Abstract
Purpose
Microbiota-derived metabolites could alter the brain tissue toward the neurodegeneration disease. This study aims to select the genes associated with Propionic acid (PPA) and compromise Alzheimer’s disease (AD) to find the possible roles of PPA in AD pathogenesis.
Methods
Microbiota-derived metabolites could alter the brain tissue toward the neurodegeneration disease. This study aims to select the genes associated with Propionic acid (PPA) and compromise Alzheimer’s disease (AD) to find the possible roles of PPA in AD pathogenesis.
Results
Amongst all genes associated with PPA and AD, 284 genes to be shared by searching databases and were subjected to further analysis. AD-PPA genes mainly involved in cancer, bacterial and virus infection, and neurological and non-neurological diseases. Gene Ontology and pathway analysis covered the most AD hallmark, such as amyloid formation, apoptosis, proliferation, inflammation, and immune system. Network analysis revealed hub and bottleneck genes. MCODE analysis also indicated the seed genes represented in the significant subnetworks. ICAM1 and CCND1 were the hub, bottleneck, and seed genes.
Conclusions
PPA interacted genes implicated in AD act through pathways initiate neuronal cell death. In sum up, AD-PPA shared genes exhibited evidence that supports the idea PPA secreted from bacteria could alter brain physiology toward the emerging AD signs. This idea needs to confirm by more future investigation in animal models.
Electronic supplementary material
The online version of this article (10.1007/s40200-020-00564-7) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer’s disease, Propionic acid, Microbiota, Bioinformatics, System biology
Introduction
Alzheimer’s disease (AD) is known as the most common neurodegenerative disorder that its primitive hallmarks are the accumulation of beta-amyloid peptide (Aβ) and neurofibrillary tangles [1]. In the recent decade, another hypothesis explains AD pathogenesis that named the infection hypothesis. This hypothesis explains that microorganisms might have an essential role in AD progression [2]. This hypothesis reconstructed by pioneering work of Itzhaki’s group, who showed that plaques contain residues of HSV-1 viral DNA and some experimental data intimate that other viruses, such as CMV, may also be involved in the pathogenesis of AD [3, 4]. The epidemiological studies show that the presence of antiviral antibodies has been correlated with the long-term development of AD [5]. Other groups of investigators suggested a role for spirochetes in the pathogenesis of AD or the presence of Borrelia burgdorferi in the post-mortem brains of many AD patients [6, 7]. Cutibacterium acnes(propionibacterium acnes) was detected in the cortex of patients with AD [8]. Scientists at Bristol University also used 16S ribosomal NGS to assess the bacterial component of the microbiome in post-mortem tissue from the temporal cortex of AD and control. This study suggests that AD brains tend to have large bacterial loads compared to controls. They reported phyla such as Firmicutes and most consistently Actinobacteria, especially Cutibacterium acnes [9]. Cutibacterium acnes is a Gram-positive and anaerobic human skin commensal that involved in the pathogenesis of acne. It was formerly named Propionibacterium acnes for its ability to generate propionic acid (PPA) [10, 11]. In addition to microbiota found in post-mortem brain tissue of AD and related them to neurological pathologies [9],we, therefore, hypothesized the microbiota-derived metabolites such as PPA might be a significant risk factor for AD.
PPA is found in the gut, along with other short-chain fatty acids, such as acetate and butyrate, which are major metabolic products of enteric bacteria, following fermentation of dietary carbohydrates and some amino acids [12, 13]. Many bacteria existing in the oral mucosa also produce PPA [14–16]. PPA is taken up by neuroglia and neurons and enters the CNS, where it is thought to comprise a significant energy source in cellular metabolism, particularly during early brain development [17, 18]. Besides, PPA changes several physiological processes such as neurotransmitter synthesis and release, cell signaling, free radical production, mitochondrial function, lipid metabolism, immune function, gap junction gating, intracellular pH maintenance, and modulation of gene expression through phosphorylation and histone acetylation [19, 18, 20–22]. PPA inoculation induced abnormal neural cell organization, which may have led to autism-like neurobehaviors [23, 24]. Studies showed that following PPA administration in rats elevates levels of microglia (CD68 positive) and neurotoxic cytokines, including interleukin (IL)-6, tumor necrosis factor (TNF)-α, and interferon-γ [25–27].
Tanzi and Moir have produced a compelling piece of evidence demonstrating amyloid β that accumulated in the AD brain is an antimicrobial peptide [28]. These observations were supported by the in vivo testing of the antimicrobial activity of Aβ [29]. Antimicrobial activity of Aβ in the presence of pathogens or pathogen-derived metabolites like PPA could be a crucial trigger of starting AD pathogenesis. Multiple studies focus on the positive effect of PPA on treated cells in animal models [25–27], or determined its association as an environmental contributor in autism spectrum disorders [23, 24, 30]. Since there is no investigation directly shows the AD pathology and neuronal toxicity of PPA in experimental models, this study used bioinformatic analysis to identify PPA complications link to emerge AD pathophysiology and signs. Herein, we aimed to combine the multi-source PPA-related data in a meaningful manner as retrieved from multiple databases to uncover the mechanism associated with these critical genes interacted with PPA and involved in AD. This study will improve our ability to understand and diagnose the possible microbiota-derived PPA and consequences of AD pathology and it will help to prevent long-term complications.
Methodology
Study design and prioritization of AD and PPA genes
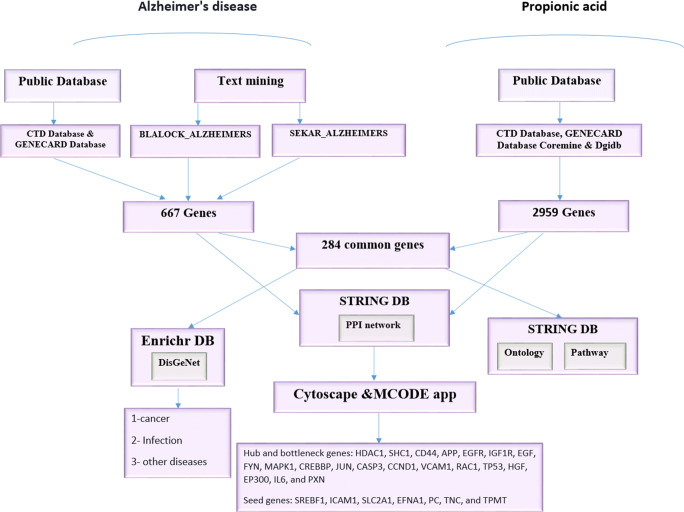
To find the possible relationship of PPA secreted by microbiota from variable resources and AD pathogenesis, we selected the genes shared in PPA and AD for further functional and structural analysis. There are different public resources that contain the interactions of genes with chemicals, drugs, and other different agents. These kinds of data emerged from experimental [31, 32] and computational knowledge [32, 33]. Besides, database repositories of high throughput gene expression, including Gene Expression Omnibus (GEO) [34] and ArrayExpress [35]contain data, have been produced from the direct effect of drugs and chemicals on cell lines, animal models, or the result of human clinical trials. In this study, collected genes associated with PPA and AD separately from several of the following resources are explained in data gathering part and selected the shared genes (named AD-PPA shared genes or PPA-AD shared genes) for the next analysis. Used Gene Ontology (GO) and Pathway enrichment tools to find the mechanism related to these genes as the functional analysis of data and applied network analysis as the structural analysis of data to find the crucial genes. An outline of the workflow of this study was summarized in Fig. 1. To prioritize the AD-PPA genes, first focused on the data gathering from gene expression databases and compared significant data with data obtained from public databases [36]. Public databases generally collected data from different molecular databases and, based on unique algorithms designed for their analysis; report the output with significant value. In this study, it was almost selected output from different databases with significant value for both AD and PPA keywords. The other prioritization of AD-PPA genes, analysis of AD, and PPA network separately and compare the output with the network of AD-PPA genes. For analysis gene-diseases association, gene ontology, and KEGG pathway, used genes that are shared in AD and PPA.
Fig. 1.
The workflow of data gathering related to AD and PPA and their functional and structural analysis
Data gathering related to Alzheimer disease and propionic acid
The parent terms “Alzheimer disease” was used to find the genes and proteins. The AD data was collected from public databases (CTD, and GeneCards) and gene expression data from text mining. Comparative Toxicogenomics Database (CTD, http://ctdbase.org) is a public resource that carries information about interactions between gene products and environmental chemicals. Over 15 million toxicogenomic relationships in CTD provide a user-friendly database that helps to find the effect of these interactions on human diseases [36]. In CTD, the input is a gene list or gene name, drug, chemical agent, and disease term. The output will be chemicals interact with genes, genes interact with the drug and chemical agent, and genes involved in the disease. We searched the keyword “Alzheimer’s disease” in the search box related to keyword queries and selected Alzheimer’s disease between 2 queries obtained from the input. Then we opened the gene box in Alzheimer’s disease page to download the genes associated with AD. We used all data retrieved from this database without considering the inference score. For prioritizing inferences of the different database used in CTD, CTD provides a statistic named “Inference Score” that it reflects the degree of similarity between CTD chemical–gene-disease networks and a similar scale-free random network [36].
GeneCards (https://www.genecards.org/) includes comprehensive useful and annotative information of known and predicted human genes that categorized in different terms including, genetic loci, gene clusters, RNA genes, pseudogenes, and protein coding. These data integrate automatically from over 125 data sources [37]. In this database, also search for genes, biological process, or human disease and output will be all genes linked to the searching term. To find the gene associated with Alzheimer’s disease, search this keyword as input in the search box of the GeneCards database and exported all data related to Alzheimer’s disease. For selecting the genes in this database without considering scoring, applied all data as output. GeneCards used GIFtS score linked to the GIFtS algorithm that reflected the degree of a gene’s functionality. [37].
Gene expression data were obtained from text mining. We found papers that analyzed the microarray and RNAseq data [38, 39]. Brandon L. Pearson et al. used previously published neurodegenerative disease data, including these two data named BLALOCK_ALZHEIMERS and SEKAR_ALZHEIMERS. An Excel list of brain disorders gene sets and genes was accessible in Supplementary Data 2 [40].We picked up BLALOCK_ALZHEIMERS and SEKAR_ALZHEIMERS data (were statistically significant (FDR < 0.1)) and combined these two AD datasets and introduced them as “text mining data.“
Blalock EM et al. have been generated microarray data that used snap-frozen hippocampal specimens of 31 subjects at different levels of ADs that assessed with Braak stages, adjusted Mini-mental Status Exam (MMSE) test and neurofibrillary tangle (NFT) count. The control tissues were matched for age, and neurodegenerative disorders (MMSE > 25, < Braak stage II). The samples were provided by Brain Bank of the Alzheimer’s Disease Research Center at the University of Kentucky. To generate gene expression data used, human GeneChips (HG-U133A) and MICROARRAY SUITE 5 (MAS5; 50) [38].
Sekar Sh. et al. in a study with dbGaP accession NO. phs000745.v1.p1 also has been used ten post-mortem brain samples from late-onset AD subjects(Braak stages ranging from IV to VI) and ten healthy elderly control subjects(Braak stages ranging from I to IV). They were collected at the Banner Sun Health Research Institute’s Brain and Body Donation Program. Library pools were sequenced by synthesis on the Illumina HiSeq2000 using Illumina’s Truseq PE Cluster Kit v3 and Illumina’s TruSeq SBS Kits v3 for paired 83 bp read lengths [39].
The final gene list related to AD was obtained from the genes that were shared between CTD and GeneCards databases and text mining.The parent term “propionic acid” was used to find the genes that interact with propionic acid. In GEO or ArrayExpress databases, there was not found high-throughput data that have been directly produced from the effect of PPA on cell line or animal models, so to collect PPA data used the public databases, CTD and GeneCards, DGIDB, and Coremine search engine. In CTD, searched propionic acid in keyword queries and selected propionic acid between 316 results to find the genes associated with propionic acid. Then opened the gene box in the propionic acid page and downloaded all the genes as output. GeneCards also searched the propionic acid in the keyword search box and exported all genes associated with propionic acid as output. Here for CTD and GeneCards, selected all genes without considering a specific threshold for scoring. The Drug Gene Interaction Database (DGIDB, www.dgidb.org), is an open research resource for drug-gene interaction. This database applies 30 disparate databases and web resources to analyze information associated with the interaction of genes and drugs [33]. In DGIDB databases, input was the propionic acid in search category box and selected drug sheet to search all genes interact with propionic acid but did not use filter related to the Clinically Actionable, Druggable Genome and Drug Resistance. All genes retrieved from this database used for further analysis. Coremine search engine is a free Internet service for searching that developed by PubGene Company (www.coremine.com). Coremine Medical is a domain for sharing medical information. In the Coremine Medical search engine, propionic acid was explored as input, then selected the gene/protein in extracted association part and copied the genes that were highlighted based on the significant level and shown by at least one blue rectangle.Final gene list of PPA was the combination of all genes obtained from databases, CTD, GeneCards, and DGIDB, and Coremine search engine.
AD genes and genes interacted PPA were downloaded in December 2019.
AD-PPA shared genes
To determine the shared AD-related genes from databases and text mining applied Venn diagram software (http://bioinformatics.psb.ugent.be/webtools/Venn/). To find the shared genes between AD and PPA (shared genes) also used Venn diagram software. All of these shared genes of AD and PPA were selected for further analysis.
Gene-disease association analysis
In order to find the comorbidity disease-related to a gene set, the Enrichr database use several disease databases, OMIM, DSigDB, dbGaP, MSigDB, GeneSigDB, to analyze genes for association with diseases. The input of this database is a gene set, and output will be a table of significant diseases and associated genes [41]. To identify the disease possibly related to the AD-PPA genes used Enrichr database (http://amp.pharm.mssm.edu/Enrichr). That is an open-source database that used different libraries to enrich a gene list [41]. For gene-disease association analysis, selected the DisGeNet database in Enrichr database. We insert the AD-PPA genes as input to the search box, then used the DisGeNet database between disease databases in the disease page of Enrichr and exported the table contain different statistical analyses for the relationship between genes and diseases. We selected the diseases that their P-values < 0.05.
Gene ontology and KEGG pathway analysis
Functional analysis was performed using the String database (version 11.0) (https://string-db.org). This database determined the protein-protein interaction and held the enrichment tools to analyze the ontologies and pathways. The Gene Ontology (GO) categories are GO Biological Process, GO Molecular Function, and GO Cellular Component. String database connected to the KEGG database to analyze genes for the enriching pathway. We inserted the AD-PPA genes as input to search box related to multiple proteins and selected homo sapience for the organism. The output contains network and several characteristics information linked to genes in the network. Then analysis box opened and find the different enrichment analyses that we selected the Gene Ontology and KEGG pathway analysis and downloaded. Enrichment analyses in the String database contain only significant terms with P-value < 0.05, and it is not necessary to select significant output manually.
Protein-protein interaction network construction and topological analysis
Network topology indicates the topological structure of a network [42]. In protein-protein interaction network is a layout of the physical connections between proteins [43]. In the analysis of biological networks used topological structure analysis to identify some aspects such as hubs and bottleneck nodes or modules as the groups of nodes with high topological overlap. A biological network is a scale-free network contains a few highly connected nodes (hubs) which link the rest of the less connected nodes and are sensitive to deletion hubs. Also, hub nodes in the protein-protein interaction network are more likely to be essential for a vital cellular process [43]. All genes related to PPA, AD, and AD-PPA shared genes were fed into the String database to construct the protein-protein interaction networks for each gene set. Then, the protein-protein interaction network (tsv file) was imported to Cytoscape software (version 3.5.1) (http://www.cytoscape.org/) to decipher the crucial protein from the analysis of the network. The Cytoscape is open-source software that constructs and analysis network [44]. In order to introduce the hub and bottleneck genes, it was employed network analysis that assesses the top 10% of degree and betweenness-centrality of every node, respectively. Hub genes are defined as highly connected nodes in the protein-protein interaction network. The network is sensitive to delete the hub genes [45]. Betweenness-centrality is a measure of centrality in a network based on shortest paths. In network analysis, the genes with higher betweenness-centrality are crucial nodes contain valuable information that passes through these nodes, and whole network connectivity depends on these nodes [46]. Hub and bottleneck genes play essential roles in the characteristics and the development of the disease [45]. To find the seed gene used the MCODE app in Cytoscape. MCODE used the topological feature to find highly interconnected regions in a network and introduce numerous clusters and seed genes. The seed genes are interest genes in identified subnetworks and defined as the highest-scoring gene in a gene cluster [47]. The input for analysis in MCODE was selecting every network and then separately run the MCODE. The output was that the sub-networks of each network were significant and contained a score. MCODE parameter was selected to analyze data included, Node Score Cutoff: 0.2, Haircut: true, Fluff: false, K-Core: 2, Max. Depth from Seed: 100. The clusters with the highest score selected for functional analysis (GO and KEGG pathway analysis). Consequently, a protein-protein interaction network is composed of many significant genes (hub and bottleneck genes) and the seed genes allocated to the subnetwork analysis. In this study, constructed networks from AD, PPA, and AD-PPA shared genes, determined the hub and bottleneck nodes in each network, then by MCODE determined the subnetworks and compared the seed genes between all networks. The data selected based on hub-bottleneck-seed genes were listed for functional analysis by GO and pathway analysis.
Statistical analysis
All data with P-values of < 0.05 were considered to be statistically significant.
Results
Genes associated with AD; genes associated with PPA and shared genes between AD and PPA
There is no data that explain the role of microbiota-derived PPA in AD pathophysiology. Here, by using available data related to the AD and PPA, try to find how genes interacted with PPA could affect and change the brain cells to promote AD. In order to meet this aim applied several filters to prioritize genes and mechanisms. First, it was in data gathering that integrated both gene expression data and data obtained from public databases. In order to find the accurate genes related to AD, we selected the expressed genes available in text mining data produced from BLALOCK_ALZHEIMERS (1218 genes) and SEKAR_ALZHEIMERS (226 genes), and genes available in public databases, CTD (22668 genes), and GeneCards (8605 genes) databases. All shared genes were obtained by Venn Diagram software (Supplementary Data S1 and Fig. 2a). It was determined 667-shared genes in AD. To find the interaction of the genes with PPA, we integrated data available in CTD (17 genes), DGIDB (3 genes), and GeneCards (2719 genes) databases and 513 genes found significantly in the Coremine search engine. It was collected 2959 genes interact with PPA in different databases in which found 284 shared genes with AD (Fig. 2b). These genes named AD-PPA shared genes that contain genes associated with both AD and PPA. All detected genes related to AD and PPA and AD-PPA shared genes were listed in Supplementary Data S1. The second step in prioritization was the analysis of AD, PPA, and AD-PPA genes separately, particularly network analysis. AD network constructed from 667 genes whereas the PPA network constructed from 2959 genes.
Fig. 2.

Venn Diagram of genes associated with AD (a) and the common genes detected in AD and PPA (b)
Gene-disease association
Enrichr database was used for gene-disease association analysis and selected DisGeNet database to determine the possible disease enriched for 284 AD-PPA shared genes. The results of Enrichr analysis within the DisGeNet database indicated that AD-PPA shared genes are related to some diseases that organized in three categories, infection, cancer, and other diseases (represented in Table 1 and Supplementary data S2). The most significant disease enriched was allocated to cancer progression and metastatic process and represented these genes are dysregulated in several cancer types. The next significant category related to infection by several bacteria and viruses such as HIV, Cytomegalovirus, Respiratory syncytial virus, Helicobacter pylori, Epstein-Barr virus, Enterovirus infections, Retroviridae infections, Salmonella infections, and Herpes Simplex. Several neurocognitive disorders including, autistic disorder, parkinsonian disorders, prion diseases, meningitis, epilepsy, transient cerebral ischemia, depressive symptoms, marijuana abuse, and dementia (vascular) or disability such as intellectual disability, mental retardation, mental deficiency, and poor school performance. In addition to neurological disorders, AD-PPA shared genes enriched in non-neurological disease, including obesity, hypertensive disease, diabetes mellitus, degenerative polyarthritis, heart failure, asthma, endometriosis, and keratosis. Several recognized disorders were associated with the cellular alteration that emerged in AD condition, such as memory loss, gliosis, Tauopathies, Neural Tube Defects, protein deficiency.
Table 1.
Top significant of DisGeNet disease related to PPA-AD genes that organized in 3 categories, infections, cancer and other diseases
| Term | Adjusted P-value | Genes |
|---|---|---|
| Infections | ||
| HIV Infections | 1.26E-25 | APP;CDKN1A;MCM7;CD81;HFE;TAT;TNC;SLC2A1;GPT;ICAM1;EDNRA;… |
| Infection | 9.58E-23 | APP;SCARB1;KHDRBS1;CD81;HFE;PDGFB;SLC2A1;LPL;EGFR;ICAM1;PTGS1;… |
| Infection caused by Helicobacter pylori | 3.90E-16 | HFE;ODC1;TGFA;EGFR;ICAM1;GJA1;KHSRP;CASP3;PIM1;HMOX1;RAC1;JUN;… |
| Herpes Simplex Infections | 7.27E-15 | HDAC4;APP;ITIH4;CDKN1A;SPARC;TAT;ODC1;LEF1;SLC2A1;NOLC1;COMT;EGFR;… |
| Respiratory syncytial virus (RSV) infection in conditions classified elsewhere and of unspecified site | 1.61E-12 | ABCA1;TGFB1;VCAM1;IL15;IL18;CLU;CXCL2;EGFR;NFKB1;ICAM1;NFKBIA;IL1A;IL6;… |
| Cytomegalovirus Infections | 3.10E-12 | LRP1;ODC1;EGFR;ICAM1;EDNRA;GJA1;MAPK1;NCAM1;CD34;MICA;SREBF1;JUN;… |
| Bacterial Infections | 3.45E-11 | ITIH4;CD163;TGFB1;HFE;HGF;IL18;TGFA;PLA2G4A;CFLAR;CXCL2;EGFR;NFKB1;NFKBIA… |
| Epstein-Barr Virus Infections | 6.07E-11 | TGFB1;VCAM1;IL18;EGFR;NFKB1;DCN;ICAM1;IL1A;IL6;CCND1;BCL6;IFNG;CASP3;… |
| Cancer | ||
| Neoplasm Metastasis | 3.89E-77 | APP;SPARC;MYLK;ICAM1;AQP1;EDNRA;GJA1;PSMD4;RPS6KA1;DPYSL3;TNFSF10;… |
| Tumor Progression | 4.82E-53 | SPARC;CSF1;TNC;IRS2;FGF1;ELK1;CLU;ICAM1;IGF1R;EDNRA;GJA1;ZFP36;CCND3;… |
| Other Diseases | ||
| Rheumatoid Arthritis | 1.52E-51 | APP;TPMT;CSF1;HFE;TNC;FGF1;CLU;ICAM1;IGF1R;C4B;EDNRA;ZFP36;CCND3… |
| Atherosclerosis | 3.47E-50 | APP;SCARB1;SPARC;CSF1;HFE;TNC;IRS2;FGF1;ELK1;CLU;MYLK;ICAM1;… |
| Obesity | 2.11E-48 | APP;SCARB1;FAAH;SPARC;CSF1;CD81;HFE;INPPL1;TNC;IRS2;FGF1;… |
| Hypertensive disease | 2.16E-46 | APP;SPARC;HFE;INPPL1;TNC;IRS2;FGF1;COMT;CLU;MYLK;ICAM1;AQP1;… |
| Diabetes Mellitus | 7.58E-41 | APP;SCARB1;SPARC;HFE;INPPL1;IRS2;COMT;CLU;ICAM1;AQP1;IGF1R;EDNRA;… |
| Degenerative polyarthritis | 1.34E-38 | DDR1;APP;CDKN1A;CSF1;THRA;HFE;TNC;SLC2A1;PTPRK;FGF1;CLU;CXCL2;… |
| Heart failure | 1.01E-36 | DDR1;GSK3B;CDKN1A;CSF1;HFE;TNC;SLC2A1;GPT;ECE1;BRCA1;PRL;CXCL2;… |
| Asthma | 5.29E-35 | CDKN1A;TNC;CTSS;MYLK;ICAM1;CASP9;AKAP13;EDNRA;ZFP36;TNFSF10;… |
| Endometriosis | 1.86E-34 | CSF1;THRA;TGFB1I1;TNC;SLC2A1;IRS2;BRCA1;PRL;FGF1;COMT;ICAM1;… |
| Autistic Disorder | 4.84E-11 | APP;FAAH;TNC;PRL;COMT;AKAP1;C4B;GJA1;ERBB4;EP300;MAPK1;DMD;… |
| Parkinsonian Disorders | 5.94E-10 | PDGFRB;APP;NQO1;TBP;PDGFB;PRL;PLA2G6;COMT;SOD2;CP;GFAP;TPO;… |
| Neurocognitive Disorders | 1.63E-06 | ITIH4;ZFP36;IFNG;TAT;F2R;HMOX1;TP53;MAP3K11 |
| Intellectual Disability | 1.87E-06 | APP;THRA;TAT;ARHGAP1;INPPL1;SLC2A1;ELK1;IGF1R;EDNRA;GJA1;SPTLC2;… |
| Mental Retardation | 1.95E-06 | HDAC4;APP;THRA;TAT;ARHGAP1;SLC2A1;PLA2G6;ASCL1;IGF1R;PURA;GJA1;TPO;… |
| Depressive Symptoms | 1.95E-06 | TGFB1;TAT;BRCA1;COMT;CYP3A4;EGFR;IL6;FOLH1;DAO;MAPK1;NCAM1;DRD2;CD34 |
| Prion Diseases | 1.97E-06 | ABCA1;APP;IL1A;TGFB2;IL6;GSN;CYBB;SOD2;CLU;NFKB1 |
| Marijuana Abuse | 2.14E-06 | ABCA1;SCARB1;FAAH;EDNRA;NCAM1;COMT;DRD2;TP53;EGFR |
| Memory Loss | 2.52E-06 | PDGFRB;ABCA1;APP;IL6;PAH;HSF1;PDGFB;BCL2;COMT;DRD2;EGFR;CTSB |
| Meningitis | 6.05E-06 | C4B;IL6;TGFB1;IFNG;TNFSF10;CYBB;FLNA;RAC1;JAK3;ICAM1 |
| Dementia, Vascular | 1.65E-05 | APP;IL1A;IL6;TGFB1;EGF;MMP2;TP53;SREBF2;FGFR1 |
| Epilepsy, Temporal Lobe | 1.73E-05 | SLC2A1;NFKB1;ICAM1;PLD2;TNFRSF1A;IL1A;GJA1;CASP3;TNFSF10;BCL2;MBP;FYN;TP53 |
| Transient Cerebral Ischemia | 1.75E-05 | CASP9;MMP12;GSK3B;BAD;GPX3;SOD2;TP53 |
| Autism Spectrum Disorders | 6.17E-05 | DDR1;FAAH;EGF;HFE;HGF;IL18;CASK;IRS2;PRL;SOD2;GFAP;IL1A;EDNRA;MMP16;IFNG;… |
| Keratosis | 9.53E-05 | CAST;MMP12;CCND1;BCL2;FGFR3;TP53 |
| Gliosis | 1.10E-04 | TBP;HGF;ITGA1;MAPK1;PLA2G6;EGFR;LMNB1;GFAP |
| Poor school performance | 4.44E-04 | HDAC4;APP;TAT;SLC2A1;PLA2G6;ASCL1;IGF1R;PURA;GJA1;TPO;FLNA;EP300;MAPK1;DMD;GGT1;… |
| Tauopathies | 5.76E-04 | APP;KHDRBS1;GSK3B;IL1A;TPO;IL6;HMOX1;MAPK1;TP53 |
| Neural Tube Defects | 6.58E-04 | C5;FOLH1;TCN2;IFNG;SNAI2;NCAM1;COMT;TP53;LMNB1;LRP6 |
| Basal cell carcinoma | 7.90E-04 | LATS1;IL6;TGFB1;CFLAR;TP53;EGFR;IGF1R;MCL1 |
| Mental deficiency | 9.18E-04 | HDAC4;APP;TAT;SLC2A1;PLA2G6;ASCL1;IGF1R;PURA;GJA1;TPO;FLNA;EP300;MAPK1;… |
| Protein Deficiency | 9.37E-04 | C4B;APP;SCARB1;IFNG;SLC2A1;TP53;LDLR |
Enriched Gene Ontology
Gene ontology enrichment for AD-PPA shared genes that participated in the biological processes revealed in 1997 terms that linked to the different hallmarks of AD. The biological processes related to AD represented briefly in Table 2 and completed shown in Supplementary Data S2. Hallmark of AD was selected based on Kelly N. H. Nudelman et al. study [48]. They have included AD neuropathology, cell death, proliferative signaling, growth suppressors, angiogenesis, cell adhesion, genomic instability, inflammation, immune function, and cellular energetics. GO molecular function was enriched 219 terms that represented the top 20 in Table 3 and shown Supplementary Data S2. Most significant molecular functions were protein binding (P-value = 2.27E-48), signaling receptor binding (P-value = 2.28E-26) and kinase activity (P-value = 6.97E-15). Out of them, 5 genes (CLU, INSR, LDLR, SCARB1, TGFB2) associated with amyloid-beta (P-value = 0.0114). The significant cellular components terms enriched from AD-PPA shared genes were 177 that listed the top 20 in Table 4. The main cellular components are the cytoplasmic part, extracellular region, cell surface, vesicle, Golgi, ER, mitochondrion, cytoskeleton, or various parts of the neuron (cell body, dendrite, synapse, myelin sheath). Supplementary Data S2 contains all cellular components enrichment related to AD-PPA shared genes.
Table 2.
Biological process related to common genes in PPA-AD potentially involved by Ontology with default parameters. Observed gene count indicate the number of genes from our list and background gene count indicate the number of genes allocated to the specific GO term in its library. The head title obtained from the important hallmarks of cancer that observed in AD
| Gene Onthology ID | Description | Observed gene count | Background gene count | False discovery rate |
|---|---|---|---|---|
| AD Neuropathology | ||||
| GO:1904645 | response to amyloid-beta | 5 | 29 | 0.00063 |
| GO:1900221 | regulation of amyloid-beta clearance | 4 | 10 | 0.00023 |
| GO:1902003 | regulation of amyloid-beta formation | 4 | 18 | 0.0012 |
| GO:1902947 | regulation of tau-protein kinase activity | 3 | 10 | 0.0033 |
| Cell Death | ||||
| GO:0043523 | regulation of neuron apoptotic process | 23 | 195 | 2.08E-12 |
| GO:0043067 | regulation of programmed cell death | 100 | 1516 | 4.65E-36 |
| GO:1903201 | regulation of oxidative stress-induced cell death | 7 | 65 | 0.00044 |
| GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway | 4 | 34 | 0.0078 |
| GO:2001238 | positive regulation of extrinsic apoptotic signaling pathway | 4 | 51 | 0.0238 |
| Proliferative Signaling | ||||
| GO:0048666 | neuron development | 38 | 758 | 1.17E-09 |
| GO:0031175 | neuron projection development | 31 | 616 | 5.69E-08 |
| GO:0048699 | generation of neurons | 64 | 1422 | 2.33E-14 |
| GO:0030182 | neuron differentiation | 45 | 940 | 8.53E-11 |
| GO:0042127 | regulation of cell population proliferation | 115 | 1594 | 5.60E-46 |
| GO:2000177 | regulation of neural precursor cell proliferation | 6 | 79 | 0.0056 |
| GO:0007265 | Ras protein signal transduction | 12 | 155 | 4.03E-05 |
| GO:0043406 | positive regulation of MAP kinase activity | 27 | 264 | 3.48E-13 |
| Growth Suppressors | ||||
| GO:0045787 | positive regulation of cell cycle | 31 | 376 | 7.98E-13 |
| GO:0007346 | regulation of mitotic cell cycle | 31 | 608 | 4.28E-08 |
| GO:0045786 | negative regulation of cell cycle | 19 | 517 | 0.0014 |
| GO:0007050 | cell cycle arrest | 9 | 149 | 0.0022 |
| GO:0090400 | stress-induced premature senescence | 2 | 8 | 0.0253 |
| GO:0090399 | replicative senescence | 2 | 13 | 0.0482 |
| Angiogenesis | ||||
| GO:0001568 | blood vessel development | 29 | 464 | 2.07E-09 |
| GO:0008015 | blood circulation | 22 | 373 | 7.13E-07 |
| GO:0003073 | regulation of systemic arterial blood pressure | 7 | 92 | 0.0025 |
| GO:0007596 | blood coagulation | 11 | 288 | 0.0136 |
| GO:1903589 | positive regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis | 2 | 9 | 0.0296 |
| GO:0090049 | regulation of cell migration involved in sprouting angiogenesis | 4 | 37 | 0.01 |
| GO:0001525 | angiogenesis | 17 | 297 | 2.46E-05 |
| GO:0001666 | response to hypoxia | 26 | 288 | 1.36E-11 |
| Cell adhesion | ||||
| GO:0022409 | positive regulation of cell-cell adhesion | 22 | 238 | 4.63E-10 |
| GO:0007155 | cell adhesion | 39 | 843 | 5.40E-09 |
| GO:0001952 | regulation of cell-matrix adhesion | 9 | 105 | 0.00024 |
| GO:0048041 | focal adhesion assembly | 3 | 24 | 0.0206 |
| GO:0001764 | neuron migration | 7 | 118 | 0.0081 |
| Genomic Instability | ||||
| GO:0006974 | cellular response to DNA damage stimulus | 22 | 749 | 0.0067 |
| GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator | 3 | 17 | 0.0101 |
| GO:0006268 | DNA unwinding involved in DNA replication | 2 | 8 | 0.0253 |
| GO:0006975 | DNA damage induced protein phosphorylation | 2 | 8 | 0.0253 |
| GO:0032392 | DNA geometric change | 4 | 63 | 0.0408 |
| GO:0000077 | DNA damage checkpoint | 6 | 138 | 0.0451 |
| GO:2001252 | positive regulation of chromosome organization | 9 | 160 | 0.0033 |
| Inflammation | ||||
| GO:0006954 | inflammatory response | 41 | 482 | 2.20E-17 |
| GO:0002526 | acute inflammatory response | 8 | 73 | 0.00014 |
| GO:0002248 | connective tissue replacement involved in inflammatory response wound healing | 3 | 5 | 0.00082 |
| GO:0006925 | inflammatory cell apoptotic process | 2 | 5 | 0.0138 |
| GO:0002544 | chronic inflammatory response | 2 | 13 | 0.0482 |
| GO:0001774 | microglial cell activation | 3 | 20 | 0.0138 |
| GO:0034097 | response to cytokine | 77 | 1035 | 3.86E-30 |
| GO:0019221 | cytokine-mediated signaling pathway | 55 | 655 | 2.51E-23 |
| Immune Function | ||||
| GO:0045824 | negative regulation of innate immune response | 5 | 49 | 0.0044 |
| GO:0002376 | immune system process | 105 | 2370 | 1.19E-24 |
| GO:0002250 | adaptive immune response | 24 | 280 | 2.53E-10 |
| GO:0006959 | humoral immune response | 14 | 252 | 0.0002 |
| Cellular Energetics | ||||
| GO:0043467 | regulation of generation of precursor metabolites and energy | 7 | 96 | 0.0031 |
| GO:0042593 | glucose homeostasis | 7 | 169 | 0.0367 |
| GO:0005979 | regulation of glycogen biosynthetic process | 5 | 29 | 0.00063 |
Table 3.
Top 20 of Molecular function obtain from enrichment of PPA- AD common genes. Observed gene count indicate the number of genes from our list and background gene count indicate the number of genes allocated to the specific GO term in its library
| Gene Onthology ID | Description | Observed gene count | Background gene count | False discovery rate |
|---|---|---|---|---|
| GO:0005515 | protein binding | 221 | 6605 | 2.27E-48 |
| GO:0005488 | binding | 266 | 11878 | 1.97E-34 |
| GO:0005102 | signaling receptor binding | 87 | 1513 | 2.28E-26 |
| GO:0019899 | enzyme binding | 98 | 2197 | 2.95E-22 |
| GO:0004672 | protein kinase activity | 46 | 635 | 2.48E-16 |
| GO:0016773 | phosphotransferase activity, alcohol group as acceptor | 50 | 767 | 3.44E-16 |
| GO:0044877 | protein-containing complex binding | 54 | 968 | 6.79E-15 |
| GO:0016301 | kinase activity | 50 | 835 | 6.97E-15 |
| GO:0019900 | kinase binding | 44 | 678 | 3.82E-14 |
| GO:0042802 | identical protein binding | 72 | 1754 | 8.55E-14 |
| GO:0019901 | protein kinase binding | 40 | 599 | 3.29E-13 |
| GO:0098772 | molecular function regulator | 69 | 1793 | 6.63E-12 |
| GO:0008134 | transcription factor binding | 38 | 610 | 1.05E-11 |
| GO:0004713 | protein tyrosine kinase activity | 22 | 180 | 1.08E-11 |
| GO:0003824 | catalytic activity | 140 | 5592 | 1.43E-11 |
| GO:0043167 | ion binding | 145 | 6066 | 1.44E-10 |
| GO:0043168 | anion binding | 85 | 2696 | 1.44E-10 |
| GO:0140096 | catalytic activity, acting on a protein | 74 | 2176 | 1.77E-10 |
| GO:0008144 | drug binding | 63 | 1710 | 4.28E-10 |
| GO:0097367 | carbohydrate derivative binding | 72 | 2163 | 9.22E-10 |
Table 4.
Top 20 of cellular component enrichment detected from common AD-PPA genes. Observed gene count indicate the number of genes from our list and background gene count indicate the number of genes allocated to the specific GO term in its library
| Gene Onthology ID | Description | Observed gene count | Background gene count | False discovery rate |
|---|---|---|---|---|
| GO:0044444 | cytoplasmic part | 214 | 9377 | 4.25E-18 |
| GO:0005576 | extracellular region | 97 | 2505 | 1.46E-17 |
| GO:0005737 | cytoplasm | 235 | 11238 | 2.71E-17 |
| GO:0009986 | cell surface | 49 | 690 | 2.71E-17 |
| GO:0005615 | extracellular space | 62 | 1134 | 4.48E-17 |
| GO:0044421 | extracellular region part | 68 | 1375 | 7.88E-17 |
| GO:0031982 | vesicle | 90 | 2318 | 1.44E-16 |
| GO:0044459 | plasma membrane part | 95 | 2651 | 1.71E-15 |
| GO:0005829 | cytosol | 138 | 4958 | 3.91E-15 |
| GO:0031410 | cytoplasmic vesicle | 83 | 2226 | 3.69E-14 |
| GO:0071944 | cell periphery | 139 | 5254 | 1.63E-13 |
| GO:0005886 | plasma membrane | 137 | 5159 | 2.17E-13 |
| GO:0044464 | cell part | 277 | 16244 | 3.10E-13 |
| GO:0012505 | endomembrane system | 120 | 4347 | 2.48E-12 |
| GO:0098805 | whole membrane | 63 | 1554 | 5.50E-12 |
| GO:0045121 | membrane raft | 27 | 300 | 8.19E-12 |
| GO:0044422 | organelle part | 191 | 9111 | 6.90E-11 |
| GO:0043227 | membrane-bounded organelle | 219 | 11244 | 1.20E-10 |
| GO:0044446 | intracellular organelle part | 187 | 8882 | 1.20E-10 |
| GO:0005622 | intracellular | 254 | 14286 | 2.61E-10 |
Enriched KEGG Pathway
The KEGG pathway analysis demonstrated that 175 pathways are linked to AD-PPA genes. Table 5 contains the top 20 pathways and complete result represented in Supplementary Data S2. The most important pathways are included, pathways in cancer, microRNAs in cancer, proteoglycans in cancer, MAPK signaling pathway, PI3K-Akt signaling pathway, Ras signaling pathway, HIF-1 signaling pathway, focal adhesion, EGFR tyrosine kinase inhibitor resistance, Jak-STAT signaling pathway.
Table 5.
Top 20 of KEGG pathways of common AD-PPA genes. Observed gene count indicate the number of genes from our list and background gene count indicate the number of genes allocated to the specific KEGG pathway term in its library
| KEGG- ID | Description | Observed gene count | Background gene count | False discovery rate |
|---|---|---|---|---|
| hsa05200 | Pathways in cancer | 63 | 515 | 5.19E-35 |
| hsa04010 | MAPK signaling pathway | 43 | 293 | 3.02E-26 |
| hsa05206 | MicroRNAs in cancer | 32 | 149 | 8.06E-24 |
| hsa04151 | PI3K-Akt signaling pathway | 41 | 348 | 6.08E-22 |
| hsa04014 | Ras signaling pathway | 32 | 228 | 4.91E-19 |
| hsa05167 | Kaposi’s sarcoma-associated herpesvirus infection | 28 | 183 | 1.58E-17 |
| hsa05215 | Prostate cancer | 22 | 97 | 6.66E-17 |
| hsa04066 | HIF-1 signaling pathway | 21 | 98 | 9.72E-16 |
| hsa05169 | Epstein-Barr virus infection | 26 | 194 | 3.77E-15 |
| hsa05205 | Proteoglycans in cancer | 26 | 195 | 3.80E-15 |
| hsa04380 | Osteoclast differentiation | 22 | 124 | 4.05E-15 |
| hsa04510 | Focal adhesion | 26 | 197 | 4.05E-15 |
| hsa05210 | Colorectal cancer | 19 | 85 | 1.03E-14 |
| hsa05166 | HTLV-I infection | 28 | 250 | 1.04E-14 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 18 | 78 | 3.30E-14 |
| hsa04630 | Jak-STAT signaling pathway | 23 | 160 | 3.34E-14 |
| hsa05211 | Renal cell carcinoma | 17 | 68 | 5.75E-14 |
| hsa04933 | AGE-RAGE signaling pathway in diabetic complications | 19 | 98 | 7.24E-14 |
| hsa04722 | Neurotrophin signaling pathway | 20 | 116 | 9.34E-14 |
| hsa04015 | Rap1 signaling pathway | 24 | 203 | 3.29E-13 |
Protein-protein interaction networks construction and analysis
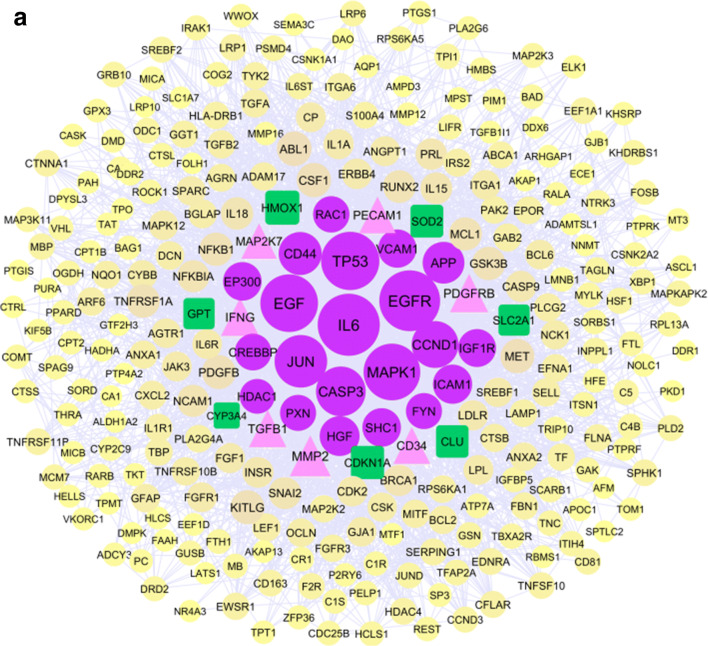
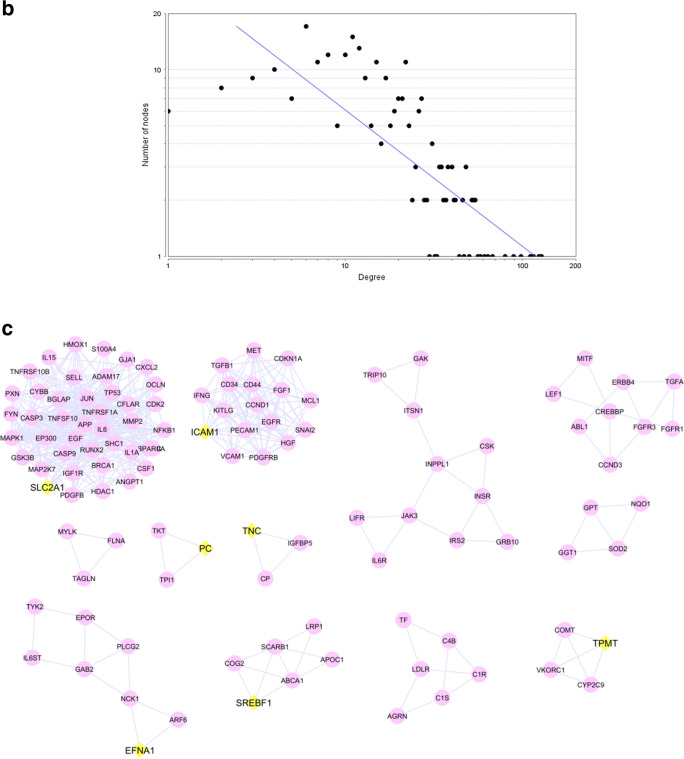
The Cytoscape software was used for network analysis. We constructed three protein-protein interaction networks, AD, PPA, and AD-PPA, then analyzed every network separately and finally compared the crucial genes related to each network. The primary protein-protein interaction network obtained from the String database was imported into Cytoscape. Constructed AD network contains 591 nodes and 5076 edges, while the PPA network contains 2768 nodes and 104202 edges. The network analysis determined hub and bottleneck genes for AD and PPA network that represented in Supplementary Table S1 and S2 respectively. We used the lists obtained from the hub, and bottleneck nodes of AD and PPA network to prioritization the genes belong to AD and PPA and then compare the list will obtain from the network of AD-PPA shared genes. The constructed network from AD-PPA shared genes (284 genes) contains 280 nodes, which are connected to 2859 edges. The overview figure of integrating 3 networks was represented in Supplementary Figure S1. All three networks were a scale-free network that included a few numbers of hub genes with the most number of interactions and many numbers of nodes with a few interactions. Figure 3a represented the protein-protein interaction network of AD-PPA genes in which hub and bottleneck nodes have a more prominent size with different colors and located in the center, and Fig. 3b represented the node degree distribution. Besides, the analysis of the AD-PPA network revealed the several parameters that are containing; the clustering coefficient is 0.402, network density is 0.073, the network diameter is 5, and the connected component is 1, the shortest paths are 78120. The network hub and bottleneck nodes were also listed in Table 6. Out of them, HDAC1, SHC1, CD44, APP, EGFR, IGF1R, EGF, FYN, MAPK1, CREBBP, JUN, CASP3, CCND1, VCAM1, RAC1, TP53, HGF, EP300, IL6, and PXN are the genes shared between hub and bottleneck and introduce as relevant genes in the protein-protein interaction network.
Fig. 3.
The protein-protein interaction networks of 284 genes are common in AD and PPA (a) that constructed by Cytoscape software. The size of nodes was determined by their degree. Hub and bottleneck genes are purple ellipse nodes. The genes that are only hub are pink triangle nodes while the genes that are only bottleneck represented as green rectangle nodes. Degree distribution related nodes in network (b) revealed the AD-PPA network is a scale free network. MCODE analysis detected 12 clusters that seed detected as diamond nodes with yellow color(c)
Table 6.
Hub genes and bottleneck related the common genes of AD and PPA network obtained from Cytoscape software
| Gene name | Degree (Hub) | Gene name | Betweenness centrality (Bottleneck) |
|---|---|---|---|
| IL6 | 128 | EGFR | 0.097698 |
| EGFR | 124 | IL6 | 0.093625 |
| TP53 | 116 | TP53 | 0.075256 |
| EGF | 114 | MAPK1 | 0.069349 |
| MAPK1 | 111 | EGF | 0.063399 |
| JUN | 98 | APP | 0.038658 |
| CASP3 | 89 | JUN | 0.038026 |
| CCND1 | 80 | CASP3 | 0.036808 |
| CD44 | 68 | CCND1 | 0.029613 |
| APP | 64 | RAC1 | 0.02678 |
| MMP2 | 61 | EP300 | 0.022104 |
| HGF | 58 | GPT | 0.022064 |
| ICAM1 | 56 | SOD2 | 0.021618 |
| EP300 | 54 | SLC2A1 | 0.019361 |
| SHC1 | 54 | PXN | 0.018438 |
| VCAM1 | 53 | HGF | 0.017373 |
| PDGFRB | 53 | CYP3A4 | 0.016507 |
| IGF1R | 52 | CD44 | 0.015733 |
| IFNG | 52 | CREBBP | 0.015148 |
| TGFB1 | 50 | SHC1 | 0.014951 |
| CREBBP | 48 | HDAC1 | 0.014795 |
| CD34 | 48 | IGF1R | 0.012673 |
| PECAM1 | 48 | HMOX1 | 0.01255 |
| RAC1 | 47 | ICAM1 | 0.012427 |
| PXN | 46 | CDKN1A | 0.012121 |
| FYN | 46 | CLU | 0.010988 |
| MAP2K7 | 44 | FYN | 0.010582 |
| HDAC1 | 42 | VCAM1 | 0.010404 |
Construction and analysis of subnetworks
The subnetworks were studied for protein-protein interaction networks, AD, PPA, and AD-PPA through MCODE app in Cytoscape to identify the seed genes. AD genes could be clustered in 14 subnetworks with seed genes, including ARL17B, ITGA6, HDAC4, CYP3A4, RUNX2, TAT, TCF3, CR1, FDFT1, SART1, and CCND1. For PPA genes, recognized 45 subnetworks with the seed genes, CCR3, CERS1, HIST2H3C, KRT10, TFAP2A, SLC22A1, MYBL2, IL2, EARS2, WNT11, VPS4B, WNK1, ACSF3, ABCG4, PIK3R2, LIPA, AQP1, PCBP2, LMX1B, BYSL, DSG2, CYP17A1, VANGL1, IL4R, COG6, SLC38A2, CCDC6, NAA30, LYST, TNFRSF10A, IFNA2, and COL2A1. MCODE analysis of AD-PPA network recognized clusters showed in Fig. 3c and Supplementary Table S3 represented the score of each cluster.
Identification and analysis of hub, bottleneck and seed nodes in AD-PPA shared genes
As depicted in Supplementary Figure S2, 80 genes of 284 AD-PPA shared genes at least had overlap with hub or bottleneck genes in one of two AD and the PPA networks. Between these overlap genes, 51 genes were shared between 3 networks. When comparing the hub or bottleneck genes in 3 networks (represented in Supplementary Figure S3), 32 genes were shared between them.
Amongst 280 AD-PPA shared genes, only seven genes, i.e., SREBF1, ICAM1, SLC2A1, EFNA1, PC, TNC, and TPMT, were identified as seed genes. When compared, the seed genes of the three networks, found no shared genes between them. As represented in the Venn diagram of Supplementary Figure S4, when compared the seed genes of AD and PPA with the all 284 shared genes of AD-PPA, found two shared genes with PPA and seven shared genes with AD. This data represented two genes; TFAP2A and AQP1 in the PPA network are crucial genes that could not distinguish as the seed genes in topological analysis of the AD-PPA subnetworks. TFAP2A was not the hub and bottleneck in the PPA network. AQP1 also was not hub and bottleneck in the PPA network but seen in the AD network. Seven genes, HDAC4, TAT, ITGA6, CCND1, CYP3A4, CR1, and RUNX2, are seed genes of AD network that could not be detected as seed genes in AD-PPA subnetwork. CCND1 seen in all three networks AD, PPA, and AD-PPA, belongs to the hub and bottleneck nodes. CCND1 was also observed in most of GO categories or KEGG pathways. CYP3A4, also seen in three networks, had high betweenness centrality. CYP3A4 was also seen in several GO categories and diseases. RUNX2 detected as hub and bottleneck nodes in the PPA network and recognized as a hub in the AD network. This gene was not a hub and bottleneck nodes in the AD-PPA network. HDAC4, ITGA6, CR1 seen in three networks but not the hub and bottleneck nodes. TAT was seen in AD but was not a hub and bottleneck nodes.
ICAM1 is a seed gene of AD-PPA subnetwork that belongs to the hub and bottleneck genes. This gene also allocated to several biological processes and diseases. SLC2A1 is a seed gene of AD-PPA subnetwork that also detected as a bottleneck gene. SLC2A1 is a part of genes attributed to biological processes, including cellular response to glucose starvation and response to peptide. Moreover, SLC2A1 was recognized in most of the diseases enriched by DisGeNet analysis but not the virus and bacterial infection. When assessed functionally 40 hubs, bottlenecks, and seed genes of AD-PPA network in the String database, these genes were found that enriched all biological process, molecular function, cellular component, and pathways obtained from all genes. The most significant GO and Pathways listed in Table 7. For two top ranked AD-PPA subnetworks obtained from MCODE (score 16.146 and score 13.125 contain 59 genes) included 29 genes had overlap with 40 hub-bottleneck-seed genes. In addition, the functional analysis in Enrichr for GO and KEGG pathway enrichments found a similar result to the analysis of 40 hub-bottlenecks-seed genes.
Table 7.
Functional analysis of 40 genes including hub, bottlenecked and seed genes in String database. Observed gene count indicate the number of genes from our list and background gene count indicate the number of genes allocated to the specific GO and KEGG pathway term in its library
| #term ID | Description | Observed gene count | Background gene count | False discovery rate | Proteins |
|---|---|---|---|---|---|
| Biological process | |||||
| GO:0007166 | cell surface receptor signaling pathway | 29 | 2198 | 6.92E-16 | APP,CASP3,CCND1,CD44,CDKN1A,CREBBP,EFNA1,EGF,EGFR,EP300,FYN,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAPK1,MMP2,PDGFRB,PXN,RAC1,SHC1,SOD2,SREBF1,TGFB1,TP53,VCAM1 |
| GO:0001934 | positive regulation of protein phosphorylation | 21 | 941 | 2.17E-14 | APP,CCND1,CD44,CDKN1A,CLU,EFNA1,EGF,EGFR,FYN,HGF,ICAM1,IFNG,IL6,JUN,MAP2K7,MAPK1,PDGFRB,RAC1,SHC1,TGFB1,TP53 |
| GO:2000377 | regulation of reactive oxygen species metabolic process | 13 | 169 | 2.46E-14 | APP,CD34,CDKN1A,CLU,EGFR,FYN,ICAM1,IFNG,PDGFRB,RAC1,SHC1,TGFB1,TP53 |
| GO:0006950 | response to stress | 31 | 3267 | 2.63E-14 | APP,CASP3,CCND1,CD34,CD44,CDKN1A,CLU,CREBBP,EGFR,EP300,FYN,HDAC1,HMOX1,ICAM1,IFNG,IL6,JUN,MAP2K7,MAPK1,MMP2,PDGFRB,PXN,RAC1,SHC1,SLC2A1,SOD2,SREBF1,TGFB1,TNC,TP53,VCAM1 |
| GO:0031401 | positive regulation of protein modification process | 22 | 1149 | 2.63E-14 | APP,CCND1,CD44,CDKN1A,CLU,EFNA1,EGF,EGFR,FYN,HGF,ICAM1,IFNG,IL6,JUN,MAP2K7,MAPK1,PDGFRB,RAC1,SHC1,SREBF1,TGFB1,TP53 |
| GO:0070887 | cellular response to chemical stimulus | 29 | 2672 | 2.63E-14 | APP,CASP3,CCND1,CD44,CDKN1A,CREBBP,CYP3A4,EGFR,EP300,FYN,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAPK1,MMP2,PDGFRB,PXN,RAC1,SHC1,SOD2,SREBF1,TGFB1,TNC,TP53,VCAM1 |
| GO:0010033 | response to organic substance | 29 | 2815 | 4.54E-14 | APP,CASP3,CCND1,CD44,CDKN1A,CLU,EGFR,EP300,FYN,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAP2K7,MAPK1,MMP2,PDGFRB,PXN,SHC1,SLC2A1,SOD2,SREBF1,TGFB1,TNC,TP53,VCAM1 |
| GO:0042221 | response to chemical | 33 | 4153 | 5.15E-14 | APP,CASP3,CCND1,CD44,CDKN1A,CLU,CREBBP,CYP3A4,EFNA1,EGFR,EP300,FYN,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAP2K7,MAPK1,MMP2,PDGFRB,PXN,RAC1,SHC1,SLC2A1,SOD2,SREBF1,TGFB1,TNC,TP53,VCAM1 |
| GO:0010941 | regulation of cell death | 24 | 1638 | 5.19E-14 | APP,CASP3,CD34,CD44,CDKN1A,CLU,CREBBP,EFNA1,EGFR,FYN,HDAC1,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAP2K7,PDGFRB,SHC1,SOD2,TGFB1,TP53 |
| Molecular function | |||||
| GO:0005515 | protein binding | 36 | 6605 | 8.21E-11 | APP,CASP3,CCND1,CD34,CDKN1A,CLU,CREBBP,CYP3A4,EFNA1,EGF,EGFR,EP300,FYN,HDAC1,HGF,HMOX1,ICAM1,IFNG,IGF1R,IL6,JUN,MAP2K7,MAPK1,PC,PDGFRB,PECAM1,PXN,RAC1,SHC1,SLC2A1,SOD2,SREBF1,TGFB1,TNC,TP53,VCAM1 |
| GO:0019899 | enzyme binding | 23 | 2197 | 4.01E-10 | APP,CASP3,CCND1,CDKN1A,CLU,CYP3A4,EGF,EGFR,FYN,HDAC1,HGF,HMOX1,JUN,MAP2K7,MAPK1,PDGFRB,PXN,RAC1,SHC1,SLC2A1,SREBF1,TGFB1,TP53 |
| GO:0005102 | signaling receptor binding | 19 | 1513 | 2.91E-09 | APP,CASP3,CLU,EFNA1,EGF,EGFR,EP300,FYN,HGF,ICAM1,IFNG,IGF1R,IL6,PDGFRB,PXN,SHC1,TGFB1,TP53,VCAM1 |
| GO:0019901 | protein kinase binding | 11 | 599 | 1.39E-06 | CCND1,CDKN1A,EGFR,MAP2K7,MAPK1,PDGFRB,PXN,RAC1,SHC1,SREBF1,TP53 |
| GO:0004672 | protein kinase activity | 11 | 635 | 2.19E-06 | CCND1,CDKN1A,EGF,EGFR,FYN,HGF,IGF1R,MAP2K7,MAPK1,PDGFRB,RAC1 |
| GO:0070851 | growth factor receptor binding | 6 | 131 | 1.19E-05 | APP,EGF,FYN,IL6,PDGFRB,SHC1 |
| GO:0005178 | integrin binding | 5 | 122 | 0.00011 | APP,EGFR,ICAM1,PXN,VCAM1 |
| GO:0005088 | Ras guanyl-nucleotide exchange factor activity | 6 | 243 | 0.00019 | EGF,EGFR,FYN,HGF,PDGFRB,SHC1 |
| GO:0033613 | activating transcription factor binding | 4 | 69 | 0.00025 | CREBBP,EP300,HDAC1,JUN |
| GO:0002039 | p53 binding | 4 | 73 | 0.0003 | CREBBP,EP300,HDAC1,TP53 |
| Cellular component | |||||
| GO:0045121 | membrane raft | 9 | 300 | 2.98E-06 | APP,CASP3,EGFR,FYN,HMOX1,ICAM1,MAPK1,PECAM1,SLC2A1 |
| GO:0044421 | extracellular region part | 15 | 1375 | 3.09E-06 | APP,CD34,CLU,EGF,EGFR,HGF,HMOX1,ICAM1,IFNG,IL6,MMP2,PECAM1,TGFB1,TNC,VCAM1 |
| GO:0044444 | cytoplasmic part | 35 | 9377 | 1.02E-05 | APP,CASP3,CCND1,CD34,CD44,CDKN1A,CLU,CYP3A4,EGF,EGFR,EP300,FYN,GPT,HDAC1,HGF,HMOX1,IL6,JUN,MAP2K7,MAPK1,MMP2,PC,PDGFRB,PECAM1,PXN,RAC1,SHC1,SLC2A1,SOD2,SREBF1,TGFB1,TNC,TP53,TPMT,VCAM1 |
| GO:0031982 | vesicle | 16 | 2318 | 0.00018 | APP,CD44,CLU,EGF,EGFR,FYN,HGF,ICAM1,MAPK1,PDGFRB,PECAM1,RAC1,SLC2A1,SREBF1,TGFB1,VCAM1 |
| GO:0009986 | cell surface | 9 | 690 | 0.00019 | APP,CD34,CD44,CLU,EGFR,ICAM1,PDGFRB,TGFB1,VCAM1 |
| GO:0097386 | glial cell projection | 2 | 18 | 0.0047 | APP,FYN |
| GO:0030425 | dendrite | 5 | 531 | 0.0212 | APP,CLU,FYN,MAPK1,RAC1 |
| GO:0000123 | histone acetyltransferase complex | 2 | 76 | 0.0412 | CREBBP,EP300 |
| GO:0005739 | mitochondrion | 8 | 1531 | 0.0417 | CLU,FYN,MAPK1,MMP2,PC,SHC1,SOD2,TP53 |
| GO:0070062 | extracellular exosome | 2 | 80 | 0.0435 | EGF,ICAM1 |
| KEGG pathways | |||||
| hsa05200 | Pathways in cancer | 21 | 515 | 1.54E-20 | CASP3,CCND1,CDKN1A,CREBBP,EGF,EGFR,EP300,HDAC1,HGF,HMOX1,IFNG,IGF1R,IL6,JUN,MAPK1,MMP2,PDGFRB,RAC1,SLC2A1,TGFB1,TP53 |
| hsa05206 | MicroRNAs in cancer | 14 | 149 | 7.73E-18 | CASP3,CCND1,CD44,CDKN1A,CREBBP,EGFR,EP300,HDAC1,HMOX1,MAPK1,PDGFRB,SHC1,TNC,TP53 |
| hsa04066 | HIF-1 signaling pathway | 11 | 98 | 8.67E-15 | CDKN1A,CREBBP,EGF,EGFR,EP300,HMOX1,IFNG,IGF1R,IL6,MAPK1,SLC2A1 |
| hsa04510 | Focal adhesion | 13 | 197 | 8.67E-15 | CCND1,EGF,EGFR,FYN,HGF,IGF1R,JUN,MAPK1,PDGFRB,PXN,RAC1,SHC1,TNC |
| hsa05205 | Proteoglycans in cancer | 13 | 195 | 8.67E-15 | CASP3,CCND1,CD44,CDKN1A,EGFR,HGF,IGF1R,MAPK1,MMP2,PXN,RAC1,TGFB1,TP53 |
| hsa04068 | FoxO signaling pathway | 11 | 130 | 9.14E-14 | CCND1,CDKN1A,CREBBP,EGF,EGFR,EP300,IGF1R,IL6,MAPK1,SOD2,TGFB1 |
| hsa04010 | MAPK signaling pathway | 13 | 293 | 3.56E-13 | CASP3,EFNA1,EGF,EGFR,HGF,IGF1R,JUN,MAP2K7,MAPK1,PDGFRB,RAC1,TGFB1,TP53 |
| hsa04151 | PI3K-Akt signaling pathway | 13 | 348 | 1.81E-12 | CCND1,CDKN1A,EFNA1,EGF,EGFR,HGF,IGF1R,IL6,MAPK1,PDGFRB,RAC1,TNC,TP53 |
| hsa04360 | Axon guidance | 4 | 173 | 0.00086 | EFNA1,FYN,MAPK1,RAC1 |
| hsa05100 | Bacterial invasion of epithelial cells | 3 | 72 | 0.00088 | PXN,RAC1,SHC1 |
Discussion
The infection hypothesis is an interesting theory in Alzheimer’s disease onset and explains the role of microbiota in inducing AD [49]. In addition to microbiota, the microbiota-derived metabolites play important roles in various neurological pathologies. Propionic acid(propionate) is one of the most short-chain fatty acids metabolites produced by a variety of microbiota [12, 50]. Since there is no experimental data related to the effect of PPA in AD pathology here used the system biology approaches and bioinformatics analysis to find the association of PPA and AD. The system biology approach integrates different levels of molecular biology to deeply interpret the pathological origin of a multifactorial disease [51].
In the present study, we combined the genes involved in PPA available in CTD, DISEASES, and Gene Cards databases and the Coremine search engine by Venn diagram software. As a result, according to the Fig. 1, 284 PPA genes were found to be shared with AD genes (extracted from text mining, CTD, and GeneCards databases). We used network and enrichment analysis to uncover critical molecular mechanisms and relationships between PPA and AD. DisGeNet enriched diseases that arranged in categories related to infection, cancer, and neurological and non-neurological diseases. As depicted in Table 1, several bacteria and virus infections were disclosed. Out of them, Salmonella typhimurium able to generate PPA during fermentation [52]. Prior microbiome researches have also shown oral and genital herpes, Epstein Barr virus, cytomegalovirus, HIV, gut bacteria, liver bacteria, Helicobacter pylori, periodontal pathogens, and Chlamydophila pneumonia present in AD pathogenesis [49, 53, 54]. These pathogens may cross the blood-brain barrier or brain-CSF barrier and attack the CNS [55, 56]. Also, it was determined that microbiotas influence CNS by microbiota-derived metabolites and inflammation [49, 55, 57–60]. Since we used the AD-PPA genes to detect the diseases, this data suggests that PPA can induce the same effect provided by microbiota. The most significant diseases were related to cancer progression and metastatic processes that also validated by the KEGG pathway analysis and previously were mentioned as familiar hallmarks of cancer and AD [48]. In the analysis of biological process also enriched cancer as the first gene ontology. Our results are supporting this evidence that PPA could able to promote brain tissue toward AD pathology like seen in cancer. AD-PPA genes also identified disease-related Neurocognitive Disorders and several disabilities. It has been reported excessive PPA has implied disadvantageous effects such as propionic acidemia, a neurodevelopmental metabolic disorder, that identified by elevation of PPA levels in the blood, cerebrospinal fluid, and neurons [61, 62]. For instance, several studies showed that intraventricular inoculation of PPA created behavioral and brain abnormalities in rats similar to autism spectrum disorder [53–56].
Dysregulation in the cell cycle was another significant KEGG pathway that previously has been contributed to surviving neuron cells and accumulating the amyloid fibril, which eventually undergoes apoptosis [63]. Dysregulation in proliferative signaling and evade growth suppressor have been identified as an important hallmark of AD [64]. The shared genes of AD-PPA enriched the biological processes related to neuron generation, development, projection, and differentiation. Besides, they contain genes that describe the positive and negative regulation of proliferation. MAP kinase pathway that enriched in our data is the primary signaling activity in neuron and glial cells that promote phosphorylation of tau deposit [65]. Also, another proliferative signaling is RAS that the pathway-related this GTPase also enriched and previously suggested in tau phosphorylation [66]. It has been determined that neurodegeneration is associated with improper cell cycle progression that increases neuropathological processes and finally leads to apoptosis [67]. Out of genes, 19 genes found the negative cell cycle regulators while there existed 31 genes with function in positive regulation of cell cycle that most of them were shared in apoptosis and proliferative process. In AD patients, Aβ42 causes hyperactivation of MEK-ERK signaling lead to cell death by possibly mediating Tau hyperphosphorylation. In addition, the Aβ42-mediated aberrant MEK-ERK signaling pathway may promote S-phase cell cycle reentry through inducing expression of cyclin D1(CCND1) and neuronal cell death [68, 69]. CCND1 as a crucial gene in cell cycle reentry of postmitotic neurons was an important hub and bottleneck gene in three networks AD, PPA, and AD-PPA. Other impressive gene ontology enrichment results had a direct link to the amyloid formation and tau pathology. This finding was also confirmed by gene-disease association analysis. Tauopathies, Neural Tube Defects, Protein Deficiency were recognized in DisGeNet analysis. These results confirm the role of PPA in AD as a risk factor could able to enhance the process, leading to cell cycle deregulation and contribute to neuronal loss and neurodegeneration observed in the AD brains. This finding was also supported by the enrichment output of AD-PPA genes that associated with apoptosis activated by the p53 signaling pathway or cell death through both intrinsic and extrinsic apoptosis signaling pathways. Apoptosis activated by the p53 signaling pathway has implicated by the treatment of neuronal cells by PPA in vitro [70]. Lobzhanidze G et al. reported that a low level of PPA could change amygdala cells toward apoptosis [30]. Previously indicated enhanced Caspase-3 mRNA expression and inhibited Bcl-2 mRNA expression in the brain of rat pups that had been exposed to PPA [71].
Another mediator of brain injury in AD is Oxidative stress [72] that regulation of oxidative stress-induced cell death was the significant biological processes detected by AD-PPA genes. Increased reactive oxygen species (ROS) production may lead to apoptosis [73]. Alteration of PPA treatment in the enzymatic antioxidant capacity in rat brains has been represented a significant decrease in superoxide dismutase (SOD) and catalase(CAT) activities [71, 74]. According to Table 6, SOD2 is the bottleneck genes distinguished in network analysis, play role as a critical antioxidant. Accumulating evidence disclosed that ROS produced by various enzymatic reactions and chemical processes had implicated the pathogenesis of neurodegenerative disorders such as AD [75]. ROS involved in the expression of well-defined inflammatory mediators, including MMPs, cPLA2, COX-2, and adhesion molecules. We recognized several pro-inflammatory genes and the protein response to this event, including APP, CASP3, EGF, EGFR, EP300, HMOX1, ICAM1, IL6, JUN, MMP2, PDGFRB, and VCAM1 that were belonged to the hub- bottleneck- seed genes. The previous research on natural products such as resveratrol, curcumin, berberine has been shown that they could able to elicit anti-cancer and antiaging by intracellular signaling mechanisms. Many of the beneficial effects have been attributed to their anti-inflammatory properties [76]. Between the genes underlying these effects, we detected APP, CASP3, GPT, HMOX1, ICAM1, IL6, JUN, MAPK1, TGFB1, TP53, and VCAM1 that are significant genes of our gene list. As mentioned, they belong to inflammatory genes that also targeted in antiaging investigations. Within this list, ICAM1 was the only gene distinguished as a hub-bottleneck-seed node. Several studies have been emphasized the importance of ICAM1 gene polymorphism and its expression levels in AD pathogenesis [77–79].
Animal studies showing that oxidative damage to proteins may be involved in the pathophysiology of PPA [80]. The alterations of protein structure by oxidants may affect the function of receptors, enzymes, and transport proteins, resulting in a partial or complete loss of protein functionality [81]. The most crucial molecular function enrichments link to protein binding and kinase activity that this oxidation could change the protein interaction, and unfavorable results would contain alteration seen in the protein-protein interaction network. Previously determined, the increase of phosphorylation promotes pathogenesis-related TAU protein [1]. In AD-PPA genes enriched protein binding as the most significant molecular function. It has been previously indicated the aberrant protein binding in AD [63]. In addition to protein oxidation, PPA animates lipid peroxidation in the rat brain and the plasma of patients with propionic academia that are finally resulting in cell damage [82, 83]. PPA inhibits the antioxidant enzyme activities and induces Malondialdehyde, which may have happened in mitochondrial dysfunction [84, 85]. Mitochondrial hypometabolism characterizes brain aging and AD. Redox dysregulation and chronic neuroinflammation are observed in brain aging and AD that linked to energy metabolism and inflammatory responses. The metabolic-inflammatory axis describes the dynamic interaction of these systems in the brain [86]. Changing in brain metabolism and cellular energy are the hallmark of AD that increased levels of PPA affect several processes related to energy metabolism [87]. Here, we detected a few genes of our list implicated in glucose hemostasis. One of the critical genes in our gene list was SLC2A1 that was a bottleneck-seed gene. This gene provides instructions for producing GLUT1 that acts as a mediator for transporting glucose at the blood-brain barrier. GLUT1 diminutions worsen cerebrovascular degeneration in AD [88].
PPA induced not only lipid damage but also DNA damage [71]. In this study, AD- PPA genes enrich the process response to DNA damage, DNA geometric change, DNA damage checkpoint, or activated apoptotic pathway in response to DNA damage and positive regulation of chromosome organization. Previously determine the association of genomic instability observed in AD that one of evidence is emerging early-onset AD in a patient with down syndrome [89].
Inflammation process and response to acute and chronic inflammation or cytokine production by the immune system are processes that enriched AD-PPA genes. Inflammation in glia cells “Gliosis” is the hallmark of AD that enhances the amyloidogenic process [90] and detected by analysis of our genes in DisGeNet. Innate and adaptive immune response or humoral immune system enriched by 284 genes. The immune system plays an essential role in the progression, or maybe it is a risk factor for AD [91]. Lobzhanidze G et al. have been recognized the significant structural alteration in the amygdala beyond the administration of PPA in adolescent rats. They reported the Glial alterations, the activation of astrocytes and microglia, and axons demyelination [30]. Microglia plays an essential role in the clearance of tau oligomers and the actin cytoskeleton for phagocytosis in AD [92]. In addition, the pathway-related proliferation immune system response and inflammation induced by PPA noticed in autism spectrum disorder [25].
There was the amount of biological process linked to angiogenesis blood circulating and allocated the 26 genes to the hypoxia-induced response. Previous studies have indicated the vascular dysfunctions in the early year increase regionally blood flow as a compensatory mechanism while observed decrease eventually in the later stage of AD. The capillary amyloid angiopathy induced by hypoxia through activating β- and γ-secretases can be attributed to AD pathology [93]. Hypoxia condition modulates hypometabolism in several regions of the brain by overexpression of amyloid precursor protein and decreases the clearance of Aβ. This event promotes inflammation and, ultimately, neuronal cell death [94]. HIF-1 signaling pathway supports cellular adaptation in hypoxic conditions that were seen in the KEGG pathway of AD-PPA genes.
Cell adhesion that detected in AD-PPA genes enriched biological processes related to cell-cell adhesion and cell-matrix adhesion. It has been determined that the genes with pleiotropic roles in cell adhesion highlighted in AD pathogenesis [95] ] and PPA also had been prepared induced extensive alterations in gene expression, including neuronal cell adhesion molecules [90].
Cellular component analysis enriched the critical part of a cell that promotes AD pathogenesis. Cytoskeleton change in neuron cell that enriched by AD-PPA genes effect on the synapse. Rush T et al. reported that Aβ oligomers induce aberrant actin stabilization and synaptic loss and impairment [96]. Dysfunctionality was observed in several parts of the neuron, i.e., synapse or cellular secretion [90, 97]. Elevated PPA could also produce sensitivity to oxidative stress and, in turn, increase the damage caused by other toxic environmental factors such as metals or infectious agents [87]. Mitochondria also implicated alteration specific in function, size, and form by PPA treatment in vivo [30, 90, 98].
Conclusions
AD-PPA genes analysis unveiled the comorbidity with diseases that surprisingly were related to the effect of bacteria and virus infection or enriched the neurological disease that previously reported in PPA intervention, such as meningitis or autism disorder. It could be pinpointed that AD-PPA genes carry biological processes that cover almost all of AD pathogenesis hallmarks. Functional analysis of hub-bottleneck-seed genes represented the role of these crucial genes in redox signaling, neuroinflammation, and cell cycle, and cell death. Since it has not been produced high throughput data directly obtained from the effect of PPA on cell line or animal models, our analysis opens the view of the possible effect of PPA on AD pathogenesis. Therefore, it is necessary to design further empirical investigations to attain deep insights into the PPA metabolite secreted by microbiota and implicated in AD pathogenesis.
Electronic supplementary material
(DOCX 871 kb)
(XLSX 467 kb)
(XLSX 102 kb)
Acknowledgements
This study is a part of the Ph.D. thesis of Morteza Aliashrafi.
Author contributions
Morteza Aliashrafi design the study carried out data collection and statistical analysis drafted the manuscript and interpreted the data; Mohammad Nasehi designed and supervised the study; Mohammad-Reza Zarrindast designed and supervised the study; Mohammad Taghi Joghataei design the study; Hakimeh Zali carried out data collection and statistical analysis; Seyed Davar Siadat designed and supervised the study. All authors read and approved the final manuscript.
Funding information
The Proteomics Research Centre of Shahid Beheshti University of Medical Sciences supported this study.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mohammad Nasehi, Email: nasehi@iricss.org.
Hakimeh Zali, Email: hakimehzali@gmail.com, Email: h.zali@sbmu.ac.ir.
References
- 1.Jack CR, Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimer Dement. 2018;14(4):535–62. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemark DK, Allen SJ. The microbiome and disease: reviewing the links between the oral microbiome, aging, and Alzheimer’s disease. J Alzheimer Dis. 2015;43(3):725–38. doi: 10.3233/JAD-141170. [DOI] [PubMed] [Google Scholar]
- 3.Itzhaki RF. Herpes and Alzheimer’s disease: subversion in the central nervous system and how it might be halted. J Alzheimer Dis. 2016;54(4):1273–81. doi: 10.3233/JAD-160607. [DOI] [PubMed] [Google Scholar]
- 4.Lövheim H, Olsson J, Weidung B, Johansson A, Eriksson S, Hallmans G, et al. Interaction between cytomegalovirus and herpes simplex virus type 1 associated with the risk of Alzheimer’s disease development. J Alzheimer Dis. 2018;61(3):939–45. doi: 10.3233/JAD-161305. [DOI] [PubMed] [Google Scholar]
- 5.Lövheim H, Gilthorpe J, Adolfsson R, Nilsson L-G, Elgh F. Reactivated herpes simplex infection increases the risk of Alzheimer’s disease. Alzheimer Dement. 2015;11(6):593–9. doi: 10.1016/j.jalz.2014.04.522. [DOI] [PubMed] [Google Scholar]
- 6.Miklossy J. Are there spirochetes in the brain in more than 90 percent of Alzheimer’s Disease cases? J Neuroinflammation. 2011;8(1):90. [Google Scholar]
- 7.Miklossy J. Bacterial amyloid and DNA are important constituents of senile plaques: Further evidence of the spirochetal and biofilm nature of senile plaques. Handbook of Infection and Alzheimer’s Disease. 2017;5:89. [DOI] [PMC free article] [PubMed]
- 8.Kornhuber H. Propionibacterium acnes in the cortex of patients with Alzheimer’s disease. Eur Arch Psychiatry Clin NeuroSci. 1996;246(2):108–9. doi: 10.1007/BF02274902. [DOI] [PubMed] [Google Scholar]
- 9.Emery DC, Shoemark DK, Batstone TE, Waterfall CM, Coghill JA, Cerajewska TL, et al. 16S rRNA next generation sequencing analysis shows bacteria in Alzheimer’s post-mortem brain. Front Aging Neurosci. 2017;9:195. doi: 10.3389/fnagi.2017.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirschbaum J, Kligman A. The pathogenic role of Corynebacterium acnes in acne vulgaris. Arch Dermatol. 1963;88(6):832–3. doi: 10.1001/archderm.1963.01590240156026. [DOI] [PubMed] [Google Scholar]
- 11.Douglas H, Gunter SE. The taxonomic position of Corynebacterium acnes. J Bacteriol. 1946;52(1):15. [PMC free article] [PubMed] [Google Scholar]
- 12.Sa’ad H, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta Mol Cell Biol Lipids. 2010;1801(11):1175–83. [DOI] [PubMed]
- 13.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time? Nutr Clin Pract. 2006;21(4):351–66. doi: 10.1177/0115426506021004351. [DOI] [PubMed] [Google Scholar]
- 14.Zouboulis C, Eady A, Philpott M, Goldsmith L, Orfanos C, Cunliffe W, et al. What is the pathogenesis of acne? Exp Dermatol. 2005;14(2):143. [DOI] [PubMed]
- 15.Borgström M, Edwardsson S, Svensäter G, Twetman S. Acid formation in sucrose-exposed dental plaque in relation to caries incidence in schoolchildren. Clin Oral Investig. 2000;4(1):9–12. doi: 10.1007/s007840050106. [DOI] [PubMed] [Google Scholar]
- 16.Niederman R, Zhang J, Kashket S. Short-chain carboxylic-acid-stimulated, PMN-mediated gingival inflammation. Crit Rev Oral Biol Med. 1997;8(3):269–90. doi: 10.1177/10454411970080030301. [DOI] [PubMed] [Google Scholar]
- 17.MacFabe DF. Short-chain fatty acid fermentation products of the gut microbiome: implications in autism spectrum disorders. Microb Ecol Health Dis. 2012;23(1):19260. doi: 10.3402/mehd.v23i0.19260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakao S, Moriya Y, Furuyama S, Niederman R, Sugiya H. Propionic acid stimulates superoxide generation in human neutrophils. Cell Biol Int. 1998;22(5):331–7. doi: 10.1006/cbir.1998.0263. [DOI] [PubMed] [Google Scholar]
- 19.DeCastro M, Nankova BB, Shah P, Patel P, Mally PV, Mishra R, et al. Short chain fatty acids regulate tyrosine hydroxylase gene expression through a cAMP-dependent signaling pathway. Mol Brain Res. 2005;142(1):28–38. doi: 10.1016/j.molbrainres.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 20.Rörig B, Klausa G, Sutor B. Intracellular acidification reduced gap junction coupling between immature rat neocortical pyramidal neurones. J Physiol. 1996;490(1):31–49. doi: 10.1113/jphysiol.1996.sp021125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiber J, Chapman KA, Summar ML, Mew NA, Sutton VR, MacLeod E, et al. Neurologic considerations in propionic acidemia. Mol Genet Metab. 2012;105(1):10–5. doi: 10.1016/j.ymgme.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Nyhan WL, Bay C, Beyer EW, Mazi M. Neurologic nonmetabolic presentation of propionic acidemia. Arch Neurol. 1999;56(9):1143–7. doi: 10.1001/archneur.56.9.1143. [DOI] [PubMed] [Google Scholar]
- 23.MacFabe DF, Cain DP, Rodriguez-Capote K, Franklin AE, Hoffman JE, Boon F, et al. Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res. 2007;176(1):149–69. doi: 10.1016/j.bbr.2006.07.025. [DOI] [PubMed] [Google Scholar]
- 24.Thomas RH, Foley KA, Mepham JR, Tichenoff LJ, Possmayer F, MacFabe DF. Altered brain phospholipid and acylcarnitine profiles in propionic acid infused rodents: further development of a potential model of autism spectrum disorders. J Neurochem. 2010;113(2):515–29. doi: 10.1111/j.1471-4159.2010.06614.x. [DOI] [PubMed] [Google Scholar]
- 25.El-Ansary AK, Bacha AB, Kotb M. Etiology of autistic features: the persisting neurotoxic effects of propionic acid. J Neuroinflamm. 2012;9(1):74. doi: 10.1186/1742-2094-9-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacFabe DF, Cain NE, Boon F, Ossenkopp K-P, Cain DP. Effects of the enteric bacterial metabolic product propionic acid on object-directed behavior, social behavior, cognition, and neuroinflammation in adolescent rats: relevance to autism spectrum disorder. Behav Brain Res. 2011;217(1):47–54. doi: 10.1016/j.bbr.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Shultz SR, MacFabe DF, Ossenkopp K-P, Scratch S, Whelan J, Taylor R, et al. Intracerebroventricular injection of propionic acid, an enteric bacterial metabolic end-product, impairs social behavior in the rat: implications for an animal model of autism. Neuropharmacology. 2008;54(6):901–11. doi: 10.1016/j.neuropharm.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 28.Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, et al. The Alzheimer’s disease-associated amyloid β-protein is an antimicrobial peptide. PloS One. 2010;5(3). [DOI] [PMC free article] [PubMed]
- 29.Kumar DKV, Choi SH, Washicosky KJ, Eimer WA, Tucker S, Ghofrani J, et al. Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci Transl Med. 2016;8(340):340ra72-ra72. doi: 10.1126/scitranslmed.aaf1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lobzhanidze G, Lordkipanidze T, Zhvania M, Japaridze N, Pochkidze N, Gasimov E, et al. Effect of propionic acid on the morphology of the amygdala in adolescent male rats and their behavior. Micron. 2019;125:102732. doi: 10.1016/j.micron.2019.102732. [DOI] [PubMed] [Google Scholar]
- 31.Davis A, Murphy C, Johnson R, Lay J. ea Lennon-Hopkins. The Comparative Toxicogenomics Database: update 2013. Nucleic Acids Res 41:D1104-14. [DOI] [PMC free article] [PubMed]
- 32.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010. [DOI] [PMC free article] [PubMed]
- 33.Cotto KC, Wagner AH, Feng Y-Y, Kiwala S, Coffman AC, Spies G, et al. DGIdb 3.0: a redesign and expansion of the drug–gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–73. [DOI] [PMC free article] [PubMed]
- 34.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parkinson H, Kapushesky M, Shojatalab M, Abeygunawardena N, Coulson R, Farne A, et al. ArrayExpress—a public database of microarray experiments and gene expression profiles. Nucleic Acids Res. 2007;35(suppl_1):D747–50. [DOI] [PMC free article] [PubMed]
- 36.Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The Comparative Toxicogenomics Database (CTD) Environ Health Perspect. 2003;111(6):793–5. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebhan M, Chalifa-Caspi V, Prilusky J, Lancet D. GeneCards: integrating information about genes, proteins and diseases. Trends Genet. 1997;13(4):163. doi: 10.1016/s0168-9525(97)01103-7. [DOI] [PubMed] [Google Scholar]
- 38.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, Landfield PW. Incipient Alzheimer’s disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci. 2004;101(7):2173–8. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sekar S, McDonald J, Cuyugan L, Aldrich J, Kurdoglu A, Adkins J, et al. Alzheimer’s disease is associated with altered expression of genes involved in immune response and mitochondrial processes in astrocytes. Neurobiol Aging. 2015;36(2):583–91. doi: 10.1016/j.neurobiolaging.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearson BL, Simon JM, McCoy ES, Salazar G, Fragola G, Zylka MJ. Identification of chemicals that mimic transcriptional changes associated with autism, brain aging and neurodegeneration. Nat Commun. 2016;7(1):1–12. doi: 10.1038/ncomms11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knabe JF. Computational Genetic Regulatory Networks: Evolvable, Self-organizing Systems. Berlin: Springer; 2012.
- 43.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4(1). [DOI] [PubMed]
- 44.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Yun GH, Zhu J, Zhang C, Niu Y-M. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 2019;10:1081. doi: 10.3389/fphys.2019.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mantrach A, Yen L, Callut J, Francoisse K, Shimbo M, Saerens M. The sum-over-paths covariance kernel: A novel covariance measure between nodes of a directed graph. IEEE Trans Pattern Anal Mach Intell. 2009;32(6):1112–26. doi: 10.1109/TPAMI.2009.78. [DOI] [PubMed] [Google Scholar]
- 47.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nudelman KN, McDonald BC, Lahiri DK, Saykin AJ. Biological hallmarks of cancer in Alzheimer’s disease. Mol Neurobiol. 2019;56(10):7173–87. doi: 10.1007/s12035-019-1591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Liu C-C, Zheng H, Huang TY. Amyloid, tau, pathogen infection and antimicrobial protection in Alzheimer’s disease–conformist, nonconformist, and realistic prospects for AD pathogenesis. Transl Neurodegener. 2018;7(1):34. doi: 10.1186/s40035-018-0139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wong JM, De Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–43. doi: 10.1097/00004836-200603000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Karns R, Tabar S, Bardes E, Jegga A, Aronow B. How Do Bioinformatics approaches apply to the analysis and understanding of disease pathology? 2014.
- 52.Fülöp T, Itzhaki RF, Balin BJ, Miklossy J, Barron AE. Role of microbes in the development of Alzheimer’s disease: state of the art–An International Symposium Presented at the 2017 IAGG Congress in San Francisco. Front Genet. 2018;9:362. doi: 10.3389/fgene.2018.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fülöp T, Itzhaki RF, Balin BJ, Miklossy J, Barron AE. Role of microbes in the development of Alzheimer’s disease: state of the art–An International Symposium Presented at the 2017 IAGG Congress in San Francisco. Front Genet. 2018;9:362. doi: 10.3389/fgene.2018.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doulberis M, Kotronis G, Thomann R, Polyzos SA, Boziki M, Gialamprinou D, et al. Impact of Helicobacter pylori on Alzheimer’s disease: What do we know so far? Helicobacter. 2018;23(1):e12454. doi: 10.1111/hel.12454. [DOI] [PubMed] [Google Scholar]
- 55.Kristensson K, Masocha W, Bentivoglio M. Mechanisms of CNS invasion and damage by parasites. Handbook of clinical neurology. Amsterdam: Elsevier; 2013. p. 11–22. [DOI] [PubMed]
- 56.Dando SJ, Mackay-Sim A, Norton R, Currie BJ, John JAS, Ekberg JA, et al. Pathogens penetrating the central nervous system: infection pathways and the cellular and molecular mechanisms of invasion. Clin Microbiol Rev. 2014;27(4):691–726. doi: 10.1128/CMR.00118-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dahm T, Rudolph H, Schwerk C, Schroten H, Tenenbaum T. Neuroinvasion and inflammation in viral central nervous system infections. Mediat Inflamm. 2016;2016. [DOI] [PMC free article] [PubMed]
- 58.Aguayo S, Schuh CMAP, Vicente B, Aguayo LG. Association between Alzheimer’s Disease and oral and gut microbiota: are pore forming proteins the missing link? J Alzheimer Dis. 2018;65(1):29–46. doi: 10.3233/JAD-180319. [DOI] [PubMed] [Google Scholar]
- 59.Martins IJ. Bacterial lipopolysaccharides change membrane fluidity with relevance to phospholipid and amyloid beta dynamics in Alzheimer’s disease. 2016.
- 60.Zhao Y, Jaber V, Lukiw WJ. Secretory products of the human GI tract microbiome and their potential impact on Alzheimer’s disease (AD): detection of lipopolysaccharide (LPS) in AD hippocampus. Front Cell Infect Microbiol. 2017;7:318. doi: 10.3389/fcimb.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu F, Fan T, Duan J, Chen D. Clinical analysis of organic acidemia in neonates from neonatal intensive care units. Zhongguo dang dai er ke za zhi = Chinese. J Contemp Pediatr. 2012;14(5):336–9. [PubMed] [Google Scholar]
- 62.Feliz B, Witt DR, Harris BT. Propionic acidemia: a neuropathology case report and review of prior cases. Arch Pathol Lab Med. 2003;127(8):e325-e8. doi: 10.5858/2003-127-e325-PAANCR. [DOI] [PubMed] [Google Scholar]
- 63.Lorenzo A, Yuan M, Zhang Z, Paganetti PA, Sturchler-Pierrat C, Staufenbiel M, et al. Amyloid β interacts with the amyloid precursor protein: a potential toxic mechanism in Alzheimer’s disease. Nat Neurosci. 2000;3(5):460–4. doi: 10.1038/74833. [DOI] [PubMed] [Google Scholar]
- 64.Keeney JT, Swomley AM, Harris JL, Fiorini A, Mitov MI, Perluigi M, et al. Cell cycle proteins in brain in mild cognitive impairment: insights into progression to Alzheimer disease. Neurotox Res. 2012;22(3):220–30. doi: 10.1007/s12640-011-9287-2. [DOI] [PubMed] [Google Scholar]
- 65.Ferrer I, Blanco R, Carmona M, Puig B. Phosphorylated mitogen-activated protein kinase (MAPK/ERK-P), protein kinase of 38 kDa (p38-P), stress-activated protein kinase (SAPK/JNK-P), and calcium/calmodulin-dependent kinase II (CaM kinase II) are differentially expressed in tau deposits in neurons and glial cells in tauopathies. J Neural Transm. 2001;108(12):1397–415. doi: 10.1007/s007020100016. [DOI] [PubMed] [Google Scholar]
- 66.Cavallini A, Brewerton S, Bell A, Sargent S, Glover S, Hardy C, et al. An unbiased approach to identifying tau kinases that phosphorylate tau at sites associated with Alzheimer disease. J Biol Chem. 2013;288(32):23331–47. doi: 10.1074/jbc.M113.463984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Absalon S, Kochanek DM, Raghavan V, Krichevsky AM. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J Neurosci. 2013;33(37):14645–59. doi: 10.1523/JNEUROSCI.1327-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Modi PK, Komaravelli N, Singh N, Sharma P. Interplay between MEK-ERK signaling, cyclin D1, and cyclin-dependent kinase 5 regulates cell cycle reentry and apoptosis of neurons. Mol Biol Cell. 2012;23(18):3722–30. doi: 10.1091/mbc.E12-02-0125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirouac L, Rajic AJ, Cribbs DH, Padmanabhan J. Activation of Ras-ERK signaling and GSK-3 by amyloid precursor protein and amyloid beta facilitates neurodegeneration in Alzheimer’s disease. Eneuro. 2017;4(2). [DOI] [PMC free article] [PubMed]
- 70.Yoshida T, Haga S, Numata Y, Yamashita K, Mikami T, Ogawa T, et al. Disruption of the p53–p53r2 DNA repair system in ulcerative colitis contributes to colon tumorigenesis. Int J Cancer. 2006;118(6):1395–403. doi: 10.1002/ijc.21538. [DOI] [PubMed] [Google Scholar]
- 71.Khalil SR, Abd-Elhakim YM, Selim ME, Al-Ayadhi LY. Apitoxin protects rat pups brain from propionic acid-induced oxidative stress: the expression pattern of Bcl-2 and Caspase-3 apoptotic genes. Neurotoxicology. 2015;49:121–31. doi: 10.1016/j.neuro.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 72.Cheignon C, Tomas M, Bonnefont-Rousselot D, Faller P, Hureau C, Collin F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018;14:450–64. doi: 10.1016/j.redox.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta Mol Cell Res. 2016;1863(12):2977–92. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 74.El-Ansary AK, Al-Daihan S, Ben Bacha A, Shaker GH, Al-Ayadhi LY. Comparative study on the protective effect of carnosine and carnitine against pro inflammatory/pro-oxidant effects of clindamycin and propionic acid administrations to hamsters. Afr J Microbiol Res. 2013;7(2):103–14. [Google Scholar]
- 75.Hsieh H-L, Yang C-M. Role of redox signaling in neuroinflammation and neurodegenerative diseases. BioMed Res Int. 2013;2013. [DOI] [PMC free article] [PubMed]
- 76.McCubrey JA, Lertpiriyapong K, Steelman LS, Abrams SL, Yang LV, Murata RM, et al. Effects of resveratrol, curcumin, berberine and other nutraceuticals on aging, cancer development, cancer stem cells and microRNAs. Aging. 2017;9(6):1477. doi: 10.18632/aging.101250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shawkatová I, Javor J, Párnická Z, Minárik G, Vašečková B, Králová M, et al. ICAM1 gene polymorphism in late-onset Alzheimer’s disease. Biologia. 2019;74(11):1531–8. [Google Scholar]
- 78.Wennström M, Nielsen HM. Cell adhesion molecules in Alzheimer’s disease. Degener Neurol Neuromuscul Dis. 2012;2:65. doi: 10.2147/DNND.S19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bronzuoli MR, Iacomino A, Steardo L, Scuderi C. Targeting neuroinflammation in Alzheimer’s disease. J Inflamm Res. 2016;9:199. doi: 10.2147/JIR.S86958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rigo FK, Pasquetti L, Malfatti CRM, Fighera MR, Coelho RC, Petri CZ, et al. Propionic acid induces convulsions and protein carbonylation in rats. Neurosci Lett. 2006;408(2):151–4. doi: 10.1016/j.neulet.2006.08.075. [DOI] [PubMed] [Google Scholar]
- 81.Halliwell B, Whiteman M. Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? Br J Pharmacol. 2004;142(2):231–55. doi: 10.1038/sj.bjp.0705776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Al-Dbass AM. N-Acetylcysteine reduces the neurotoxic effects of propionic acid in rat pups. J King Saud Univ-Sci. 2014;26(4):254–60. [Google Scholar]
- 83.Ribas GS, Manfredini V, de Mari JF, Wayhs CY, Vanzin CS, Biancini GB, et al. Reduction of lipid and protein damage in patients with disorders of propionate metabolism under treatment: a possible protective role of L-carnitine supplementation. Int J Dev Neurosci. 2010;28(2):127–32. doi: 10.1016/j.ijdevneu.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Frye RE, Rose S, Chacko J, Wynne R, Bennuri S, Slattery J, et al. Modulation of mitochondrial function by the microbiome metabolite propionic acid in autism and control cell lines. Transl Psychiatry. 2016;6(10):e927-e. doi: 10.1038/tp.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.MacFabe DF, Rodríguez-Capote K, Hoffman JE, Franklin AE, Mohammad-Asef Y, Taylor AR, et al. A novel rodent model of autism: intraventricular infusions of propionic acid increase locomotor activity and induce neuroinflammation and oxidative stress in discrete regions of adult rat brain. Am J Biochem Biotechnol. 2008;4(2):146–66. [Google Scholar]
- 86.Yin F, Sancheti H, Patil I, Cadenas E. Energy metabolism and inflammation in brain aging and Alzheimer’s disease. Free Radic Biol Med. 2016;100:108–22. doi: 10.1016/j.freeradbiomed.2016.04.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wajner M, Latini A, Wyse A, Dutra-Filho C. The role of oxidative damage in the neuropathology of organic acidurias: insights from animal studies. J Inherit Metab Dis. 2004;27(4):427–48. doi: 10.1023/B:BOLI.0000037353.13085.e2. [DOI] [PubMed] [Google Scholar]
- 88.Winkler EA, Nishida Y, Sagare AP, Rege SV, Bell RD, Perlmutter D, et al. GLUT1 reductions exacerbate Alzheimer’s disease vasculo-neuronal dysfunction and degeneration. Nat Neurosci. 2015;18(4):521. doi: 10.1038/nn.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yurov YB, Vorsanova SG, Iourov IY. The DNA replication stress hypothesis of Alzheimer’s disease. Sci World J. 2011;11:2602–12. doi: 10.1100/2011/625690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nankova BB, Agarwal R, MacFabe DF, La Gamma EF. Enteric bacterial metabolites propionic and butyric acid modulate gene expression, including CREB-dependent catecholaminergic neurotransmission, in PC12 cells-possible relevance to autism spectrum disorders. PloS One. 2014;9(8). [DOI] [PMC free article] [PubMed]
- 91.McManus RM, Heneka MT. Role of neuroinflammation in neurodegeneration: new insights. Alzheimer Res Ther. 2017;9(1):14. doi: 10.1186/s13195-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Das R, Balmik AA, Chinnathambi S. Phagocytosis of full-length Tau oligomers by Actin-remodeling of activated microglia. J Neuroinflamm. 2020;17(1):1–15. doi: 10.1186/s12974-019-1694-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salminen A, Kauppinen A, Kaarniranta K. Hypoxia/ischemia activate processing of Amyloid Precursor Protein: impact of vascular dysfunction in the pathogenesis of Alzheimer’s disease. J Neurochem. 2017;140(4):536–49. doi: 10.1111/jnc.13932. [DOI] [PubMed] [Google Scholar]
- 94.Ashok BS, Ajith TA, Sivanesan S. Hypoxia-inducible factors as neuroprotective agent in Alzheimer’s disease. Clin Exp Pharmacol Physiol. 2017;44(3):327–34. doi: 10.1111/1440-1681.12717. [DOI] [PubMed] [Google Scholar]
- 95.Ramanan VK, Kim S, Holohan K, Shen L, Nho K, Risacher SL, et al. Genome-wide pathway analysis of memory impairment in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort implicates gene candidates, canonical pathways, and networks. Brain Imaging Behav. 2012;6(4):634–48. doi: 10.1007/s11682-012-9196-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rush T, Martinez-Hernandez J, Dollmeyer M, Frandemiche ML, Borel E, Boisseau S, et al. Synaptotoxicity in Alzheimer’s disease involved a dysregulation of actin cytoskeleton dynamics through cofilin 1 phosphorylation. J Neurosci. 2018;38(48):10349–61. doi: 10.1523/JNEUROSCI.1409-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee S, Mankhong S, Kang J-H. Extracellular vesicle as a source of Alzheimer’s biomarkers: Opportunities and challenges. Int J Mol Sci. 2019;20(7):1728. doi: 10.3390/ijms20071728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brass EP, Beyerinck RA. Effects of propionate and carnitine on the hepatic oxidation of short-and medium-chain-length fatty acids. Biochem J. 1988;250(3):819–25. doi: 10.1042/bj2500819. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 871 kb)
(XLSX 467 kb)
(XLSX 102 kb)