoters.
FIGURE 8.
Ca2+ (A) and Mg2+ (B) reduce expression of PhoP-activated genes, but do not affect PhoP-repressed ones. For qPCR measurement, RNA was isolated from the wild type (wt) or phoP mutant Pve cells grown in MMN with either 10 μM or 10 mM of the specified ion. Mean values of relative expression levels with 95% confidence intervals are shown.
Unexpectedly, divalent cation concentrations had little effect on PhoP-repressed genes. Expression levels of the three genes tested (citM, pelI, and OA04_29000) differed less than twofold between the wild type and phoP mutant strains and these differences were not statistically significant (Figure 8). We conclude that (i) PhoP amounts present in the wild type Pve cells grown at 10 mM Ca2+ or Mg2+ are sufficient to cause repression of the two transcriptional units involved and (ii) phosphorylation may not be required for PhoP to bind its operator sites. The last conclusion is in line with the previous report of both dimerisation and DNA binding of PhoP being independent of its phosphorylation (Perron-Savard et al., 2005). The ability of reduced PhoP amounts to repress transcription of pelI and the citM-OA04_29000 operon is not surprising, as regulatory regions of both transcriptional units have multiple high scoring (and presumably high-affinity) PhoP binding sites.
PhoP Regulon Expression in planta
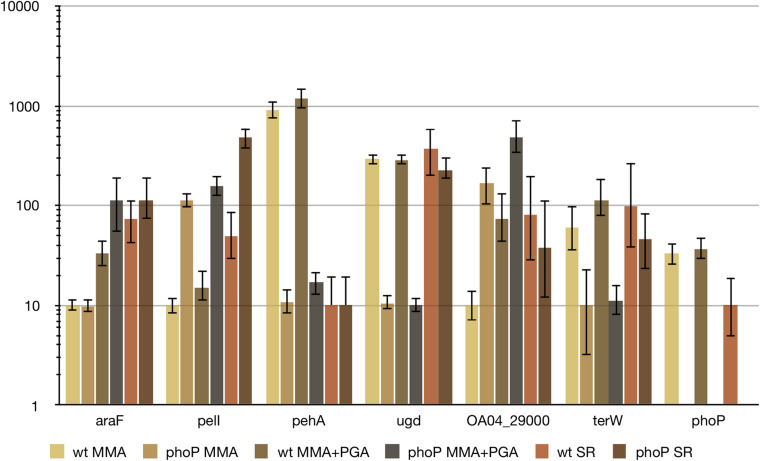
Due to autoregulation, PhoP involvement in the adaptation to the plant environment must be accompanied by changes in its expression level. We have previously noticed decreased phoP expression in another example of Pectobacterium-plant interaction. A 3.5- and 5.2-fold phoP repression was observed for P. atrosepticum strain SCRI 1043 in asymptomatic and necrotic zones of infected tobacco stems (Gorshkov et al., 2018). This reduction of phoP expression correlated well with expression level changes of PhoP controlled genes: e.g., pehA expression was reduced while pelI and ara genes expression was increased.
In the current investigation, phoP expression decreased about 4 times in macerated potato tuber tissues (Figure 9). In the bacterial cells isolated from the rotten tuber tissues, expression level differences of PhoP regulon members between the wild type and mutant strains were much less pronounced than in vitro or even absent (Figure 9). This could be attributed to the partial inactivation of the PhoPQ system in planta as described in the Discussion section. A single notable exception was the pelI gene, which had similar differences of expression levels between the wild type and phoP mutant strains in planta and in vitro. A large number of high-affinity PhoP binding sites in front of pelI may permit even reduced amounts of PhoP to still cause pelI repression.
FIGURE 9.
Relative expression levels of PhoP regulon members in vitro and in planta. For qPCR measurement, RNA was isolated from the wild type (wt) or phoP mutant Pve cells grown in MMA, MMA with 0.5% sodium polypectate (MMA + PGA) or from the cells of rotten potato tissues (SR).
Two genes that show significantly higher expression in phoP mutant cells within potato tubers than in vitro are pelI and ugd. Regulatory regions of both genes contain binding sites for FNR, a key transcriptional activator under anaerobiosis. As the tubers were incubated in anaerobic conditions after inoculation, FNR-dependent transcription activation could be responsible for the observed higher in planta expression of pelI and ugd.
Discussion
Our experimental results and in silico analysis show that plant pathogenesis-specific part constitutes the majority of PhoP-controlled genes in Pve. As a result, the PhoPQ two-component system can be considered a major modulator of Pve gene expression that ensures the levels of certain proteins are appropriate for particular stages of plant colonisation.
The major functional categories of PhoP-controlled proteins include stress resistance factors, hydrolases of plant cell wall polysaccharides, transporters and enzymes necessary for the utilisation of plant-derived products (Figure 10).
FIGURE 10.
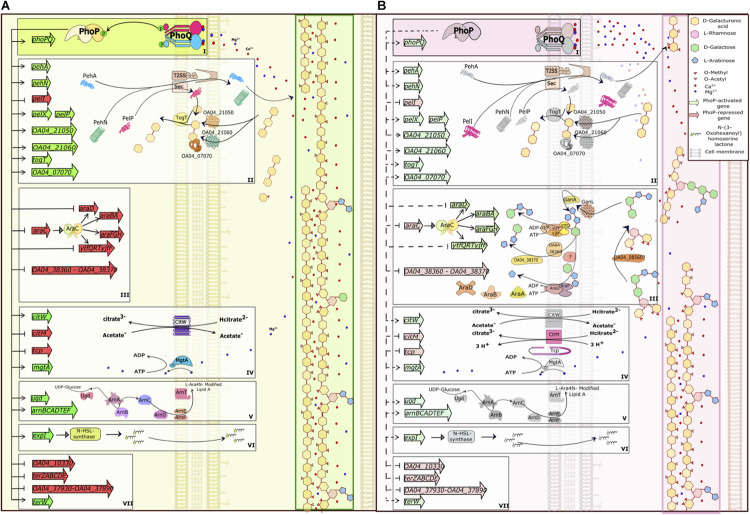
PhoPQ-dependent switch of virulence gene expression in Pve. (A) PhoQ kinase phosphorylates PhoP at low concentrations of calcium and magnesium ions (early infection) leading to maximal activation and repression of its targets. (B) As the infection progresses, extensive plant cell wall degradation releases vast amounts of Ca2+ and Mg2+ leading to inhibition of PhoQ kinase activity and PhoP dephosphorylation. This prevents PhoP-dependent activation, but still allows repression of target genes that have high-affinity PhoP operators. The drawing tries to link the expression of PhoPQ-controlled genes to plant cell wall degradation but does not take into the account other regulators that might influence the same genes. The picture shows the majority of PhoP-repressed genes and a selection of PhoP-activated genes from the same functional categories. The categories are framed and labelled I (the PhoPQ TCS), II (polygalacturonic acid degradation and utilisation), III (arabinogalactan degradation and utilisation), IV (transporters), V (LPS modification), VI (quorum sensing) and VII (tellurite resistance). In the categories II and III, the T2SS and Sec systems, two galactanases and the GanL porin not controlled by PhoP are added for clarity; these proteins are induced in planta and complete the full pathway of degradation, transport and utilisation of pectin components. To simplify the picture, categories IV and V are shown only partially; arabinogalactan degradation products are simplified (fewer sugar residues are drawn). Activation/repression are shown as pointed/blunt arrows. Activated/repressed genes are shaded green/red. PhoP-activated gene products are greyed on (B) to mark their presence in reduced amounts.
phoP transcription in Pve decreased within rotten tissues of potato tuber. We had previously seen a similar decrease in phoP transcription in a slightly different pathosystem (Gorshkov et al., 2018). In these two cases, the decrease of phoP transcription occurs at the late stages of infection. Recent work with P. brasiliense has not only reported decreased phoP expression at late infection stage but also registered an increase of phoP transcript amounts shortly after tuber inoculation (Bellieny-Rabelo et al., 2020). Since phoP is autoregulated, we think the changes of phoP expression level reflect the state of the PhoPQ two-component system: induced at the onset of the infection and repressed at later stages. The switch between the two states occurs due to the detection of plant-derived stimuli and helps the pathogen to adjust its properties, mostly cell envelope-related, that are required for proper interaction with the host plant.
Of the stimuli known to be detected by PhoQ in other bacteria, we were able to register strong response only to divalent cation concentrations. The pectobacterial PhoPQ system was originally described in P. parmentieri as responding to Ca2+ ions (Flego et al., 1997, 2000) while we have observed similar PhoPQ response to both Ca2+ and Mg2+ in Pve. In plant cells, Ca2+ was shown to be mostly cell wall-bound but it is released when the cell wall is degraded by pectobacteria (Park et al., 2004). Mg2+ concentration in plant apoplast was also reported to be quite low (Haque et al., 2008). On the other hand, the release of all ions bound in plant cells would result in concentrations of about 10 mM of Ca2+ and Mg2+ (Pagel and Heitefuss, 1989; Brown et al., 2012). Therefore, PhoPQ expression can be expected to be high at the onset of infection, resulting in the activation of the pehA gene expression. High polygalacturonase activity at the first stage of infection might change the availability of other cell wall components and would release cell wall-bound Ca2+. The combined action of divalent cations would lead to the repression of phoPQ transcription.
PelI is highly active in the presence of high Ca2+ concentrations, is known to be highly expressed in planta and is thought to be especially effective at the degradation of the calcium-rich middle lamella (Shevchik et al., 1997; Hugouvieux-Cotte-Pattat et al., 2014). High pelI expression, observed in potato tubers at late infection stages, may have a major effect on tissue maceration. Therefore, even higher PelI amounts in the phoP mutant can be the reason for its greater maceration ability during the artificial tuber inoculation.
The switch from PehA to PelI as the major pectinolytic enzyme correlates with the most likely sequence of events during plant cell wall polygalacturonate degradation and utilisation by a soft rot pathogen (Reverchon et al., 2016). Polygalacturonases have an acidic pH optimum and are therefore required at early stages of infection, as apoplastic fluid is slightly acidic. In contrast, PelI is active at the alkaline pH (Shevchik et al., 1997) and is relevant at the later stages of infection when the alkalinisation of the infection area occurs (Nachin and Barras, 2000). Release of further amounts of Ca2+ and Mg2+ by PelI may cause an even stronger decrease of phoPQ expression, resulting in the final stabilisation of PhoP regulon members expression at the levels required for the late stages of infection.
Contrary to PhoP-dependent transcription activation, repression does not seem to require phosphorylated PhoP and hence can still occur in the presence of high Ca2+ and/or Mg2+ concentrations, especially at promoter regions with multiple and/or high-affinity PhoP binding sites. However, as PhoP amounts are somewhat decreased in these conditions, the outcomes for different PhoP-repressed genes might vary: from partial derepression of genes with single and/or low-affinity PhoP binding site(s) to weak or almost no derepression of genes with multiple and/or high-affinity PhoP binding site(s).
An interesting aspect of PhoP-dependent repression links metabolic pathways of galacturonic acid-containing and arabinose-containing cell wall component utilisation. PhoP represses uptake and degradation of arabinose-containing cell wall components only in the presence of PGA. Although the exact mechanism of PGA involvement is not certain, the physiological consequences of such regulation are quite clear: galacturonic acid-containing cell wall breakdown products are preferred over arabinose-containing ones. This preference is relieved in the soft rot area (which could be due to exhausting PGA amounts) allowing the pathogen to utilise additional nutrients.
Many transporters are controlled by PhoP at least to some extent and most of them are activated. High transporter activity at the onset of infection would increase intracellular nutrient availability for the pathogen. Increased external nutrient availability at the later stages of infection justifies the reduction of transporter gene expression. Overall, transporter-related genes constitute approximately 1/3 of the DEGs with two largest groups of 7 genes each encoding iron and arabinose transporters. Citrate transporters seem to be a special case since one of them (CitM) is repressed while the other one (CitW) is activated by PhoP. The switch from CitW to CitM during plant colonisation may ensure continuous uptake of the abundant nutrient. Additional tri- and dicarboxylate transporters are likely to be controlled by PhoP too, but have been poorly expressed under conditions used in our work, possibly due to the missing inducers.
PhoP is also involved in the control of several stress resistance genes. The two largest groups of these genes defend Pve from cationic peptides and tellurite. Plants produce a rich arsenal of mostly CAMPs (Benko-Iseppon et al., 2010). The ugd-arnBCADTEF locus is strongly activated by PhoP and should protect against CAMPs from the early stages of infection when Pve cells may face high CAMP concentrations in the apoplast. In contrast, a large group of poorly characterised “tellurite resistance” genes are PhoP repressed and should be better expressed at the later stages of infection. PhoP-dependent coregulation of the ter genes with well-characterised virulence factors hints at their involvement in host adaptation. A possible contribution of these genes to oxidative resistance (Chasteen et al., 2009) may help plant pathogen to overcome host-generated oxidative burst. Since tellurite resistance genes are among the few that uniquely distinguish Pve from other pectobacteria, their detailed study may help to better understand the pathogenic strategy of this species.
It has long been established that anaerobic conditions are required for successful plant infection by soft rot bacteria (see Perombelon, 2002, for a review). This requirement was attributed to both the impairment of oxygen-dependent host resistance systems and increase of pathogen virulence. RNA-seq data show the involvement of PhoP in the regulation of 15 genes involved in anaerobic metabolism while in silico analysis suggests even more potential anaerobiosis-related PhoP targets. Since neither of these genes was reported to be controlled by PhoP in animal pathogens, their addition to PhoP regulon of Pve constitutes plant pathogen-specific extension of PhoP regulon. Some, but not all of the PhoP-controlled anaerobiosis-related genes are co-regulated by the main anaerobiosis regulator FNR, creating possibilities for complex regulatory effects. An interesting example is provided by cationic peptide resistance genes ugd and arnBCADTEF. Our results suggest that PhoP maintains a high expression level of these two transcription units in aerobic conditions in vitro while high expression level in phoP mutant cells within macerated potato tuber tissues can be explained by FNR-dependent transcription activation in anaerobic conditions. Other cases of Pve-specific PhoP and FNR operator co-occurrence include the ter operon and several pectinolysis genes.
Our results give additional hints to PhoPQ position in the regulatory network of Pve. Direct regulation of expI transcription links PhoP to the quorum sensing system. This link may result in a significant impact of PhoPQ on the secondary metabolism (including virulence factors production) via the well-established signal chain consisting of N-acylhomoserine lactone (AHL) produced by ExpI, AHL-responsive transcription factors ExpR (ExpR1) and VirR (ExpR2), and the global posttranscriptional regulator RsmA (Cui et al., 2006; Sjöblom et al., 2006). However, due to several autoregulatory mechanisms within the quorum sensing system and its intricate connections with other regulators, the exact outcome of increasing expI expression in a particular condition (especially in planta) is hard to predict. These mechanisms include autorepression of at least one of the AHL-responsive TFs (Castang et al., 2006) and interference between convergently transcribed expI and expR (Gogoleva et al., 2014). Additional complexity arises due to RsmA being targeted by multiple other regulators, including plant-responsive ones (Mukherjee et al., 2000; Hyytiainen et al., 2001). Of these, KdgR is the most important in the context of our work. KdgR directly represses transcription of RsmB, the regulatory RNA antagonising RsmA (Liu et al., 1999). In addition to this global regulatory mode, KdgR is also the major repressor of multiple pectinolysis genes (Liu et al., 1999). We have previously located KdgR binding sites within a pectobacterial genome (Nikolaichik and Damienikan, 2016) and now observed frequent co-occurrence of KdgR and PhoP binding sites in the regulatory regions of pectinolysis-related genes in Pve. This creates possibilities for delicate regulation of virulence properties, since both KdgR and PhoPQ can be inactivated in planta by their corresponding ligands, 2-keto-3-deoxygluconate (Nasser et al., 1992) and Ca2+/Mg2+. The mode of regulation is opposite for the two regulators (repression by KdgR and mostly activation by PhoP), hence the resulting changes in gene expression would depend on relative timing and extent of inactivation of the two regulators. Due to the overall complexity of the interaction network in soft rot pathogens, PhoP involvement in regulation of pectinolysis and anaerobiosis deserves dedicated studies.
We finally note that the PhoP regulon in Pve is likely to include many more genes apart from the DEGs found in this work. We believe that many of the additional PhoP binding sites found by the in silico approach are the true ones and can result in PhoP-dependent expression level changes in conditions different from the ones studied here. Taking into account numerous potential PhoP targets, high proportion of virulence-related and regulatory genes among them and likely interactions with other regulators, the possibility of PhoP involvement should be routinely considered when studying the expression of virulence factors in Pve and related plant pathogens.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA627079.
Author Contributions
UK: phoP mutant construction and preliminary characterisation, assessment of polymyxin B and tellurite resistance, virulence tests, gene expression measurement with qRT-PCR, visualisation, and writing, reviewing, and editing of the manuscript. NG: cDNA library preparation, sequencing, data quality control, and filtering. NK: gene expression measurement with qRT-PCR. AK: RNA-seq data analysis. YD: construction of the pPQ plasmid and complementation tests. YG: conceptualisation of the study, funding acquisition, resources, supervision, and reviewing and editing of the manuscript. YN: conceptualisation of the study, funding acquisition, resources, supervision, writing, reviewing, and editing of the manuscript, RNA isolation, virulence tests, structure analysis, TFBS inference, PhoP regulon analysis, and genome annotation correction. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Alexander Ageichik for critical reading of the manuscript. A preprint of this paper was published (Kravchenko et al., 2020).
Funding. This work was supported by grants from Belarusian Republican Foundation for Basic Research (project no. B18R-117), Russian Foundation for Basic Research (project no. 18-54-00021), State Programme for Scientific Research “Biotechnology” (project no. 2.44), and BSU Rector grant. The RNA-Seq analysis was supported by the Russian Science Foundation (project no. 17-14-01363). The cDNA-library preparation was worked out at the financial support of the Ministry of Science and Higher Education of the Russian Federation (grant no. 075-15-2019-1881). DNA sequencing was performed within the frameworks of the government assignment for FRC Kazan Scientific Center of RAS. The study was carried out by using the equipment of the CSF-SAC FRC KSC RAS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.621391/full#supplementary-material
References
- Abbott D. W., Boraston A. B. (2008). Structural biology of pectin degradation by Enterobacteriaceae. Microbiol. Mol. Biol. Rev. 72 301–316. 10.1128/MMBR.00038-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott D. W., Hrynuik S., Boraston A. B. (2007). Identification and characterization of a novel periplasmic polygalacturonic acid binding protein from Yersinia enterolitica. J. Mol. Biol. 367 1023–1033. 10.1016/j.jmb.2007.01.030 [DOI] [PubMed] [Google Scholar]
- Anantharaman V., Iyer L. M., Aravind L. (2012). Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. Biosyst. 8 3142–3165. 10.1039/c2mb25239b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader M. W., Sanowar S., Daley M. E., Schneider A. R., Cho U., Xu W., et al. (2005). Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122 461–472. 10.1016/j.cell.2005.05.030 [DOI] [PubMed] [Google Scholar]
- Bellieny-Rabelo D., Nkomo N. P., Shyntum D. Y., Moleleki L. N. (2020). Horizontally acquired quorum-sensing regulators recruited by the phop regulatory network expand the host adaptation repertoire in the phytopathogen Pectobacterium brasiliense. mSystems 5 e619–e650. 10.1128/mSystems.00650-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benko-Iseppon A. M., Lins Galdino S., Calsa T., Jr., Akio Kido E., Tossi A., Carlos Belarmino L., et al. (2010). Overview on plant antimicrobial peptides. Curr. Protein Pept. Sci. 11 181–188. 10.2174/138920310791112075 [DOI] [PubMed] [Google Scholar]
- Bera A., Herbert S., Jakob A., Vollmer W., Götz F. (2004). Why are pathogenic staphylococci so lysozyme resistant? the peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus: lysozyme resistance of S. aureus. Mol. Microbiol. 55 778–787. 10.1111/j.1365-2958.2004.04446.x [DOI] [PubMed] [Google Scholar]
- Berman H. M. (2000). The protein data bank. Nucleic Acids Res. 28 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott M. (1997). Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167 78–88. 10.1007/s002030050419 [DOI] [PubMed] [Google Scholar]
- Bowman S. E. J., Backman L. R. F., Bjork R. E., Andorfer M. C., Yori S., Caruso A., et al. (2019). Solution structure and biochemical characterization of a spare part protein that restores activity to an oxygen-damaged glycyl radical enzyme. J. Biol. Inorg. Chem. 24 817–829. 10.1007/s00775-019-01681-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N. L., Pimentel H., Melsted P., Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brown C. R., Haynes K. G., Moore M., Pavek M. J., Hane D. C., Love S. L., et al. (2012). Stability and broad-sense heritability of mineral content in potato: calcium and magnesium. Am. J. Potato Res. 89 255–261. 10.1007/s12230-012-9240-9 [DOI] [Google Scholar]
- Bullock W. O. (1987). XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with beta-galactosidase selection. Bio. Tecch. 5 376–379. [Google Scholar]
- Burr T., Barnard A. M. L., Corbett M. J., Pemberton C. L., Simpson N. J. L., Salmond G. P. C., et al. (2006). Identification of the central quorum sensing regulator of virulence in the enteric phytopathogen, erwinia carotovora: the VirR repressor. Mol. Microbiol. 59 113–125. 10.1111/j.1365-2958.2005.04939.x [DOI] [PubMed] [Google Scholar]
- Castang S., Reverchon S., Gouet P., Nasser W. (2006). Direct evidence for the modulation of the activity of the erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J. Biol. Chem. 281 29972–29987. 10.1074/jbc.M601666200 [DOI] [PubMed] [Google Scholar]
- Chasteen T. G., Fuentes D. E., Tantaleán J. C., Vásquez C. C. (2009). Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol. Rev. 33 820–832. 10.1111/j.1574-6976.2009.00177.x [DOI] [PubMed] [Google Scholar]
- Chatterjee A., Cui Y., Chaudhuri S., Chatterjee A. K. (2002). Identification of regulators of hrp/hop genes of Erwinia carotovora ssp carotovora and characterization of HrpLEcc (SigmaLEcc), an alternative sigma factor. Mol. Plant Pathol. 3 359–370. 10.1046/j.1364-3703.2002.00128.x [DOI] [PubMed] [Google Scholar]
- Coornaert A., Chiaruttini C., Springer M., Guillier M. (2013). Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 9:e1003156. 10.1371/journal.pgen.1003156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie M. J., Groisman E. A. (2010). Promoter and Riboswitch Control of the Mg2+ Transporter MgtA from Salmonella enterica. J. Bacteriol. 192 604–607. 10.1128/JB.01239-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Chatterjee A., Chatterjee A. K. (2001). Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant Microbe. Interact. 14 516–526. 10.1094/mpmi.2001.14.4.516 [DOI] [PubMed] [Google Scholar]
- Cui Y., Chatterjee A., Hasegawa H., Chatterjee A. K. (2006). Erwinia carotovora subspecies produce duplicate variants of ExpR, LuxR homologs that activate rsmA transcription but differ in their interactions with N-acylhomoserine lactone signals. J. Bacteriol. 188 4715–4726. 10.1128/JB.00351-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Chatterjee A., Hasegawa H., Dixit V., Leigh N., Chatterjee A. K., et al. (2005). ExpR, a LuxR homolog of Erwinia carotovora subsp. carotovora, activates transcription of rsmA, which specifies a global regulatory RNA-binding protein. J. Bacteriol. 187 4792–4803. 10.1128/JB.187.14.4792-4803.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Chatterjee A., Yang H., Chatterjee A. K. (2008). Regulatory network controlling extracellular proteins in erwinia carotovora subsp. carotovora: FlhDC, the master regulator of flagellar genes, activates rsmb regulatory RNA production by affecting gacA and hexA (lrhA) expression. J. Bacteriol. 190 4610–4623. 10.1128/JB.01828-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic-Attree I., Toussaint B., Garin J., Vignais P. M. (1997). Cloning, sequence and mutagenesis of the structural gene of Pseudomonas aeruginosa CysB, which can activate algD transcription. Mol. Microbiol. 24 1275–1284. 10.1046/j.1365-2958.1997.4121799.x [DOI] [PubMed] [Google Scholar]
- Eddy S. R. (2011). Accelerated profile HMM searches. PLoS Comput. Biol. 7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow J. M., Hudson L. L., Wells G., Coleman J. P., Pesci E. C. (2015). CysB negatively affects the transcription of pqsR and Pseudomonas quinolone signal production in Pseudomonas aeruginosa. J. Bacteriol. 197 1988–2002. 10.1128/JB.00246-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flego D., Marits R., Eriksson A. R., Koiv V., Karlsson M. B., Heikinheimo R., et al. (2000). A two-component regulatory system, pehR-pehS, controls endopolygalacturonase production and virulence in the plant pathogen erwinia carotovora subsp. carotovora. Mol. Plant Microbe. Interact. 13 447–455. 10.1094/mpmi.2000.13.4.447 [DOI] [PubMed] [Google Scholar]
- Flego D., Pirhonen M., Saarilahti H., Palva T. K., Palva E. T. (1997). Control of virulence gene expression by plant calcium in the phytopathogen Erwinia carotovora. Mol. Microbiol. 25 831–838. 10.1111/j.1365-2958.1997.mmi501.x [DOI] [PubMed] [Google Scholar]
- Fujimoto Z., Ichinose H., Maehara T., Honda M., Kitaoka M., Kaneko S., et al. (2010). Crystal Structure of an Exo-1,5-α-l-arabinofuranosidase from Streptomyces avermitilis provides insights into the mechanism of substrate discrimination between Exo- and Endo-type Enzymes in glycoside hydrolase family 43. J. Biol. Chem. 285 34134–34143. 10.1074/jbc.M110.164251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garriga X., Eliasson R., Torrents E., Jordan A., Barbé J., Gibert I., et al. (1996). nrdD and nrdG genes are essential for strict anaerobic growth of Escherichia coli. Biochem. Biophys. Res. Commun. 229 189–192. 10.1006/bbrc.1996.1778 [DOI] [PubMed] [Google Scholar]
- Gogoleva N., Kravchenko U., Nikolaichik Y., Gogolev Y. (2020). Transcriptomic dataset of wild type and phoP mutant Pectobacterium versatile. Data Brief 32:106123. 10.1016/j.dib.2020.106123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogoleva N. E., Shlykova L. V., Gorshkov V. Y., Daminova A. G., Gogolev Y. V. (2014). Effect of topology of quorum sensing-related genes in Pectobacterium atrosepticum on their expression. Mol. Biol. 48 583–589. 10.1134/S0026893314040049 [DOI] [PubMed] [Google Scholar]
- Gorshkov V., Gubaev R., Petrova O., Daminova A., Gogoleva N., Ageeva M., et al. (2018). Transcriptome profiling helps to identify potential and true molecular switches of stealth to brute force behavior in Pectobacterium atrosepticum during systemic colonization of tobacco plants. Eur. J. Plant Pathol. 152 957–976. 10.1007/s10658-018-1496-6 [DOI] [Google Scholar]
- Groisman E. A., Hollands K., Kriner M. A., Lee E.-J., Park S.-Y., Pontes M. H., et al. (2013). Bacterial Mg 2+ homeostasis, transport, and virulence. Annu. Rev. Genet. 47 625–646. 10.1146/annurev-genet-051313-051025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque M. M., Nahar K., Rahim M. A., Gomes I., Tsuyumu S. (2008). PhoP-PhoQ Two-component system required for colonization leading to virulence of dickeya dadantii 3937 in planta. Bangladesh J. Microbiol. 25 36–40. 10.3329/bjm.v25i1.4853 [DOI] [Google Scholar]
- Haque M. M., Tsuyumu S. (2005). Virulence, resistance to magainin II, and expression of pectate lyase are controlled by the PhoP-PhoQ two-component regulatory system responding to pH and magnesium in Erwinia chrysanthemi 3937. J. Gen. Plant Pathol. 71 47–53. 10.1007/s10327-004-0158-z [DOI] [Google Scholar]
- Harari O., Park S.-Y., Huang H., Groisman E. A., Zwir I. (2010). Defining the plasticity of transcription factor binding sites by deconstructing DNA consensus sequences: the PhoP-binding sites among gamma/enterobacteria. PLoS Comput. Biol. 6:e1000862. 10.1371/journal.pcbi.1000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. J., Shih Y. L., Bentley S. D., Salmond G. P. (1998). The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol. Microbiol. 28 705–717. 10.1046/j.1365-2958.1998.00825.x [DOI] [PubMed] [Google Scholar]
- He X., Wang L., Wang S. (2016). Structural basis of DNA sequence recognition by the response regulator PhoP in Mycobacterium tuberculosis. Sci. Rep. 6:24442. 10.1038/srep24442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horler R. S. P., Müller A., Williamson D. C., Potts J. R., Wilson K. S., Thomas G. H., et al. (2009). Furanose-specific sugar transport characterization of a bacterial galactofuranose-binding protein. J. Biol. Chem. 284 31156–31163. 10.1074/jbc.M109.054296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Condemine G., Shevchik V. E. (2014). Bacterial pectate lyases, structural and functional diversity: bacterial pectate lyases. Environ. Microbiol. Rep. 6 427–440. 10.1111/1758-2229.12166 [DOI] [PubMed] [Google Scholar]
- Hyytiainen H., Montesano M., Palva E. T. (2001). Global regulators ExpA (GacA) and KdgR modulate extracellular enzyme gene expression through the RsmA-rsmB system in Erwinia carotovora subsp. carotovora. Mol. Plant Microbe. Interact. 14 931–938. 10.1094/mpmi.2001.14.8.931 [DOI] [PubMed] [Google Scholar]
- Imperi F., Tiburzi F., Fimia G. M., Visca P. (2010). Transcriptional control of the pvdS iron starvation sigma factor gene by the master regulator of sulfur metabolism CysB in Pseudomonas aeruginosa. Environ. Microbiol. 12 1630–1642. 10.1111/j.1462-2920.2010.02210.x [DOI] [PubMed] [Google Scholar]
- Iwadate Y., Kato J. (2019). Identification of a formate-dependent uric acid degradation pathway in Escherichia coli. J. Bacteriol. 201 e518–e573. 10.1128/JB.00573-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafra S., Figura I., Hugouvieux-Cotte-Pattat N., Lojkowska E. (1999). Expression of erwinia chrysanthemi pectinase genes pelI, pelL, and pelZ during infection of potato tubers. Mol. Plant-Microbe. Interact. 12 845–851. 10.1094/mpmi.1999.12.10.845 [DOI] [Google Scholar]
- Kästner C. N., Schneider K., Dimroth P., Pos K. M. (2002). Characterization of the citrate/acetate antiporter CitW of Klebsiella pneumoniae. Arch. Microbiol. 177 500–506. 10.1007/s00203-002-0420-8 [DOI] [PubMed] [Google Scholar]
- Kotake T., Yamanashi Y., Imaizumi C., Tsumuraya Y. (2016). Metabolism of L-arabinose in plants. J. Plant Res. 129 781–792. 10.1007/s10265-016-0834-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko U., Gogoleva N., Kolubako A., Kruk A., Diubo J., Gogolev Y., et al. (2020). The PhoPQ two-component system is the major regulator of cell surface properties, stress responses and plant-derived substrate utilisation during development of Pectobacterium versatile -host plant pathosystem. bioRxiv 2020:060806 10.1101/2020.04.24.060806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy I. V., Boeva V. A., Favorov A. V., Makeev V. J. (2010). Deep and wide digging for binding motifs in ChIP-Seq data. Bioinforma. Oxf. Engl. 26 2622–2623. 10.1093/bioinformatics/btq488 [DOI] [PubMed] [Google Scholar]
- Linares-Pastén J. A., Falck P., Albasri K., Kjellström S., Adlercreutz P., Logan D. T., et al. (2017). Three-dimensional structures and functional studies of two GH43 arabinofuranosidases from Weissella sp. strain 142 and Lactobacillus brevis. FEBS J. 284 2019–2036. 10.1111/febs.14101 [DOI] [PubMed] [Google Scholar]
- Liu H., Coulthurst S. J., Pritchard L., Hedley P. E., Ravensdale M., Humphris S., et al. (2008). Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathog. 4:e1000093. 10.1371/journal.ppat.1000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chatterjee A., Chatterjee A. K. (1994). Nucleotide sequence, organization and expression of rdgA and rdgB genes that regulate pectin lyase production in the plant pathogenic bacterium Erwinia carotovora subsp. carotovora in response to DNA-damaging agents. Mol. Microbiol. 14 999–1010. 10.1111/j.1365-2958.1994.tb01334.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Jiang G., Cui Y., Mukherjee A., Ma W. L., Chatterjee A. K., et al. (1999). KdgREcc negatively regulates genes for pectinases, cellulase, protease, HarpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J. Bacteriol. 181 2411–2421. 10.1128/jb.181.8.2411-2421.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., Henrissat B. (2014). The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42 D490–D495. 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjurul Haque M., Hirata H., Tsuyumu S. (2012). Role of PhoP–PhoQ two-component system in pellicle formation, virulence and survival in harsh environments of Dickeya dadantii 3937. J. Gen. Plant Pathol. 78 176–189. 10.1007/s10327-012-0372-z [DOI] [Google Scholar]
- Matsuo N., Kaneko S., Kuno A., Kobayashi H., Kusakabe I. (2000). Purification, characterization and gene cloning of two α-L-arabinofuranosidases from Streptomyces chartreusis GS901. Biochem. J. 346 9–15. 10.1042/bj3460009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClean K. H., Chhabra S. R., Camara M., Daykin M., Swift S., Bycroft B. W., et al. (1997). Quorum sensing and Chromobacteriurn violaceum:exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143 3703–3711. 10.1099/00221287-143-12-3703 [DOI] [PubMed] [Google Scholar]
- McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. 10.1093/nar/gkt444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf W. W., Jiang W., Wanner B. L. (1994). Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6K gamma origin plasmids at different copy numbers. Gene 138 1–7. 10.1016/0378-1119(94)90776-5 [DOI] [PubMed] [Google Scholar]
- Meyer S., De Angeli A., Fernie A. R., Martinoia E. (2010). Intra- and extra-cellular excretion of carboxylates. Trends Plant Sci. 15 40–47. 10.1016/j.tplants.2009.10.002 [DOI] [PubMed] [Google Scholar]
- Moreira C. G., Herrera C. M., Needham B. D., Parker C. T., Libby S. J., Fang F. C., et al. (2013). Virulence and stress-related periplasmic protein (VisP) in bacterial/host associations. Proc. Natl. Acad. Sci. U S A. 110 1470–1475. 10.1073/pnas.1215416110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Cui Y., Ma W., Liu Y., Chatterjee A. K. (2000). hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum-sensing signal, N-(3-oxohexanoyl)- l-homoserine lactone. Environ. Microbiol. 2 203–215. 10.1046/j.1462-2920.2000.00093.x [DOI] [PubMed] [Google Scholar]
- Myamin V. E., Pesnyakevich A. G., Nikolaichik E. A., Prokulevich V. A. (2004). Genetic regulation of pathogenicity and virulence factors in bacteria erwinia carotovora subsp. atroseptica: identification of gene kduD. Russ. J. Genet. 40 970–976. 10.1023/B:RUGE.0000041374.14133.20 [DOI] [PubMed] [Google Scholar]
- Nachin L., Barras F. (2000). External pH: an environmental signal that helps to rationalize pel gene duplication in Erwinia chrysanthemi. Mol. Plant Microbe. Interact. 6 882–886. 10.1094/mpmi.2000.13.8.882 [DOI] [PubMed] [Google Scholar]
- Nasser W., Reverchon S., Robert-Baudouy J. (1992). Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol. Microbiol. 65 257–265. 10.1111/j.1365-2958.1992.tb02007.x [DOI] [PubMed] [Google Scholar]
- Needham B. D., Trent M. S. (2013). Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 11 467–481. 10.1038/nrmicro3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D. L., Kennedy E. P. (1971). Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J. Biol. Chem. 246 3042–3049. [PubMed] [Google Scholar]
- Nikolaichik Y., Damienikan A. U. (2016). SigmoID: a user-friendly tool for improving bacterial genome annotation through analysis of transcription control signals. PeerJ. 4:e2056. 10.7717/peerj.2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novichkov P. S., Kazakov A. E., Ravcheev D. A., Leyn S. A., Kovaleva G. Y., Sutormin R. A., et al. (2013). RegPrecise 3.0 – A resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 14:745. 10.1186/1471-2164-14-745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel W., Heitefuss R. (1989). Calcium content and cell wall polygalacturonans in potato tubers of cultivars with different susceptibilities to Erwinia carotovora subsp. atroseptica. Physiol. Mol. Plant Pathol. 35 11–21. 10.1016/0885-5765(89)90003-9 [DOI] [Google Scholar]
- Park S.-W., Hwang B.-H., Kim W.-Y., Kim J. (2004). Changes in cell wall carbohydrates composition and Ca distribution of Brassica campestris ssp. pekinesis in relation to Erwinia polygalacturonase production during soft rot development. Hortic. Environ. Biotechnol. 45 223–232. [Google Scholar]
- Pelosi L., Vo C.-D.-T., Abby S. S., Loiseau L., Rascalou B., Hajj Chehade M., et al. (2019). Ubiquinone biosynthesis over the entire O 2 range: characterization of a conserved O 2 -independent pathway. mBio 10:e1319. 10.1128/mBio.01319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold R. J., Pemberton J. M. (1992). An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118 145–146. 10.1016/0378-1119(92)90263-o [DOI] [PubMed] [Google Scholar]
- Perez J. C., Shin D., Zwir I., Latifi T., Hadley T. J., Groisman E. A., et al. (2009). Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:1000428. 10.1371/journal.pgen.1000428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perombelon M. C. M. (2002). Potato diseases caused by soft rot erwinias: an overview of pathogenesis. Plant Pathol. 51 1–12. 10.1046/j.0032-0862.2001.short [DOI] [Google Scholar]
- Perron-Savard P., De Crescenzo G., Moual H. L. (2005). Dimerization and DNA binding of the Salmonella enterica PhoP response regulator are phosphorylation independent. Microbiology 151 3979–3987. 10.1099/mic.0.28236-0 [DOI] [PubMed] [Google Scholar]
- Pfaffl M. W., Horgan G. W., Dempfle L. (2002). Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. 10.1093/nar/30.9.e36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier P., Pédron J., Taghouti G., Fischer-Le Saux M., Caullireau E., Bertrand C., et al. (2019). Elevation of pectobacterium carotovorum subsp. odoriferum to species level as pectobacterium odoriferum sp. nov., proposal of pectobacterium brasiliense sp. nov. and pectobacterium actinidiae sp. nov., emended description of pectobacterium carotovorum and description of pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 69 3207–3216. 10.1099/ijsem.0.003611 [DOI] [PubMed] [Google Scholar]
- Prost L. R., Daley M. E., Le Sage V., Bader M. W., Le Moual H., Klevit R. E., et al. (2007). Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26 165–174. 10.1016/j.molcel.2007.03.008 [DOI] [PubMed] [Google Scholar]
- Raghavan R., Groisman E. A., Ochman H. (2011). Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 21 1487–1497. 10.1101/gr.119370.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S., Muskhelisvili G., Nasser W. (2016). “Virulence Program of a Bacterial Plant Pathogen: The Dickeya Model,” in Progress in Molecular Biology and Translational Science. Amsterdam: Elsevier; 51–92. [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahota G., Stormo G. D. (2010). Novel sequence-based method for identifying transcription factor binding sites in prokaryotic genomes. Bioinformatics 26 2672–2677. 10.1093/bioinformatics/btq501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleif R. (2010). AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 34 779–796. 10.1111/j.1574-6976.2010.00226.x [DOI] [PubMed] [Google Scholar]
- Sharma C. M., Papenfort K., Pernitzsch S. R., Mollenkopf H.-J., Hinton J. C. D., Vogel J. (2011). Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA: GcvB regulon. Mol. Microbiol. 81 1144–1165. 10.1111/j.1365-2958.2011.07751.x [DOI] [PubMed] [Google Scholar]
- Shevchik V. E., Boccara M., Vedel R., Hugouvieux-Cotte-Pattat N. (1998). Processing of the pectate lyase PelI by extracellular proteases of Erwinia chrysanthemi 3937. Mol. Microbiol. 29 1459–1469. [DOI] [PubMed] [Google Scholar]
- Shevchik V. E., Robert-Baudouy J., Hugouvieux-Cotte-Pattat N. (1997). Pectate lyase PelI of Erwinia chrysanthemi 3937 belongs to a new family. J. Bacteriol. 179 7321–7330. 10.1128/jb.179.23.7321-7330.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirshikov F. V., Korzhenkov A. A., Miroshnikov K. K., Kabanova A. P., Barannik A. P., Ignatov A. N., et al. (2018). Draft genome sequences of new genomospecies “Candidatus Pectobacterium maceratum” strains, which cause soft rot in plants. Genome Announc. 6 e218–e260. 10.1128/genomeA.00260-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöblom S., Brader G., Koch G., Palva E. T. (2006). Cooperation of two distinct ExpR regulators controls quorum sensing specificity and virulence in the plant pathogen Erwinia carotovora. Mol. Microbiol. 60 1474–1489. 10.1111/j.1365-2958.2006.05210.x [DOI] [PubMed] [Google Scholar]
- Slauch J. M., Lee A. A., Mahan M. J., Mekalanos J. J. (1996). Molecular characterization of the oafA locus responsible for acetylation of Salmonella typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J. Bacteriol. 178 5904–5909. 10.1128/jb.178.20.5904-5909.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer A. M., Currenti S., Bonocora R. P., Baranowski C., Petrone B. L., Palumbo M. J., et al. (2014). Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J. Bacteriol. 196 660–671. 10.1128/JB.01007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson N. R., Nasser W., McGowan S., Sebaihia M., Salmond G. P. (1999). Erwinia carotovora has two KdgR-like proteins belonging to the IciR family of transcriptional regulators: identification and characterization of the RexZ activator and the KdgR repressor of pathogenesis. Microbiology 145 1531–1545. 10.1099/13500872-145-7-1531 [DOI] [PubMed] [Google Scholar]
- Toth I. K., Birch P. R. (2005). Rotting softly and stealthily. Curr. Opin. Plant Biol. 8 424–429. 10.1016/j.pbi.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Touzé T., Blanot D., Mengin-Lecreulx D. (2008a). Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J. Biol. Chem. 283 16573–16583. 10.1074/jbc.M800394200 [DOI] [PubMed] [Google Scholar]
- Touzé T., Tran A. X., Hankins J. V., Mengin-Lecreulx D., Trent M. S. (2008b). Periplasmic phosphorylation of lipid a is linked to the synthesis of undecaprenyl phosphate. Mol. Microbiol. 67 264–277. 10.1111/j.1365-2958.2007.06044.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsers I., Gorshkov V., Gogoleva N., Parfirova O., Petrova O., Gogolev Y. (2020). Plant soft rot development and regulation from the viewpoint of transcriptomic profiling. Plants 9:1176. 10.3390/plants9091176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A., Cutcutache I., Koressaar T., Ye J., Faircloth B. C., Remm M., et al. (2012). Primer3—new capabilities and interfaces. Nucleic Acids Res. 40:e115. 10.1093/nar/gks596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbany C., Neuhaus H. E. (2008). Citrate uptake into pectobacterium atrosepticum is critical for bacterial virulence. Mol. Plant. Microbe Interact. 21 547–554. 10.1094/MPMI-21-5-0547 [DOI] [PubMed] [Google Scholar]
- Valková D., Valkovičová L., Vávrová S., Kováčová E., Mravec J., Turňa J. (2007). The contribution of tellurite resistance genes to the fitness of Escherichia coli uropathogenic strains. Open Life Sci. 2 182–191. 10.2478/s11535-007-0019-9 [DOI] [Google Scholar]
- Valkovicova L., Valkova D., Vavrova S., Alekhina O., Hoang V. P., Jezna M., et al. (2011). The role of TerW protein in the tellurite resistance of uropathogenic Escherichia coli. Biologia 66 565–573. 10.2478/s11756-011-0075-5 [DOI] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Véscovi E. G., Soncini F. C., Groisman E. A. (1996). Mg2+ as an Extracellular Signal: Environmental Regulation of Salmonella Virulence. Cell 84 165–174. 10.1016/S0092-8674(00)81003-X [DOI] [PubMed] [Google Scholar]
- Wagner A. F. V., Schultz S., Bomke J., Pils T., Lehmann W. D., Knappe J. (2001). YfiD of Escherichia coli and Y06I of Bacteriophage T4 as autonomous Glycyl radical cofactors reconstituting the catalytic center of oxygen-fragmented pyruvate formate-lyase. Biochem. Biophys. Res. Commun. 285 456–462. 10.1006/bbrc.2001.5186 [DOI] [PubMed] [Google Scholar]
- Whelan K. F., Colleran E., Taylor D. E. (1995). Phage inhibition, colicin resistance, and tellurite resistance are encoded by a single cluster of genes on the IncHI2 plasmid R478. J. Bacteriol. 177 5016–5027. 10.1128/jb.177.17.5016-5027.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan K. F., Sherburne R. K., Taylor D. E. (1997). Characterization of a region of the IncHI2 plasmid R478 which protects Escherichia coli from toxic effects specified by components of the tellurite, phage, and colicin resistance cluster. J. Bacteriol. 179 63–71. 10.1128/jb.179.1.63-71.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojnowska M., Walker D. (2019). FusB energises import across the outer membrane through direct interaction with its ferredoxin substrate. Microbiology 11 e2020–e2081. 10.1101/749960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T. L., et al. (2012). Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Jin F., Glatter T., Sourjik V. (2017). Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. U S A. 114 E10792–E10798. 10.1073/pnas.1717272114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanegina O., Kirsanov D., Baulin E., Karyagina A., Alexeevski A., Spirin S., et al. (2016). An updated version of NPIDB includes new classifications of DNA–protein complexes and their families. Nucleic Acids Res. 44 D144–D153. 10.1093/nar/gkv1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA627079.