Abstract
Background
Type 2 diabetes, as the most prevalent metabolic disorder, is one of the leading causes of death worldwide. Recent studies showed a significant association between intestinal microbiota and type 2 diabetes. These studies have shared evidences that alteration in the composition of intestinal microbiota can disrupt the balance of the host homeostasis and lead to metabolic disorders such as type 2 diabetes. In the present study, we compared the intestinal microbiota composition in three groups of type 2 diabetes patients, pre-diabetic patients and healthy individuals of Iranian population.
Methods
After obtaining informed consent, stool samples were collected from 90 individuals of three studied groups. The DNA was extracted using column-based method. Intestinal microbiota composition was evaluated by quantitative real-time PCR using specific bacterial 16S rRNA primers. The difference of bacterial load was compared between three groups.
Results
The prevalence of Akkermansia muciniphila and Bifidobacteria species in healthy group was higher than type 2 diabetes group (P Value 0.006 and 0.001, respectively). In contrast, the load of Lactobacillus (P Value 0.044), Escherichia coli (P Value 0.005), and Bacteroides fragilis (P Value 0.017) in type 2 diabetes group, and the frequency of E. coli (P Value 0.001) and Bacteroides fragilis (P Value 0.004) in pre-diabetic group was significantly higher than healthy group. Moreover, the frequency of Faecalibacterium prausnitzii in healthy group was significantly higher compared to two other groups (P Value 0.005).
Conclusion
There is a correlation between intestinal microbiota composition and type 2 diabetes. Determination and restoration of this microbiota composition pattern may have a possible role in prevention and control of type 2 diabetes in a certain population.
Keywords: microbiota, gut, type 2 diabetes, pre-diabetes
Background
Gut flora, also known as gut microbiota, differs widely among individuals. It is estimated that there are about 100 trillion bacteria in human’s gastrointestinal tract, while 70 to 80% of them cannot be cultured [1]. Gastrointestinal flora composition also changes with diet [2], age [3], and other human factors [4]. The role of microbes in human health is not fully understood. But in recent years, studies have shown that microbes play an important role in the health, disease, and even treatment [5]. Intestinal microbiota is considered as a key factor in metabolism of carbohydrates, lipids, and amino acids and thus influences several metabolic disorders, including obesity; type 2 diabetes, and atherosclerosis [6, 7]. At the same time, Intestinal microbiota regulate inflammatory status by producing short-chain fatty acids (SCFAs) via carbohydrate fermentation and Lipopolysaccharide (LPS) of microbial membranes [8, 9].
Diabetes mellitus is a chronic and debilitating disorder characterized by persistent hyperglycemia. It may be due to dysfunction in insulin secretion (type 1 diabetes) or resistance to peripheral actions of insulin (type 2 diabetes). Type 2 diabetes is a metabolic disease with multiple predisposing factors, including obesity, genetic, physical inactivity and stress [10, 11]. A number of studies found out that intestinal microbiota and its metabolites can regulate the inflammatory responses, energy homeostasis, and lipid and glucose metabolism [12–14]. In this concern, it is documented that stimulation of glucagon-like peptide-1 release is a potential mediating mechanism for the effects of SCFAs on glucose homeostasis (SCFA-producing bacteria, such as Bifidobacterium). Hence, strategies have usually focused on a type 2 diabetes intestinal bacteria with anti-inflammatory properties and propensity to produce SCFAs. The clinical trials indicate that the modulation of the intestinal microbiota could be effective in diabetes management [15]. There is also evidences that intestinal microbiota has a significant role in triggering the chronic, low-grade inflammation that underpins insulin resistance and type 2 diabetes [16–18].
Numerous Factors like diet, hygiene and antibiotic consumption may alter composition of microbiota [19] and Contrary to studies carried out recently to explore the gut microbiota in diabetic patients and to identify the probable connection between type 2 diabetes and microbiota, there is still need for evidence supporting this hypothesis. Therefore, in the present study we tried to identify and compare the intestinal microbiota composition in three groups of type 2 diabetes patients, pre-diabetic patients and healthy individuals in Iranian population.
Materials and Methods
Study Design and Population
Ninety individuals referred to university hospitals in Tehran, Iran in 2018 were included in this case-control study. The inclusion criteria were age between 40 to 60 years. The exclusion criteria were suffering from chronic diseases (including inflammatory bowel diseases, liver cirrhosis, and chronic kidney diseases), smoking, alcohol consumption, cancer, current infectious diseases, receiving antibiotics and corticosteroids within the last month. Total sample size considering the standard effect size and normal approximation using the Z statistic, derived from published studies.
The subjects recruited into three groups. Group 1) thirty type 2 diabetes patients with A1c Hemoglobin less than 10%; group 2) thirty pre-diabetic patients identified with high fasting glucose (100 ≤ IFG < 126 mg/dl) or increased glucose within two hours after glucose injection (140 ≤ IGT < 200 mg/dl) and with A1c Hemoglobin less than 6.4; and group 3) thirty healthy individuals.
Method
After obtaining a written informed consent, the stool samples collected in sterile laboratory dishes containing ice and transferred to the Tuberculosis and Lung Research Department in Pasteur Institute, Tehran Iran. The samples preserved in −20 °C until analysis. DNA from the 200 mg of each stool sample was extracted using QIAamp® DNA Stool Mini Kit according to the manufacturer’s instructions method [20]. The concentration and purity of extracted DNA evaluated by a Nanodrop spectrophotometer (Thermo scientific, USA). The extracted DNA samples were stored at −20 for further analysis.
Quantitative real-time PCR was used in order to determine and quantify different bacterial populations. Genus-specific sequences of primers targeted the bacterial 16SrRNA genes used in the current study. Specificity of the primers was evaluated using the nucleotide BLAST in NCBI. The specific primers sequences are listed in Table 1 [19–27]. PCR reactions were performed in duplicate using Roche LightCycler® 96 system (Roche, Switzerland). Each 20 μl PCR reaction contained 1 ul DNA, 10 ul syber green master mix (Takara, Japan), 8 ul distilled water, and 0.5 μl of forward and reverse primers (10 pmol/L). The mixture was heated for 1 min at 95 °C, followed by 40 amplification cycles: denaturation (95 °C for 5 s), annealing (55 °C for 30 s), and extension (72 °C for 30 s). After amplification, the final melting curve analysis was performed by cooling down PCR product from 95 °C to 60 °C.
Table 1.
16S rRNA specific primers used in this study
| Bacteria | Forward | Reverse | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Akkermansia | CAGCACGTGAAGGTGGGGAC | CCTTGCGGTTGGCTTCAGAT | 327 | [19] |
| Bacteroides | CTGAACCAGCCAAGTAGCG | CCGCAAACTTTCACAACTGACTTA | 230 | [20, 21] |
| Lactobacillus | AGCAGTAGGGAATCTTCCA | CACCGCTACACATGGAG | 341 | [22, 23] |
| E. coli | CATTGACGTTACCCGCAGAAGAAGC | CTCTACGAGACTCAAGCTTGC | 195 | [24, 25] |
| Bifidobacterium | TCGCGTCYGGTGTGAAAG | CCACATCCAGCRTCCAC | 128 | [25, 26] |
| Faecalibacterium | GGAGGAAGAAGGTCTTCGG | AATTCCGCCTACCTCTGCACT | 248 | [25, 27] |
To calculate the concentration of bacteria, 10 fold serial dilutions of extracted DNA from standard strain E. coli were prepared. This standard curve allowed calculating DNA concentration of each bacterium from stool samples.
Statistical Analysis
The normality of data distribution was evaluated using Kolmogorov-Smirnov test. For data with normal distribution, One-way ANOVA and Fisher’s Least Significant Difference Test (LSD) were used. For non-normally distributed data, non-parametric Kruskal-Wallis test and Man-Whitney Test were applied. Mann-Whitney Test was used to analyze the correlation between gender and bacterial abundance. P Value less than 0.05 considered statistically significant. Statistical analyses performed using SPSS version 25.0 (SPSS Inc., Chicago, IL, USA).
Results
The mean age of patients with type 2 diabetes (53.3 years) was significantly higher than pre-diabetics (48.63 years) and healthy controls (51.63 years) (P Value = 0.024). Gender, Weight and BMI were not significantly different between studied groups (P Value >0.05). The concentration of FBS (mean ± SD) was significantly higher in pre-diabetic (100 ± 13.3) and type 2 diabetes patients (136 ± 17.03) compared to healthy controls (85.6 ± 9.9) (P Value ˂0.001).
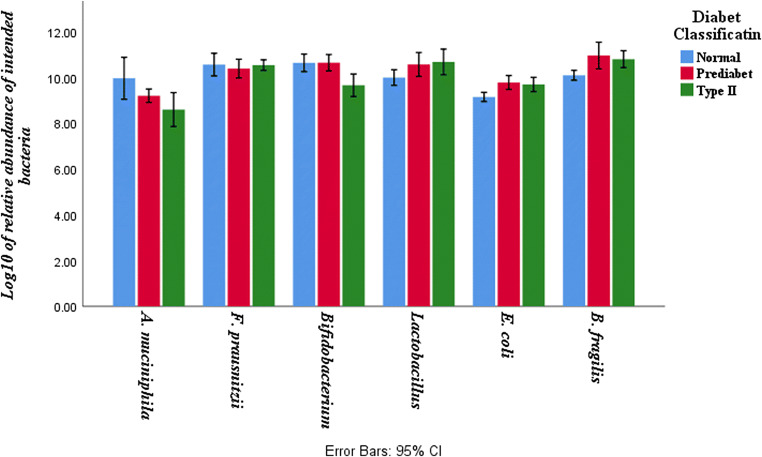
The load of the studied bacteria differs between the studied groups (Fig. 1). The bacterial load of E. coli was higher in fecal samples of pre-diabetic and type 2 diabetic patients compared to healthy controls (P-Values 0.001 and 0.005, respectively). The quantity of Bacteroides fragilis was also higher in pre-diabetic and type 2 diabetes patients compared to healthy controls (P Values 0.004 and 0.017, respectively).
Fig. 1.
Bacterial load of fecal samples in pre-diabetic, type 2 diabetic patients and healthy controls. Values are calculated based on Log10 CFU/g stool and are presented as mean ± SD
Faecalibacterium prausnitzii, a butyrate-producing genus, was significantly higher in healthy volunteers compared to pre-diabetic (P- Value 0.05) and type 2 diabetes patients (P Value 0.05).
The difference between the load of Akkermansia muciniphila in healthy group compared to pre-diabetic group was not significant but it was statistically significant compared to type 2 diabetes patients (P Values 0.11 and 0.006, respectively). In contrast, the bacterial load of lactobacillus was higher in type 2 diabetic patients than healthy group (P Value 0.006). The difference between pre-diabetic group and healthy controls was not statistically significant (P Value >0.05).
The number of Bifidobacterium was significantly higher in healthy subjects compared to type 2 diabetic patients (P Value 0.001). The pre-diabetic group also had a higher load of Bifidobacterium compared to type 2 diabetic patients (P Value 0.001).
The study further explored the correlation between subjects’ gender and bacterial abundance. The load of Akkermansia muciniphila, Faecalibacterium prausnitzii, E. coli, and Bacteroides fragilis was higher in male subjects whereas female participants had a larger number of Bifidobacterium and Lactobacillus but the results were not statistically significant (P Value 0.1).
Discussion
Previous studies showed that gut microbiota composition is different in populations with different ethnicities. It is hypothesized that gut microbiota including Akkermansia muciniphila, Faecalibacterium prausnitzii, Bifidobacterium, Lactobacillus, E. coli and Bacteroides fragilis has a role on regulating metabolic profile and immune system and in triggering the chronic, low-grade inflammation which leads to insulin resistance and type 2 diabetes [21, 22].
In the present study we showed that the frequency of Akkermansia muciniphila and Bifidobacterium in healthy group was higher than that in type 2 diabetes group. In contrast, the load of Lactobacillus, E. coli, and Bacteroides fragilis in type 2 diabetes group and the load of E. coli and Bacteroides fragilis in pre-diabetes group were significantly higher than healthy group. Also, the frequency of Faecalibacterium prausnitzii in the healthy group was significantly greater than the other two groups.
Karlsson et al. study as one of the most comprehensive studies on the gut microbiota composition, indicated lower abundance of butyrate-producing bacteria in type 2 diabetes patients [23]. In another similar study, Qin et al. found that the frequency of Faecalibacterium prausnitzii decreased in patients with type 2 diabetes, while opportunistic pathogens such as Bacteroides and E.coli increased. They also reported that the prevalence of Akkermansia muciniphila in patients with Type 2 diabetes was higher than healthy individuals [24]. In the present study, we also showed a significant decrease in the frequency of Faecalibacterium prausnitzii bacteria in patients with type 2 diabetes compared to the control group. Furthermore, we showed that the prevalence of Bacteroides fragilis and E. coli was significantly higher in pre-diabetic and type 2 diabetes patients than in normal subjects. Interestingly, in contrast to Qin et al., the results of our studies showed that the frequency of Akkermansia muciniphila was significantly higher in healthy individuals than in those with type 2 diabetes. Different geographical and epidemiological factors may cause these differences.
Consistent with the present results, Le et al. showed that the frequency of Lactobacillus in fecal samples of patients with type 2 diabetes was significantly higher than healthy subjects, while Bifidobacterium was lower [25].
In previous investigations, Wu et al. observed a lower load of Bifidobacterium in patient with type 2 diabetes. [26]. Also, Moghaddam et al. reported a significant decrease in frequency of Faecalibacterium prausnitzii in Iranian type 2 diabetic patients compared to control group but there was no significant difference in the number of Bifidobacterium and Bacteroides fragilis between type 2 diabetic and healthy individuals [27]. Consistent with these studies, we showed a significant decrease in the frequency of Faecalibacterium prausnitzii and Bifidobacterium in type 2 diabetes patients compared to the control group. Additionally, we showed that the frequency of Bacteroides fragilis in Pre-diabetic and type 2 diabetic patients were significantly higher than that in healthy subjects.
Sedighi et al. studied the association between frequency of Lactobacillus and Bifidobacterium and type 2 diabetes in Iranian population. They reported that in patients with type 2 diabetes the frequency of Lactobacillus was significantly higher than healthy individuals. In contrast, the frequency of Bifidobacterium in patients with type 2 diabetes was significantly lower than healthy individuals [28]. In contrast, Le et al. and Halawa and colleagues, reported significantly lower Lactobacillus in stool sample among type 2 diabetic patients when compared to control group [25, 29]. The present study indicated that the prevalence of Lactobacillus bacteria in the gastrointestinal tract of patients with type 2 diabetes was significantly higher than healthy individuals. Our study also confirmed the reduction of Bifidobacterium load in type 2 diabetes patients and showed that there was a significant correlation between Bifidobacterium frequency and type 2 diabetes.
Ejtahed et al. reported that bacterial load of Bifidobacterium and Bacteroides was higher in healthy control group in Iranian population [30]. Since long-term diet affects Bacteroides, It has been reported that Bacteroides was associated with a diet rich in protein and animal fat [31].
In the present study we found that the bacterial load of Akkermansia muciniphila and Faecalibacterium prausnitzii in healthy group is higher than type 2 diabetes group. In contrast with the present evidences, Remely et al. reported that the frequency of Akkermansia muciniphila and Faecalibacterium prausnitzii also increased in type 2 diabetes group [32].
Controversial results regarding the gut microbiota composition in type 2 diabetes can be seen in different studies. These controversies may be due to the affecting co-factors including, ethnicity, different locations, life style, various diet, drug consumption history and even the methodology to evaluate the microbiota.
In this study we were able to evaluate the association between gut microbiota and type 2 diabetes in Iranian population. Because of time and financial limitations, we only selected several bacteria to evaluate in this research. Conducting a multi-centric national study on individuals from most parts of Iran and evaluation of more bacterial genera in gut microbiota (more representative sample) could increase the reliability of these findings.
The considered time of cessation of antibiotic prior to sampling in this study was one month. We were not able to consider longer time (three months). We were not also able to rule out all situations that may lead to chronic inflammation and use them as exclusion criteria.
Conclusion
The correlation between gut microbiota and different medical conditions like type 2 diabetes has been widely established in several studies, with differences in findings and attributable strains. It is important that possible interventions for changing the composition of intestinal microbiota in order to prevent or control inflammatory conditions should be based on regional habits and dietary factors.
Acknowledgements
This work was supported by the National Institute for Medical Research Development (NIMAD), Tehran, Iran (942995). The authors would like to thank the colleagues at Mycobacteriology and Pulmonary Research Department and Microbiology Research Center at Pasteur Institute of Iran, Tehran, Iran.
Abbreviations
- IFG
Impaired Fasting Glucose
- IGT
Impaired Glucose Tolerance
- A1c
Glycated Hemoglobin
- PCR
Polymerase Chain Reaction
- DNA
Deoxyribonucleic Acid
- CFU
Colony Forming Unit
Compliance with Ethical standards
Conflict of interests
The authors declare that they have no conflict of interest.
Informed consent
Prior to sampling, a written informed consent obtained from all participants.
Footnotes
The original online version of this article was revised: In the original publication of the article, the grant number in Acknowledgement Section has been processed incorrectly. It has been updated in this correction.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/15/2022
A Correction to this paper has been published: 10.1007/s40200-022-01136-7
References
- 1.Ho JT, Chan GC, Li JC. Systemic effects of gut microbiota and its relationship with disease and modulation. BMC Immunol. 2015;16:21. doi: 10.1186/s12865-015-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott KP, Gratz SW, Sheridan PO, Flint HJ, Duncan SH. The influence of diet on the gut microbiota. Pharmacol Res. 2013;69(1):52–60. doi: 10.1016/j.phrs.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 3.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hasan N, Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young VB. The intestinal microbiota in health and disease. Curr Opin Gastroenterol. 2012;28(1):63–69. doi: 10.1097/MOG.0b013e32834d61e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulange CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 2016;8(1):42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang WH, Kitai T, Hazen SL. Gut Microbiota in Cardiovascular Health and Disease. Circ Res. 2017;120(7):1183–1196. doi: 10.1161/CIRCRESAHA.117.309715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.d'Hennezel E, Abubucker S, Murphy LO, Cullen TW. Total Lipopolysaccharide from the Human Gut Microbiome Silences Toll-Like Receptor Signaling. mSystems. 2017;2(6). [DOI] [PMC free article] [PubMed]
- 10.Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115(5):1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 12.Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short-chain fatty acids, and host metabolic regulation. Nutrients. 2015;7(4):2839–2849. doi: 10.3390/nu7042839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017;18(1):2. doi: 10.1186/s12865-016-0187-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pussinen PJ, Havulinna AS, Lehto M, Sundvall J, Salomaa V. Endotoxemia is associated with an increased risk of incident diabetes. Diabetes Care. 2011;34(2):392–397. doi: 10.2337/dc10-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Adeshirlarijaney A, Gewirtz AT. Considering gut microbiota in treatment of type 2 diabetes mellitus. Gut Microbes 2020:1–12. [DOI] [PMC free article] [PubMed]
- 16.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016;15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carvalho BM, Saad MJ. Influence of gut microbiota on subclinical inflammation and insulin resistance. Mediat Inflamm. 2013;2013:986734. doi: 10.1155/2013/986734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3(4):279–288. doi: 10.4161/gmic.19625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutr. 2016;63(10):560–568. doi: 10.1016/j.endonu.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Gong J, Cottrill M, Yu H, de Lange C, Burton J, Topp E. Evaluation of QIAamp DNA Stool Mini Kit for ecological studies of gut microbiota. J Microbiol Methods. 2003;54(1):13–20. doi: 10.1016/S0167-7012(02)00260-9. [DOI] [PubMed] [Google Scholar]
- 21.D'Argenio V, Salvatore F. The role of the gut microbiome in the healthy adult status. Clin Chim Acta. 2015;451(Pt A):97–102. doi: 10.1016/j.cca.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Sommer F, Backhed F. The gut microbiota--masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 23.Karlsson FH, Tremaroli V, Nookaew I, Bergstrom G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498(7452):99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 24.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D, Peng Y, Zhang D, Jie Z, Wu W, Qin Y, Xue W, Li J, Han L, Lu D, Wu P, Dai Y, Sun X, Li Z, Tang A, Zhong S, Li X, Chen W, Xu R, Wang M, Feng Q, Gong M, Yu J, Zhang Y, Zhang M, Hansen T, Sanchez G, Raes J, Falony G, Okuda S, Almeida M, LeChatelier E, Renault P, Pons N, Batto JM, Zhang Z, Chen H, Yang R, Zheng W, Li S, Yang H, Wang J, Ehrlich SD, Nielsen R, Pedersen O, Kristiansen K, Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 25.Le KA, Li Y, Xu X, Yang W, Liu T, Zhao X, et al. Alterations in fecal Lactobacillus and Bifidobacterium species in type 2 diabetes patients in Southern China population. Front Physiol. 2012;3:496. doi: 10.3389/fphys.2012.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X, Ma C, Han L, Nawaz M, Gao F, Zhang X, Yu P, Zhao C’, Li L, Zhou A, Wang J, Moore JE, Cherie Millar B, Xu J. Molecular characterisation of the faecal microbiota in patients with type 2 diabetes. Curr Microbiol. 2010;61(1):69–78. doi: 10.1007/s00284-010-9582-9. [DOI] [PubMed] [Google Scholar]
- 27.Navab-Moghadam F, Sedighi M, Khamseh ME, Alaei-Shahmiri F, Talebi M, Razavi S, Amirmozafari N. The association of type 2 diabetes with gut microbiota composition. Microb Pathog. 2017;110:630–636. doi: 10.1016/j.micpath.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 28.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, Amirmozafari N. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microb Pathog. 2017;111:362–369. doi: 10.1016/j.micpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 29.Halawa MR, El-Salam MA, Mostafa BM, Sallout SS. The Gut Microbiome, Lactobacillus acidophilus; Relation with Type 2 Diabetes Mellitus. Curr Diabetes Rev. 2019;15(6):480–485. doi: 10.2174/1573399815666190206162143. [DOI] [PubMed] [Google Scholar]
- 30.Ejtahed H-S, Hoseini-Tavassol Z, Khatami S, Zangeneh M, Behrouzi A, Ahmadi Badi S, Moshiri A, Hasani-Ranjbar S, Soroush AR, Vaziri F, Fateh A, Ghanei M, Bouzari S, Najar-Peerayeh S, Siadat SD, Larijani B. Main gut bacterial composition differs between patients with type 1 and type 2 diabetes and non-diabetic adults. Journal of Diabetes & Metabolic Disorders. 2020;19(1):265–271. doi: 10.1007/s40200-020-00502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, Sinha R, Gilroy E, Gupta K, Baldassano R, Nessel L, Li H, Bushman FD, Lewis JD. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remely M, Hippe B, Zanner J, Aumueller E, Brath H, Haslberger AG. Gut Microbiota of Obese, type 2 diabetes Individuals is Enriched in Faecalibacterium prausnitzii, Akkermansia muciniphila and Peptostreptococcus anaerobius after Weight Loss. Endocr Metab Immune Disord Drug Targets. 2016;16(2):99–106. doi: 10.2174/1871530316666160831093813. [DOI] [PubMed] [Google Scholar]