Abstract
Positive single-strand ribonucleic acid [(+)ssRNA] viruses can cause multiple outbreaks, for which comprehensive tailored therapeutic strategies are still missing. Virus and host cell dynamics are tightly connected, generating a complex dynamics that conveys in virion assembly to ensure virus spread in the body. Starting from the knowledge of relevant processes in (+ss)RNA virus replication, transcription, translation, virions budding and shedding, and their respective energy costs, we built up a systems thinking (ST)–based diagram of the virus–host interaction, comprehensive of stocks, flows, and processes as well-described in literature. In ST approach, stocks and flows are expressed by a proxy of the energy embedded and transmitted, respectively, whereas processes are referred to the energy required for the system functioning. In this perspective, healthiness is just a particular configuration, in which stocks relevant for the system (equivalent but not limited to proteins, RNA, DNA, and all metabolites required for the survival) are constant, and the system behavior is stationary. At time of infection, the presence of additional stocks (e.g., viral protein and RNA and all metabolites required for virion assembly and spread) confers a complex network of feedbacks leading to new configurations, which can evolve to maximize the virions stock, thus changing the system structure, output, and purpose. The dynamic trajectories will evolve to achieve a new stationary status, a phenomenon described in microbiology as integration and symbiosis when the system is resilient enough to the changes, or the system may stop functioning and die. Application of external driving forces, acting on processes, can affect the dynamic trajectories adding a further degree of complexity, which can be captured by ST approach, used to address these new configurations. Investigation of system configurations in response to external driving forces acting is developed by computational analysis based on ST diagrams, with the aim at designing novel therapeutic approaches.
Keywords: systems thinking (ST), RNA-virus, virus–host interaction, dynamics, modeling, simulation – computers, evolution trajectories
Introduction
Positive single-stranded ribonucleic acid [(+)ssRNA] viruses, including picornaviruses, flaviviruses, Togaviridae, and human coronaviruses (CoVs) (Ahlquist et al., 2003; Lam et al., 2016; Scutigliani and Kikkert, 2017; Primadharsini et al., 2019), cause multiple outbreaks, for which tailored antiviral strategies are still missing (Zumla et al., 2016; Dinesh et al., 2020; Gordon et al., 2020b). (+)ssRNA viruses package their genomes as messenger sense, single-stranded RNA and replicate those genomes solely through RNA intermediates in the cytosol of the host cells (Den Boon et al., 2010). RNA-dependent RNA polymerases lack coreplicative and postreplicative fidelity-enhancing pathways; this final RNA genome copies incorporate mutations at a high rate (Lauring et al., 2013; Acevedo et al., 2014), providing the viral quasi-species with a higher probability to evolve and adapt to new environments and challenges during infection (Burch and Chao, 2000; Vignuzzi et al., 2006). The diversity is essential for both viral fitness (Wargo and Kurath, 2012) and pathogenesis because of the complex relationships among virus replication (VR), host cells, and immune system, as almost all (+)ssRNA viruses can delay antiviral innate immune response (Kobasa et al., 2007) in multiple ways (Diebold et al., 2003; Hogan et al., 2004; Meylan et al., 2005; Saito et al., 2008; Yang et al., 2015; Beachboard and Horner, 2016; Nelemans and Kikkert, 2019). Host immunogenetic factors can be sensitive to a variation in the viral load, leading to a defective response of the innate immunity, that could explain the variable clinical course of infection (Fanning et al., 2001; Nelemans and Kikkert, 2019).
Recent studies confirmed the complexity of viral dynamics, whose fitness is improved by the complex interactions with the host proteins (Bosl et al., 2019; Sruthi and Prakash, 2019), as previously described in modeling the virus–host interactions at subcell and cell levels (Dapat and Oshitani, 2016; Jonsdottir and Dijkman, 2016; Gao et al., 2017; Patzina et al., 2017; Gordon et al., 2020b). However, models that address a specific aspect of the virus–host interaction do not capture the wide range of intertwined spatial and temporal (hours to days) dynamic scales (Apweiler et al., 2018), which are related to the interactions of different concurrent hierarchical levels. For this reason, we aimed at describing the virus–host cell interaction as a dynamic system by a systems thinking (ST) approach (Northridge and Metcalf, 2016).
In ST, the behavior of a dynamic system can be described and predicted by the temporal evolution of its configurations, given by hierarchical feedback loops and self-organization. The configuration evolution then can be analytically computed by proper simulators (Odum and Odum, 2000; Tegner et al., 2009; Hassmiller Lich et al., 2017; Spill et al., 2018) to further address suitable leverage points for intervening (Meadows and Wright, 2008). In particular, dynamic models, where the temporal evolution of extensive variables (stocks) is simulated in the form of trajectories, derive their initial conditions from available and assessed evidence. Varying the key system parameters, trajectories represent the possible evolutive (structural) patterns of the system at issue, becoming abstracted with respect to local specific attributes related to single case studies. However, being stocks associated to system observables, the relation with those attributes is maintained, making it possible to compare predicted trajectories with observed data and constituting suitable counterfactuals with respect to the laboratory measurements results.
System dynamics (SD) approach is mostly used for strategic modeling, typically for ecological and socioeconomic systems, to understand the supply chain performance. In particular, results of SD modeling provide a set of alternative evolutive patterns in form of graphs, capturing the internal dynamics of a system even in lack of some experimental data to fit. These can provide alternative scenarios that, when fitting experimental evidences, indicate the most effective leverage points to control the system evolution.
In this article, we show that, by approaching the host–virus interaction as a dynamic systemic problem (Sterman, 2002; Meadows and Wright, 2008), it is possible to identify potential systemic leverage points to minimize the release of virions, so addressing effective systemic intervention strategies.
Materials and Methods
Development of a Stock-Flow Diagram
The basic SD element is the stock and flow diagram. Stocks are countable extensive variables Qi, i = 1, 2,…, n, relevant to the study at issue, that constitute an n-ple of numbers (possibly derived from experimental measurements), which at any time represents the state of the system. A stock may change its value only upon its inflows and/or its outflows, represented by arrows entering or exiting the stock. Processes are any occurrence capable to alter—either quantitatively or qualitatively—a flow, by the action of one or more of the system elements. In a stationary state of the system, stock values are either constant or regularly oscillating. Processes, which cause the stationarity or perturbation of a system, must be activated by a driver, acting on the flows where the process is located. The pattern of the feedbacks acting in the system configurations is the feature that ultimately defines the systems dynamics. Each flow depends on the state variables Qi by relationships of the kind dQi/dt = kf(Qj), i, j = 1,…, n, where n is the number of stocks in the system.
The stocks and flows inventory reported in Table 1 was based on information from existing knowledge on the biological mechanisms at issue, listing the variables and parameters necessary to set up the equations describing the system dynamics. Turnover times of stocks included in the RNA-virus–host interaction ST diagram have been reported in Table 2, derived from the available literature (Konig et al., 2008; Friedel and Haas, 2011; Pfefferle et al., 2011; Jourdan et al., 2012; Munday et al., 2012; Naji et al., 2012; Wu et al., 2012; Emmott et al., 2013; Garcia-Dorival et al., 2014; Verchot, 2014; Watanabe et al., 2014; York et al., 2014; Zheng et al., 2014; Dong et al., 2016; Kuo et al., 2016, 2018; Wang et al., 2016, 2017a, b; Gao et al., 2017; Hafirassou et al., 2017; Khamina et al., 2017; King et al., 2017; Martinez-Gil et al., 2017; Patzina et al., 2017; Coyaud et al., 2018; Iwasaki et al., 2018; Lescar et al., 2018; Ziegler et al., 2018; Bosl et al., 2019; Chakravorty et al., 2019; Garcia-Moreno et al., 2019; Rothenburg and Brennan, 2020). All stocks, flows, and processes were expressed using a common proxy unit, representing the energy embedded, transmitted, and used, respectively, during the system operation. The proxy unit was expressed as the number of ATP (and ATP-equivalent) hydrolysis events (Mahmoudabadi et al., 2017). This choice allowed calculating each parameter of the system on the basis of stocks and characteristic times of the flows derived from the literature, without further need for experimental data.
TABLE 1.
Inventory of stocks and flows depicted in the diagram of Figure 1.
| Stock | Biological meaning | Dynamic equation* | Calibration value | References |
| Q1 | Resources available for protein synthesis | dQ1/dt = J0 + J21A + J21B − J1 − J13 − J15 − J17 | 3.9 | Odum, 1996, 2002; Odum and Odum, 2000 |
| Q2A | Short-half-life proteins | dQ2A/dt = J2A − J21A − J20A | 13 | Wheatley et al., 1980; Eden et al., 2011; Siwiak and Zielenkiewicz, 2013 |
| Q2B | Long-half-life proteins | dQ2B/dt = J2B − J21B − J23 − J25 − J27 − J20B | 13 | Wheatley et al., 1980; Eden et al., 2011; Siwiak and Zielenkiewicz, 2013 |
| Q3 | Viral ss + RNA | dQ3/dt = J3 − J4 | 3 | Baccam et al., 2006 |
| Q4 | Viral proteins | dQ4/dt = J4 − J5 | 0.024 | Kummer et al., 2014 |
| Q5 | Virions | dQ5/dt = J6 − J7 | 0 | NA |
| Flow | Biological role | Phenomenological coefficients (k) | References | |
| J0 = k0 × R × (1 + Q2A) | Enter of resources allocated for protein synthesis | 3.9E-06 | Odum, 1996 | |
| J1 = k1 × Q1 | Host-cell RNA transcription and translation | 6.9E-05 | Eden et al., 2011; Kummer et al., 2014 | |
| J2A = k2A × Q1 | Short-half-life protein synthesis | 2.6E-05 | Adelman et al., 2002; Klumpp and Hwa, 2008; Eden et al., 2011 | |
| J2B = k2B × Q1 × (1 + Q4) | Long-half-life protein synthesis | 1.6E-05 | Proshkin et al., 2010; Eden et al., 2011 | |
| J3 = k3 × Q1 × Q2B × Q3 × Q5 | Virus-RNA replication | 1.0E-01 | Mahmoudabadi et al., 2017 | |
| J4 = k4 × Q3 | Viral RNA translation | 6.9E-05 | Kummer et al., 2014 | |
| J5 = k5 × Q1 × Q2B × Q4 | Recruitment of resources and host-cell protein machinery for virion assembly | 1.7E-03 | Mahmoudabadi et al., 2017 | |
| J6 = k6 × Q1 × Q2B × Q4 | Virion assembly | 3.0E-03 | Mahmoudabadi et al., 2017 | |
| J7 = k7 × Q1 × Q2B × Q5 | Virion budding | 8.3E-04 | Mahmoudabadi et al., 2017 | |
| J13 = k13 × Q2B × Q1 × Q3 × Q5 | Flow of host-cell resources diverted to let virus enter | 8.3E-02 | Mahmoudabadi et al., 2017 | |
| J15 = k15 × Q1 × Q4 × Q2B | Flow of host-cell resources diverted to let virion assembly | 1.7E-03 | Mahmoudabadi et al., 2017 | |
| J17 = k17 × Q1 × Q5 × Q2B | Flow of host-cell resources diverted to let virion shedding | 8.3E-04 | Mahmoudabadi et al., 2017 | |
| J20A = k20A × Q2A | Flow of host-cell short-half-life proteins addressed to degradation | 3.9E-06 | Eden et al., 2011; Boisvert et al., 2012 | |
| J20B = k20B × Q2B | Flow of host-cell long-half-life proteins addressed to degradation | 1.6E-06 | Eden et al., 2011; Boisvert et al., 2012 | |
| J21A = k21A × Q2A | Proteostasis mechanisms, including proteasome degradation and autophagy to re-cycle unfolded, old or not functional host-cell short-half-life proteins | 3.9E-06 | Eden et al., 2011; Boisvert et al., 2012 | |
| J21B = k21B × Q2B | Proteostasis mechanisms, including proteasome degradation and autophagy of to re-cycle unfolded, old or not functional host-cell long-half-life proteins | 3.9E-06 | Eden et al., 2011; Boisvert et al., 2012 | |
| J23 = k23 × Q1 × Q2B × Q3 × Q5 | Flow of host-cell proteins recruited to let virus enter and RNA transcription | 8.3E-02 | Mahmoudabadi et al., 2017 | |
| J25 = k25 × Q1 × Q2B × Q4 | Flow of host-cell proteins recruited to let virion assembly | 1.7E-03 | Mahmoudabadi et al., 2017 | |
| J27 = k27 × Q1 × Q2B × Q5 | Flow of host-cell proteins recruited to let virion budding | 1.7E-03 | Mahmoudabadi et al., 2017 | |
| J35 = k35 × Q4 | Flow of viral RNA to embed in the virion | 4.6E-05 | Mahmoudabadi et al., 2017 | |
| J50 = k50 × Q1 × Q2B × Q5 | Virion shedding | 4.6E-05 | Mahmoudabadi et al., 2017 | |
* Equations representing the dynamics of the stocks and calibration values are also shown. Calibration value is expressed in ×1012 ATP-eq.
TABLE 2.
Relevant flows included for the diagram and simulator development.
| Flow | Description | Value | Stock turnover time | References |
| J1 | Host protein translation | 4 h | Kummer et al., 2014 | |
| J1 + J2A | Protein synthesis rate | 10–20 aa/s | Kudva et al., 2013 | |
| J1 + J2B | Protein synthesis rate | 10–20 aa/s | Kudva et al., 2013 | |
| J2A | Host protein transcription and translation (long-half-life proteins) | 13 aa/s | Adelman et al., 2002; Klumpp and Hwa, 2008 | |
| J2A | Host protein transcription and translation (long-half-life proteins) | 2–500 (140) mRNA/h 1,000/proteins/mRNA/h 660 mRNA/h/cell | 1 h | Howard-Ashby et al., 2006; Ben-Tabou De-Leon and Davidson, 2009; Schwanhausser et al., 2011 |
| J2A | Host protein transcription and translation (long-half-life proteins) | 60,200 mRNA/h to convert in ATP eq | Pelechano et al., 2010 | |
| J2B | Host protein transcription and translation (short half-life proteins) | 42 nt/s + 14 aa/s | Proshkin et al., 2010 | |
| J2A + J2B | 15% 20% respiration rate | 0.99 μmol/kg/day | Buttgereit and Brand, 1995; Waterlow, 2006 | |
| J20 | Protein degradation | 0.08/h | Doherty et al., 2009 | |
| J20 | Turnover rate of protein | 20/hs | Boisvert et al., 2012 | |
| J13 | Virus transcription and replication | 2,500 nt/s | 12 s | Mahmoudabadi et al., 2017 |
| J3 | Virus transcription and replication | 2,500 nt/s | 12 s | Mahmoudabadi et al., 2017 |
| J4 | Viral protein translation | 4 h | Kummer et al., 2014 | |
| J5 | Virion assembly | 10 min | Mahmoudabadi et al., 2017 | |
| J6 | Virion assembly | 10 min | Mahmoudabadi et al., 2017 | |
| J7 | Virion budding | 20 min | Mahmoudabadi et al., 2017 | |
| J50 | Virion shedding | 24% in 21.6 h | 21.6 h | Murphy et al., 1980 |
| J15 | Virion assembly | 10 min | Mahmoudabadi et al., 2017 | |
| J17 | Virion budding | 20 min | Mahmoudabadi et al., 2017 | |
| J23 | Virus transcription and replication | 2,500 nt/s | 12 s | Mahmoudabadi et al., 2017 |
| J25 | Virion assembly | Mahmoudabadi et al., 2017 | ||
| J27 | Virion budding | 20 min | Mahmoudabadi et al., 2017 | |
| J21 | Protein recycling of short and long half unfolded-defective proteins | 0.08/h | Doherty et al., 2009 | |
| J3 + J4 + J5 + J6 + J7 + J50 | vRNA replication and virion shedding, virus production after infection | 6 h | Sedmak and Grossberg, 1973; Baccam et al., 2006 |
Development of the Virus–Host Interaction Systemic Simulator
After setting the initial conditions at time 0 for the stocks, system solutions were obtained using recursive computation for a relative short period of time (identified with the median life of an epithelial cell, 7 days), in order to appreciate the model dynamic behavior. The computational model based on a set of differential equations that describe the rates of change of all stocks in the ST diagram (Odum and Odum, 2000; Bossel, 2007) was developed using the open-source software package SCILAB1, which uses approximation techniques to evaluate stocks.
Given a set of initial conditions for the stocks (i.e., the initial state of the system) and a set of phenomenological coefficients k associated to flows, the set of interconnected equations was treated by a standard finite-different method, taking care of choosing a time step short enough to evidence the dynamics of any of the studied processes. The coefficients ki were calculated, on the basis of literature data, considering the dynamics of each single stock, by quantifying flows and stocks during the time interval set as simulation step, as shown in Table 1. When different flows coparticipate in a process, each coefficient gathers all the actions that concur to the intensity of the outcoming flow(s). In detail, the parameters used to run the model (i.e., the set of values for the ki coefficients), describing the reaction of each system component to a change in any other one, were derived from the stocks turnover times (Odum and Odum, 2000; Bossel, 2007). Therefore, the host–virus interaction computational model, built on experimental evidences as listed in Tables 1, 2, is not specific for a unique virus, but may represent the patterns of any virus–host interaction, in which stocks, flows, and processes are those relevant for the operation of the system at issue.
The reliability of both available data and modeling was tested by evaluating the effect of the variation of each of the most relevant input data (stocks and processes) on the system trajectories. Unfortunately, here is not a single comprehensive procedure suitable for the validation of all dynamic models, being dependent on their usefulness, in turn referred to the very purpose of the model itself (Grüne-Yanoff and Weirich, 2010). We chose the sensitivity analysis approach (Qudrat-Ullah, 2012; Hekimoğlu and Barlas, 2016), which allows to see to what extent a variation on these values can lead to alternative evaluations of the system dynamics. In particular, we applied a 50% variation (either positive or negative) to those parameters that the results were more sensitive to. As expected, while the corresponding simulations varied as well, the general patterns presented in the following remained the same, especially concerning the overall trends shown by comparing the groups of simulations, providing a model validation.
Results
Stock-Flow Diagram of (+)ssRNA Virus–Host Interaction
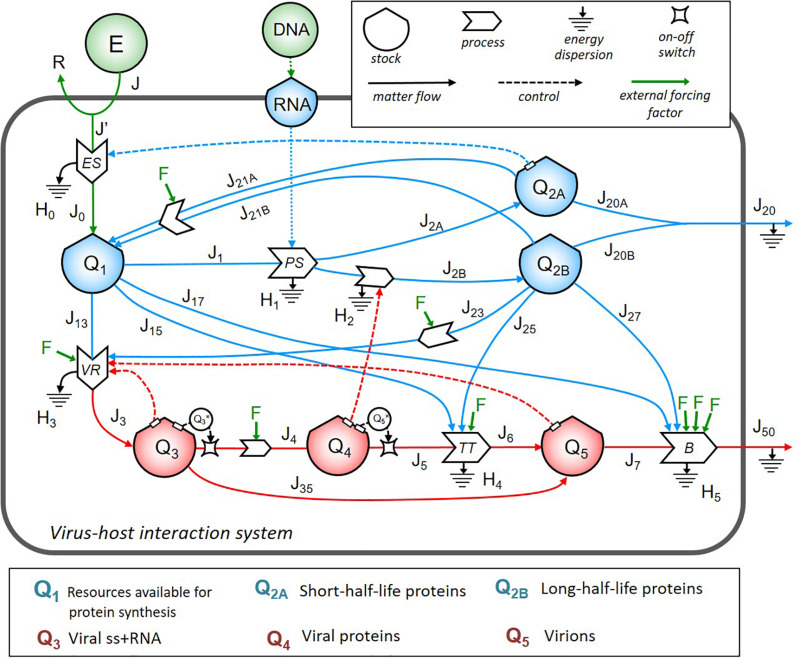
First, we identified the important structures in the system and then used to build up the stock-flow diagram of the virus–host interaction system. In Figure 1, symbols were borrowed from the energy language (Odum and Odum, 2000; Brown, 2004): shields indicate the stocks; big solid arrows indicate the processes; line arrows indicate the flows; dashed lines show the controls exerted by the stocks on the processes.
FIGURE 1.
The energy systemic diagram of a cell infected by ss+ RNA virus. Stock-flow diagram of the virus–host interaction system. In the upper right box are the meaning of symbols. The color code is as follows: blue for host cell energy stocks and relative inflows and outflows; red for virus energy stocks and relative inflows and outflows; green for external energy inputs and external driving forces F corresponding to different therapeutic strategies. The lower box lists the biological contents of the stocks, all expressed in terms of energy (ATP-equivalent units).
The dynamics of energy allocation for protein synthesis contained in the stock Q1 depended on the cell bioenergetics, e.g., the number of mitochondria, OX-PHOS activity levels, and cell cycle phase (Murayama et al., 2008; Canto et al., 2009; Li et al., 2020). In the absence of virus, the stationary configuration was given by energy required to flow from stock Q1 (via J1, J2A, and J2B flows) to stocks Q2A and Q2B to produce, respectively, short- and long-half-life proteins, which could, in turn, be recruited by VR machinery. J20A and J20B, grouped into the flow J20, represented the outflow of folded, fully functional proteins addressed to secretion or surface exposure. Based on basal proteostasis of host cell, recovery of energetic sources from proteins not addressed to leave the system could be possible via several complex processes (e.g., proteasomal degradation, and autophagy), identified by flows of materials J21A and J21B, respectively, from Q2A and Q2B back to Q1.
The viral load in the system, expressed by the stocks Q3 (identified as viral RNA content to be used for viral transcription and translation), Q4 (translated viral proteins content), and Q5 (full assembled virions to shed virus outside), diverted, at the time of infection, resources directly from Q1 (through flows J13, J15, and J17) and Q2B (through flows J23, J25, and J27). Virions shedding was represented by the flows J7 and J50 through the contribution of the host flows J17 and J27. The output flows J4 and J5 were set to be effective only if the value of the respective stock Q3 and Q5 was higher than a threshold, as represented by the two switch symbols in the diagram.
We identified four feedback loops (represented by dot lines in Figure 1): (i) the positive control of Q2A stock on the energy supply process (occurring when more structural host proteins operate to maintain the energetics homeostasis of the host cell); (ii) the positive control of Q3 stock on the VR process (highlighting that the more viral RNA is in the system, the more intensive replication can occur if host sources are available); (iii) the positive control of Q4 stock on the processes of synthesis and maturation of host proteins (highlighting that the more viral proteins are made, the more host proteins are synthesized to be recruited in the virion assembly machinery, increasing J2B); (iv) the positive control of Q5 stock on the VR process (highlighting that the more virions are produced, the more resources are diverted from the host cell to viral replication).
System Dynamics of (+)ssRNA Virus–Host Interaction
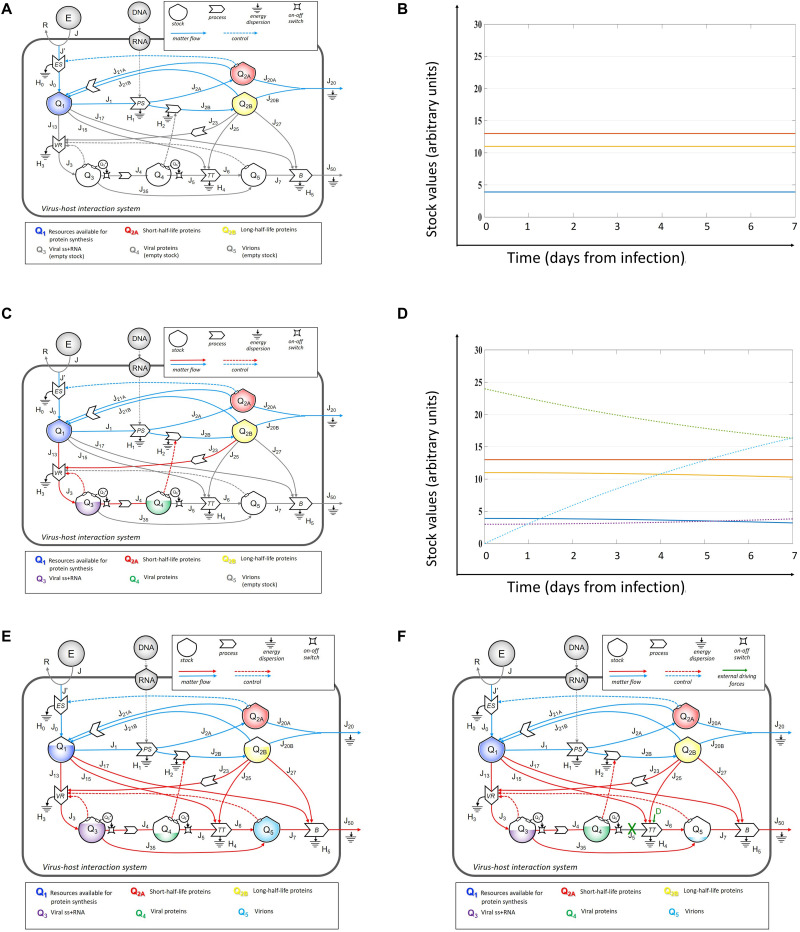
First, a computational model was derived from the stock-flow diagram shown in Figure 1 using the standardized workflow of systemic modeling (Odum and Odum, 2000; Xue et al., 2018). Figure 2 shows two different system self-organized patterns (configurations) to guide reader in the overall comprehension of the proposed approach. The virus–host interaction was represented as an evolving set of simulated trajectories, to which the positive value of Q3 stock had given access, using preexisting stocks, processes, and flows of the host cell, followed overtime by progressive filling of Q4 and Q5 stocks. In (Figure 2A) configuration, the viral load is null (the stocks Q3, Q4, and Q5 are empty), and the values of stocks Q1, Q2A, and Q2B are constant; thus, the system behavior is stationary (Figure 2B). At time of infection, the Q3 stock was fed, and its proteins could interact with the host proteome to sustain RNA replication. Based on previous works in the field (Wei et al., 1995; Adelman et al., 2002; Mohler et al., 2005; Regoes et al., 2005; De Boer et al., 2010), we identified a time delay of 2–6 h required to record changes in the Q5 stock.
FIGURE 2.
Systems configurations based on initial conditions and effects of external driver forces. In the configuration of initial null viral load (A), the value of stocks Q1, Q2A, and Q2B were constant, and the system behavior was stationary (B), with constant values of all stocks overtime. At time of infection, the network of flows and feedbacks identified a new configuration (C), to generate a not stationary pattern (D), in which stock values change overtime in response to the other elements of the system, which can evolve to maximize the virions’ stock (E). Application of external driving forces, acting on processes (identified by red cross on J5), can reduce the flows and address new configurations (F), identifying leverage points that can be explored at different magnitude and timepoints with a computational simulator.
Moreover, the value of Q5 varied over time due to changes that occurred at different timepoints in the stocks Q2B, Q3, and Q4. Thus, the network of flows and feedbacks could identify a new configuration (Figure 2C), to generate a non-stationary behavior (Figure 2D), where the values of Q3, Q4, and Q5 stocks evolved in a non-linear way (Supplementary Figure 1), to maximize the value of virions stock in the configuration (Figure 2E). We define the (Figure 2A) configuration as healthy, the (Figure 2C) configuration as early infection associated to asymptomatic disease, the (Figure 2E) configuration as late infection associated to symptomatic disease, and the (Figure 2F) configuration as symbiotic infection, consequent to any approach derived from the application of external driving forces at any time able to maintain configuration (Figure 2C) without crashing the system. The goal for any curative approach should be to recover the (Figure 2A) configuration when an infection occurs.
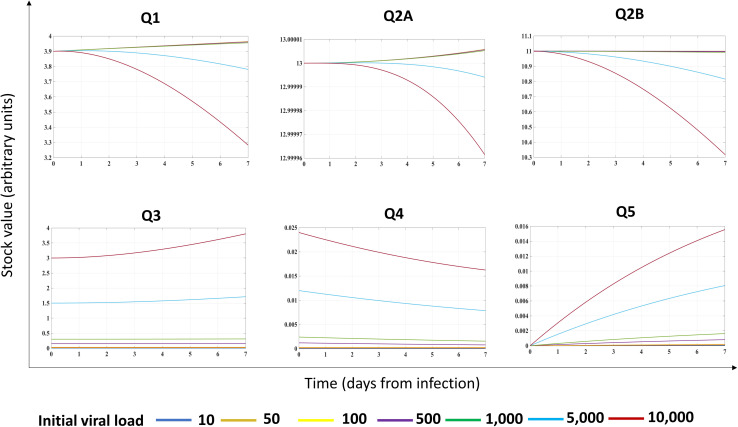
Second, we investigated the system dynamics under different initial conditions, exploring the possible role of different initial viral loads (Figure 3). Assuming different initial viral loads (10–10,000 RNA copies range), we found a threshold (at about 5,000 RNA copies) for triggering the progressive reduction of Q1 (Figure 3). Indeed, for low initial viral load (10–1,000 RNA copies), the system perturbation could be absorbed by the configuration itself (Supplementary Figure 1), without affecting the overtime stock value of Q1 and Q2B but maintaining constant Q3, Q4, and Q5.
FIGURE 3.
Effects of initial viral load on the energy dynamics of a cell infected by a (+)ssRNA virus. Stock values, expressed in ATP-eq (arbitrary units chosen as proxy), are shown from Day 0 though Day 7 as a function of different initial viral loads (indicated at the bottom with different color codes). Evolutionary pattern for each stock was not linear.
The behavior of stock values Q1, Q2A, and Q2B diverged non-linearly at the threshold value, with a progressive decrease, starting at Day 3 from infection (cyan and red lines, respectively). We found a non-linear evolution of the system output (the Q5 stock) depending on the initial conditions: for Q3 stock in the range (10–1,000 RNA copies), the Q5 was linear, whereas for higher initial viral load, the growth of Q5 was linear in the first day and non-linear in the further timeframe, associated to a progressive, unpredictable reduction of Q3–Q4 stocks, reflecting in biological terms the turnover of viral proteins required for virions budding.
The non-linear behavior of the (+)ssRNA virus–host interaction was due to the control of Q4 stock on processes of the host cell, forcing host proteins Q2B to favor the production of virions (to feed the Q5 stock value) through the increased J23, J25, and J27 flows. In biological terms, the progressive Q1 reduction reflects the metabolic rewiring of infected host cells (Chen et al., 2016), with progressive reduction of resources allocated for the maintenance of host processes, requiring a metabolic shift to less efficient but more rapid source of the available energy required for downstream processes (Thaker et al., 2019). The described system dynamics was experimentally validated for influenza virus (Mahmoudabadi et al., 2017). Results confirmed a previous theoretical assumption, showing a Gibbs free energy for virus lower than its host (Popovic and Minceva, 2020a, b), when virus and host cells are evaluated separately and not as a unique system as herein proposed.
The behavior of Q1 following different initial values of Q3 could explain both the variable incubation time in each individual subject and why infections due to (+)ssRNA viruses can occur asymptomatically in most cases. The resilience of virus–host system for a specific range of Q3 amount could, in turn, depend on intrinsic and extrinsic factors. Indeed, in response to manipulation of stocks and flows, and based on timeframe of observation, the system could evolve along different, non-linear trajectories, requiring early intervention upon infection to make the system resilient to growth of viral stocks.
System Dynamics of (+)ssRNA Virus–Host Interaction in Response to External Driving Forces Applied to Reduce Virions Outflow
Currently, the search for a therapeutic strategy is based on single target-related parameters, while we propose to identify systemic targets (i.e., polytarget) to improve host response to host–virus interaction. Starting from the pharmacodynamics of compounds currently under investigation for a typical (+)ssRNA virus (Thorlund et al., 2020), we could reclassify them, based on their systemic mechanisms of action as listed in Table 3. Their effects may be potentially simulated to establish the single-cell effect, the best time, and/or schedule of administration, as shortcut of in vitro studies, with a detail level established on the basis of the purpose of the study design. To this end, we applied the search of systemic leverage points by simulating the dynamics of multiple scenarios, upon the action of a generic external driving force (D), assuming that the minimization of the value of Q5 stock over time should limit the propagation of virions outside the cells.
TABLE 3.
Examples of drugs that could act as external forcing factors on identified systemic flow targets.
| Flow target(s) | Compound | Mechanism of action | References |
| J0, J3, J4, J21A, J21B | FK506 (tacrolimus) | FKBP15 inhibitor. ER protein quality control regulators. Bioenergetics regulators. mRNA translation inhibitor. | Wiederrecht et al., 1993; Gordon et al., 2020b |
| J0, J4, J21A, J21B | Rapamycin | Jefferies et al., 1997; Wang et al., 2014; Gordon et al., 2020b | |
| J1, J3, J21B | Valproic acid | HDAC2 inhibitor | Cong and Bacchetti, 2000; Phiel et al., 2001; Nagesh et al., 2017; Gordon et al., 2020b |
| J1, J4 | SAHA | pan HDAC inhibitors | Saha and Parks, 2019 |
| J1, J35, J2B | Selinexor | mRNA nuclear export complex inhibitor | Bardina et al., 2009; Castelló et al., 2009; Schmidt et al., 2013; Walker et al., 2013; Watters et al., 2017; Gordon et al., 2020b |
| J4, J2A, J2B | Dabrafenib | Kinase inhibitor, protein synthesis inhibitor | Burkard et al., 2015; Kindrachuk et al., 2015; Gordon et al., 2020b |
| J0, J13 | Metformin | Inhibitor of respiratory electron transport, glycolysis regulation | Fontaine et al., 2015; Gordon et al., 2020b |
| J3, J13 | Camostat, nafamostat | TMPRSS inhibitors proteolytic cleavage of viral spike protein priming to the receptor ACE2 present in human cell | Gordon et al., 2020b; Kailas et al., 2020 |
| J4, J2A, J2B | Ponatinib | Kinase inhibitor, protein synthesis inhibitor | Burkard et al., 2015; Gordon et al., 2020b |
| J3 | Ribavirin | Nucleoside inhibitor (mutagenic ribonucleoside) | Patterson and Fernandez-Larsson, 1990; Crotty et al., 2000; Gordon et al., 2020b |
| J13 | Chloramphenicol, tigecycline, linezolid | Antibiotics, able to inhibit mitochondrial ribosomes | Colberg-Poley et al., 2000; Gordon et al., 2020b |
| J21A, J21B | Chloroquine | SIGMAR1/SIGMAR2 inhibitor. Autophagy inhibitor. | Keyaerts et al., 2004; Vincent et al., 2005; Saeed et al., 2011; Cottam et al., 2014; Borba et al., 2020; Gordon et al., 2020b |
| J3, J5, J21A, J21B | Hydroxychloroquine | Autophagy inhibitor, antiviral effect | Cottam et al., 2014; Gautret et al., 2020; Gordon et al., 2020b; Liu et al., 2020 |
| J1, J4, J5 | Zotatifin, ternatin 4, tomvosertib | mRNA translation inhibitors | Gordon et al., 2020b |
| J1, J2B, J23 | Silmitasertib or TMCB | Casein kinase II inhibitors, protein synthesis inhibitor | Siddiqui-Jain et al., 2010; Reineke et al., 2017; Gordon et al., 2020b |
| J3 | Remdesivir | Nucleoside analog, interferes with RNA-dependent RNA polymerase | Sheahan et al., 2017 |
| J13 | Umifenovir | Inhibitor of the fusion between the viral envelope (surrounding the viral capsid) and the cell membrane of the target cell | Shi et al., 2007; Pécheur et al., 2016 |
| J13 | Lisinopril, losartan | ACE inhibitors, prevent the fusion between the viral envelope and the cell membrane of the target cell | Hoffmann et al., 2020 |
| J3 | Favipiravir | Selective inhibitor of RNA-dependent RNA polymerase | Cai et al., 2020; Shiraki and Daikoku, 2020 |
| J2A, J2B, J3*, J4, J7*, J21 | Macrolide antibiotics azithromycin*, clarithromycin | Inhibition of ribosomal translation Autophagy inhibition | Tenson et al., 2003; Stamatiou et al., 2009; Gielen et al., 2010; Petroni et al., 2020 |
∗Close to Js, refers to azithromycin, in the other colum, being the compount impactinng on the identified flows.
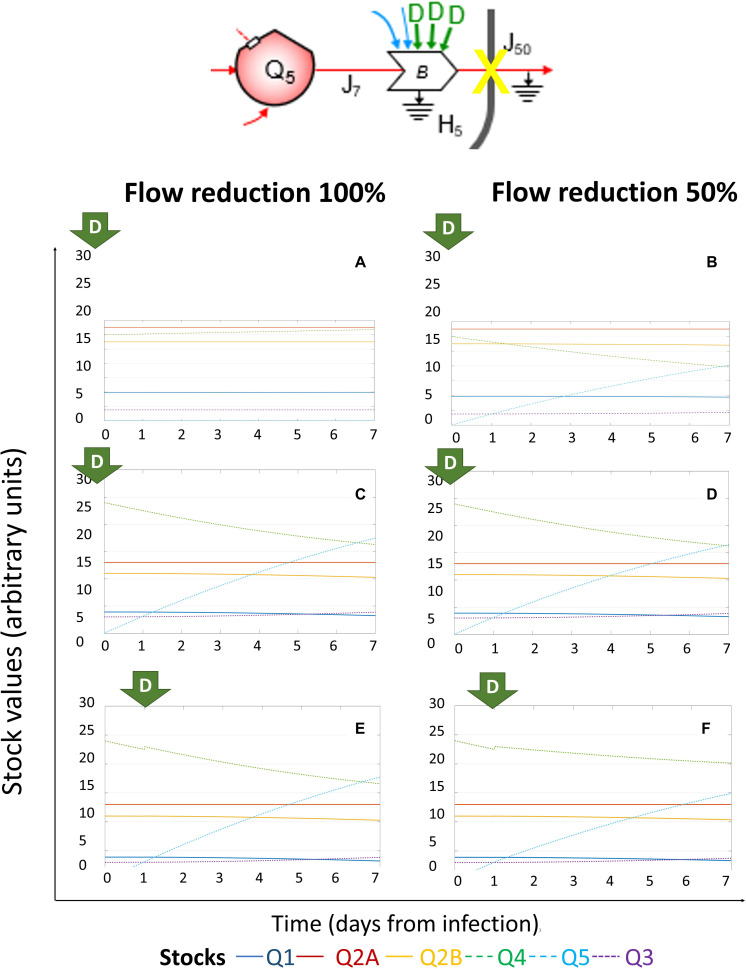
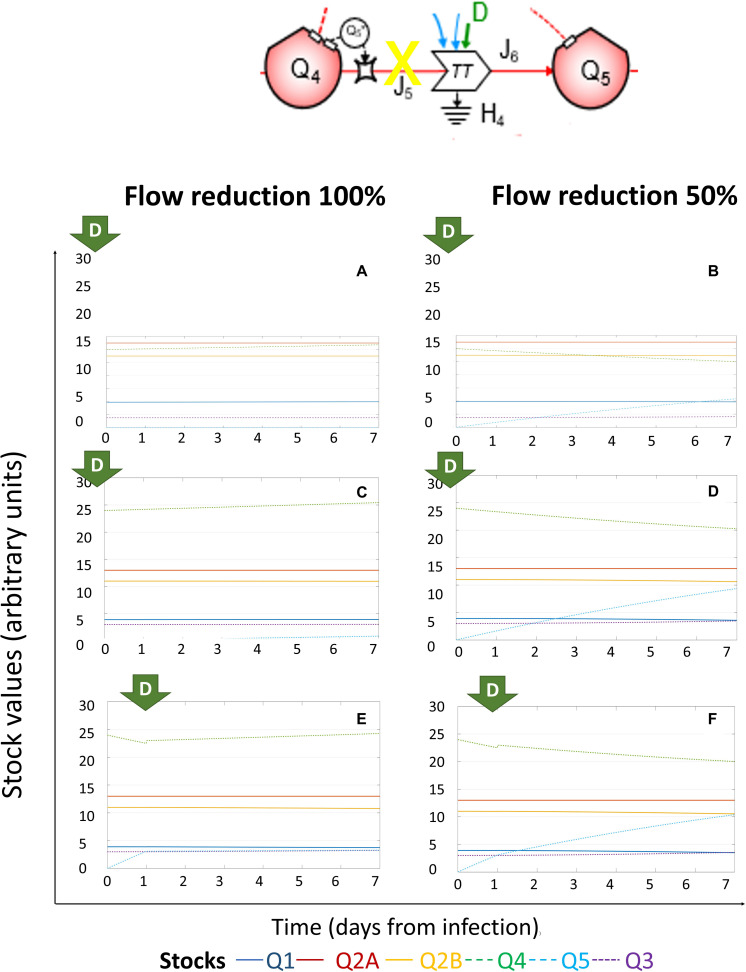
First, we explored the system configurations upon reduction of the outflows from the Q5 virions stock, via manipulation of J7 and/or J50. However, minimization of J7 was counter-effective, due to the increase of Q5 as consequence of the feedback action in the VR process (data not shown). The effects of full (100%) or partial (50%) reduction of J50 (flow of energy required for virions budding) could prevent the outflow from Q5 without stopping its growth, diverting resources from Q1 and Q2B to Q5, so further supporting viral hijacking of cellular metabolism and impairing host cell homeostasis. As shown in Figure 4, the effect of driving forces acting to modulate J50 was different based on application time, Day 0 (Figures 4A–D) versus Day 1 (Figures 4E,F), and initial viral load, as the early abrogation of J50 when the amount of Q3 was 5,000 RNA copies could restore the stationary status (Figure 4A), while halving J50 maintained homeostasis for host-cell stock, but could not prevent the growth of Q5 (Figure 4B). At increasing initial viral load, reduction of J50 applied at Day 0 (Figures 4C,D) or at Day 1 (Figures 4E,F) could not prevent the growth of Q5 and the progressive decrease of Q1 and Q2B. This systemic dynamics can explain the relationship between the time of initiation of neuraminidase inhibitors and their efficacy (Moscona, 2005), as treatment starting within the first 12 h after the onset of fever shortened the illness by more than 3 days, as compared with treatment starting at 48 h (Aoki et al., 2003).
FIGURE 4.
System dynamics of (+)ssRNA virus–host interaction in response to external driving forces applied to reduce virions outflow. Changes over time of the values of each stock of the system diagrammed in Figure 1 (for the color code, see bottom), expressed in ATP-eq, in response to reduction of J50 (flow of energy required for virions budding). Several scenarios are shown: initial viral load 5k and application of full (100%, A) or partial (50%, B) J50 reduction at Day 0; initial viral load 10k and application of full (100%, C) or partial (50%, D) J50 reduction at Day 0; initial viral load 10k and application of full (100%, E) or partial (50%, F) J50 reduction at Day 1.
System Dynamics of (+)ssRNA Virus–Host Interaction in Response to External Driving Forces Applied to Reduce Virions Assembly
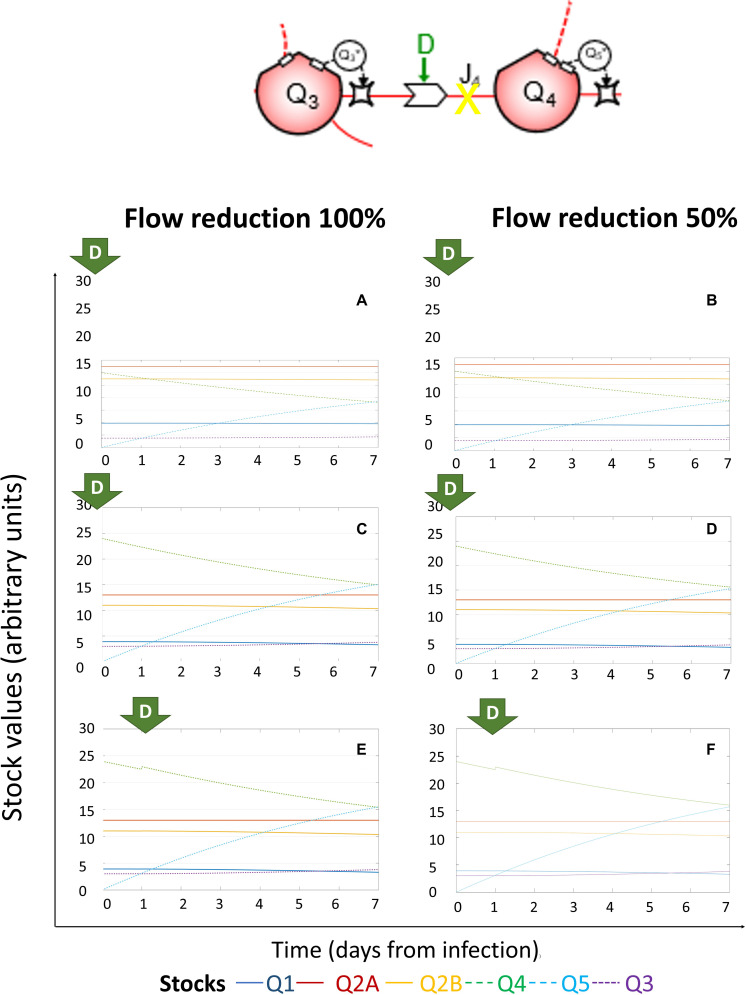
Second, we explored the systemic response to full (100%) or partial (50%) reduction of either J3 (flow of energy required for RNA replication), or J4 (flow of energy required for viral RNA translation and viral protein synthesis), or J5 (flow of energy required for virions assembly), which are involved in virions assembly and are typically dependent on intrinsic viral biological properties.
Full reduction of J5 at Day 0 could recover the systems dynamics in a stationary status, in all scenarios of initial viral load tested (5,000 RNA copies, Figure 5A; 10,000 RNA copies, Figure 5C), with constant values for all stocks. A partial reduction of J5 at Day 0 maintained a stationary status only for lower initial viral load (5,000 RNA copies, Figure 5B) associated to a reduced amount of stocks Q4 and Q5 (Figure 5D). The abrogation of J5 at Day 0 was still effective to preserve the stationary status (Figure 5E), but halving J5 at Day 1 could reduce but not prevent the growth of Q5 (Figure 5F). We also simulated the effect of applying the same external inputs at different times: after 1 (Supplementary Figure 2), 3 (Supplementary Figure 3), or 5 days (Supplementary Figure 4) from the initial infection, confirming the role of early application of external driving forces to recover the stationary status of the system.
FIGURE 5.
System dynamics of (+)ssRNA virus–host interaction in response to external driving forces applied to reduce virions assembly. Changes over time of the values of each stock of the system diagrammed in Figure 1 (for the color code, see bottom), expressed in ATP-eq, in response to reduction of J5 (flow of energy required for virions assembly). Several scenarios are shown: initial viral load 5k and application of full (100%, A) or partial (50%, B) J5 reduction at Day 0; initial viral load 10k and application of full (100%, C) or partial (50%, D) J5 reduction at Day 0; initial viral load 10k and application of full (100%, E) or partial (50%, F) J5 reduction at Day 1.
The importance of full abrogation of J5 flow for a (+)ssRNA virus–host interaction has been indirectly confirmed by the data recently published by Gordon et al., who cloned, tagged, and expressed 26 of 29 severe acute respiratory syndrome (SARS)–CoV-2 proteins individually in HEK293T cells and used mass spectrometry to measure protein–protein interactions (Gordon et al., 2020b), to identify 69 existing drugs, known to target host proteins or associated pathways, which interact with SARS-CoV-2, addressing the importance to target the host–virus interaction at the level of RNA translation.
System Dynamics of (+)ssRNA Virus–Host Interaction in Response to External Driving Forces Applied to Reduce Viral Protein Synthesis
Full (100%) or partial (50%) reduction of J4 (flow of energy required for viral RNA translation and viral protein synthesis) at Day 0 did not affect the dynamics of the system (Figure 6). In particular, for lower initial viral load (5,000 RNA copies, Figures 6A,B), the Q5 stock was always lower than Q2A, thus not affecting Q1, which remained constant. However, when Q5 stock was greater than Q2A, Q1 started to decrease, again suggesting that the effect of viral infection on host metabolism is associated to threshold values specific for each system, and not for each cell type. The observation that—based on initial viral load—the higher value of Q5 is lower than the stable quantity of Q2A and Q2B for 5k ATP-eq and higher than Q2A and Q2B for 10k ATP-eq can explain the contribution of initial viral load and configuration of host-cell stocks in the viral fitness, which depends on host cell cycle stage, addressed as initial value of Q2A and K2A (Wargo and Kurath, 2012).
FIGURE 6.
System dynamics of (+)ssRNA virus–host interaction in response to external driving forces applied to reduce viral protein synthesis. Changes over time of the values of each stock of the system diagrammed in Figure 1 (for the color code, see bottom), expressed in ATP-eq, in response to reduction of J4 (flow of energy required for viral RNA translation and viral protein synthesis). Several scenarios are shown: initial viral load 5k and application of full (100%, A) or partial (50%, B) J4 reduction at Day 0; initial viral load 10k and application of full (100%, C) or partial (50%, D) J4 reduction at Day 0; initial viral load 10k and application of full (100%, E) or partial (50%, F) J4 reduction at Day 1.
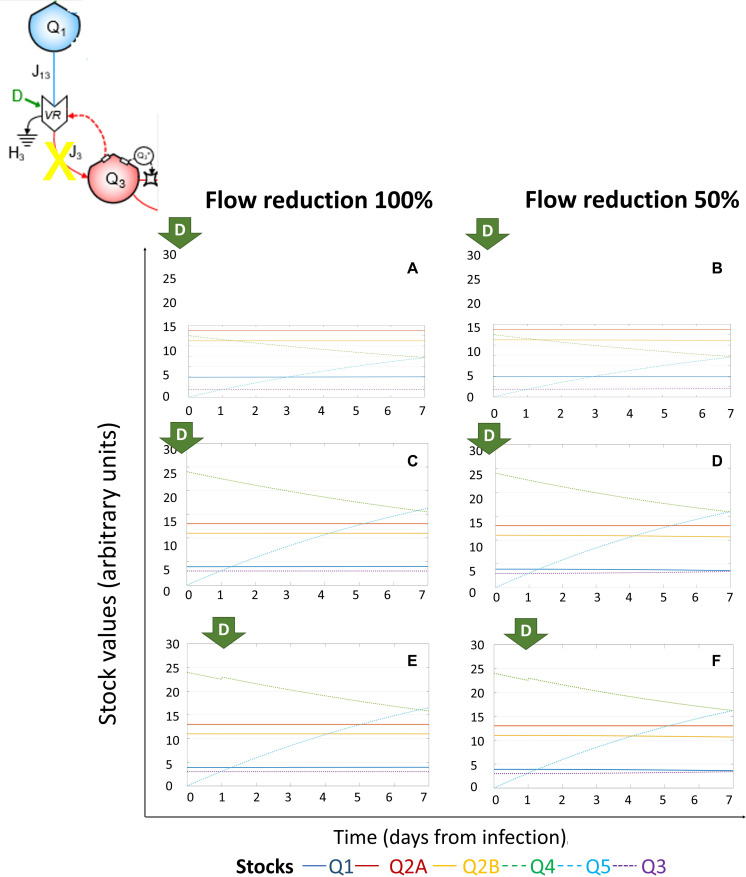
System Dynamics of (+)ssRNA Virus–Host Interaction in Response to External Driving Forces Applied to Reduce Viral RNA Replication
Full (100%) or partial (50%) reduction of J3 (flow of energy required for RNA replication) at Day 0 did not affect the dynamics of the system (Figure 7), but—differently from the previous scenario—Q1 remained constant even if Q5 was lower than the stable quantity of Q2A and Q2B, even if administered later after infection (on Day 3 or 5, as shown in Supplementary Figures 2–4). This suggests that the host cell can preserve its homeostasis upon early exposure to inhibitors of viral replication. Interestingly, the pattern in response to single external driving forces was maintained over time, with the values of each stock just shifted, based on the time of application.
FIGURE 7.
System dynamics of (+)ssRNA virus–host interaction in response to external driving forces applied to reduce viral RNA replication. Changes over time of the values of each stock of the system diagrammed in Figure 1 (for the color code, see bottom), expressed in ATP-eq, in response to reduction of J3 (flow of energy required for RNA replication). Several scenarios are shown: initial viral load 5k and application of full (100%, A) or partial (50%, B) J3 reduction at Day 0; initial viral load 10k and application of full (100%, C) or partial (50%, D) J3 reduction at Day 0; initial viral load 10k and application of full (100%, E) or partial (50%, F) J3 reduction at Day 1.
Several strategies can be used to reduce selectively virus RNA, including nucleoside analogs, which are metabolized intracellularly into their active ribonucleoside 5′-triphosphate forms and incorporated into the nascent viral RNA by error-prone viral RNA-dependent RNA polymerase (RdRps), to disrupt RNA synthesis directly via chain termination, or accumulation of deleterious mutations in the viral genome. The response to nucleoside analogs could be better tested by our computational model, adding the mutations rate and the DNA/RNA metabolism of host cell, not included in the diagram for lack of experimental data about the turnover of RNA stock, specific for each host cell type of interest.
System Configurations Dynamics in Response to Multiple External Driving Forces Applied at Different Timepoints From Infection
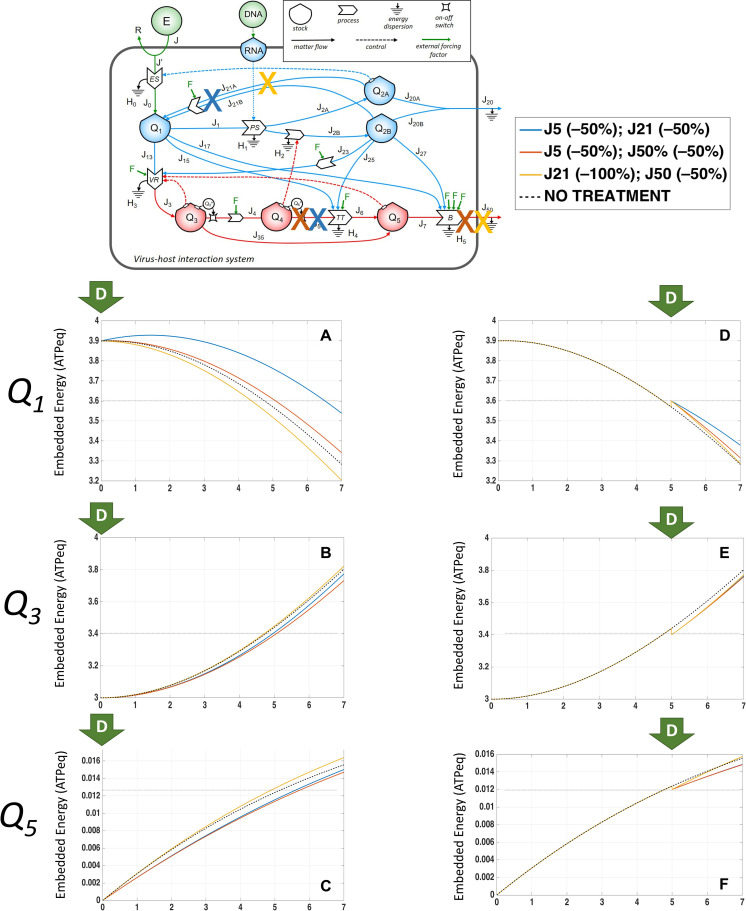
Third, we next explored the application of multiple external forcing factors, addressing the requirement of a polytarget approach (Bizzarri et al., 2020). The positive effects on Q1, Q3, and Q5 arising from targeting J5 was mitigated by the combination with reduction of J50 or J21, since the combination targeting J5 and J21 (indicated by the blue line in Figure 8) applied at Day 0 could preserve the Q1 amount better than no treatment (Figure 8A) associated to slower increase of the growth of Q3 (Figure 8B) and Q5 (Figure 8C), leading us to explore further combinations. Application of external driving forces acting on J50 and J21 (yellow line in Figure 8) increased the Q5 stock value instead of the expected decrease (Figure 8C); thus, action on a single flow was more efficient than on two of them, given the emergence of compensatory feedbacks and flows. These observations are supported by the controversial findings about the efficacy of macrolides, chloroquine, and their derivatives in the recent COVID-19 pandemic, as their systemic effects include reduction of J5 and J21 (Table 3), and could be affected by the viral load and time of application, with weak changes in virions’ spread if applied later in the clinical course of disease as shown by preliminary results of randomized trials (Derwand et al., 2020; Kashour et al., 2020; Magagnoli et al., 2020; Fiolet et al., 2021).
FIGURE 8.
System configurations dynamics in response to multiple external driving forces applied at different timepoints from infection. Changes over time of Q1 stocks, expressed in ATP-eq, in response to partial (50%) reduction of J5 and J21 (blue line), J5 and J50 (orange line), J21 and J50 (yellow line), or no application of external driving forces applied at Day 0 (A) or at Day 5 (D) of infection. Changes over time of Q3 stocks, expressed in ATP-eq, in response to partial (50%) reduction of J5 and J21 (blue line), J5 and J50 (orange line), J21 and J50 (yellow line), or no application of external driving forces applied at Day 0 (B) or at Day 5 (E) of infection. Changes over time of Q5 stocks, expressed in ATP-eq, in response to partial (50%) reduction of J5 and J21 (blue line), J5 and J50 (orange line), J21 and J50 (yellow line), or no application of external driving forces applied at Day 0 (C) or at Day 5 (F) of infection.
When the combination targeting J5 and J50 was applied later (at Day 5 from infection, orange line in Figure 8), Q1 was higher than untreated (Figure 8D), associated to slower growth of Q3 (Figure 8E) and Q5 (Figure 8F), but not leading to restore of stationarity. This behavior is a typical systemic feature, where an intervention on a specific local process may lead to counterintuitive rearrangements in the system dynamics. Thus, once the main flow to reduce was found to identify the n-ple stock associated with the desired system output, molecular insights should suggest the biological process to assess a tailored treatment.
Discussion
In this work, we approached the host–virus interaction dynamics as a systemic problem, and for the first time in the field, we used combined ST tools as a conceptual framework to build up a systemic description of the viral action and host response, critically depending on the existing metabolic environment.
The basic ST idea is to integrate the traditional bottom-up approach—which describes “local” behaviors through cause–effect chains and functional units—with a top-down approach, which points out at the global behavior of the system in terms of its operational configurations, emerging from the feedback-driven response to different driving forces, like those represented, for example, by the chemistry of new drugs (Odum, 1996). The utility of computational simulations stems from their capacity to identify structural side effects, non-linearities, and time delays, which are left unexplained by other approaches. Systems biology already recognized the relevance of complexity in the study of microbiological systems (Kaneko, 2006; Loscalzo and Barabasi, 2011), but although successfully applied in several fields ranging from hard sciences to ecology and economics (Brown, 2004; Brown and Ulgiati, 2004), the potential of ST in the study of biological systems is still underexploited. Thanks to its abstract nature, stock-flow description can be used in a wide range of different fields, realizing the conceptual bridge that connects the language of biological systems to that of ecology.
In targeting the virus–host interaction, there is an emerging need of tools that could early identify those compounds, not primarily designed for their antiviral action, identifiable by in silico approaches (Gordon et al., 2020b), which alone or in combination can provide clinical efficacy (Mina et al., 2016; Cheng et al., 2018; Bogdanow et al., 2019; Panja et al., 2019; Gordon et al., 2020b). There are known advantages of in silico modeling of the action of therapeutic agents on known diseases through agent-based modeling (Mao et al., 2018). However, the literature evidenced some intrinsic limitations on the choice of parameters, such as the size of investigated populations (Mina et al., 2016), while major problems are related to model validation (Mina et al., 2016; Donkin et al., 2017), also requiring to supplement the models with adequate formal ontologies (Kalfoglou and Schorlemmer, 2003; Gotts et al., 2019).
The proposed model was developed at the single-cell scale. However, in order to define an overall therapeutic approach, the integration with a multiscale approach would be also desirable. In particular, depending on the availability of appropriate data, a future model could focus on different scales, with a more detailed description of some components at the subcellular level, which were grouped (e.g., short- and long-half-life proteins, lipids, and vesicles trafficking) in the present study. On the other hand, the interaction between different cell populations in the host could be also developed, to represent the interaction between healthy and infected cells, and the contribution of immune system (Tan et al., 2007; Katze et al., 2008; Wu et al., 2020) or the repertoire of receptors on the surface of the host cell (Gordon et al., 2020a; Hoffmann et al., 2020; Kailas et al., 2020) to surveil and limit the size of Q3 stock at the single-cell level. Other natural system constraints could be also included, like some physiological parameters (e.g., temperature and metabolic rate), whose impact on the human body energy dynamics is already understood. The use of a multiscale hierarchical perspective is in principle already possible, as discussed in previous works adopting the same sort of system representation (Ulgiati and Brown, 2009; Ulgiati et al., 2011).
Conclusion
This work highlights the advantages of applying an ST-based approach to the study of virus–host interaction, being reflected in the possibility of extracting systemic dynamic features that would be otherwise counterintuitive. While a traditional single-target approach would address strategies targeting the viral RNA (Q3) or the replication process (J3), our results suggest that the virus growth is more vulnerable if the process of virion growth before expulsion (process TT, involving flow J5) is targeted.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
AR designed the study and collected medical knowledge. FG built up the diagram. MC built up the simulator. All authors performed simulation, analyzed data, prepared the figures, and wrote the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Luca Naso, Francesco Di Raimondo, and Maria Sanfilippo from SIES, Società Italiana di Ematologia Sperimentale for the support provided.
Funding. This work was supported by SIES, Società Italiana di Ematologia Sperimentale.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.600254/full#supplementary-material
System dynamics of (+)ssRNA virus–host interaction in response to initial viral load. Changes overtime of the values of each stock of the system diagrammed in Figure 1 (for the color code see at the bottom), expressed in ATP-eq: in absence of infection the system status was stationary (A). Upon infection initial values of Q3 stock identify the response of system, at 50 (B), 100 (C), 1,000 (D), 5,000 (E) and 10,000 (F) RNA copies, expressed in ATP-eq. For Q3 stock in the range (10–1,000 RNA copies) the trajectory of Q5 evolution was linear, while for higher initial viral load the growth of Q5 was linear in the first day and non-linear in the further timeframe.
Effects of targeting leverage point applying external driving force (D) at Day 1. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 1, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
Effects of targeting leverage point applying external driving force (D) at Day 3. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 3, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
Effects of targeting leverage point applying external driving force (D) at Day 5. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 5, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
References
- Acevedo A., Brodsky L., Andino R. (2014). Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505 686–690. 10.1038/nature12861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K., La Porta A., Santangelo T. I. J. I., Lis J. T., Roberts J. W., Wang M. D. (2002). Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc. Natl. Acad. Sci. U S A 99 13538–13543. 10.1073/pnas.212358999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist P., Noueiry A. O., Lee W.-M., Kushner D. B., Dye B. T. (2003). Host Factors in Positive-Strand RNA Virus Genome Replication. J. Virol. 77 8181–8186. 10.1128/jvi.77.15.8181-8186.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki F. Y., Macleod M. D., Paggiaro P., Carewicz O., El Sawy A., Wat C., et al. (2003). Early administration of oral oseltamivir increases the benefits of influenza treatment. J. Antimicrob. Chemother. 51 123–129. 10.1093/jac/dkg007 [DOI] [PubMed] [Google Scholar]
- Apweiler R., Beissbarth T., Berthold M. R., Blüthgen N., Burmeister Y., Dammann O., et al. (2018). Whither systems medicine? Exp. Mole. Med. 50 e453–e453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P., Beauchemin C., Macken C. A., Hayden F. G., Perelson A. S. (2006). Kinetics of influenza A virus infection in humans. J. Virol. 80 7590–7599. 10.1128/jvi.01623-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardina M. V., Lidsky P. V., Sheval E. V., Fominykh K. V., Van Kuppeveld F. J. M., Polyakov V. Y., et al. (2009). Mengovirus-Induced Rearrangement of the Nuclear Pore Complex: Hijacking Cellular Phosphorylation Machinery. J. Virol. 83 3150–3161. 10.1128/jvi.01456-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beachboard D. C., Horner S. M. (2016). Innate immune evasion strategies of DNA and RNA viruses. Curr. Opin. Microbiol. 32 113–119. 10.1016/j.mib.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Tabou De-Leon S., Davidson E. H. (2009). Modeling the dynamics of transcriptional gene regulatory networks for animal development. Dev. Biol. 325 317–328. 10.1016/j.ydbio.2008.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzarri M., Minini M., Monti N. (2020). “Revisiting the Concept of Human Disease,” in Approaching Complex Diseases, ed. Bizzarri M. (Berlin: Springer Nature; ), 1–34. 10.1007/978-3-030-32857-3_1 [DOI] [Google Scholar]
- Bogdanow B., Wang X., Eichelbaum K., Sadewasser A., Husic I., Paki K., et al. (2019). The dynamic proteome of influenza A virus infection identifies M segment splicing as a host range determinant. Nat. Commun. 10:5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F. M., Ahmad Y., Gierlinski M., Charriere F., Lamont D., Scott M., et al. (2012). A quantitative spatial proteomics analysis of proteome turnover in human cells. Mol. Cell Proteomics 11:M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borba M. G. S., Val F. F. A., Sampaio V. S., Alexandre M. A. A., Melo G. C., Brito M., et al. (2020). Effect of High vs Low Doses of Chloroquine Diphosphate as Adjunctive Therapy for Patients Hospitalized With Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Infection: A Randomized Clinical Trial. JAMA Netw. Open 3:e208857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl K., Ianevski A., Than T. T., Andersen P. I., Kuivanen S., Teppor M., et al. (2019). Common Nodes of Virus-Host Interaction Revealed Through an Integrated Network Analysis. Front. Immunol. 10:2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossel H. (2007). Systems and models: complexity, dynamics, evolution, sustainability. Norderstedt: Books on Demand GmbH. [Google Scholar]
- Brown M. T. (2004). A picture is worth a thousand words: energy systems language and simulation. Ecol. Model. 178 83–100. 10.1016/j.ecolmodel.2003.12.008 [DOI] [Google Scholar]
- Brown M. T., Ulgiati S. (2004). Energy quality, emergy, and transformity: H.T. Odum’s contributions to quantifying and understanding systems. Ecol. Model. 178 201–213. 10.1016/j.ecolmodel.2004.03.002 [DOI] [Google Scholar]
- Burch C. L., Chao L. (2000). Evolvability of an RNA virus is determined by its mutational neighbourhood. Nature 406 625–628. 10.1038/35020564 [DOI] [PubMed] [Google Scholar]
- Burkard C., Verheije M. H., Haagmans B. L., Van Kuppeveld F. J., Rottier P. J., Bosch B. J., et al. (2015). ATP1A1-mediated Src signaling inhibits coronavirus entry into host cells. J. Virol. 89 4434–4448. 10.1128/jvi.03274-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F., Brand M. D. (1995). A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J. 312(Pt 1), 163–167. 10.1042/bj3120163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. (2020). Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study. Engineering 6 1192–1198. 10.1016/j.eng.2020.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C., Gerhart-Hines Z., Feige J. N., Lagouge M., Noriega L., Milne J. C., et al. (2009). AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458 1056–1060. 10.1038/nature07813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló A., Izquierdo J. M., Welnowska E., Carrasco L. (2009). RNA nuclear export is blocked by poliovirus 2A protease and is concomitant with nucleoporin cleavage. J. Cell Sci. 122 3799–3809. 10.1242/jcs.055988 [DOI] [PubMed] [Google Scholar]
- Chakravorty S., Yan B., Wang C., Wang L., Quaid J. T., Lin C. F., et al. (2019). Integrated Pan-Cancer Map of EBV-Associated Neoplasms Reveals Functional Host-Virus Interactions. Cancer Res. 79 6010–6023. 10.1158/0008-5472.can-19-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I. T., Lee D. Y., Huang Y. T., Kou G. H., Wang H. C., Chang G. D., et al. (2016). Six Hours after Infection, the Metabolic Changes Induced by WSSV Neutralize the Host’s Oxidative Stress Defenses. Sci. Rep. 6:27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Desai R. J., Handy D. E., Wang R., Schneeweiss S., Barabasi A. L., et al. (2018). Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 9:2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colberg-Poley A. M., Patel M. B., Erezo D. P., Slater J. E. (2000). Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 81 1779–1789. 10.1099/0022-1317-81-7-1779 [DOI] [PubMed] [Google Scholar]
- Cong Y. S., Bacchetti S. (2000). Histone deacetylation is involved in the transcriptional repression of hTERT in normal human cells. J. Biol. Chem. 275 35665–35668. 10.1074/jbc.c000637200 [DOI] [PubMed] [Google Scholar]
- Cottam E. M., Whelband M. C., Wileman T. (2014). Coronavirus NSP6 restricts autophagosome expansion. Autophagy 10 1426–1441. 10.4161/auto.29309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E., Ranadheera C., Cheng D., Goncalves J., Dyakov B. J. A., Laurent E. M. N., et al. (2018). Global Interactomics Uncovers Extensive Organellar Targeting by Zika Virus. Mol. Cell Proteom. 17 2242–2255. 10.1074/mcp.tir118.000800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S., Maag D., Arnold J. J., Zhong W., Lau J. Y. N., Hong Z., et al. (2000). The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6 1375–1379. 10.1038/82191 [DOI] [PubMed] [Google Scholar]
- Dapat C., Oshitani H. (2016). Novel insights into human respiratory syncytial virus-host factor interactions through integrated proteomics and transcriptomics analysis. Exp. Rev. Anti. Infect. Ther. 14 285–297. 10.1586/14787210.2016.1141676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer R. J., Ribeiro R. M., Perelson A. S. (2010). Current estimates for HIV-1 production imply rapid viral clearance in lymphoid tissues. PLoS Comput. Biol. 6:e1000906. 10.1371/journal.pcbi.1000906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Boon J. A., Diaz A., Ahlquist P. (2010). Cytoplasmic viral replication complexes. Cell Host Microb. 8 77–85. 10.1016/j.chom.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derwand R., Scholz M., Zelenko V. (2020). COVID-19 outpatients: early risk-stratified treatment with zinc plus low-dose hydroxychloroquine and azithromycin: a retrospective case series study. Int. J. Antimicrob. Agents 56:106214. 10.1016/j.ijantimicag.2020.106214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold S. S., Montoya M., Unger H., Alexopoulou L., Roy P., Haswell L. E., et al. (2003). Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424 324–328. 10.1038/nature01783 [DOI] [PubMed] [Google Scholar]
- Dinesh D. C., Tamilarasan S., Rajaram K., Boura E. (2020). Antiviral Drug Targets of Single-Stranded RNA Viruses Causing Chronic Human Diseases. Curr. Drug Targets 21 105–124. 10.2174/1389450119666190920153247 [DOI] [PubMed] [Google Scholar]
- Doherty M. K., Hammond D. E., Clague M. J., Gaskell S. J., Beynon R. J. (2009). Turnover of the human proteome: determination of protein intracellular stability by dynamic SILAC. J. Proteome. Res. 8 104–112. 10.1021/pr800641v [DOI] [PubMed] [Google Scholar]
- Dong S., Liu L., Wu W., Armstrong S. D., Xia D., Nan H., et al. (2016). Determination of the interactome of non-structural protein12 from highly pathogenic porcine reproductive and respiratory syndrome virus with host cellular proteins using high throughput proteomics and identification of HSP70 as a cellular factor for virus replication. J. Proteomic. 146 58–69. 10.1016/j.jprot.2016.06.019 [DOI] [PubMed] [Google Scholar]
- Donkin E., Dennis P., Ustalakov A., Warren J., Clare A. (2017). Replicating complex agent based models, a formidable task. Environ. Model. Soft. 92 142–151. 10.1016/j.envsoft.2017.01.020 [DOI] [Google Scholar]
- Eden E., Geva-Zatorsky N., Issaeva I., Cohen A., Dekel E., Danon T., et al. (2011). Proteome half-life dynamics in living human cells. Science 331 764–768. 10.1126/science.1199784 [DOI] [PubMed] [Google Scholar]
- Emmott E., Munday D., Bickerton E., Britton P., Rodgers M. A., Whitehouse A., et al. (2013). The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 87 9486–9500. 10.1128/jvi.00321-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning L. J., Levis J., Kenny-Walsh E., Whelton M., O’sullivan K., Shanahan F. (2001). HLA class II genes determine the natural variance of hepatitis C viral load. Hepatology 33 224–230. 10.1053/jhep.2001.20642 [DOI] [PubMed] [Google Scholar]
- Fiolet T., Guihur A., Rebeaud M. E., Mulot M., Peiffer-Smadja N., Mahamat-Saleh Y., et al. (2021). Effect of hydroxychloroquine with or without azithromycin on the mortality of coronavirus disease 2019 (COVID-19) patients: a systematic review and meta-analysis. Clin. Microbiol. Infect. 27 19–27. 10.1016/j.cmi.2020.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine K. A., Sanchez E. L., Camarda R., Lagunoff M. (2015). Dengue virus induces and requires glycolysis for optimal replication. J. Virol. 89 2358–2366. 10.1128/jvi.02309-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel C. C., Haas J. (2011). Virus-host interactomes and global models of virus-infected cells. Trends Microbiol. 19 501–508. 10.1016/j.tim.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Gao Z., Hu J., Liang Y., Yang Q., Yan K., Liu D., et al. (2017). Generation and Comprehensive Analysis of Host Cell Interactome of the PA Protein of the Highly Pathogenic H5N1 Avian Influenza Virus in Mammalian Cells. Front. Microbiol. 8:739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Dorival I., Wu W., Dowall S., Armstrong S., Touzelet O., Wastling J., et al. (2014). Elucidation of the Ebola virus VP24 cellular interactome and disruption of virus biology through targeted inhibition of host-cell protein function. J. Proteome Res. 13 5120–5135. 10.1021/pr500556d [DOI] [PubMed] [Google Scholar]
- Garcia-Moreno M., Noerenberg M., Ni S., Jarvelin A. I., Gonzalez-Almela E., Lenz C. E., et al. (2019). System-wide Profiling of RNA-Binding Proteins Uncovers Key Regulators of Virus Infection. Mol. Cell 74 196.e–211.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautret P., Lagier J. C., Parola P., Hoang V. T., Meddeb L., Mailhe M., et al. (2020). Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 56:105949. 10.1016/j.ijantimicag.2020.105949 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gielen V., Johnston S. L., Edwards M. R. (2010). Azithromycin induces anti-viral responses in bronchial epithelial cells. Eur. Respir. J. 36 646–654. 10.1183/09031936.00095809 [DOI] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020a). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. E., Jang G. M., Bouhaddou M., Xu J., Obernier K., White K. M., et al. (2020b). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotts N. M., Van Voorn G. A. K., Polhill J. G., Jong E. D., Edmonds B., Hofstede G. J., et al. (2019). Agent-based modelling of socio-ecological systems: Models, projects and ontologies. Ecol. Compl. 40:100728 10.1016/j.ecocom.2018.07.007 [DOI] [Google Scholar]
- Grüne-Yanoff T., Weirich P. (2010). The Philosophy and Epistemology of Simulation: A Review. Simulation Gaming 41 20–50. 10.1177/1046878109353470 [DOI] [Google Scholar]
- Hafirassou M. L., Meertens L., Umana-Diaz C., Labeau A., Dejarnac O., Bonnet-Madin L., et al. (2017). A Global Interactome Map of the Dengue Virus NS1 Identifies Virus Restriction and Dependency Host Factors. Cell Rep. 21 3900–3913. 10.1016/j.celrep.2017.11.094 [DOI] [PubMed] [Google Scholar]
- Hassmiller Lich K., Urban J. B., Frerichs L., Dave G. (2017). Extending systems thinking in planning and evaluation using group concept mapping and system dynamics to tackle complex problems. Eval. Prog. Plann 60 254–264. 10.1016/j.evalprogplan.2016.10.008 [DOI] [PubMed] [Google Scholar]
- Hekimoğlu M., Barlas Y. (2016). Sensitivity analysis for models with multiple behavior modes: a method based on behavior pattern measures. Syst. Dyn. Rev. 32 332–362. 10.1002/sdr.1568 [DOI] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., et al. (2020). SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 181 271.e–280.e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan R. J., Gao G., Rowe T., Bell P., Flieder D., Paragas J., et al. (2004). Resolution of primary severe acute respiratory syndrome-associated coronavirus infection requires Stat1. J. Virol. 78 11416–11421. 10.1128/jvi.78.20.11416-11421.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard-Ashby M., Materna S. C., Brown C. T., Chen L., Cameron R. A., Davidson E. H. (2006). Identification and characterization of homeobox transcription factor genes in Strongylocentrotus purpuratus, and their expression in embryonic development. Dev. Biol. 300 74–89. 10.1016/j.ydbio.2006.08.039 [DOI] [PubMed] [Google Scholar]
- Iwasaki M., Minder P., Cai Y., Kuhn J. H., Yates J. R., III, Torbett B. E., et al. (2018). Interactome analysis of the lymphocytic choriomeningitis virus nucleoprotein in infected cells reveals ATPase Na+/K+ transporting subunit Alpha 1 and prohibitin as host-cell factors involved in the life cycle of mammarenaviruses. PLoS Pathog. 14:e1006892. 10.1371/journal.ppat.1006892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferies H. B., Fumagalli S., Dennis P. B., Reinhard C., Pearson R. B., Thomas G. (1997). Rapamycin suppresses 5’TOP mRNA translation through inhibition of p70s6k. Embo J. 16 3693–3704. 10.1093/emboj/16.12.3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir H. R., Dijkman R. (2016). Coronaviruses and the human airway: a universal system for virus-host interaction studies. Virol J. 13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan S. S., Osorio F., Hiscox J. A. (2012). An interactome map of the nucleocapsid protein from a highly pathogenic North American porcine reproductive and respiratory syndrome virus strain generated using SILAC-based quantitative proteomics. Proteomics 12 1015–1023. 10.1002/pmic.201100469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kailas S., Sagar S. B., Maruti J. D., Shailesh R. W., Naiem H. N., Subodh A. K., et al. (2020). Homology Modeling and Docking Studies of TMPRSS2 with Experimentally Known Inhibitors Camostat Mesylate, Nafamostat and Bromhexine Hydrochloride to Control SARS-Coronavirus-2. ChemRxiv. Preprint 10.26434/chemrxiv.12162360.v1 [DOI] [Google Scholar]
- Kalfoglou Y., Schorlemmer M. (2003). Ontology mapping: the state of the art. Know. Eng. Rev. 18 1–31. 10.1017/s0269888903000651 [DOI] [Google Scholar]
- Kaneko K. (2006). Life: An Introduction to Complex Systems Biology. Berlin: Springer. [Google Scholar]
- Kashour Z., Riaz M., Garbati M. A., Aldosary O., Tlayjeh H., Gerberi D., et al. (2020). Efficacy of chloroquine or hydroxychloroquine in COVID-19 patients: a systematic review and meta-analysis. J. Antimicrob. Chemother. 76 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katze M. G., Fornek J. L., Palermo R. E., Walters K.-A., Korth M. J. (2008). Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat. Rev. Immunol. 8 644–654. 10.1038/nri2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. (2004). In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 323 264–268. 10.1016/j.bbrc.2004.08.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamina K., Lercher A., Caldera M., Schliehe C., Vilagos B., Sahin M., et al. (2017). Characterization of host proteins interacting with the lymphocytic choriomeningitis virus L protein. PLoS Pathog. 13:e1006758. 10.1371/journal.ppat.1006758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindrachuk J., Ork B., Hart B. J., Mazur S., Holbrook M. R., Frieman M. B., et al. (2015). Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 59 1088–1099. 10.1128/aac.03659-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King B. R., Hershkowitz D., Eisenhauer P. L., Weir M. E., Ziegler C. M., Russo J., et al. (2017). A Map of the Arenavirus Nucleoprotein-Host Protein Interactome Reveals that Junin Virus Selectively Impairs the Antiviral Activity of Double-Stranded RNA-Activated Protein Kinase (PKR). J. Virol. 91 e763–e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S., Hwa T. (2008). Stochasticity and traffic jams in the transcription of ribosomal RNA: Intriguing role of termination and antitermination. Proc. Natl. Acad. Sci. 105 18159–18164. 10.1073/pnas.0806084105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D., Jones S. M., Shinya K., Kash J. C., Copps J., Ebihara H., et al. (2007). Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445 319–323. 10.1038/nature05495 [DOI] [PubMed] [Google Scholar]
- Konig R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., et al. (2008). Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell 135 49–60. 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudva R., Denks K., Kuhn P., Vogt A. (2013). gative bacteria: the Sec and Tat dependent protein transport pathways. Res. Microbiol. 164 505–534. 10.1016/j.resmic.2013.03.016 [DOI] [PubMed] [Google Scholar]
- Kummer S., Flöttmann M., Schwanhäusser B., Sieben C., Veit M., Selbach M., et al. (2014). Alteration of protein levels during influenza virus H1N1 infection in host cells: a proteomic survey of host and virus reveals differential dynamics. PLoS One 9:e94257. 10.1371/journal.pone.0094257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo R. L., Chen C. J., Tam E. H., Huang C. G., Li L. H., Li Z. H., et al. (2018). Interactome Analysis of NS1 Protein Encoded by Influenza A H7N9 Virus Reveals an Inhibitory Role of NS1 in Host mRNA Maturation. J. Proteome Res. 17 1474–1484. 10.1021/acs.jproteome.7b00815 [DOI] [PubMed] [Google Scholar]
- Kuo R. L., Li Z. H., Li L. H., Lee K. M., Tam E. H., Liu H. M., et al. (2016). Interactome Analysis of the NS1 Protein Encoded by Influenza A H1N1 Virus Reveals a Positive Regulatory Role of Host Protein PRP19 in Viral Replication. J. Proteome Res. 15 1639–1648. 10.1021/acs.jproteome.6b00103 [DOI] [PubMed] [Google Scholar]
- Lam T. T., Zhu H., Guan Y., Holmes E. C. (2016). Genomic Analysis of the Emergence, Evolution, and Spread of Human Respiratory RNA Viruses. Annu. Rev. Genomics Hum. Genet. 17 193–218. 10.1146/annurev-genom-083115-022628 [DOI] [PubMed] [Google Scholar]
- Lauring A. S., Frydman J., Andino R. (2013). The role of mutational robustness in RNA virus evolution. Nat. Rev. Microb. 11 327–336. 10.1038/nrmicro3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescar J., Soh S., Lee L. T., Vasudevan S. G., Kang C., Lim S. P. (2018). The Dengue Virus Replication Complex: From RNA Replication to Protein-Protein Interactions to Evasion of Innate Immunity. Adv. Exp. Med. Biol. 1062 115–129. 10.1007/978-981-10-8727-1_9 [DOI] [PubMed] [Google Scholar]
- Li A., Wu Q., Warnick G., Li S., Libby E. N., Garcia D. A., et al. (2020). The incidence of thromboembolism for lenalidomide versus thalidomide in older patients with newly diagnosed multiple myeloma. Ann. Hematol. 99 121–126. 10.1007/s00277-019-03860-2 [DOI] [PubMed] [Google Scholar]
- Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., et al. (2020). Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 6:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscalzo J., Barabasi A. L. (2011). Systems biology and the future of medicine. Wiley Interdiscip Rev. Syst. Biol. Med. 3 619–627. 10.1002/wsbm.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magagnoli J., Narendran S., Pereira F., Cummings T. H., Hardin J. W., Sutton S. S., et al. (2020). Outcomes of Hydroxychloroquine Usage in United States Veterans Hospitalized with COVID-19. medRxiv. Preprint 10.1101/2020.04.16.20065920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoudabadi G., Milo R., Phillips R. (2017). Energetic cost of building a virus. Proc. Natl. Acad. Sci. 114 E4324–E4333. 10.1073/pnas.1701670114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X., Mcmanaway S., Jaiswal J. K., Patel P. B., Wilson W. R., Hicks K. O., et al. (2018). An agent-based model for drug-radiation interactions in the tumour microenvironment: Hypoxia-activated prodrug SN30000 in multicellular tumour spheroids. PLoS Comput. Biol. 14:e1006469. 10.1371/journal.pcbi.1006469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Gil L., Vera-Velasco N. M., Mingarro I. (2017). Exploring the Human-Nipah Virus Protein-Protein Interactome. J. Virol. 91 e1461–e1417. 10.1128/JVI.01461-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows D. H., Wright D. (2008). Thinking in systems : a primer. [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., et al. (2005). Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437 1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Mina P., Tsaneva-Atanasova K., Bernardo M. D. (2016). Entrainment and Control of Bacterial Populations: An in Silico Study over a Spatially Extended Agent Based Model. ACS Synthetic Biol. 5 639–653. 10.1021/acssynbio.5b00243 [DOI] [PubMed] [Google Scholar]
- Mohler L., Flockerzi D., Sann H., Reichl U. (2005). Mathematical model of influenza A virus production in large-scale microcarrier culture. Biotechnol. Bioeng. 90 46–58. 10.1002/bit.20363 [DOI] [PubMed] [Google Scholar]
- Moscona A. (2005). Neuraminidase inhibitors for influenza. N. Engl. J. Med. 353 1363–1373. 10.1056/NEJMra050740 [DOI] [PubMed] [Google Scholar]
- Munday D. C., Surtees R., Emmott E., Dove B. K., Digard P., Barr J. N., et al. (2012). Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics 12 666–672. 10.1002/pmic.201100488 [DOI] [PubMed] [Google Scholar]
- Murayama A., Ohmori K., Fujimura A., Minami H., Yasuzawa-Tanaka K., Kuroda T., et al. (2008). Epigenetic Control of rDNA Loci in Response to Intracellular Energy Status. Cell 133 627–639. 10.1016/j.cell.2008.03.030 [DOI] [PubMed] [Google Scholar]
- Murphy B. R., Rennels M. B., Douglas R. G., Jr., Betts R. F., Couch R. B., Cate T. R., Jr., et al. (1980). Evaluation of influenza A/Hong Kong/123/77 (H1N1) ts-1A2 and cold-adapted recombinant viruses in seronegative adult volunteers. Infect Immun. 29 348–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagesh P. T., Hussain M., Galvin H. D., Husain M. (2017). Histone Deacetylase 2 Is a Component of Influenza A Virus-Induced Host Antiviral Response. Front. Microbiol. 8:1315. 10.3389/fmicb.2017.01315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naji S., Ambrus G., Cimermancic P., Reyes J. R., Johnson J. R., Filbrandt R., et al. (2012). Host cell interactome of HIV-1 Rev includes RNA helicases involved in multiple facets of virus production. Mol. Cell Proteom. 11 M111.015313. 10.1074/mcp.M111.015313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelemans T., Kikkert M. (2019). Viral Innate Immune Evasion and the Pathogenesis of Emerging RNA Virus Infections. Viruses 11:916. 10.20944/preprints201909.0212.v1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northridge M. E., Metcalf S. S. (2016). Enhancing implementation science by applying best principles of systems science. Health Res. Policy Syst. 14:74. 10.1186/s12961-016-0146-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odum H. T. (1996). Environmental accounting : EMERGY and environmental decision making. New York, NY: Wiley. [Google Scholar]
- Odum H. T. (2002). Explanations of ecological relationships with energy systems concepts. Ecol. Model. 158 201–211. 10.1016/s0304-3800(02)00232-6 [DOI] [Google Scholar]
- Odum H. T., Odum E. C. (2000). Modeling for all scales : an introduction to system simulation. San Diego: Academic Press. [Google Scholar]
- Panja A. S., Sarkar A., Biswas R., Bandyopadhyay B., Bandopadhyay R. (2019). Modification of drug-binding proteins associated with the efflux pump in MDR-MTB in course of evolution: an unraveled clue based on in silico approach. J. Antibiot. 72 282–290. 10.1038/s41429-019-0146-3 [DOI] [PubMed] [Google Scholar]
- Patterson J. L., Fernandez-Larsson R. (1990). Molecular Mechanisms of Action of Ribavirin. Rev. Infect. Dis. 12 1139–1146. 10.1093/clinids/12.6.1139 [DOI] [PubMed] [Google Scholar]
- Patzina C., Botting C. H., Garcia-Sastre A., Randall R. E., Hale B. G. (2017). Human interactome of the influenza B virus NS1 protein. J. Gen. Virol. 98 2267–2273. 10.1099/jgv.0.000909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pécheur E. I., Borisevich V., Halfmann P., Morrey J. D., Smee D. F., Prichard M., et al. (2016). The Synthetic Antiviral Drug Arbidol Inhibits Globally Prevalent Pathogenic Viruses. J. Virol. 90 3086–3092. 10.1128/jvi.02077-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V., Chavez S., Perez-Ortin J. E. (2010). A complete set of nascent transcription rates for yeast genes. PLoS One 5:e15442. 10.1371/journal.pone.0015442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroni G., Bagni G., Iorio J., Duranti C., Lottini T., Stefanini M., et al. (2020). Clarithromycin inhibits autophagy in colorectal cancer by regulating the hERG1 potassium channel interaction with PI3K. Cell Death Dis. 11:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferle S., Schopf J., Kogl M., Friedel C. C., Muller M. A., Carbajo-Lozoya J., et al. (2011). The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 7:e1002331. 10.1371/journal.ppat.1002331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phiel C. J., Zhang F., Huang E. Y., Guenther M. G., Lazar M. A., Klein P. S. (2001). Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276 36734–36741. 10.1074/jbc.m101287200 [DOI] [PubMed] [Google Scholar]
- Popovic M., Minceva M. (2020a). Thermodynamic insight into viral infections 2: empirical formulas, molecular compositions and thermodynamic properties of SARS, MERS and SARS-CoV-2 (COVID-19) viruses. Heliyon 6:e04943. 10.1016/j.heliyon.2020.e04943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic M., Minceva M. (2020b). A thermodynamic insight into viral infections: do viruses in a lytic cycle hijack cell metabolism due to their low Gibbs energy? Heliyon 6:e03933. 10.1016/j.heliyon.2020.e03933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primadharsini P. P., Nagashima S., Okamoto H. (2019). Genetic Variability and Evolution of Hepatitis E Virus. Viruses 127 216–228. 10.1016/j.virusres.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proshkin S., Rahmouni A. R., Mironov A., Nudler E. (2010). Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328 504–508. 10.1126/science.1184939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qudrat-Ullah H. (2012). On the validation of system dynamics type simulation models. Telecommun. Syst. 51 159–166. 10.1007/s11235-011-9425-4 [DOI] [Google Scholar]
- Regoes R. R., Crotty S., Antia R., Tanaka M. M. (2005). Optimal replication of poliovirus within cells. Am. Nat. 165 364–373. 10.2307/3473412 [DOI] [PubMed] [Google Scholar]
- Reineke L. C., Tsai W. C., Jain A., Kaelber J. T., Jung S. Y., Lloyd R. E. (2017). Casein Kinase 2 Is Linked to Stress Granule Dynamics through Phosphorylation of the Stress Granule Nucleating Protein G3BP1. Mol. Cell Biol. 37 e596–e516. 10.1128/MCB.00596-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S., Brennan G. (2020). Species-Specific Host-Virus Interactions: Implications for Viral Host Range and Virulence. Trends Microbiol. 28 46–56. 10.1016/j.tim.2019.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed M., Suzuki R., Watanabe N., Masaki T., Tomonaga M., Muhammad A., et al. (2011). Role of the Endoplasmic Reticulum-associated Degradation (ERAD) Pathway in Degradation of Hepatitis C Virus Envelope Proteins and Production of Virus Particles. J. Biol. Chem. 286 37264–37273. 10.1074/jbc.M111.259085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B., Parks R. J. (2019). Histone Deacetylase Inhibitor Suberoylanilide Hydroxamic Acid Suppresses Human Adenovirus Gene Expression and Replication. J. Virol. 93 e88–e19. 10.1128/JVI.00088-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr. (2008). Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454 523–527. 10.1038/nature07106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., Braggio E., Kortuem K. M., Egan J. B., Zhu Y. X., Xin C. S., et al. (2013). Genome-wide studies in multiple myeloma identify XPO1/CRM1 as a critical target validated using the selective nuclear export inhibitor KPT-276. Leukemia 27 2357–2365. 10.1038/leu.2013.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473 337–342. 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- Scutigliani E. M., Kikkert M. (2017). Interaction of the innate immune system with positive-strand RNA virus replication organelles. Cytokine Growth Fact. Rev. 37 17–27. 10.1016/j.cytogfr.2017.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmak J. J., Grossberg S. E. (1973). Interferon bioassay: reduction in yield of myxovirus neuraminidases. J. Gen. Virol. 21 1–7. 10.1099/0022-1317-21-1-1 [DOI] [PubMed] [Google Scholar]
- Sheahan T. P., Sims A. C., Graham R. L., Menachery V. D., Gralinski L. E., Case J. B., et al. (2017). Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 9:eaal3653. 10.1126/scitranslmed.aal3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Xiong H., He J., Deng H., Li Q., Zhong Q., et al. (2007). Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo. Arch. Virol. 152 1447–1455. 10.1007/s00705-007-0974-5 [DOI] [PubMed] [Google Scholar]
- Shiraki K., Daikoku T. (2020). Favipiravir, an anti-influenza drug against life-threatening RNA virus infections. Pharm. Therap. 209:107512 10.1016/j.pharmthera.2020.107512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui-Jain A., Drygin D., Streiner N., Chua P., Pierre F., O’brien S. E., et al. (2010). CX-4945, an orally bioavailable selective inhibitor of protein kinase CK2, inhibits prosurvival and angiogenic signaling and exhibits antitumor efficacy. Cancer Res. 70 10288–10298. 10.1158/0008-5472.CAN-10-1893 [DOI] [PubMed] [Google Scholar]
- Siwiak M., Zielenkiewicz P. (2013). Transimulation - protein biosynthesis web service. PLoS One 8:e73943. 10.1371/journal.pone.0073943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spill F., Bakal C., Mak M. (2018). Mechanical and Systems Biology of Cancer. Comput. Struct. Biotechnol. J 16 237–245. 10.1016/j.csbj.2018.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sruthi C. K., Prakash M. K. (2019). Statistical characteristics of amino acid covariance as possible descriptors of viral genomic complexity. Sci. Rep. 9:18410. 10.1038/s41598-019-54720-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatiou R., Paraskeva E., Boukas K., Gourgoulianis K. I., Molyvdas P.-A., Hatziefthimiou A. A. (2009). Azithromycin has an antiproliferative and autophagic effect on airway smooth muscle cells. Eur. Respir. J. 34 721–730. 10.1183/09031936.00089407 [DOI] [PubMed] [Google Scholar]
- Sterman J. D. (2002). All models are wrong: reflections on becoming a systems scientist. Syst. Dyn. Rev. 18 501–531. 10.1002/sdr.261 [DOI] [Google Scholar]
- Tan S.-L., Ganji G., Paeper B., Proll S., Katze M. G. (2007). Systems biology and the host response to viral infection. Nat. Biotechnol. 25 1383–1389. 10.1038/nbt1207-1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegner J. N., Compte A., Auffray C., An G., Cedersund G., Clermont G., et al. (2009). Computational disease modeling - fact or fiction? BMC Syst. Biol. 3:56. 10.1186/1752-0509-3-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T., Lovmar M., Ehrenberg M. (2003). The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J. Mol. Biol. 330 1005–1014. 10.1016/S0022-2836(03)00662-4 [DOI] [PubMed] [Google Scholar]
- Thaker S. K., Ch’ng J., Christofk H. R. (2019). Viral hijacking of cellular metabolism. BMC Biol. 17:59. 10.1186/s12915-019-0678-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorlund K., Dron L., Park J., Hsu G., Forrest J., I, Mills E. J. (2020). A real-time dashboard of clinical trials for COVID-19. Lancet Digit. Health 2 e286–e287. 10.1016/S2589-7500(20)30086-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulgiati S., Brown M. T. (2009). Emergy and ecosystem complexity. Commun. Nonlinear Sci. Numerical. Simul. 14 310–321. 10.1016/j.cnsns.2007.05.028 [DOI] [Google Scholar]
- Ulgiati S., Ascione M., Zucaro A., Campanella L. (2011). Emergy-based complexity measures in natural and social systems. Ecol. Indic. 11 1185–1190. 10.1016/j.ecolind.2010.12.021 [DOI] [Google Scholar]
- Verchot J. (2014). The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 5:66. 10.3389/fpls.2014.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M., Stone J. K., Arnold J. J., Cameron C. E., Andino R. (2006). Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439 344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M. J., Bergeron E., Benjannet S., Erickson B. R., Rollin P. E., Ksiazek T. G., et al. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol J. 2:69. 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E. J., Younessi P., Fulcher A. J., Mccuaig R., Thomas B. J., Bardin P. G., et al. (2013). Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS One 8:e71316. 10.1371/journal.pone.0071316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. H., Chung F. T., Lin S. M., Huang S. Y., Chou C. L., Lee K. Y., et al. (2014). Adjuvant treatment with a mammalian target of rapamycin inhibitor, sirolimus, and steroids improves outcomes in patients with severe H1N1 pneumonia and acute respiratory failure. Crit. Care Med. 42 313–321. 10.1097/CCM.0b013e3182a2727d [DOI] [PubMed] [Google Scholar]
- Wang L., Fu B., Li W., Patil G., Liu L., Dorf M. E., et al. (2017a). Comparative influenza protein interactomes identify the role of plakophilin 2 in virus restriction. Nat. Commun. 8:13876. 10.1038/ncomms13876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li Q., Liu R., Zheng M., Wen J., Zhao G. (2016). Host cell interactome of PA protein of H5N1 influenza A virus in chicken cells. J. Proteomics 136 48–54. 10.1016/j.jprot.2016.01.018 [DOI] [PubMed] [Google Scholar]
- Wang Q., Li Y., Dong H., Wang L., Peng J., An T., et al. (2017b). Identification of host cellular proteins that interact with the M protein of a highly pathogenic porcine reproductive and respiratory syndrome virus vaccine strain. Virol J. 14:39. 10.1186/s12985-017-0700-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo A. R., Kurath G. (2012). Viral fitness: definitions, measurement, and current insights. Curr. Opin. Virol. 2 538–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kawakami E., Shoemaker J. E., Lopes T. J., Matsuoka Y., Tomita Y., et al. (2014). Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microb. 16 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterlow J. C. (2006). Protein turnover. Cambridge, MA: CABI Pub. [Google Scholar]
- Watters K., Inankur B., Gardiner J. C., Warrick J., Sherer N. M., Yin J., et al. (2017). Differential Disruption of Nucleocytoplasmic Trafficking Pathways by Rhinovirus 2A Proteases. J. Virol. 91 e2472–e2416. 10.1128/JVI.02472-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X., Ghosh S. K., Taylor M. E., Johnson V. A., Emini E. A., Deutsch P., et al. (1995). Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373 117–122. 10.1038/373117a0 [DOI] [PubMed] [Google Scholar]
- Wheatley D. N., Giddings M. R., Inglis M. S. (1980). Kinetics of degradation of ‘short-’ and ‘long-lived’ proteins in cultured mammalian cells. Cell Biol. Int. Rep. 4 1081–1090. 10.1016/0309-1651(80)90045-4 [DOI] [PubMed] [Google Scholar]
- Wiederrecht G., Lam E., Hung S., Martin M., Sigal N. (1993). The mechanism of action of FK-506 and cyclosporin A. Ann. N. Y. Acad. Sci. 696 9–19. 10.1111/j.1749-6632.1993.tb17137.x [DOI] [PubMed] [Google Scholar]
- Wu N. C., Otwinowski J., Thompson A. J., Nycholat C. M., Nourmohammad A., Wilson I. A. (2020). Major antigenic site B of human influenza H3N2 viruses has an evolving local fitness landscape. Nat. Commun. 11:1233. 10.1038/s41467-020-15102-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W., Tran K. C., Teng M. N., Heesom K. J., Matthews D. A., Barr J. N., et al. (2012). The interactome of the human respiratory syncytial virus NS1 protein highlights multiple effects on host cell biology. J. Virol. 86 7777–7789. 10.1128/JVI.00460-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Liu G., Casazza M., Ulgiati S. (2018). Development of an urban FEW nexus online analyzer to support urban circular economy strategy planning. Energy 164 475–495. 10.1016/j.energy.2018.08.198 [DOI] [Google Scholar]
- Yang Y., Ye F., Zhu N., Wang W., Deng Y., Zhao Z., et al. (2015). Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 5:17554. 10.1038/srep17554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York A., Hutchinson E. C., Fodor E. (2014). Interactome analysis of the influenza A virus transcription/replication machinery identifies protein phosphatase 6 as a cellular factor required for efficient virus replication. J. Virol. 88 13284–13299. 10.1128/JVI.01813-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L. L., Li C., Ping J., Zhou Y., Li Y., Hao P. (2014). The domain landscape of virus-host interactomes. Biomed. Res. Int. 2014:867235. 10.1155/2014/867235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C. M., Eisenhauer P., Kelly J. A., Dang L. N., Beganovic V., Bruce E. A., et al. (2018). A Proteomics Survey of Junin Virus Interactions with Human Proteins Reveals Host Factors Required for Arenavirus Replication. J. Virol. 92 e1565–e1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Chan J. F. W., Azhar E. I., Hui D. S. C., Yuen K.-Y. (2016). Coronaviruses — drug discovery and therapeutic options. Nat. Rev. Drug Discov. 15 327–347. 10.1038/nrd.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
System dynamics of (+)ssRNA virus–host interaction in response to initial viral load. Changes overtime of the values of each stock of the system diagrammed in Figure 1 (for the color code see at the bottom), expressed in ATP-eq: in absence of infection the system status was stationary (A). Upon infection initial values of Q3 stock identify the response of system, at 50 (B), 100 (C), 1,000 (D), 5,000 (E) and 10,000 (F) RNA copies, expressed in ATP-eq. For Q3 stock in the range (10–1,000 RNA copies) the trajectory of Q5 evolution was linear, while for higher initial viral load the growth of Q5 was linear in the first day and non-linear in the further timeframe.
Effects of targeting leverage point applying external driving force (D) at Day 1. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 1, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
Effects of targeting leverage point applying external driving force (D) at Day 3. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 3, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
Effects of targeting leverage point applying external driving force (D) at Day 5. Time change of the stocks values (for the color code see at the bottom), expressed in ATP-eq, were depicted over-time, from infection (Day 0, Time 0) through the next 7 days, upon application, at Day 5, of generic external driving forces (D) able to reduce the target flows (schemed on the left) of 100% or 50%, respectively.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.