Abstract
Key message
Eleven new major resistance genes for lettuce downy mildew were introgressed from wild Lactuca species and mapped to small regions in the lettuce genome.
Abstract
Downy mildew, caused by the oomycete pathogen Bremia lactucae Regel, is the most important disease of lettuce (Lactuca sativa L.). The most effective method to control this disease is by using resistant cultivars expressing dominant resistance genes (Dm genes). In order to counter changes in pathogen virulence, multiple resistance genes have been introgressed from wild species by repeated backcrosses to cultivated lettuce, resulting in numerous near-isogenic lines (NILs) only differing for small chromosome regions that are associated with resistance. Low-pass, whole genome sequencing of 11 NILs was used to identify the chromosome segments introgressed from the wild donor species. This located the candidate chromosomal positions for resistance genes as well as additional segments. F2 segregating populations derived from these NILs were used to genetically map the resistance genes to one or two loci in the lettuce reference genome. Precise knowledge of the location of new Dm genes provides the foundation for marker-assisted selection to breed cultivars with multiple genes for resistance to downy mildew.
Electronic supplementary material
The online version of this article (10.1007/s00122-020-03711-z) contains supplementary material, which is available to authorized users.
Introduction
Downy mildew, caused by the oomycete Bremia lactucae, is the most important disease in lettuce (Lactuca sativa L.) worldwide. B. lactucae is a biotrophic pathogen that primarily infects the foliar tissue, reducing yield and decreasing the quality of the marketable portion of the crop. The use of cultivars carrying dominant resistant genes (Dm genes) is the most effective strategy to control this disease. However, B. lactucae rapidly evolves to new virulence phenotypes that defeat individual Dm genes. For this reason, lettuce breeding programs continually seek new sources of resistance to downy mildew and new resistance genes (Crute 1992; Lebeda et al. 2014; Parra et al. 2016).
For over fifty years, breeding for resistance to B. lactucae has relied on the introgression of new genes from wild species (Crute 1992; Beharav et al. 2006; Parra et al. 2016). Of the 100 wild Lactuca species described thus far, only 14 are known to be natural hosts of B. lactucae (Lebeda et al. 2002). L. serriola and L. saligna have been used as sources of resistance to downy mildew in multiple breeding programs. L. virosa also possesses race-specific resistance to B. lactucae, but its use in breeding programs has been restricted due to infertility barriers between L. virosa and L. sativa; however, interspecific crosses followed by embryo rescue have occasionally allowed the transfer of resistance from L. virosa to L. sativa (Maisonneuve et al. 1999; Maisonneuve 2003). Introgression of resistance from L. serriola, L. saligna, and L. virosa through repeated backcrosses to L. sativa has been carried out by several public and commercial breeding programs resulting in a large increase in the number of resistance genes being deployed (Michelmore et al. 2013).
The genome of L. sativa cv. Salinas has been sequenced and assembled (Reyes-Chin-Wo et al. 2017). The latest assembly covers 2.4 Gb of the total estimated 2.7 Gb lettuce genome; these sequences are ordered in nine chromosomal pseudomolecules (Reyes-Chin-Wo et al. 2017). The 27 known Dm genes are located in major resistance clusters (MRC) that contain genes encoding nucleotide-binding leucine-rich repeats (NLRs) (Christopoulou et al. 2015a, b). Currently, over 50 genes for resistance to lettuce downy mildew have been reported and genetically characterized to varying extents (reviewed in Parra et al. 2016). While 27 Dm genes have been mapped to individual clusters in the reference genome, another 23 resistance factors remain uncharacterized (Parra et al. 2016). The constant advances in genome sequencing technologies have increased the amount of sequence data generated, while the cost of sequencing continues to decrease; this enables the use of whole-genome sequencing (WGS) as a tool to compare different genotypes, find introgressed regions with high divergence, and indicate candidate positions of resistance genes.
In this study, we used WGS of 11 near isogenic lines (NILs) and recurrent genotypes to identify the chromosome segments introgressed from wild donor species. Segregating populations derived from these NILs were used to position the new resistances on the reference genome of lettuce. The knowledge of the position of these new genes for resistance to lettuce downy mildew will facilitate marker-assisted selection to pyramid multiple Dm genes into cultivars, which will increase the evolutionary hurdle for the pathogen to become virulent and provide more durable disease resistance.
Methods
Plant materials
Multiple advanced breeding lines have been generated during the last four decades as products of the lettuce breeding program at UC Davis. The 11 NILs used in this study had been previously generated from crosses of wild Lactuca accessions and L. sativa cv. Salinas, followed by six backcrosses to cv. Salinas (Table 1). F2 populations were derived from crosses of each NIL (male) with L. sativa cv. Salinas (female).
Table 1.
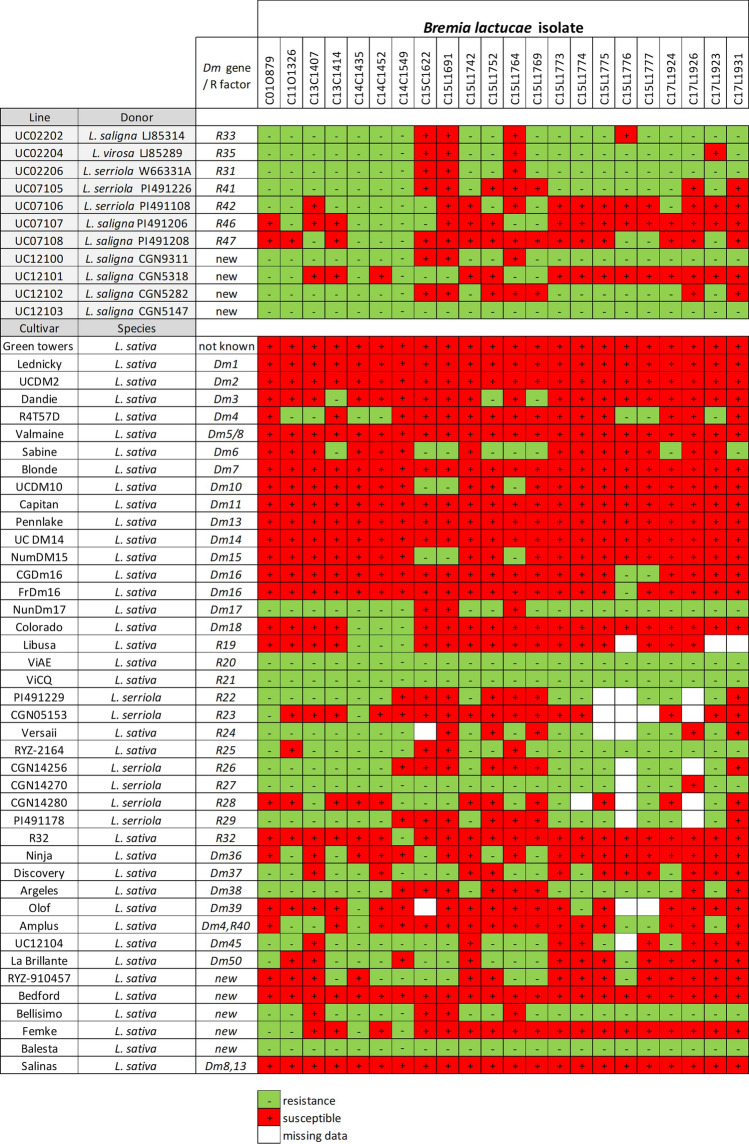
Resistance specificities to multiple isolates of B. lactucae in near isogenic lines (NILs) with introgressed chromosomal segments from wild species of Lactuca
Green/negative = resistance; red/plus = susceptible. L. sativa cv. Salinas (recurrent parent) is susceptible to all isolates used in this study
Low-pass whole genome sequencing
DNA was extracted from seedlings of each NIL using GeneElute™ Plant DNA extraction miniprep kit (Sigma-Aldrich, MO, USA). DNA was fragmented using a COVARISR sonicator in order to obtain an average fragment size of 350 bp. Genomic DNA libraries for individual NILs were made using KAPA hyper prep kit for Illumina sequencing (KAPA Biosystems, MA, USA). Libraries were size selected for a range of 300–400 bp using BluePippin (Sage Science, MA, USA), then barcoded, multiplexed, and sequenced using an Illumina Hi-seq 2500 platform (Illumina Inc., CA, USA).
Determination of introgressed region
Read mapping, variant detection, and visualization of SNP density was done using CLC Genomics Workbench 11.0 (https://www.qiagenbioinformatics.com/). Reads from each NIL were mapped into the Salinas reference genome v8 (Reyes-Chin-Wo et al. 2017), using a minimum cut-off of 90% similarity. Single nucleotide polymorphisms (SNPs) were only called for regions with a minimum coverage of three reads and when present in 90% of the total number of reads at each position. To avoid non-specific variants, SNPs generated by mapping cv. Salinas reads against the cv. Salinas reference genome were filtered out. Illumina reads from cv. Salinas 88 (cv. Salinas + mo gene introgressed from L. serriola PI 251245) were used to set the optimal parameters for mapping and variant detection against the cv. Salinas reference genome.
Tests for resistance to Bremia lactucae
NILs were evaluated using seedling-based tests for resistance to multiple isolates of B. lactucae from California. All isolates were grown on seedlings of L. sativa cv. Green Towers, which expresses no known genes for resistance to downy mildew. Spores were collected in distilled water after 7 days and used immediately for inoculations. Seedlings from NILs and 42 lettuce cultivars (Table 1) were grown in a growth chamber at 17 °C for 7 days and sprayed with a ~ 104 spores/mL suspension of B. lactucae. Lines/cultivars with sporulation on more than 50% of the seedlings were considered susceptible; lines/cultivars with no sporulation were considered resistant. For the phenotyping of F2 individuals, cotyledons from 7-day-old seedlings were detached and placed over wet filter paper in 48-well cell culture plates and then inoculated with ~ 25 µL of a suspension of B. lactucae spores. Cotyledons with full sporulation covering the whole leaf area were considered susceptible, while cotyledons with no sporulation were considered resistant. Cotyledons with sparse sporulation (only a few such cotyledons were observed) were not considered in the analysis. For the phenotyping of F3 families derived from F2 individuals, seedlings were inoculated following the same protocol used for the NILs. Resistance phenotypes for both seedlings and detached cotyledons were recorded 7 and 14 days post-inoculation.
Genotyping and mapping of R-genes
A subset of the F2 populations derived from each NIL were genotyped using genotyping-by-sequencing (GBS) as described in Macias-González et al. (2019) (Table 2). DNA was extracted from ~ 50 seeds from each F2 plant, digested with AvaII, ligated to barcoded adapters, and multiplexed in pools ranging from 250 to 300 individuals, with cv. Salinas included in each set. Pools were size-selected to ~ 350 bp using BluePippin and paired-end sequenced using an Illumina Hi-seq 2500 platform (Illumina Inc., CA, USA). After demultiplexing, TASSEL3 was used to obtain individual genotypes (Glaubitz et al. 2014). Visualization and selection of SNPs was done using Microsoft Excel. The SNP genotypes were converted to alleles A, H, or B, where A was homozygous for the susceptible parent, B was homozygous for the resistant parent, and H was heterozygous. For the lines UC07106 and UC07108, standard interval mapping was performed using R-QTL (Broman et al. 2003). The significance threshold for the logarithm (base 10) of odds scores (LOD) for QTLs was calculated by 1000 permutations at the significance thresholds of α < 0.05 and α < 0.01.
Table 2.
F2 segregation ratios and chromosomal positions of R-genes present in NILs
| Line | Isolate (F2 phenotyping) | No. plants (R/S) | Seg. ratio | Chi-square | Genetics | Isolate (F3 phenotypimg) | No. of sequenced | R-gene position* | Dm gene |
|---|---|---|---|---|---|---|---|---|---|
| UC02202 | C11O1326 | 105R/35S | 3:1 | 0 | 1 Dominant | C15L1742 | 65 | Chr1 | Dm33 |
| UC02204 | C11O1326 | 107R/36S | 3:1 | 0.002 | 1 Dominant | C15L1777, C17L1926 | 72 | Chr1 | Dm35 |
| UC02206 | C11O1326 | 109R/35S | 3:1 | 0.037 | 1 Dominant | C17L1926 | 72 | Chr1 | R31 |
| UC07105 | C11O1326 | 105R/31S | 3:1 | 0.352 | 1 Dominant | C15L1777 | 67 | MRC4 | R41 |
| UC07106 | C11O1326 | 90R/44S | ~3:1 | 3.59 | 1 Dominant | C15L1752 | 96 | MRC1 | R42 |
| UC07107 | C11O1326 | 32R/97S | 1:3 | 0.002 | 1 Recessive | C1101326 | 116 | MRC1 | Dm46 |
| UC07108 | C13O1407 | 68R/56S | 9:7 | 0.1 | 2 Dominant | C15L1777 | 106 | MRC2, MRC4 | R47 |
| UC12100 | C11O1326 | 104R/32S | 3:1 | 0.157 | 1 Dominant | C15L1742, C17L1926 | 72 | Chr1 | s.f.d (R60) |
| UC12101 | C11O1326 | 81R/63S | 9:7 | 0 | 2 Dominant | C15L1691 | 143 | MRC1, MRC2 | s.f.d (R64) |
| UC12102 | C11O1326 | 109R/35S | 3:1 | 0.037 | 1 Dominant | C15L1777 | 72 | MRC4 | s.f.d (R61) |
| UC12103 | C11O1326 | 111R/33S | 3:1 | 0.333 | 1 Dominant | C17L1926 | 72 | Chr1 | s.f.d (R62) |
| UC12103 | 22R/10S | 3:1 | 0.666 | 1 Dominant | C15L1691 | 32 | MRC1 | s.f.d (R63) |
Fragments co-located with MRC clusters are shown in bold letters (*based on the reference genome of cv. Salinas). R/S resistant/susceptible. Chi-square calculated according to Σ (observed value-expected value)2/expected value. s.f.d: submitted for denomination
Availability of data and materials
Phenotypic and genotypic datasets for the progenies are available upon request.
Results
Screening of NILs for resistance to multiple B. lactucae isolates from California
Eleven NILs and another 42 lettuce cultivars carrying known Dm genes were evaluated using seedling-based tests for resistance to 25 isolates of B. lactucae from California. Seven of these NILs were resistant to most of the isolates tested, while the other four NILs were resistant to at least ten B. lactucae isolates (Table 1). Several NILs showed patterns of resistance consistent with the expression of new Dm resistance genes. Interestingly, the NILs UC02206 and UC12100 displayed the same resistance specificity as the line NunDm17, which carries Dm17 (Table 1), and the NILs UC07105 and UC12102 exhibited the same resistance profile. UC12103 was resistant to all tested isolates. Only one lettuce cultivar, Balesta, was resistant to all tested isolates (Table 1). Two lines with resistance from L. virosa, ViAE and ViCQ, were also resistant to all isolates tested.
Determination of introgressed regions in NILs using low-pass whole genome shotgun sequencing data
L. sativa cv. Salinas that was used as a recurrent parent for the generation of the NILs was also sequenced to assemble the reference genome (Reyes-Chin-Wo et al. 2017). This facilitated the identification of introgressed regions in the NILs by visualizing chromosomal regions with high densities of SNPs when comparing reads of the NILs aligned against the reference genome. This approach was validated using cv. Salinas 88, which contains the mo gene for resistance to lettuce mosaic virus introgressed from L. serriola PI251245 (Ryder 1991; Mikel 2007) into cv. Salinas. Salinas 88 reads were aligned against the Salinas reference genome and a single small (16 Mb) introgressed segment was identified on Chromosome (Chr) 4, which had previously been shown genetically to contain the mo gene (Fig. 1; Irwin et al. 1999). Salinas 88 was sequenced to 2.2×; analysis with different levels of coverage determined that the minimal average read coverage needed to accurately detect the size of the introgressed segment in cv. Salinas 88 was 0.5×.
Fig. 1.
Detection of introgression in Salinas 88. Detection of SNP variants against the Lactuca sativa cv. Salinas reference genome shows a highly polymorphic small region in Chromosome 4 (red bars). The mo gene is located in the middle of the introgressed region
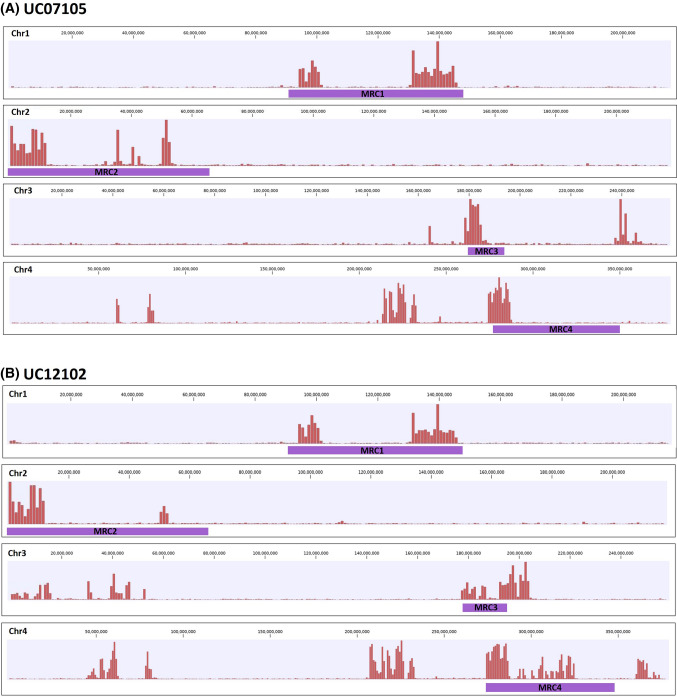
We were able to identify introgressed regions in all of the NILs. The NILs used in this study had been sequenced to an average read coverage of 1.1 × when mapped against the cv. Salinas reference genome. Some breeding lines had only a few separate segments with a high density of SNPs, while others contained multiple polymorphic regions (Supplementary Fig. 1). All of the NILs showed potential introgressions of regions encoding one or more MRCs (Supplementary Fig. 1). Most of the lines had introgressions of MRC1 and MRC2; only a few NILs had introgressions of MRC3 and MRC4 (Fig. 2). The SNP profiles are similar among some lines at MRC1 or MRC2; the reason for this is unclear but may reflect uneven recombination across the genome following interspecific crosses.
Fig. 2.
Example of introgressions in near isogenic lines (NILs) that are co-located with MRCs. a Introgressions for line UC07105; b introgressions for line UC12102. SNP density is indicated with red bars. Purple bars indicate the MRC regions in the reference genome
Genetic mapping of new genes for resistance to downy mildew in lettuce
To identify which of the introgressed segments in the NILs are involved in DM resistance, F2 populations derived from ten NILs (UC02202, UC02204, UC02206, UC07105, UC07106, UC07107, UC12100, UC12101, UC12102, and UC12103) were screened for resistance to B. lactucae isolate C11O1326. The F2 population derived from UC07108 was screened for resistance to B. lactucae isolate C13C1407 rather than C11O1326 because the latter isolate was virulent on UC07108. The F2 populations derived from UC02202, UC02204, UC02206, UC07105, UC07106, UC12100, UC12102, and UC12103 showed a segregation ratio of 3:1 between resistant and susceptible individuals, consistent with one dominant resistant gene conferring resistance to downy mildew. The F2 populations derived from UC07108 and UC12101 showed a segregation ratio of 9:7, consistent with two dominant genes being required for resistance. The population derived from UC07107 had a segregation ratio of 1:3 between resistant and susceptible individuals, consistent with the presence of a recessive resistance gene.
F2 populations were genotyped using GBS and resistance genes were positioned on the reference genome (Table 2, Fig. 3, and Supplementary Table 1). From the nine populations with simple Mendelian segregation (3:1), six had resistance that mapped to Chr 1, while two populations had resistance that mapped to Chr 4. The recessive resistance to B. lactucae isolate C11O1326 present in UC07107 was positioned inside MRC1, while the resistance to the same isolate present in UC07105 and UC12102 was positioned inside MRC4. For the resistance genes present in the F2 populations derived from UC07108 and UC12101, which had segregation ratios of 9:7, the resistances were mapped to MRC2 and MRC4, and MRC1 and MRC2, respectively. The resistances to C11O1326 present in UC02202, UC02204, UC02206, UC12100, and UC12103 mapped outside the MRC regions (Fig. 3). For line UC07106, a region in Chr 1 was significantly associated with resistance.
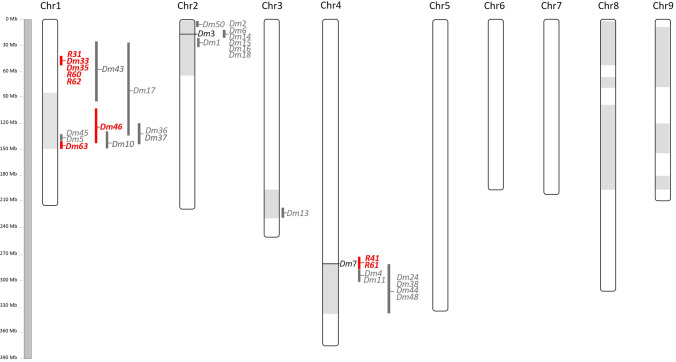
Fig. 3.
Graphic representation of lettuce Chromosomes 1, 2, 3, and 4 with the genomic location of new and known resistance genes. Genes in red letters were mapped in this study. Gray regions show the size intervals of the major resistance clusters MRC1, MRC2, MRC3, and MRC4
Phenotyping seedlings of F3 families allowed us to narrow down the mapping interval for resistance genes from NILs UC07105, UC07106, UC12100, and UC12102 (due to the ability to phenotypically distinguish homozygous and heterozygous F2 resistant lines) to under half of the size detected in the F2. Additionally, by using F3 families we were able to test the resistances found in the F2 populations against another four B. lactucae isolates (C15L1742, C15L1752, C15L1777, C17L1926, and C15L1691). We found that the resistance genes detected in nine NILs can also confer resistance to at least one additional B. lactucae isolate. Moreover, we found an additional resistance gene in UC12103 that confers resistance to C15L1691, which was positioned inside MRC1.
Denomination of the new Dm genes
The resistance identified in UC02202, UC02204, UC07105, UC07106, UC07107, and UC07108 was derived from donors with R-factors previously described in Parra et al. (2016); therefore, the Dm genes in these lines were named according to the R-factor denominations previously assigned to these donors (Table 2). However, until the uniqueness of the resistance specificity in UC02206 and UC07105 has been proven, the R-genes in these lines will remain R-factors. The same criteria was applied to the resistance genes in UC12100, UC12102, and the two new R genes in UC12103, which have been submitted to the International Bremia Evaluation Board (IBEB) for denomination as R60, R61, R62, and R63, respectively (Table 2). Genes in UC07108 and UC12101 will be kept as resistance factors (R), until we know which of the two genes in each line is the actual R-gene.
Discussion
The genetic mapping of resistance genes in lettuce at high resolution can be challenging. Previously, Dm genes have been mapped using segregating populations, mostly F2 populations (Hulbert and Michelmore 1985; Farrara et al. 1987; Maisonneuve et al. 1994; McHale et al. 2009; Jeuken et al. 2009; Simko et al. 2015), allowing the determination of the map position for 27 Dm genes (Fig. 3) (Parra et al. 2016). However, the mapping resolution for most of these genes comprises windows from 10 to 70 Mb (McHale et al. 2009, Christopoulou et al. 2015b). In this study, we mapped an additional 11 Dm genes to different positions in the L. sativa cv. Salinas reference genome, six of them to genomic intervals no larger than 6 Mb (Fig. 3, Supplementary Table 1). The highest resolution was the localization of the resistance gene in UC12102 to a 3.4 Mb window.
Several NILs used in this study had unique resistance profiles against the range of B. lactucae isolates tested. The line UC12103 showed resistance to all 25 isolates tested. The lines Balesta, VIAE, and VICQ were also resistant to all tested isolates. However, the genomic positions of the resistances present in these lines have not been reported, which makes it impossible to determine whether they share the same gene(s), or whether the observed phenotype is due to a combination of several Dm genes that recognize multiple Avr proteins from these isolates of B. lactucae. Interestingly, the NILs UC02206 and UC12100 displayed the same resistance profile as NunDm17, which carries Dm17, despite the fact their resistances were introgressed from different wild sources (L. serriola W66331A, L. saligna CGN9311, and L. serriola LS102, respectively). Both resistances present in these lines mapped to Chromosome 1, overlapping the genomic region where Dm17 had been previously mapped (Maisonneuve et al. 1994; Mc Hale et al. 2009) (Fig. 3). Similarly, the lines UC07105 and UC12102 had identical resistance specificities to each other and their resistances mapped to the same region on Chromosome 4, despite both resistances being introgressed from different Lactuca species (L. serriola PI491226 and L. saligna CGN5282, respectively). Different lettuce genotypes with the same resistance specificity to isolates of B. lactucae have been observed before. An example of this is Dm5 in L. serriola PI167150 and Dm8 in L. serriola PI104584. Dm5 and Dm8 confer resistance to the same spectrum of isolates and map to the same genetic position (Norwood and Crute 1984; Hulbert and Michelmore 1985). Future cloning of the genes present in the NILs will clarify whether these lines carry the same Dm gene or different Dm genes with the same downy mildew resistance specificity.
Genome introgressions from wild Lactuca spp.
Whole genome sequencing and variant detection is a successful and low-cost method to identify introgressions from wild species (Severin et al. 2010). However, the polymorphic regions visualized on the cv. Salinas reference genome may not necessarily represent the real size of the introgressed segments from the wild donors in the NILs. Alignment to a reference genome limits the possibility of displaying regions with extremely high divergence or regions that have been deleted/inserted in the chromosomal segment of the wild donor. Some introgressed segments were very small in the NILs, as expected in lines that have been backcrossed multiple times to the recurrent parent. However, some lines had extremely large chromosomal segments introgressed from the wild parent. Low recombination rates can be a product of highly divergent regions between cultivated and wild type Lactuca spp. (Liharska et al. 1996).
For UC07106, a ~ 40 Mb region in MRC1 was significantly associated with resistance to isolate C11O1326 (Supplementary Fig. 2), although not all the susceptible individuals had a homozygous susceptible genotype, indicative of incomplete penetrance of this resistance. Phenotyping of F3 families followed by QTL mapping narrowed down the region to 31 Mb and confirmed the discrepancy between the susceptible genotype and phenotype; F3 data also demonstrated cosegregation of resistance to an additional B. lactucae isolate, C15L1752 (Supplementary Fig. 2). Segregation in the Salinas x UC07106 F2 population deviated from simple Mendelian expected ratios; however, the closest fit was to a 3:1 segregation ratio, consistent with the presence of a dominant resistance gene. According to the SNP density profiles, the UC07106 line shows an introgression in Chromosome 1 that replaced more than half of the chromosome, including the MRC1 (Supplementary Fig. 1). This could indicate high divergence in this region between cv. Salinas and L. serriola PI491108 (recurrent and resistant donor for this NIL, respectively). Therefore, we hypothesize that the resistance gene in line UC07106 is located in a chromosomal region that is not present in the reference genome. Rearrangement of Chromosome 1 has been observed in L. sativa cv. Tizian, which showed a large region that is either missing or highly divergent from MRC1 in the cv. Salinas reference genome (Verwaaijen et al. 2018).
Different genetic basis for resistance to B. lactucae
Two NILs, UC07108 and UC12101, had an F2 segregation ratio that indicates that two dominant genes are required for the resistance phenotype. This could be indicative of the presence of one NLR R-gene that requires an additional gene for either recognition of pathogen effector or signaling of the defense response (Warren et al. 1999). One possibility is that two NLR genes are acting as a pair, with one of the NLR genes acting as a sensor NLR and the other as executor NLR (Jubic et al. 2019). However, a characteristic feature of NLR pairs in other species is that both members are often neighbors in inverse orientation in the genome and therefore genetically co-segregate (Oyuyama et al., 2011; Cesari et al., 2013; Baggs et al., 2017; Ortiz et al., 2017). The two genes in both lines are genetically unlinked, making it unlikely for them to be NLR pairs. For line UC12101, one gene was mapped to Chr1 with multiple markers to support this location. However, the second gene was mapped to Chr2, with only a few markers to support this location. Meticulous observation of mapping reads at the MRC2 disproved the lack of polymorphism as a reason for the low number of markers found in this region. This could indicate that the wild lettuce donor and the recurrent parent had extremely divergent regions at the beginning of the MRC2, which complicated the selection of SNP markers in this region.
Only one of the NILs used in this study (UC07107) showed an F2 segregation ratio of 1:3, resistance to susceptibility, indicating the presence of a recessive gene. This ratio was observed regardless of the inoculum concentration or temperature; therefore, this phenotype was not due to incomplete penetrance. The selection of resistance individuals at each backcross generation during the generation of the NILs makes the selection of a recessive gene improbable; this line may have a linked dominant gene for resistance to other isolates that were used to test backcross progeny, resulting in linkage drag of the recessive resistance gene. The isolate used for phenotyping will directly influence the R-gene identification. A clear example of this was line UC12103, where we were able to map two dominant resistance genes to different regions of Chr1, depending on the isolate used for the phenotyping of the F2 Salinas x UC12103 population (Table 2, Supplementary Fig. 3).
Implications for breeding and future directions
The resistance genes detected in the lines UC02202, UC02204, UC02206, UC07105, UC07106, UC07107, UC07108, UC12100, UC12101, UC12102, and UC12103 confer resistance to races Bl:7US, Bl:8US, Bl:9US, and three isolates with novel virulence phenotypes (Fig. 4), representing some of the most prevalent virulence phenotypes in California over the past 10 years. These lines have been made available for lettuce breeding programs. However, multiple Dm genes should be combined to maximize the evolutionary hurdle for the pathogen to become virulent, to generate more durable resistance against B. lactucae. This will require multiple intercrosses and the use of molecular markers that identify the genomic regions identified in this study.
Fig. 4.
Virulence phenotypes and races of isolates of B. lactucae detected by the resistance genes identified in this study
Most of the resistance genes identified in this study were mapped to small genomic intervals in different NILs. Sequencing the repertoire of NLR-encoding genes from these lettuce lines will allow us to find candidate genes for resistance to multiple isolates of B. lactucae. Cloning of the Dm genes present in these lines will provide the foundation for the development of stacks of Dm genes at single genomic positions using genome editing technologies. This will overcome the challenge of combining resistance genes derived from different sources that map to the same region. Stacking multiple Dm genes will greatly simplify breeding for more durable disease resistance because multiple Dm genes will be inherited as a single Mendelian locus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank Elizabeth Georgian for her edits to the final manuscript.
Author contributions statement
LP designed the experiments, carried out the DNA extractions, library constructions, analysis of sequencing data, lettuce crosses, phenotyping and genotyping of F2 populations and wrote the manuscript draft. AS and KN helped in the phenotyping of F2 and F3 populations and maintaining isolates of B. lactucae used for phenotyping. MT and OO generated and evaluated the NILs for DM resistance. RM planned the study and made significant contributions to the final manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by a Becas Chile for a Scholarship from the National Commission for Science Research and Technology (CONICYT, Chile) to L. Parra. Additional financial support was provided by grants to RWM from USDA NIFA SCRI #s 2010–51181-21631 and 2017–67013-26211.
Compliance with ethical standards
Conflicts of interest
The authors declare that they have no conflicts of interest.
Consent for publication
All authors have read the paper and agreed to publication.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Baggs E, Dagdas G, Krasileva K. NLR diversity, helpers and integrated domains: making sense of the NLR Identity. Curr Opin Plant Biol. 2017;38:59–67. doi: 10.1016/j.pbi.2017.04.012. [DOI] [PubMed] [Google Scholar]
- Beharav A, Lewinsohn D, Lebeda A, Nevo E. New wild Lactuca genetic resources with resistance against Bremia lactucae. Genet Resour Crop Evol. 2006;53:467–474. doi: 10.1007/s10722-004-1932-7. [DOI] [Google Scholar]
- Broman KW, Wu H, Sen Ś, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel J, Fournier E, Tharreau D, Terauchi R, Kroj T. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christopoulou M, McHale LK, Kozik A, Reyes-Chin-Wo S, Wroblewski T, Michelmore RW. Dissection of two complex clusters of resistance genes in lettuce (Lactuca sativa) Mol Plant Microbe Interact. 2015;28:751–765. doi: 10.1094/MPMI-06-14-0175-R. [DOI] [PubMed] [Google Scholar]
- Christopoulou M, Wo SRC, Kozik A, McHale LK, Truco MJ, Wroblewski T, Michelmore R. Genome-wide architecture of disease resistance genes in lettuce. Genes Genom Genet. 2015;5(12):2655–2669. doi: 10.1534/g3.115.020818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute IR. The role of resistance breeding in the integrated control of downy mildew (Bremia lactucae) in protected lettuce. Euphytica. 1992;63:95–102. doi: 10.1007/BF00023915. [DOI] [Google Scholar]
- Farrara BF, Illot TW, Michelmore RW. Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in species of lettuce (Lactuca sativa and L. serriola) Plant Pathol. 1987;36:499–514. doi: 10.1111/j.1365-3059.1987.tb02267.x. [DOI] [Google Scholar]
- Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES. TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE. 2014 doi: 10.1371/journal.pone.0090346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Michelmore RW. Linkage analysis of genes for resistance to downy mildew (Bremia lactucae) in lettuce (Lactuca sativa) Theor Appl Genet. 1985;70:520–528. doi: 10.1007/BF00305985. [DOI] [PubMed] [Google Scholar]
- Irwin SW, Kesseli RV, Waycott W, Ryder EJ, Cho JJ. Identification of PCR-based markers flanking the recessive LMV resistance gene mo1 in an intraspecific cross in lettuce. Genome. 1999;42:982–986. doi: 10.1139/g99-055. [DOI] [Google Scholar]
- Jeuken MJW, Zhang NW, McHale LK, Pelgrom K, Den Boer E, Lindhout P, Michelmore RW, Visser RGF, Niks RE. Rin4 causes hybrid necrosis and race-specific Resistance in an Interspecific lettuce hybrid. Plant Cell. 2009;21:3368–3378. doi: 10.1105/tpc.109.070334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubic LM, Saile S, Furzer OJ, El Kasmi F, Dangl JL. Help wanted: helper NLRs and plant immune responses. Curr Opin Plant Biol. 2019;50:82–94. doi: 10.1016/j.pbi.2019.03.013. [DOI] [PubMed] [Google Scholar]
- Lebeda A, Pink DAC, Astley D. Aspects of the interactions between wild Lactuca spp. and related genera and lettuce downy mildew (Bremia lactucae) In: SpencerPhillips PTN, Gisi U, Lebeda A, editors. Advances in downy mildew research. Dordrecht: Kluwer Academic Publishers; 2002. pp. 85–117. [Google Scholar]
- Lebeda A, Křístkova E, Kitner M, Mieslerová B, Jemelková M, Pink DAC. Wild Lactuca species, their genetic diversity, resistance to diseases and pests, and exploitation in lettuce breeding. Eur J Plant Pathol. 2014;138:597–640. doi: 10.1007/s10658-013-0254-z. [DOI] [Google Scholar]
- Liharska T, Wordragen M, Kammen A, Zabel P, Koornneef M. Tomato chromosome 6: effect of alien chromosomal segments on recombinant frequencies. Genome. 1996;39:485–491. doi: 10.1139/g96-062. [DOI] [PubMed] [Google Scholar]
- Macias-González M, Truco MJ, Bertier L, Jenni S, Simko I, Hayes RJ, Michelmore RW. Genetic architecture of tipburn resistance in lettuce. Theor Appl Genet. 2019;132(8):2209–2222. doi: 10.1007/s00122-019-03349-6. [DOI] [PubMed] [Google Scholar]
- Maisonneuve B, Bellec Y, Anderson P, Michelmore RW. Rapid mapping of two genes for resistance to downy mildew from Lactuca serriola to existing clusters of resistance genes. Theor Appl Genet. 1994;89:96–10. doi: 10.1007/BF00226989. [DOI] [PubMed] [Google Scholar]
- Maisonneuve B, Bellec Y, Souche S, Lot H (1999) New resistance against downy mildew and lettuce mosaic potyvirus in wild Lactuca spp. In: Lebeda A, Kristkova E (eds) EUCARPIA leafy vegetables 1999, Proceedings of the EUCARPIA meeting on leafy vegetables genetics and breeding, Olomouc, 8–11 June 1999, pp 191–197
- Maisonneuve B (2003) Lactuca virosa, a source of disease resistance genes for lettuce breeding: results and difficulties for gene introgression. In: Van Hintum TJL, Lebeda A, Pink DA, Schut JW (eds.) EUCARPIA leafy vegetables, Noordwijkerhout, 19–21 Mar 2003, pp 61–67
- Mc Hale LK, Truco MJ, Kozik A, Wroblewski T, Ochoa OE, Lahre KA, Knapp SJ, Michelmore RW. The genomic architecture of disease resistance in lettuce. Theor Appl Genet. 2009;118:565–580. doi: 10.1007/s00122-008-0921-1. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Truco MJ, Ochoa OE (2013) Breeding crisphead and leafy lettuce. California leafy greens research program. https://calgreens.org/wp-content/uploads/2013/07/Michelmore-Lettuce-Breeding.pdf
- Mikel MA. Genealogy of contemporary North American lettuce. HortScience. 2007;42:489–493. doi: 10.21273/HORTSCI.42.3.489. [DOI] [Google Scholar]
- Norwood JM, Crute IR. The genetic control and expression of specificity in Bremia lactucae (lettuce downy mildew) Plant Pathol. 1984;33:385–400. doi: 10.1111/j.1365-3059.1984.tb01334.x. [DOI] [Google Scholar]
- Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H, Fujibe T, Matsumura H, Shenton M, Clark-Galam D, Undan J, Ito A, Sone T, Terauchi R. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66(3):467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- Ortiz D, de Guillen K, Cesari S, Chalvon V, Gracy J, Padilla A, Kroj T. Recognition of the Magnaporthe oryzae effector AVR-Pia by the Decoy domain of the rice NLR immune receptor RGA5. Plant Cell. 2017;29(1):156–168. doi: 10.1105/tpc.16.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra L, Maisonneuve B, Lebeda A, Schut J, Christopoulou M, Jeuken M, McHale L, Truco MJ, Crute I, Michelmore RW. Rationalization of genes for resistance to Bremia lactucae in lettuce. Euphytica. 2016;210:309–326. doi: 10.1007/s10681-016-1687-1. [DOI] [Google Scholar]
- Reyes-Chin-Wo S, Wang Z, Yang X, Kozik A, Arikit S, Song C, Xia L, Froenicke L, Lavelle DO, Truco MJ, Xia R, Zhu S, Xu C, Xu H, Xu X, Cox K, Korf I, Meyers BC, Michelmore RW. Genome assembly with in vitro proximity ligation data and whole genome triplication in lettuce. Nat Commun. 2017;8:14953. doi: 10.1038/ncomms14953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder EJ. “Salinas 88” lettuce. HortScience. 1991;26(4):439–440. doi: 10.21273/HORTSCI.26.4.439. [DOI] [Google Scholar]
- Severin A, Peiffer G, Xu W, Hyten D, Bucciarelli B, O’Rourke J, Bolon Y, Grant D, Farmer A, May G, Vance C, Shoemaker R, Stupar R. An integrative approach to genomic introgression mapping. Plant Physiol. 2010;154:3–12. doi: 10.1104/pp.110.158949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simko I, Ochoa OE, Pel MA, Tsuchida C, Font Forcada C, Hayes RJ, Truco MJ, Antonise R, Galeano CH, Michelmore RW. Resistance to downy mildew in lettuce cv. La Brillante is conferred by Dm50 gene and multiple QTLs. Phytopathology. 2015;105:1220–1228. doi: 10.1094/PHYTO-02-15-0057-R. [DOI] [PubMed] [Google Scholar]
- Verwaaijen B, Wibberg D, Nelkner J, Gordin M, Rupp O, Winkler A, Bremges A, Blom J, Grosch R, Pühler A, Schlüter A. Assembly of the Lactuca sativa, L. cv. Tizian draft genome sequence reveals differences within major resistance complex 1 as compared to the cv. Salinas reference genome J Biotechnol. 2018;267:12–18. doi: 10.1016/j.jbiotec.2017.12.021. [DOI] [PubMed] [Google Scholar]
- Warren RF, Merritt PM, Holub E, Innes RW. Identification of three putative signal transduction genes involved in R gene-specified disease resistnce in Arabidopsis. Genetics. 1999;152(1):401–412. doi: 10.1093/genetics/152.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Phenotypic and genotypic datasets for the progenies are available upon request.