Abstract
Objective
Medium-chain fatty acids (MCFAs) play an increasing role in human nutrition. In the liver, one fraction is used for synthesis of MCFA-containing triacylglycerol (MCFA-TG), and the rest is used for oxidative energy production or ketogenesis. We investigated which enzymes catalyse the synthesis of MCFA-TG and how inhibition of MCFA-TG synthesis or fatty acid (FA) oxidation influences the metabolic fate of the MCFAs.
Methods
FA metabolism was followed by time-resolved tracing of alkyne-labelled FAs in freshly isolated mouse hepatocytes. Quantitative data were obtained by mass spectrometry of several hundred labelled lipid species. Wild-type hepatocytes and cells from diacylglycerol acyltransferase (DGAT)1−/− mice were treated with inhibitors against DGAT1, DGAT2, or FA β-oxidation.
Results
Inhibition or deletion of DGAT1 resulted in a reduction of MCFA-TG synthesis by 70%, while long-chain (LC)FA-TG synthesis was reduced by 20%. In contrast, DGAT2 inhibition increased MCFA-TG formation by 50%, while LCFA-TG synthesis was reduced by 5–25%. Inhibition of β-oxidation by the specific inhibitor teglicar strongly increased MCFA-TG synthesis. In contrast, the widely used β-oxidation inhibitor etomoxir blocked MCFA-TG synthesis, phenocopying DGAT1 inhibition.
Conclusions
DGAT1 is the major enzyme for hepatic MCFA-TG synthesis. Its loss can only partially be compensated by DGAT2. Specific inhibition of β-oxidation leads to a compensatory increase in MCFA-TG synthesis, whereas etomoxir blocks both β-oxidation and MCFA-TG synthesis, indicating a strong off-target effect on DGAT1.
Keywords: MCFA, Metabolic tracing, Etomoxir, Teglicar
Graphical abstract
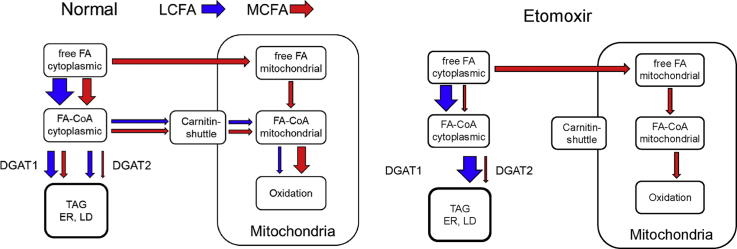
Medium-chain fatty acids (red) are oxidized or incorporated into triglycerides by DGAT1 in the liver. The popular experimental drug etomoxir inhibits the oxidation and, unexpectedly, blocks triglyceride formation by DGAT1.
1. Introduction
With globally increasing prevalence of diet-induced obesity and its associated metabolic disorders, such as non-alcoholic fatty liver disease (NAFLD), type 2 diabetes mellitus and hypertension [1,2], the interest in healthier, alternative nutrients is also increasing. Synthetic medium-chain triacylglycerols (MCT) with fatty acid (FA) chain length of 8 and/or 10 carbon atoms have long been used in parenteral nutrition [3] and as freely available food supplements (“MCT-Oil”). Further, triacylglycerols originating from coconut- or palm kernel oil and milk fat [4] that contain medium-chain fatty acids (MCFA-TGs) with mostly 12 carbon atoms are becoming an increasingly important part of general nutrition [5]. Several possible positive associations suggest MCFA-TGs as a healthy alternative to normal long-chain FA-containing TGs (LCFA-TGs) [6,7]. However, whether MCFA-TGs are healthy for the general population remains a matter of debate [8,9]. MCFA-TGs contain saturated MCFAs with a range of 8–12 carbon atoms. Globally, MCFAs are subject to a rapid oxidative metabolism, at least partially independent of facilitated transport that is otherwise necessary to metabolize LCFAs [10]. Yet, several aspects of MCFA metabolism are insufficiently understood, including the question of how the liver synthesises MCFA-TGs. While several older studies have described the ability of perfused liver [11] or freshly isolated hepatocytes [12,13] to incorporate MCFAs into TGs, the enzyme that catalyses this activity remains unknown. The major candidates are the two mammalian diacylglycerol acyltransferases, DGAT1 and DGAT2 [[14], [15], [16]].
Mammalian DGAT1 has its highest expression in the small intestine, adipose tissue and the mammary gland, but is also expressed in the liver [[16], [17], [18]]. Mammalian DGAT2 is highly expressed in the liver, adipose tissue, mammary gland and the intestine [17,19,20]. Both DGATs are located at the ER [19,[21], [22], [23]]; DGAT2 additionally localises to lipid droplets (LDs) in mammalian cells [24,25] and diytyostelium [26]. The two DGATs have distinct metabolic functions, as indicated by the phenotypes of global knockout mouse models. Loss of DGAT2 in mice is not compatible with life [27], whereas DGAT1 knockout (−/−) mice are viable, show a delayed intestinal fat absorption, have a higher energy expenditure and are resistant to high-fat diet-induced obesity, insulin resistance and hepatic steatosis [[28], [29], [30]]. Using specific inhibitors in murine hepatocytes that were labelled with various radiolabelled precursors, Qi et al. [31] concluded that DGAT1 and 2 cannot compensate for each other in LCFA-TG synthesis. They proposed that DGAT1 preferentially esterifies exogenously supplied FAs and DGAT2 endogenous FAs in the de novo pathway. In a similar setup, Li et al. [18] found that the two DGATs quantitatively compensate for each other, but TGs synthesized by DGAT1 are preferentially used for β-oxidation, whereas TGs synthetised by DGAT2 are destined for very low–density lipoprotein assembly. In addition, Wurie et al. [23] showed that DGAT2 utilises nascent diacylglycerol with de novo synthesized FAs in HepG2 cells and proposed that DGAT1 acted downstream of DGAT2 by re-acylation of DGs formed by lipolysis. Regarding MCFAs as substrates, studies in plants have shown more frequent MCFA usage by DGAT1 [[32], [33], [34]] than by DGAT2 [35]. In cows, quantitative trait loci for high MCFA content in milk fat are associated with the DGAT1 locus [36], but a direct link to DGAT function in MCFA esterification is elusive. Triggered by the phenotype of DGAT1−/− mice, numerous specific DGAT1 inhibitors have been developed in the past years [37]. Some of these have become widely used research tools [31,[38], [39], [40], [41], [42], [43]]. Etomoxir [44] has been previously shown to suppress DGAT activity in H9c2 cells [45]. Since etomoxir is probably the most frequently used experimental inhibitor of FA β-oxidation and a drug candidate itself [46], we also studied DGAT inhibition by etomoxir with a focus on DGAT specificity and a possible connection to MCFA metabolism. The major tool for both aspects of this study is time-resolved tracing of alkyne-labelled FAs. We first developed this technology with lipid class resolution and a fluorescent read-out [47] and very recently with lipid species resolution that employs quantitative mass spectrometry [48].
2. Materials and methods
2.1. Preface: nomenclature
Lipids that contain an alkyne group as a terminal triple bond play a central role in this study. Their systematic names are difficult to read and will be replaced by abbreviations. For fatty acids (FAs), the widely used short nomenclature indicates the number of C-atoms and of double bonds, i.e., oleic acid becomes FA 18:1. In the chemical sum composition, an alkyne group is equivalent to two double bonds, so an oleic acid with an additional terminal triple bond becomes an FA 18:3, which normally is used to abbreviate linolenic acid with three double bonds. In the following, we will treat the triple bond as a functionalisation of the FA and indicate it with a suffix “;Y”. This follows the strategy of the LipidMaps nomenclature [49], which does not yet have a system to indicate triple bonds in the most recent update [50]. The Y makes the triple bond visible in the abbreviation and keeps the correct number of double bonds in the abbreviation. In line with LipidMaps nomenclature, this results in FA 17:0;Y for the terminal alkyne FA with 17 C-atoms, which is used as a palmitic acid equivalent. By that, both the functionalization with a triple bond and the biochemical equivalence to palmitic acid is easily accessible. Our alkyne equivalent to oleic acid, i.e., the alkyne FA with 19C and one double bond, will then become FA 19:1;Y. For lipid classes, we will also use the suffix Y, i.e., PC;Y and TG;Y indicate phosphatidylcholine (PC) and triacylglycerol (TG) that contain an alkyne FA (FA;Y), respectively. The presence of two FA;Y in one TG would yield TG;Y2, accordingly. In glycerolipid species names, the “Y” will be placed after the double bond number, e.g., TG 51:2;Y is a frequent product of labelling with FA 17:0;Y. Accordingly, if the identity of the other FA in the TG;Y is known, the molecule would become TG 17:0;Y_16:0_18:2, if positions are known TG 17:0;Y/18:2/16:0.
2.2. Materials
Fatty acids: FA 11:0;Y (10-undecynoic acid, TCI Deutschland GmbH), FA 17:0;Y (16-heptadecynoic acid, alkyne-palmitate [47], FA 19:1;Y (nonadec-9-cis-en-18-ynoic acid, previously named alkyne-oleate [47] and FA 10:0 (decanoic acid, Merck) were used for labelling experiments as stock solutions (14–20 mM in 80% EtOH). Small molecule inhibitors: etomoxir (Eto), 30 mM in ddH2O from Cayman Chemical; A922500 (DGAT1 inhibitor (D1i)), 10 mM in DMSO (Sigma–Aldrich); JNJ-DGAT2A (DGAT2 inhibitor (D2i)), 10 mM in DMSO (Tocris); Teglicar (Tegli), 10 mM in DMSO (Avanti Polar Lipids). Twenty four-well plates and 100-μm cell strainer were purchased from Sarstedt AG. Willliams medium E was purchased from PAN Biotech. All other cell culture reagents (FCS, 200 mM of glutamine stock and 10,000 U of penicillin-streptomycin solution, Hank's Balanced Salt Solution) were purchased from Invitrogen or Sigma–Aldrich.
2.3. Methods
2.3.1. Isolation of hepatocytes
The following procedure for isolation of primary hepatocytes by collagenase perfusion followed the literature [51,52] with minor adaptations (permission LANUV NRW, 84-02.04.2015.A381). Eight-week-old male C57BL/6N mice, or DGAT1−/− mice [28] and their respective littermate controls (C57BL/6J), were injected with heparin-sodium (430 U in 100 μl, Ratiopharm) and anaesthetised with a ketamin/xylazin mixture. After cannulation (Neoflon™ 26 G 0.6 × 19 mm cannula, BD Biosciences) of the portal vein, the liver was perfused at 4 ml min−1 sequentially with preperfusion buffer (Hanks' balanced salt solution with 5 mM of EGTA (pH 7.4) and 50 μl of heparin-sodium) for 2 min, followed by collagenase buffer (Williams medium E, 0.125 mM of CaCl2, 0.5 mg ml−1 of collagenase NB46, Serva electrophoresis, Sigma cat. no. 17465) for 5 min. The liver was kept at 37 °C using a 150 W heat lamp and a thermometer (ATC2000 from World Precision Instruments). Cells were released into 50 ml of hepatocyte medium (Williams medium E, 10% FCS, 2 mM of l-glutamine, 100 U ml−1 penicillin, 100 μg of ml−1 streptomycin), and the cell suspension was centrifuged for 2 min at 20 g. The pellet, containing the primary hepatocytes, was re-suspended in fresh hepatocyte medium, filtered through a 100-μm nylon cell strainer and plated out in collagenated 24-well dishes at a density of 75,000 cells per well (12-well dishes for 150,000 cells per well, 6-well dishes for 350,000 cells per well).
2.3.2. Lipid labelling experiments
For pulse labelling experiments, hepatocytes were pre-incubated with either small-molecule inhibitors or the respective vehicle as a negative control in 400 μl of pre-incubation medium (Williams medium E, 10% FCS, 2 mM of l-glutamine, 100 U ml−1 of penicillin, 100 μg ml−1 of streptomycin and, if indicated, small molecule inhibitors: etomoxir as indicated in figure legends, 3 μM of D1i, 15 μM of D2i and teglicar as indicated, for 1 h. Pulse labelling was performed in 100 μl of pulse medium (Williams medium E, 10% FCS, 2 mM of l-glutamine, 100 U ml−1 of penicillin, 100 μg ml−1 of streptomycin with combinations of 50 μM of alkyne-labelled FAs and decanoic acid (final FA concentration: 100 μM)) for various time periods as indicated in the figure legends. After the labelling, the cells were washed once with cold hepatocyte medium and once with either ice-cold phosphate-buffered saline (PBS, TLC samples) or 100 mM of ice-cold ammonium acetate (MS samples), depending on the lipid extraction procedure. After the last washing step, liquid was removed completely, and the plates were either stored at −80 °C until lipid extraction or extracted directly thereafter.
For pulse-chase labelling experiments, hepatocytes were preincubated and pulse-labelled as described above. The pulse medium was discarded, and the cells were washed rapidly with 500 μl of prewarmed hepatocyte medium. Then, 250 μl of prewarmed chase medium, containing 100 μM of decanoic acid and the respective inhibitor combination, was added to each well. Cellular metabolism was stopped at different time points by washing the cells once with ice-cold hepatocyte medium and once with 100 mM of ice-cold ammonium acetate, taking care to remove as much liquid as possible after the last washing step. Cells were then frozen at −80 °C until lipid extraction.
2.3.3. Extraction and detection of alkyne-labelled lipids
2.3.3.1. Fluorescent TLC detection
Analysis was performed as described [47]. Briefly, lipids were extracted by sonication of multi-well plates in MeOH/CHCl3 5/1 with subsequent removal of precipitated proteins and two-phase separation of lipids. Lipid extracts (10 μl) were incubated with 40 μl of 3-azido-7-hydroxycoumarin reaction mixture (10 μl 3-azido-7-hydroxycoumarin (2 mg ml−1 in acetonitrile, 250 μl of 10 mM Cu(I)BF4 in acetonitrile, 850 μl of EtOH) overnight at 43 °C in a heating block. As standards for TLC detection, 50-pmol aliquots of synthetic alkyne-labelled lipids in CHCl3 were incubated directly in 30 μl of click reaction mixture. After the click reaction, samples were separated by TLC (solvent I: CHCl3/MeOH/H2O/acetic acid 65/25/4/1, Solvent II: isohexane/ethlyacetate 1/1)), dried and soaked in 4% N,N-diisopropylethylamine in isohexane). The fluorescent reporter clicked to the lipids was imaged with excitation at 420 nm and emission at 494/20 (coumarin fluorescence signal) and 528/28 (noise signal) bandpass emission filter set. Images were acquired, and final quantification was performed using the GelPro analyser software.
2.3.3.2. Multiplexed MS detection
Analyses were performed using the standard protocol as recently described in detail [48]. Briefly, lipids were extracted as above, and the extracts were spiked with an internal standard mix containing 10 different deuterium-labelled alkyne lipid standards. To each sample, 40 μl of reaction mixture (10 μl of 100 mM of C175 in 50% MeOH, 100 μl of 10 mM Cu(I)-TFB in acetonitrile, 900 μl 100% EtOH) containing one of the azido reporters C175-73, −75, −76 or −77 was added followed by incubation at 40 °C for 16 h. After incubation, the samples for multiplexing (4 samples, each reacted with one of the different C175 azido reporters) were pooled, 2-phase-extracted with chloroform and water, chloroform phases dried, lipids dissolved in spray buffer and analysed by MS, using the Thermo Scientific Q Exactive Plus Hybrid quadrupole-orbitrap mass spectrometer. The samples were sprayed at a flow rate of 10 μl/min using the following parameters: sheath gas 4, aux gas 2, sweep gas 0, gas heating off, spray voltage positive mode 4.0 kV, ion transfer capillary temperature 280 °C. Because the lipids clicked with the C175-XX reporter molecules result only in positive ions, the mass acquisition was done in positive ion mode. For all samples from cellular extracts, MS1 spectra were recorded from 300-1,400 m/z in 100 m/z windows for 1.2 min, followed by MS2 scans by data independent acquisition using inclusion lists from m/z 305.373–1400.119 in intervals with m/z 1.0006 to reflect the typical mass defect of lipids at the respective masses. Scan parameters were as follows: for MS1: automatic gain control (AGC) target 3 × 106, maximum ion time 800 ms, resolution 280,000, peak mode centroid, for MS2: automatic gain control (AGC) target 2 × 105, maximum ion time 700 ms, resolution 140,000, no spectral multiplexing, dynamic first mass, isolation window m/z 1.0, stepped normalised collision energy (NCE) 10, 30, 35, spectrum data type centroid. In addition, a second scan for double-charged species was performed in the scan range of m/z 300–700 with MS2 scans from m/z 300.8052–700.0648 at intervals of m/z 0.5002 and an isolation window of m/z 0.7. To identify the labelled lipids, the generated raw files were first converted to .mzml files using the MSconvert program. Files were then analysed using the programme LipidXplorer [53] with MFQL scripts to identify the species by the presence of a peak corresponding to the expected masses of the labelled lipid class combined with the characteristic neutral loss.
2.4. Statistical analysis and data evaluation
If not stated otherwise, the data are presented as mean values ± standard deviation (SD). Within one biological replica, all values were obtained from triplicate determinations. For all experimental sets, 3 biological replicas were performed. Raw data analysis was performed using Microsoft Excel and final statistical analysis was done using the GraphPad software (Prism 6 or 7). For comparison of two groups, an unpaired t-test was used. For comparisons of more than two groups within one experimental data set, a one-way analysis of variance (ANOVA) with Dunnetts's multiple comparison test was applied. When multiple comparisons were done within two or more experimental data sets, a two-way ANOVA with Sidak's multiple comparison test was applied. ∗ stands for p ≤ 0.0332, ∗∗ for p ≤ 0.0021, ∗∗∗ for p ≤ 0.0002, ∗∗∗∗ for p ≤ 0.0001 and ns stands for not significant.
3. Results
3.1. Incorporation of alkyne-MCFA into TG is inhibited by DGAT1 inhibitors and etomoxir but not by DGAT2 inhibitors
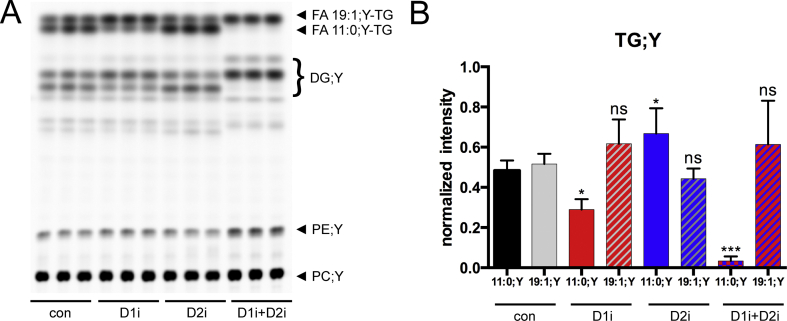
To determine the acyl-CoA chain length preference of the hepatic DGAT enzymes, we conducted 1-h labelling experiments with simultaneous addition of long-chain (FA 19:1;Y) and medium-chain (FA 11:0;Y) alkyne FAs and measured LCFA-TG and MCFA-TG synthesis in the absence or presence of DGAT inhibitors. In the negative control, both alkyne FAs were equally incorporated into TGs (Figure 1). When DGAT1 was inhibited, FA 11:0;Y-TG formation was reduced by approximately 50% (Figure 1, D1i), while DGAT2 inhibition resulted in slightly elevated FA 11:0;Y-TG formation (Figure 1, D2i). A combination of both DGAT1- and DGAT2-inhibitors resulted in a nearly complete reduction of FA 11:0;Y-TG formation (Figure 1, D1i + D2i). The formation of FA 19:1;Y-TG did not change significantly throughout all conditions (Figure 1, C19). Yet, an antiparallel effect on FA 11:0;Y-TG and FA 19:1;Y-TG formation could be observed: Upon DGAT1 inhibition, FA 11:0;Y-TG was reduced and FA 19:1;Y-TG was slightly (but not significantly) elevated. Upon DGAT2 inhibition, this effect was reversed. However, these changes were only significant for FA 11:0;Y-TG but not for FA 19:1;Y-TG.
Figure 1.
Inhibition of DGAT enzymes differentially affects MCFA or LCFA incorporation into TG. Hepatocytes (75,000 per well) were preincubated with either vehicle (con) or 3 μM of DGAT1-inhibitor (D1i), 15 μM of DGAT2-inhibitor (D2i) or a combination of both inhibitors (D1i + D2i) for 1 h. Cells were then incubated for 1 h with a combination of 50 μM of FA 11:0;Y and 50 μM of FA 19:1;Y. The cells were washed, and lipids were extracted and analysed by click reaction with azidocoumarin followed by (A) TLC separation and fluorescent imaging. (B) Quantification of fluorescent intensities of FA 11:0;Y-TG and FA 19:1;Y-TG upon either D1i (red), D2i (blue) and a combination of both (blue/red) inhibitors in comparison to the negative controls. Note that the region labelled “DG;Y” contains both 1,2- and 1,3-isomers that separate on the plate as well as some free FAs. It is not possible to unequivocally assign the bands to the respective species. The data represent mean ± SD for n = 3 biological replicates. ∗p ≤ 0.0332, ∗∗p ≤ 0.0021, ∗∗∗p ≤ 0.0002. ns, not significant.
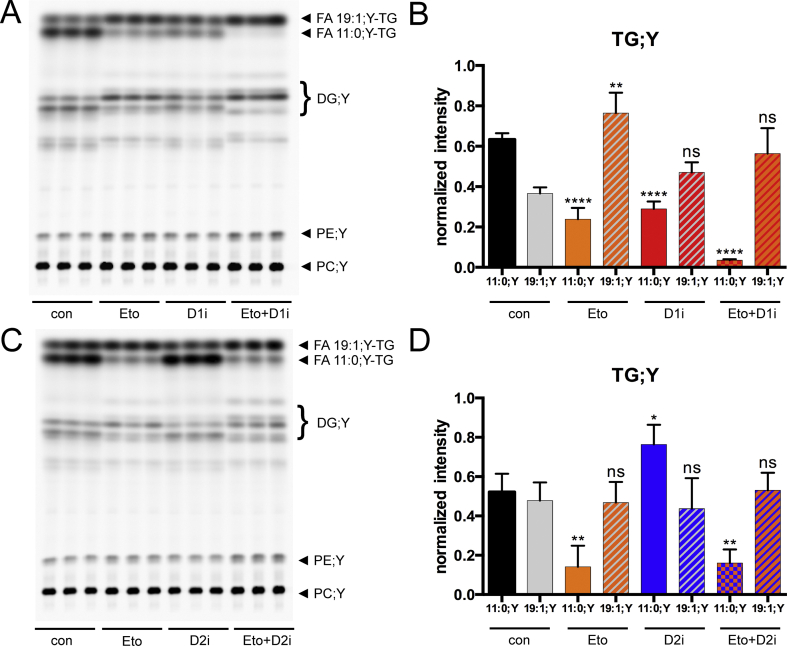
The same experimental setup was used to examine the influence of the widely used carnitine palmitoyl transferase 1 (CPT1) inhibitor etomoxir [54] on incorporation of alkyne MCFA and LCFA into TGs. We expected an increased incorporation of either of the FAs due to reduced β-oxidation. While this indeed was observed for the FA 19:1;Y FA, the treatment with etomoxir led to decreased FA 11:0;Y-TG levels similar to the treatment with the DGAT1-inhibitor (Figure 2A,B, Eto and D1i). Upon combinaton of etomoxir with the DGAT1-inhibitor, FA 11:0;Y-TG levels were strongly reduced, whereas FA 19:1;Y-TG levels were slightly elevated (Figure 2A+ B, Eto + D1i), similar to the combination of both DGAT-inhibitors (Figure 1). A combination of etomoxir and the DGAT2-inhibitor (Figure 2C,D) resulted in an inhibition of FA 11:0;Y incorporation similar to etomoxir alone.
Figure 2.
Etomoxir and DGAT1 inhibition have similar effects on FA 11:0;Y-TG formation. Hepatocytes (75,000 per well) were pre-incubated with either vehicle (con) or 3 μM of DGAT1 inhibitor (D1i), 15 μM of DGAT2 inhibitor (D2i), 50 μM of etomoxir (Eto) or a combination of both inhibitors with etomoxir (D1i + Eto or D2i + Eto) for 1 h. Cells were then incubated for 1 h with a combination of 50 μM of FA 11:0;Y and 50 μM of FA 19:1;Y. The cells were washed, and lipids were extracted and analysed by click reaction with azidocoumarin followed by (A, C) TLC separation and fluorescent imaging. (B, D) Normalised quantification of fluorescent intensities from FA 11:0;Y-TG (filled bars) and FA 19:1;Y-TG (grey hatched bars) upon either etomoxir (orange), D1i (red), D2i (blue) and a combination of both inhibitors (orange/red or orange/blue) in comparison to the negative controls (black: FA 11:0;Y, grey: FA 19:1;Y). The data represent mean ± SD for n = 3 biological replicates. ∗p ≤ 0.0332, ∗∗p ≤ 0.0021, ∗∗∗∗p ≤ 0.0001. ns = not significant.
3.2. DGAT1−/− hepatocytes show reduced synthesis of MCFA-TG
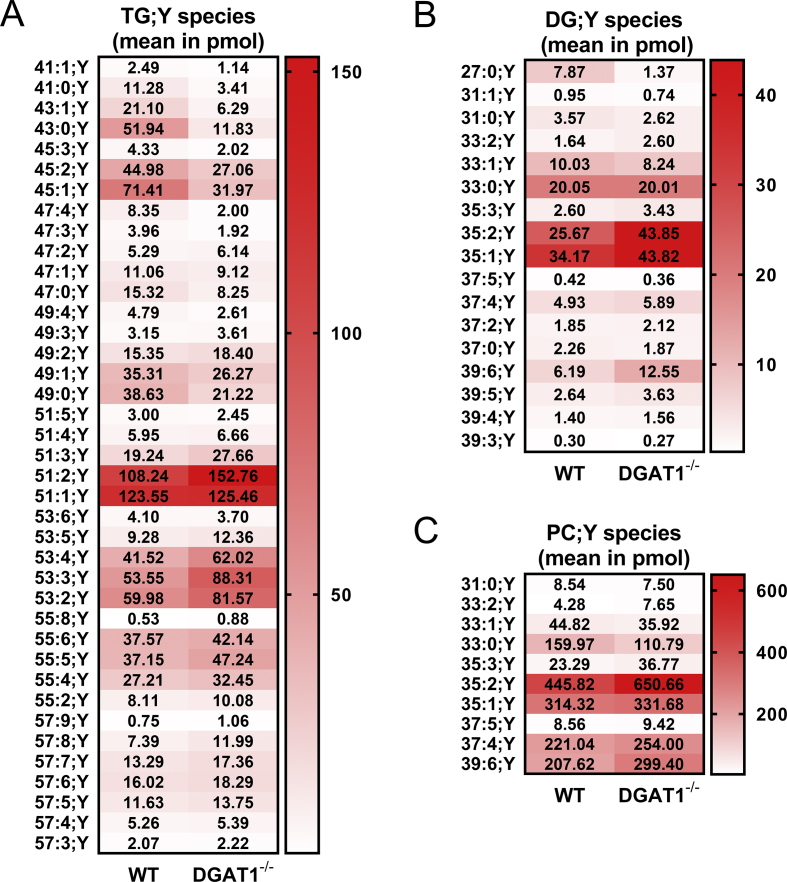
For further analyses, we applied our recent MS-based labelling technology [48], which allows unambiguous identification and absolute quantification of labelled lipid species. First, we used the alkyne palmitate analogue FA 17:0;Y together with unlabelled FA 10:0 for labelling of TG;Y species in WT and DGAT1−/− hepatocytes and analysed the absolute amounts in pmol of labelled lipid species in six major lipid classes.
In the sum of long- and medium-chain species, DGAT1−/− hepatocytes had moderately elevated PC;Y amounts, whereas TG;Y, DG;Y and PA;Y were comparable to WT cells (Suppl. Fig. 1A). Treatment with DGAT inhibitors or etomoxir or combinations thereof did not lead to significant changes of total incorporation of the labelled FAs into the cellular lipids (Suppl. Fig. 1B). On the single species level, however, major differences in TG;Y became observable. The DGAT1−/− hepatocytes had significantly lower amounts of shorter TG;Y species (Figure 3A, TG 41:1;Y - 45:1;Y). These species contain the unlabelled MCFA 10:0 (e.g., TG 43:0;Y = TG 17:0;Y_16:0_10:0), which can be seen in the MS2 fragmentation pattern (not shown). This decrease was compensated by an increase in TG;Y species with longer acyl chains (Figure 3A, TG 51:5;Y - 57:5Y), resulting in overall similar amounts in DGAT1−/− and WT hepatocytes.
Figure 3.
Analyses of alkyne-labelled lipid species from wild-type (WT) or DGAT1−/−hepatocytes, incubated with alkyne-palmitate and decanoic acid. Hepatocytes (75,000 per well) were co-incubated with 50 μM of FA 17:0;Y and 50 μM of decanoic acid (FA 10:0) for 1 h. Lipids were extracted, and alkyne-labelled species were identified and quantified by multiplexed click-MS analysis as described in the Materials and Methods section. Absolute amounts of the lipid species within the lipid classes of (A) TG;Y, (B) DG;Y and (C) PC;Y are shown in pmol. The data represent mean values for n = 3 biological replicates.
Within DG;Y (Figure 3B), three major species, namely DG 35:2;Y, 35:1;Y and 39:6;Y showed significantly higher concentrations in the DGAT1−/− hepatocytes, whereas all other species were either equally concentrated or slightly underrepresented. Only minor amounts of DG;Y that contained unlabelled FA 10:0 (i.e., DG 27:0;Y) were detected. The increased concentration of PC;Y in DGAT1−/− hepatocytes originated mainly from two species, namely PC 35:2;Y and 39:6;Y (Figure 3C), i.e., the combination of FA 17:0;Y with linoleic and docosahexaenoic acid, respectively.
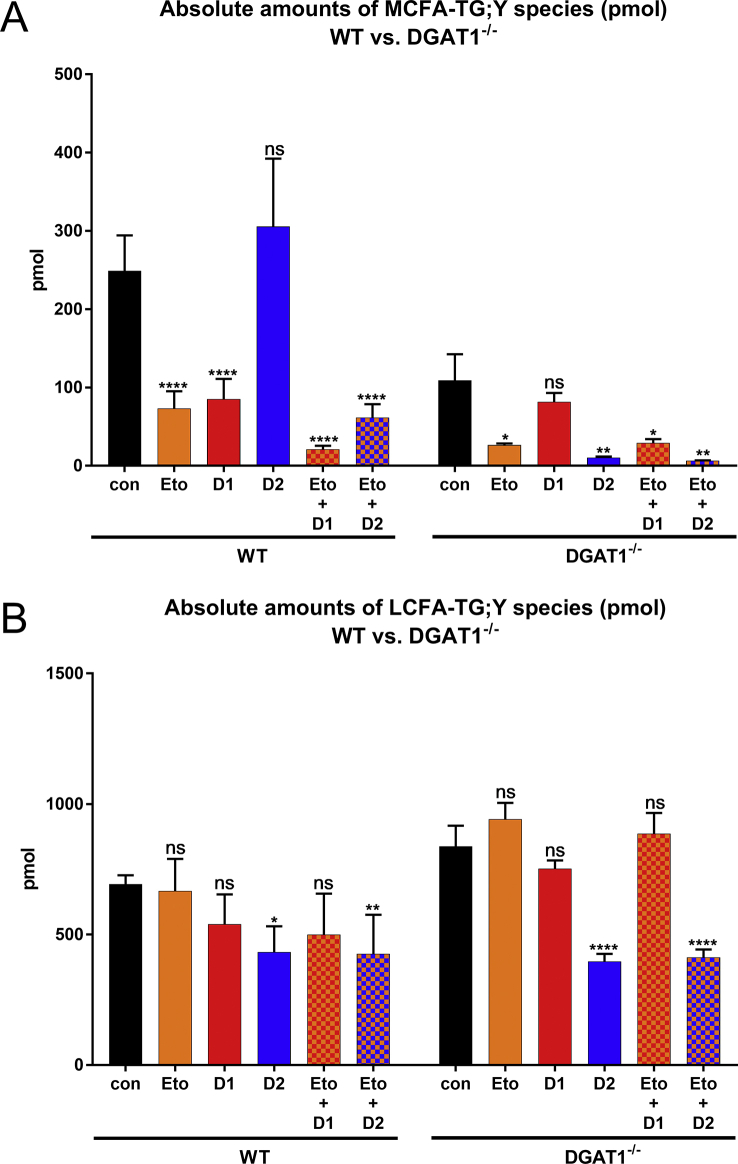
Next, we measured the influence of DGAT inhibitors on the formation of TG;Y containing MCFA or LCFA upon labelling with FA 17:0;Y and unlabelled FA 10:0 in WT and DGAT1−/− hepatocytes. In WT cells (Figure 4A left), MCFA incorporation was strongly reduced by the DGAT1 inhibitor and by etomoxir, but not by the DGAT2 inhibitor, confirming the observations in Figure 1, Figure 2. In DGAT1−/− cells (Figure 4A, right), there was a >50% reduction of MCFA-TGs that was, as expected, not further inhibited by the DGAT1 inhibitor. The DGAT2 inhibitor had a strong effect on the residual MCFA incorporation, which was further reduced by etomoxir. For the LCFA-TG synthesis in WT cells (Figure 4B, left), etomoxir and the DGAT1 inhibitor showed a tendency to reduce overall incorporation; the DGAT2 inhibitor caused a significant reduction. In DGAT1−/− cells (Figure 4B, right), there was a clear reduction of LCFA-TG synthesis by the DGAT2 inhibitor but not by etomoxir or the DGAT1 inhibitor.
Figure 4.
Analyses of alkyne-labelled TG species from wildtype (WT) or DGAT1−/−hepatocytes, treated with inhibitor combinations and incubated with alkyne-palmitate and decanoic acid. Hepatocytes (75,000 per well) were pre-incubated with either vehicle (con) or 50 μM of etomoxir (Eto), 3 μM of DGAT1-inhibitor (D1i), 15 μM of DGAT2-inhibitor (D2i) or combinations of etomoxir with each DGAT-inhibitor (Eto + D1i, Eto + D2i). Cells were then co-incubated with 50 μM of FA 17:0;Y and 50 μM of decanoic acid (FA 10:0) for 1 h. Lipids were extracted, and alkyne-labelled species were identified and quantified by multiplexed click-MS analysis as described in the Materials and Methods sections. (A) MCFA-TG;Y (∑C41–C47) and (B) LCFA-TG;Y (∑C49-57). The data represent mean ± SD for n = 3 biological replicates. ∗p ≤ 0.0332, ∗∗p ≤ 0.0021 and ∗∗∗∗p ≤ 0.0001. ns, not significant.
3.3. Normalised species spectra reveal details of DGAT inhibitor effects
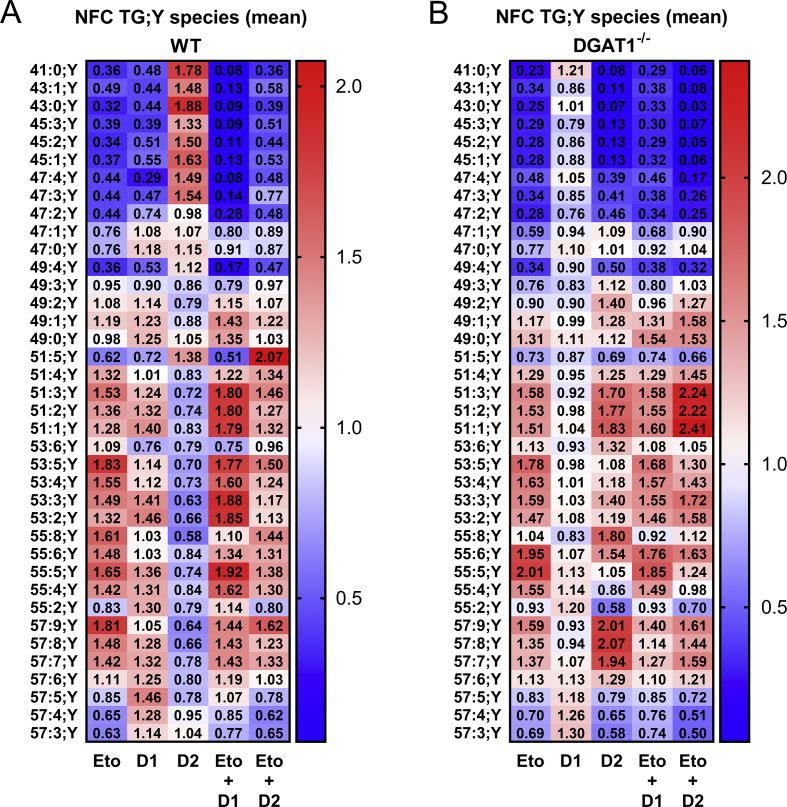
To take full advantage of the information provided by the species resolution in the data, we performed further analysis of the entire species spectrum of the samples. Systematic comparison of lipid species spectra is hampered by the large range of relative abundance of species within one class and by the change of total class abundance. To represent spectral changes normalised for both of these variations, pmol amounts of each species were divided by the corresponding control to obtain -fold changes (FCs), normalising for the relative abundance of a species within the class. Species FC values of each sample were then averaged to obtain the average FC of the lipid class. In the next step, individual FCs were divided by the class average FC to obtain normalised FC values (NFC). These NFC arrays indicate the distortion of the species spectrum of a class independent of the changes in total abundance of the class and independent of the relative abundance of the species within the class. The average NFC within one class is always 1, allowing comparison of spectral changes between different classes and samples. An accumulation of species is indicated by numbers above one (red colour) and downregulated species by numbers below one (blue colour) (Figure 5).
Figure 5.
Detailed normalised fold-change (NFC) analyses of alkyne-labelled TG species from wild-type (WT) or DGAT1−/−hepatocytes treated with inhibitor combinations and labelled with alkyne-palmitate and decanoic acid. Hepatocytes (75,000 per well) were pre-incubated with either vehicle (con) or 50 μM of etomoxir (Eto), 3 μM of DGAT1-inhibitor (D1i), 15 μM of DGAT2-inhibitor (D2i) or combinations of etomoxir with each DGAT-inhibitor (Eto + D1i, Eto + D2i). Cells were then co-incubated with 50 μM of FA 17:0;Y and 50 μM of decanoic acid (FA 10:0) for 1 h. Lipids were extracted, and alkyne-labelled species were identified and quantified by multiplexed click-MS analysis as described in the Materials and Methods section. Normalized fold changes (NFC) in concentrations of each lipid species for each inhibitor treatment of (A) WT and (B) ΔDGAT1 hepatocytes were calculated by normalisation to the negative control and to the average class fold change. The data represent mean values for n = 3 biological replicates. Additionally, values are represented by colours; the scale is indicated by the bar at the right.
The NFC pattern of TG;Y species in WT mice (Figure 5A) confirmed the previous TLC data. Inhibition of DGAT1 (D1i) or treatment with etomoxir (Eto) lead to a spectral shift from labelled MCFA-TGs (C41–C47) to LCFA-TGs (C49–C57), while inhibition of DGAT2 (D2i) resulted in the opposite species distribution. A combination of etomoxir and DGAT1 inhibitor resulted in an even stronger suppression of MCFA-TG synthesis (Eto + D1i). In the combination of etomoxir and DGAT2 inhibitor (Eto + D2i), the effect of etomoxir was dominant over the DGAT2 inhibition, resulting in a pattern that strongly resembled that of etomoxir alone. In support of the TLC-based labelling experiments, a combination of both DGAT1 and DGAT2 inhibitor resulted in a strong downregulation of MCFA-TGs (Suppl. Fig. 2, panel A, lanes labelled D1i + D2i), with a pattern very similar to the combination of etomoxir + DGAT2 inhibitor (Figure 5A).
In the DGAT1−/− hepatocytes, we observed almost no effect of the DGAT1 inhibitor on the species pattern (Figure 5 B, D1i), as expected for a specific inhibitor in a knockout of its target. Despite the lack of DGAT1, etomoxir treatment (Figure 5B, Eto) led to further MCFA-TG reduction, comparable to WT cells. This pattern was also observed upon DGAT2 inhibition and upon combination of etomoxir with either DGAT1 inhibitor or DGAT2 inhibitor. The latter combination showed a particularly strong suppression of MCFA-TG formation, along with a marked upregulation within the main labelled TGs 51:3;Y - 51:1;Y, with values >2.2.
3.4. The CPT1 inhibitors teglicar and etomoxir have opposite effects on MCFA-TG synthesis
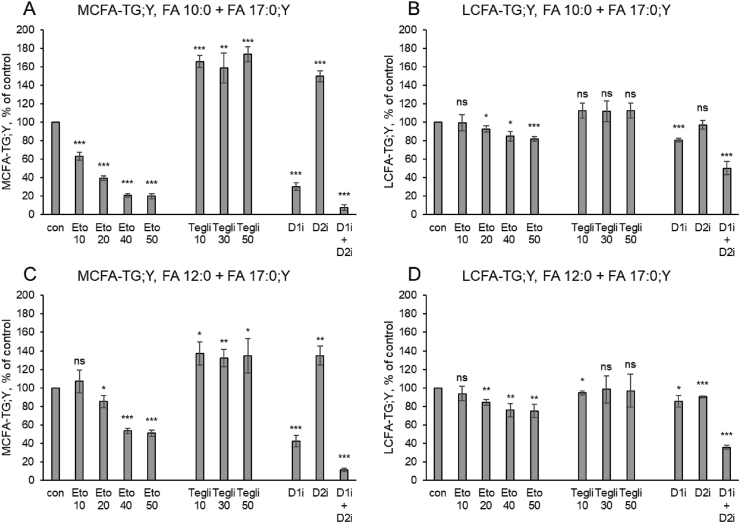
Next, we assessed whether the effects of etomoxir on MCFA-TG formation were direct, by an effect on an enzyme in the TG synthetic pathway, or indirect, i.e., connected to its genuine inhibitory action on CPT1. To answer this question, we compared the alkyne-labelled TG species profile of WT cells treated with etomoxir or teglicar [55]. This substance is a non-cleavable analogue of palmitoyl-carnitine and inhibits CPT1 by a mechanism different from the irreversible covalent modification caused by etomoxir. Both inhibitors showed opposing effects on TG synthesis. Figure 6A shows the amounts of MCFA-TG;Y that were detected after labelling with FA 17:0;Y + FA 10:0, normalised to control conditions. While etomoxir showed a concentration-dependent suppression, teglicar at any of the three tested concentrations led to a strong increase of MCFA-TG;Y. As a control, the DGAT inhibitors caused the characteristic strong decrease with D1i, increase with D2i and nearly complete suppression with D1i + D2i. In contrast, the labelled LCFA-TG;Y in the same samples (Figure 6B) were nearly unaffected by etomoxir and only weakly elevated by teglicar. In a combined etomoxir + teglicar treatment (Suppl. Fig. 3), the negative etomoxir effect on the MCFA-TG;Y species was dominant over the positive teglicar effect.
Figure 6.
MCFA-TG and LCFA-TG formation upon incubation with alkyne-palmitate and FA 10:0 or 12:0 in the presence of various inhibitors. Hepatocytes (75,000 per well) were pre-incubated with either vehicle (con) or etomoxir (Eto), teglicar (Tegli) or DGAT inhibitors (D1i, D2i). Numbers indicate inhibitor concentrations in μM. Cells were co-incubated with (A,B) 50 μM of FA 17:0;Y and 50 μM of decanoic acid (FA 10:0) or (C,D) 50 μM of FA 17:0;Y and 50 μM of FA 12:0 for 1 h in the presence of the respective inhibitors. Lipids were extracted, and alkyne-labelled species were identified and quantified by multiplexed click-MS analysis as described in the Materials and Methods section. (A,C) MCFA-TG;Y and (B,D) LCFA-TG;Y were quantified and are expressed as % of control. The data represent mean ± SD for n = 3 biological replicates. ∗p ≤ 0.05, ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001. ns, not significant.
While synthetic MCT oils mostly contain FA 8:0 and 10:0, MCFA from natural sources are dominated by FA 12:0. We performed a comparative analysis, replacing FA 10:0 by FA 12:0 in the above set of labelling experiments. Incorporation of FA 12:0 into MCFA-TG;Y (Figure 6C) was affected by the inhibitors in the same pattern, but slightly less pronounced, as that of FA 10:0. Again, the LCFA-TG;Y in the same samples was only weakly affected by inhibitors (Figure 6D). Significantly, the fraction of MCFA-TG;Y of the total TG;Y was larger for FA 12:0 than for FA 10:0 (32.6 +/− 0.4% vs. 16.2 +/− 0.9%, p = 2.1 E−5).
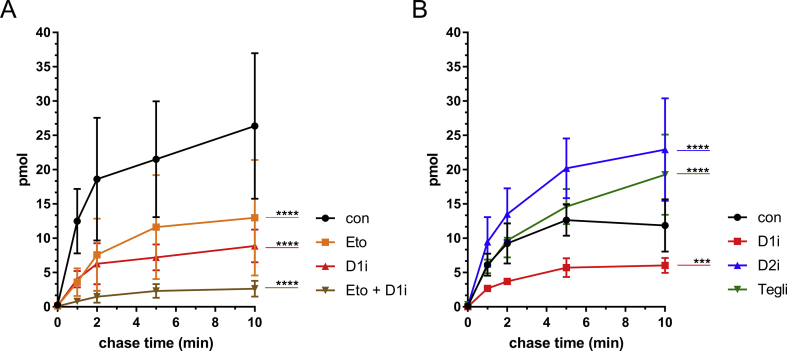
Finally, we conducted a pulse-chase experiment, in which WT hepatocytes were pulse-labelled for 2 min with FA 17:0;Y, generating a pool of FA 17:0;Y-labelled early metabolites of the glycerol phosphate pathway, namely PA and DG. During the chase times of 0–10 min, they were converted to endpoint metabolites like PC and TG. By addition of unlabelled decanoic acid (FA 10:0) and the inhibitors to the chase medium, the MCFA-TG formation by acylation of DGs with decanoic acid was directly studied. In line with the previous findings, etomoxir and the DGAT1 inhibitor as well as their combination effectively decreased the formation of the four major MCFA-TG species (TG 43:1;Y, 43:0;Y, 45:2;Y, 45:1;Y), originating from the incorporation of decanoic acid into the main four labelled DGs 33:1;Y, 33:0;Y 35:2;Y, 35:1;Y (Figure 7A and Suppl. Fig. 4). Treatment with teglicar and the DGAT2 inhibitor, however, resulted in an increased formation of the major MCFA-TG species (Figure 7B and Suppl. Fig. 5).
Figure 7.
Pulse-chase analyses of MCFA-TG formation in WT primary hepatocytes, incubated with alkyne-palmitate and chased with decanoic acid upon inhibitory treatment. After isolation, 7.5 × 104 primary mouse hepatocytes were plated in 24-well plates per well. (A) Cells were pre-incubated with either vehicle (con) or 50 μM of etomoxir (Eto), 3 μM of DGAT1-inhibitor (D1i) and a combination of both inhibitors (Eto + D1i). (B) Cells were preincubated with either vehicle (con) or 3 μM of DGAT1-inhibitor (D1i), 15 μM of DGAT2 inhibitor (D2i) or 50 μM of teglicar (Tegli). Cells were then incubated with 100 μM of FA 17:0;Y for 2 min and chased with 100 μM of decanoic acid (FA 10:0) for either 0, 1, 2, 5 or 10 min in the presence of the respective inhibitor. Lipids were extracted, and alkyne-labelled species were identified and quantified by multiplexed click-MS analysis as described in the Materials and Methods section. Absolute amounts of the lipid species within the lipid class of MCFA-TG;Y (Σ of a43:1, a43:0, a45:2, a45:1) are shown in pmol, and statistical evaluation of changes upon inhibitory treatment in comparison to the respective negative control (con) are shown for the last chase time (t = 10 min). The data represent mean ± SD for n = 3 biological replicates. ∗∗∗p ≤ 0.0002, ∗∗∗∗p ≤ 0.0001.
4. Discussion
4.1. DGAT1 is the dominant DGAT for incorporation of MCFAs into TG
To incorporate an exogenous MCFA into a MCFA-TG, a cell needs efficient (i) uptake of the MCFA, (ii) activation to the MCFA-CoA and (iii) acylation of DG to MCFA-TG by a DGAT enzyme. Consistent with the key role of DGAT enzymes in MCFA-TG synthesis, we found an almost complete suppression of MCFA-TG synthesis by a combination of DGAT1+DGAT2 inhibitors in WT hepatocytes or by inhibition of DGAT2 in DGAT1−/− hepatocytes. We did not detect relevant DGAT-independent incorporation of MCFAs into MCFA-TGs, confirming the results of a previous study using radiolabelled octanoate [12]. Regarding the preference of the mammalian DGATs for substrate chain length or saturation, both enzymes efficiently use several different unsaturated and saturated LCFA-CoAs [19]. A preference for MCFA incorporation has been established for some plant DGATs from species with major MCFA-TG synthesis [[33], [34], [35]]. To approach this question in mammalian cells, we used the very specific inhibitors that have become available in recent years due to the pharmacological interest in DGATs as drug targets in metabolic diseases.
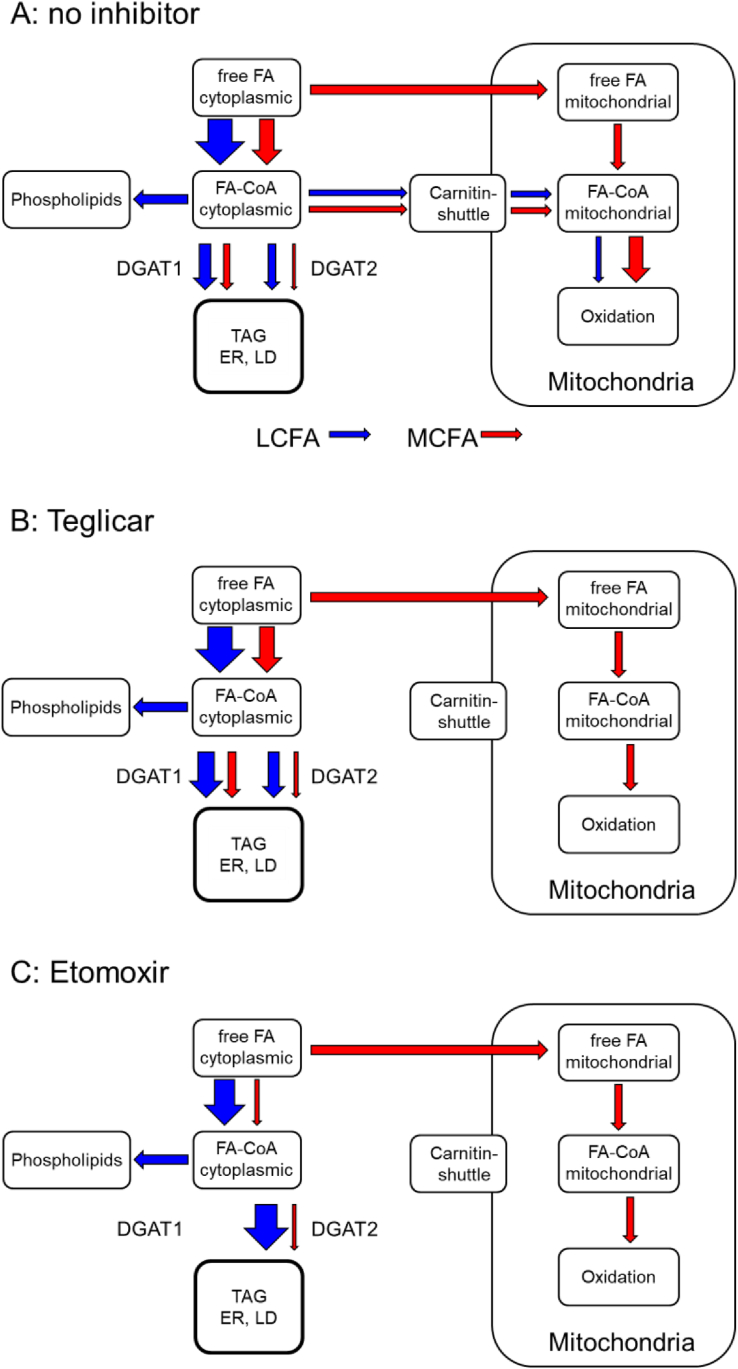
Our data clearly demonstrate that the major enzyme for MCFA-TG synthesis is DGAT1, while DGAT2 plays a minor role. This was shown both for acylation of unlabelled DGs with the labelled FA 11:0;Y (TLC-based analysis, Figure 1 + 2) and for acylation of alkyne-labelled LCFA-DGs with unlabelled decanoic acid (MS-based analysis, Figure 3, Figure 4, Figure 5, Figure 6), ruling out an influence of the terminal triple bond on FA specificity of DGATs. Given the amazing speed of lipid metabolism in hepatocytes [48], our experimental approach (1 h incubation of cells with labelled FA, Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6) reflects a mixture of biosynthesis and steady state distribution. This means that apart from direct effects of inhibitors on DGAT activities, changes in deacylation-reacylation cycles and other secondary effects due to increased levels of DG might also play a role. This was ruled out by pulse-chase experiments with a 2-min time resolution that directly showed the acylation of the major labelled long-chain DG species with unlabelled decanoic acid (Figure 7). While inhibition of DGAT1 leads to decreased MCFA-TG formation, inhibition of DGAT2 results in enhanced MCFA-TG formation, likely by providing a larger fraction of the labelled DG pool to the remaining DGAT1 with its preference for MCFA-CoAs (Figure 8A).
Figure 8.
Schematic drawing of major fluxes in FA metabolism. Fluxes of LCFA and MCFA are symbolised by blue and red arrows, respectively. (A) Baseline conditions in the absence of inhibitors, (B) in the presence of the specific CPT1 inhibitor teglicar, and (C) in the presence of etomoxir, which inhibits CPT1 and DGAT1.
4.2. Inhibition of CPT1 promotes MCFA-TG formation
Teglicar is a non-cleavable substrate analogue acting as a reversible inhibitor of liver CPT1 [55,56]. It promotes formation of both MCFA-TG and LCFA-TG (Figure 6A+ B); the effect is stronger for MCFA-TG. The general increase can be explained by a block of β-oxidation with a compensatory increase in TG synthesis from the affected FA. The stronger effect on MCFA relative to LCFA can be explained by the known preference of MCFA for hepatic β-oxidation [11]. In comparison between FA 10:0 and 12:0, the longer FA shows a smaller increase in MCFA-TG formation upon CPT1 inhibition, is better acylated to TG and is less dependent on DGAT1. Together, these findings indicate that the metabolic similarity to LCFA increases with the chain length of the MCFA. More studies will be necessary to establish a solid relationship between chain length and metabolic behaviour.
4.3. Inhibition of DGAT activity by etomoxir
Etomoxir was originally described as an irreversible inhibitor of mitochondrial LCFA β-oxidation targeting CPT1 [54]. By shifting the cellular energy utilisation from FAs to glucose, it gained interest as a potential anti-diabetic drug improving hyperglycaemia, hyperketonaemia, and hypertriglyceridaemia [[57], [58], [59]]. With an increasing understanding of the role of β-oxidation in tumour growth and cancer metabolism, CPT1 inhibitors received rejuvenated attention as possible anti-cancer drugs [60,61]. However, etomoxir has never been approved in clinical trials due to severe systemic side effects [[62], [63], [64], [65], [66]]. Etomoxir has been used as an experimental inhibitor of β-oxidation in hundreds of published studies and is present in several commercial β-oxidation assay kits. For a correct interpretation of data obtained with etomoxir, more information about possible off-target effects [16,61,67] is urgently needed. In accordance with our data, etomoxir (40 μM) inhibited DGAT activity in H9c2 cardiac myoblast cells, resulting in a 60% reduced [14C]palmitic acid incorporation into TGs [45]. When cellular subfractions from etomoxir-treated H9c2 cells were assayed for lipid metabolic activities, DGAT activity was reduced by 80%, whereas other enzymatic activities were unaffected. Additionally, the authors showed that etomoxiroyl-CoA inhibited DGAT activity in vitro, establishing a direct inhibitory action on DGAT activity. The study did not address the specificity for DGAT1 or DGAT2 or FA preferences in this etomoxir off-target effect.
In our study, DGAT inhibition by etomoxir in WT hepatocytes largely phenocopied the results observed by inhibition of DGAT1 but not of DGAT2. The TLC-based analysis showed a strong inhibition of MCFA-TG formation and a weak inhibition of LCFA-TG formation. The combination of etomoxir + DGAT1-inhibitor displayed an enhanced DGAT1 inhibitory effect, whereas etomoxir + DGAT2-inhibitor revealed comparable results as the combination of DGAT1 + DGAT2 inhibitor. These observations strongly indicate that etomoxir is a specific DGAT1-inhibitor with little effect on DGAT2. In DGAT1−/− cells, however, there was an effect on total MCFA-TG formation not explainable by DGAT1 inhibition, both by etomoxir alone and in combination with the DGAT2-inhibitor (Figure 4A). A detailed analysis of TG;Y species spectra also showed subtle differences between etomoxir and DGAT1 pattern. In each combination of etomoxir and DGAT inhibitors, etomoxir was dominant in both WT and DGAT1−/− cells. This indicates an additional inhibitory action of etomoxir with specificity for MCFA upstream of DGAT1.
4.4. Limitations
This study relies to a large extent on enzyme inhibitors, which can have off-target effects. It would be desirable to corroborate some aspects of this study with complementary data obtained by corresponding siRNA or gene deletion experiments. siRNA studies in hepatocytes are hampered by the rapid de-differentiation of freshly isolated hepatocytes. Gene deletion experiments beyond the DGAT1−/− mice are hampered by the fact that both DGAT2 [27] and CPT1a [70] deletions in mice are lethal. Regarding the DGAT1 and DGAT2 inhibitor experiments that we performed, we can be confident about our conclusions because we have confirmed their effects on TG synthesis using other specific DGAT inhibitors with virtually identical results (Supplementary Figures 6 and 7). Obviously, this is not the case for the CPT1 inhibitors etomoxir and teglicar with their opposing effects on MCFA-TG synthesis. While we conclude that etomoxir most likely is an inhibitor of DGAT1, we cannot exclude that also teglicar has off-target effects that could contribute to the phenotype that we observed. It should also be noted that the inhibition of DGAT1 by etomoxir is concentration-dependent (Figure 6). The data indicate that at concentrations below 10 μM, etomoxir might still be used as an efficient CPT1 inhibitor without a major impact on DGAT1. At higher concentrations, a significant effect on DGAT1 must be taken into account.
5. Outlook
More data are needed for a complete model that describes the chain length dependence of MCFA acylation, CPT1-dependent and -independent mitochondrial uptake and oxidation. Future studies must include a complete series of MCFAs from C6 to C12 with parallel measurement of acylation into TG, acyl-carnitine concentrations and CO2 release in the same samples.
Regarding medical aspects, the preference of DGAT1 for MCFA might also contribute to the mild to fatal diarrhoea in patients with rare homozygous DGAT1 mutations [68,69]. Since DGAT2 can mostly compensate for the LCFA-TG, but not for the MCFA-TG synthesis of missing DGAT1, these patients might benefit from food with reduced content of MCFA.
Funding
CT received funding from the DFG under Germany's Excellence Strategy – EXC2151 – 390873048 (Immunosensation). DK received funding from the Austrian Science Fund (FWF) (DK-MCD W1226, SFB F73) and the PhD programme “Molecular Medicine” of the Medical University of Graz.
Acknowledgements
The authors thank I. Hindler and A. Absenger (Medical University of Graz) for mouse care. Many thanks to Drs. C. Ejsing, K. Ekroos, E. Fahy, G. Liebisch and F. Spener for the fruitful discussion on alkyne lipid shorthand nomenclature.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.101150.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Botchlett R., Wu C. Diet composition for the management of obesity and obesity-related disorders. Journal of Diabetes Mellitus and Metabolic Syndrome. 2018;3(1):10–25. doi: 10.28967/jdmms.2018.01.18002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roden M., Shulman G.I. The integrative biology of type 2 diabetes. Nature. 2019;576(7785):51–60. doi: 10.1038/s41586-019-1797-8. [DOI] [PubMed] [Google Scholar]
- 3.Bach A.C., Babayan V.K. Medium-chain triglycerides: an update. American Journal of Clinical Nutrition. 1982;36(5):950–962. doi: 10.1093/ajcn/36.5.950. [DOI] [PubMed] [Google Scholar]
- 4.Marten B., Pfeuffer M., Schrezenmeir J. Medium-chain triglycerides. International Dairy Journal. 2006;16(11):1374–1382. doi: 10.1016/j.idairyj.2006.06.015. [DOI] [Google Scholar]
- 5.Mumme K., Stonehouse W. Effects of medium-chain triglycerides on weight loss and body composition: a meta-analysis of randomized controlled trials. Journal of the Academy of Nutrition and Dietetics. 2015;115(2):249–263. doi: 10.1016/j.jand.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 6.McCarty M.F., DiNicolantonio J.J. Lauric acid-rich medium-chain triglycerides can substitute for other oils in cooking applications and may have limited pathogenicity. Open Heart. 2016;3(2) doi: 10.1136/openhrt-2016-000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Croteau E., Castellano C.-A., Richard M.A., Fortier M., Nugent S., Lepage M. Ketogenic medium chain triglycerides increase brain energy metabolism in Alzheimer's disease. Journal of Alzheimer's Disease. 2018;64(2):551–561. doi: 10.3233/JAD-180202. [DOI] [PubMed] [Google Scholar]
- 8.Neelakantan N., Seah J.Y.H., van Dam R.M. The effect of coconut oil consumption on cardiovascular risk factors: a systematic review and meta-analysis of clinical trials. Circulation. 2020;141(10):803–814. doi: 10.1161/CIRCULATIONAHA.119.043052. [DOI] [PubMed] [Google Scholar]
- 9.Hoeks J., Mensink M., Hesselink M.K.C., Ekroos K., Schrauwen P. Long- and medium-chain fatty acids induce insulin resistance to a similar extent in humans despite marked differences in muscle fat accumulation. The Journal of Clinical Endocrinology & Metabolism. 2012;97(1):208–216. doi: 10.1210/jc.2011-1884. [DOI] [PubMed] [Google Scholar]
- 10.Papamandjaris A.A., MacDougall D.E., Jones P.J. Medium chain fatty acid metabolism and energy expenditure: obesity treatment implications. Life Sciences. 1998;62(14):1203–1215. doi: 10.1016/s0024-3205(97)01143-0. [DOI] [PubMed] [Google Scholar]
- 11.Lieber C.S., Lefèvre A., Spritz N., Feinman L., DeCarli L.M. Difference in hepatic metabolism of long- and medium-chain fatty acids: the role of fatty acid chain length in the production of the alcoholic fatty liver. Journal of Clinical Investigation. 1967;46(9):1451–1460. doi: 10.1172/JCI105637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayorek N., Bar-Tana J. Medium chain fatty acids as specific substrates for diglyceride acyltransferase in cultured hepatocytes. Journal of Biological Chemistry. 1983;258(11):6789–6792. [PubMed] [Google Scholar]
- 13.Brandes R., Mayorek N., Berry E., Arad R., Bar-Tana J. The specificity of triacylglycerol synthesis for medium-chain fatty acids in rat and human adipose preparations. Biochimica et Biophysica Acta (BBA) - Lipids and Lipid Metabolism. 1985;836(1):63–66. doi: 10.1016/0005-2760(85)90220-6. [DOI] [PubMed] [Google Scholar]
- 14.Cases S., Smith S.J., Zheng Y.-W., Myers H.M., Lear S.R., Sande E. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proceedings of the National Academy of Sciences. 1998;95(22):13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cases S., Stone S.J., Zhou P., Yen E., Tow B., Lardizabal K.D. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. Journal of Biological Chemistry. 2001;276(42):38870–38876. doi: 10.1074/jbc.M106219200. [DOI] [PubMed] [Google Scholar]
- 16.Yao C.-H., Liu G.-Y., Wang R., Moon S.H., Gross R.W., Patti G.J. Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation. PLoS Biology. 2018;16(3) doi: 10.1371/journal.pbio.2003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yen C.-L.E., Nelson D.W., Yen M.-I. Intestinal triacylglycerol synthesis in fat absorption and systemic energy metabolism. The Journal of Lipid Research. 2015;56(3):489–501. doi: 10.1194/jlr.R052902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C., Li L., Lian J., Watts R., Nelson R., Goodwin B. Roles of acyl-CoA:diacylglycerol acyltransferases 1 and 2 in triacylglycerol synthesis and secretion in primary hepatocytes. Arteriosclerosis, Thrombosis, and Vascular Biology. 2015;35(5):1080–1091. doi: 10.1161/ATVBAHA.114.304584. [DOI] [PubMed] [Google Scholar]
- 19.Yen C.-L.E., Stone S.J., Koliwad S., Harris C., Farese R.V. Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. The Journal of Lipid Research. 2008;49(11):2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLaren D.G., Han S., Murphy B.A., Wilsie L., Stout S.J., Zhou H. DGAT2 inhibition alters aspects of triglyceride metabolism in rodents but not in non-human primates. Cell Metabolism. 2018;27(6):1236–1248. doi: 10.1016/j.cmet.2018.04.004. e6. [DOI] [PubMed] [Google Scholar]
- 21.Stone S.J., Levin M.C., Farese R.V. Membrane topology and identification of key functional amino acid residues of murine acyl-CoA:diacylglycerol acyltransferase-2. Journal of Biological Chemistry. 2006;281(52):40273–40282. doi: 10.1074/jbc.M607986200. [DOI] [PubMed] [Google Scholar]
- 22.Stone S.J., Levin M.C., Zhou P., Han J., Walther T.C., Farese R.V. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. Journal of Biological Chemistry. 2009;284(8):5352–5361. doi: 10.1074/jbc.M805768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wurie H.R., Buckett L., Zammit V.A. Diacylglycerol acyltransferase 2 acts upstream of diacylglycerol acyltransferase 1 and utilizes nascent diglycerides and de novo synthesized fatty acids in HepG2 cells. FEBS Journal. 2012;279(17):3033–3047. doi: 10.1111/j.1742-4658.2012.08684.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuerschner L., Moessinger C., Thiele C. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 2008;9(3):338–352. doi: 10.1111/j.1600-0854.2007.00689.x. [DOI] [PubMed] [Google Scholar]
- 25.McFie P.J., Banman S.L., Kary S., Stone S.J. Murine diacylglycerol acyltransferase-2 (DGAT2) can catalyze triacylglycerol synthesis and promote lipid droplet formation independent of its localization to the endoplasmic reticulum. Journal of Biological Chemistry. 2011;286(32):28235–28246. doi: 10.1074/jbc.M111.256008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du X., Herrfurth C., Gottlieb T., Kawelke S., Feussner K., Rühling H. Dictyostelium discoideum Dgat2 can substitute for the essential function of Dgat1 in triglyceride production but not in ether lipid synthesis. Eukaryotic Cell. 2014;13(4):517–526. doi: 10.1128/EC.00327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stone S.J., Myers H.M., Watkins S.M., Brown B.E., Feingold K.R., Elias P.M. Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. Journal of Biological Chemistry. 2004;279(12):11767–11776. doi: 10.1074/jbc.M311000200. [DOI] [PubMed] [Google Scholar]
- 28.Smith S.J., Cases S., Jensen D.R., Chen H.C., Sande E., Tow B. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking Dgat. Nature Genetics. 2000;25(1):87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- 29.Buhman K.K., Smith S.J., Stone S.J., Repa J.J., Wong J.S., Knapp F.F. DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. Journal of Biological Chemistry. 2002;277(28):25474–25479. doi: 10.1074/jbc.M202013200. [DOI] [PubMed] [Google Scholar]
- 30.Chen H.C., Smith S.J., Ladha Z., Jensen D.R., Ferreira L.D., Pulawa L.K. Increased insulin and leptin sensitivity in mice lacking acyl CoA:diacylglycerol acyltransferase 1. Journal of Clinical Investigation. 2002;109(8):1049–1055. doi: 10.1172/JCI14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi J., Lang W., Geisler J.G., Wang P., Petrounia I., Mai S. The use of stable isotope-labeled glycerol and oleic acid to differentiate the hepatic functions of DGAT1 and -2. The Journal of Lipid Research. 2012;53(6):1106–1116. doi: 10.1194/jlr.M020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rigouin C., Croux C., Borsenberger V., Ben Khaled M., Chardot T., Marty A. Increasing medium chain fatty acids production in Yarrowia lipolytica by metabolic engineering. Microbial Cell Factories. 2018;17(1):451. doi: 10.1186/s12934-018-0989-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aymé L., Jolivet P., Nicaud J.-M., Chardot T. Molecular characterization of the elaeis guineensis medium-chain fatty acid diacylglycerol acyltransferase DGAT1-1 by heterologous expression in Yarrowia lipolytica. PloS One. 2015;10(11) doi: 10.1371/journal.pone.0143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iskandarov U., Silva J.E., Kim H.J., Andersson M., Cahoon R.E., Mockaitis K. A specialized diacylglycerol acyltransferase contributes to the extreme medium-chain fatty acid content of Cuphea seed oil. Plant Physiology. 2017;174(1):97–109. doi: 10.1104/pp.16.01894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lardizabal K.D., Mai J.T., Wagner N.W., Wyrick A., Voelker T., Hawkins D.J. DGAT2 is a new diacylglycerol acyltransferase gene family: purification, cloning, and expression in insect cells of two polypeptides from Mortierella ramanniana with diacylglycerol acyltransferase activity. Journal of Biological Chemistry. 2001;276(42):38862–38869. doi: 10.1074/jbc.M106168200. [DOI] [PubMed] [Google Scholar]
- 36.Stoop W.M., Schennink A., Visker M.H.P.W., Mullaart E., van Arendonk J.A.M., Bovenhuis H. Genome-wide scan for bovine milk-fat composition. I. Quantitative trait loci for short- and medium-chain fatty acids. Journal of Dairy Science. 2009;92(9):4664–4675. doi: 10.3168/jds.2008-1966. [DOI] [PubMed] [Google Scholar]
- 37.Naik R., Obiang-Obounou B.W., Kim M., Choi Y., Lee H.S., Lee K. Therapeutic strategies for metabolic diseases: small-molecule diacylglycerol acyltransferase (DGAT) inhibitors. ChemMedChem. 2014;9(11):2410–2424. doi: 10.1002/cmdc.201402069. [DOI] [PubMed] [Google Scholar]
- 38.King A.J., Segreti J.A., Larson K.J., Souers A.J., Kym P.R., Reilly R.M. Diacylglycerol acyltransferase 1 inhibition lowers serum triglycerides in the Zucker fatty rat and the hyperlipidemic hamster. Journal of Pharmacology and Experimental Therapeutics. 2009;330(2):526–531. doi: 10.1124/jpet.109.154047. [DOI] [PubMed] [Google Scholar]
- 39.King A.J., Segreti J.A., Larson K.J., Souers A.J., Kym P.R., Reilly R.M. In vivo efficacy of acyl CoA: diacylglycerol acyltransferase (DGAT) 1 inhibition in rodent models of postprandial hyperlipidemia. European Journal of Pharmacology. 2010;637(1–3):155–161. doi: 10.1016/j.ejphar.2010.03.056. [DOI] [PubMed] [Google Scholar]
- 40.Qi J., Lang W., Giardino E., Caldwell G.W., Smith C., Minor L.K. High-content assays for evaluating cellular and hepatic diacylglycerol acyltransferase activity. The Journal of Lipid Research. 2010;51(12):3559–3567. doi: 10.1194/jlr.D008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuda N., Kumadaki S., Higashi C., Ozawa M., Shinozaki M., Kato Y. Intestine-targeted DGAT1 inhibition improves obesity and insulin resistance without skin aberrations in mice. PloS One. 2014;9(11) doi: 10.1371/journal.pone.0112027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sachdev V., Leopold C., Bauer R., Patankar J.V., Iqbal J., Obrowsky S. Novel role of a triglyceride-synthesizing enzyme: DGAT1 at the crossroad between triglyceride and cholesterol metabolism. Biochimica et Biophysica Acta. 2016;1861(9 Pt A):1132–1141. doi: 10.1016/j.bbalip.2016.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Irshad Z., Dimitri F., Christian M., Zammit V.A. Diacylglycerol acyltransferase 2 links glucose utilization to fatty acid oxidation in the brown adipocytes. The Journal of Lipid Research. 2017;58(1):15–30. doi: 10.1194/jlr.M068197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turnbull D.M., Bartlett K., Younan S.I.M., Sherratt H.S.A. The effects of 2{5(4-chlorophenyl) pentyl}oxirane-2-carbonyl-CoA on mitochondrial oxidations. Biochemical Pharmacology. 1984;33(3):475–481. doi: 10.1016/0006-2952(84)90243-0. [DOI] [PubMed] [Google Scholar]
- 45.Xu F.Y., Taylor W.A., Hurd J.A., Hatch G.M. Etomoxir mediates differential metabolic channeling of fatty acid and glycerol precursors into cardiolipin in H9c2 cells. The Journal of Lipid Research. 2003;44(2):415–423. doi: 10.1194/jlr.M200335-JLR200. [DOI] [PubMed] [Google Scholar]
- 46.Schlaepfer I.R., Joshi M. CPT1A-mediated fat oxidation, mechanisms, and therapeutic potential. Endocrinology. 2020;161(2):267. doi: 10.1210/endocr/bqz046. [DOI] [PubMed] [Google Scholar]
- 47.Thiele C., Papan C., Hoelper D., Kusserow K., Gaebler A., Schoene M. Tracing fatty acid metabolism by click chemistry. ACS Chemical Biology. 2012;7(12):2004–2011. doi: 10.1021/cb300414v. [DOI] [PubMed] [Google Scholar]
- 48.Thiele C., Wunderling K., Leyendecker P. Multiplexed and single cell tracing of lipid metabolism. Nature Methods. 2019;16(11):1123–1130. doi: 10.1038/s41592-019-0593-6. [DOI] [PubMed] [Google Scholar]
- 49.Liebisch G., Vizcaíno J.A., Köfeler H., Trötzmüller M., Griffiths W.J., Schmitz G. Shorthand notation for lipid structures derived from mass spectrometry. The Journal of Lipid Research. 2013;54(6):1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liebisch G., Fahy E., Aoki J., Dennis E.A., Durand T., Ejsing C. Update on LIPID MAPS classification, nomenclature and shorthand notation for MS-derived lipid structures. The Journal of Lipid Research. 2020 doi: 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaytor E.N., Shih H.-m., Towle H.C. Carbohydrate regulation of hepatic gene expression. Journal of Biological Chemistry. 1997;272(11):7525–7531. doi: 10.1074/jbc.272.11.7525. [DOI] [PubMed] [Google Scholar]
- 52.Stoeckman A.K., Towle H.C. The role of SREBP-1c in nutritional regulation of lipogenic enzyme gene expression. Journal of Biological Chemistry. 2002;277(30):27029–27035. doi: 10.1074/jbc.M202638200. [DOI] [PubMed] [Google Scholar]
- 53.Herzog R., Schwudke D., Schuhmann K., Sampaio J.L., Bornstein S.R., Schroeder M. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biology. 2011;12(1):R8. doi: 10.1186/gb-2011-12-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiorpes T.C., Hoerr D., Ho W., Weaner L.E., Inman M.G., Tutwiler G.F. Identification of 2-tetradecylglycidyl coenzyme A as the active form of methyl 2-tetradecylglycidate (methyl palmoxirate) and its characterization as an irreversible, active site-directed inhibitor of carnitine palmitoyltransferase A in isolated rat liver mitochondria. Journal of Biological Chemistry. 1984;259(15):9750–9755. [PubMed] [Google Scholar]
- 55.Conti R., Mannucci E., Pessotto P., Tassoni E., Carminati P., Giannessi F. Selective reversible inhibition of liver carnitine palmitoyl-transferase 1 by teglicar reduces gluconeogenesis and improves glucose homeostasis. Diabetes. 2011;60(2):644–651. doi: 10.2337/db10-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Giannessi F., Pessotto P., Tassoni E., Chiodi P., Conti R., de Angelis F. Discovery of a long-chain carbamoyl aminocarnitine derivative, a reversible carnitine palmitoyltransferase inhibitor with antiketotic and antidiabetic activity. Journal of Medicinal Chemistry. 2003;46(2):303–309. doi: 10.1021/jm020979u. [DOI] [PubMed] [Google Scholar]
- 57.Ashrafian H., Horowitz J.D., Frenneaux M.P. Perhexiline. Cardiovascular Drug Reviews. 2007;25(1):76–97. doi: 10.1111/j.1527-3466.2007.00006.x. [DOI] [PubMed] [Google Scholar]
- 58.Selby P.L., Sherratt H.S.A. Substituted 2-oxiranecarboxylic acids: a new group of candidate hypoglycaemic drugs. Trends in Pharmacological Sciences. 1989;10(12):495–500. doi: 10.1016/0165-6147(89)90049-7. [DOI] [PubMed] [Google Scholar]
- 59.Wolf H.P. Possible new therapeutic approach in diabetes mellitus by inhibition of carnitine palmitoyltransferase 1 (CPT1) Hormone and Metabolic Research Supplement Series. 1992;26:62–67. [PubMed] [Google Scholar]
- 60.Qu Q., Zeng F., Liu X., Wang Q.J., Deng F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death & Disease. 2016;7(5) doi: 10.1038/cddis.2016.132. e2226-e2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng S., Wang G., Wang Y., Cai L., Qian K., Ju L. Fatty acid oxidation inhibitor etomoxir suppresses tumor progression and induces cell cycle arrest via PPARγ-mediated pathway in bladder cancer. Clinical Science. 2019;133(15):1745–1758. doi: 10.1042/CS20190587. [DOI] [PubMed] [Google Scholar]
- 62.Yotsumoto T., Naitoh T., Kitahara M., Tsuruzoe N. Effects of carnitine palmitoyltransferase I inhibitors on hepatic hypertrophy. European Journal of Pharmacology. 2000;398(2):297–302. doi: 10.1016/s0014-2999(00)00288-0. [DOI] [PubMed] [Google Scholar]
- 63.Merrill C.L., Ni H., Yoon L.W., Tirmenstein M.A., Narayanan P., Benavides G.R. Etomoxir-induced oxidative stress in HepG2 cells detected by differential gene expression is confirmed biochemically. Toxicological Sciences. 2002;68(1):93–101. doi: 10.1093/toxsci/68.1.93. [DOI] [PubMed] [Google Scholar]
- 64.Cabrero A., Merlos M., Laguna J.C., Carrera M.V. Down-regulation of acyl-CoA oxidase gene expression and increased NF-kappaB activity in etomoxir-induced cardiac hypertrophy. The Journal of Lipid Research. 2003;44(2):388–398. doi: 10.1194/jlr.M200294-JLR200. [DOI] [PubMed] [Google Scholar]
- 65.Carracedo A., Cantley L.C., Pandolfi P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nature Reviews Cancer. 2013;13(4):227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Connor R.S., Guo L., Ghassemi S., Snyder N.W., Worth A.J., Weng L. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Scientific Reports. 2018;8(1):27. doi: 10.1038/s41598-018-24676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Divakaruni A.S., Hsieh W.Y., Minarrieta L., Duong T.N., Kim K.K.O., Desousa B.R. Etomoxir inhibits macrophage polarization by disrupting CoA homeostasis. Cell Metabolism. 2018;28(3):490–503. doi: 10.1016/j.cmet.2018.06.001. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Haas J.T., Winter H.S., Lim E., Kirby A., Blumenstiel B., DeFelice M. DGAT1 mutation is linked to a congenital diarrheal disorder. Journal of Clinical Investigation. 2012;122(12):4680–4684. doi: 10.1172/JCI64873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Rijn J.M., Ardy R.C., Kuloğlu Z., Härter B., van Haaften-Visser D.Y., van der Doef H.P.J. Intestinal failure and aberrant lipid metabolism in patients with DGAT1 deficiency. Gastroenterology. 2018;155(1):130–143. doi: 10.1053/j.gastro.2018.03.040. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nyman L.R., Cox K.B., Hoppel C.L., Kerner J., Barnoski B.L., Hamm D.A. Homozygous carnitine palmitoyltransferase 1a (liver isoform) deficiency is lethal in the mouse. Molecular Genetics and Metabolism. 2005;86(1–2):179–187. doi: 10.1016/j.ymgme.2005.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.