Abstract
To identify signaling pathways activated by oxycodone self-administration (SA), Sprague–Dawley rats self-administered oxycodone for 20 days using short—(ShA, 3 h) and long-access (LgA, 9 h) paradigms. Animals were euthanized 2 h after SA cessation and dorsal striata were used in post-mortem molecular analyses. LgA rats escalated their oxycodone intake and separated into lower (LgA-L) or higher (LgA-H) oxycodone takers. LgA-H rats showed increased striatal protein phosphorylation of ERK1/2 and MSK1/2. Histone H3, phosphorylated at serine 10 and acetylated at lysine 14 (H3S10pK14Ac), a MSK1/2 target, showed increased abundance only in LgA-H rats. RT-qPCR analyses revealed increased AMPA receptor subunits, GluA2 and GluA3 mRNAs, in the LgA-H rats. GluA3, but not GluA2, mRNA expression correlated positively with changes in pMSK1/2 and H3S10pK14Ac. These findings suggest that escalated oxycodone SA results in MSK1/2-dependent histone phosphorylation and increases in striatal gene expression. These observations offer potential avenues for interventions against oxycodone addiction.
Subject terms: Molecular neuroscience, Psychiatric disorders, Addiction
Introduction
The opioid epidemic remains a public health crisis1,2. This is related, in part, to the over-prescription of the opioid agonist, oxycodone, for pain management3–6. Its illicit abuse has also contributed to the high number of overdose-related deaths7,8. Other complications of oxycodone use disorder include moderate to severe withdrawal symptoms1 and repeated episodes of relapses during attempts to quit through psychological or pharmacological interventions9. Chronic use of opioid drugs is also accompanied by cognitive deficits10 and post-mortem evidence of neuropathological abnormalities in the brain11. These biopsychosocial complications make the development of effective treatment of paramount importance.
Pharmacological approaches to treat opioid use disorders (OUDs) have mainly included the use of agents that interact with opioids receptors3,12,13. Upon activation, opioid receptors transmit signals to the nucleus via intracellular events that involve modulation of some kinase cascades14–16, with consequent changes in gene expression17,18. Because of several of these experiments had done using in vitro systems, it was important to address the potential neurobiological impact of repeated oxycodone self-administration in humans by using model systems that better mimic human conditions. Therefore, we decided to use the model of escalated oxycodone self-administration (SA) in rats19,20 to identify potential biochemical and molecular pathways that might be perturbed by this drug.
Herein, we used that model to measure alterations in various proteins that may impact the flow of intracellular signals from the mu opioid receptor consequent to its repeated interactions with oxycodone during a drug SA experiment. The rat dorsal striatum was dissected and processed for biochemical and molecular analyses because this structure is thought to play essential roles in the manifestation of habitual drug taking behaviors21–23. Thus, we report that the mitogen-activated protein kinase (MAPK)/mitogen- and stress-activated protein kinase (MSK) signaling cascade is activated preferentially in rats that consume large quantities of oxycodone over a period of 20 days. This was manifested by increased phosphorylation of extracellular signal-regulated kinases 1/2 (ERK1/2), MSK1/2, and increased abundance of histone H3 phosphorylated at serine 10 and acetylated at lysine 14 (H3S10pK14Ac). Altogether, these findings implicate the role of MAPK/MSK pathway and histone H3 phosphoacetylation in opioid use disorder.
Materials and methods
Intravenous surgery
We used male Sprague Dawley rats (Charles River, Raleigh, NC, USA), weighing 350–400 g before surgery and housed on a 12 h reversed light/dark cycle with food and water freely available. All procedures followed the guidelines outlined in the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals (eighth edition, https://guide-for-the-care-and-use-of-laboratory-animals.pdf) as approved by the NIDA (National Institute of Drug Abuse) Animal Care and Use Committee at the Intramural Research Program (IRP). Catheter implantations were performed as previously described24. Briefly, we anesthetized the rats with ketamine (50 mg/kg) and xylazine (5 mg/kg). Polyurethane catheters (SAI Infusion Technologies, Lake Villa, IL) were inserted into the jugular vein. The other end of the catheter was attached to a modified 22-gauge cannula (Plastics One, Roanoke, VA) that was mounted to the back of each rat. The modified cannulas, which served as infusion ports for the catheters, were connected to a fluid swivel (Instech, Plymouth, PA) via polyethylene-50 tubing that was protected by a metal spring. When the infusion ports were not used they were sealed using dust caps (PlasticOne, Roanoke, VA). Thereafter, the catheters were flushed every 48 h with gentamicin (0.05 mg/kg, Henry Schein, Melville, NY) in sterile saline to maintain patency. Intraperitoneal injection of buprenorphine (0.1 mg/kg) was used post-surgery to relieve pain.
Apparatus
Rats were trained in Med Associates SA chambers located inside sound-attenuated cabinets and controlled by a Med Associates System (Med Associates, St Albans, VT) as previously described19. In brief, each chamber was equipped with two levers located 8.5 cm above the grid floor. Presses on the retractable active lever activated the infusion pump and tone-light cue. Presses on the inactive lever had no reinforced consequences.
Training phase
Rats (n = 42) were randomly assigned to either saline (Sal) (n = 8) or oxycodone (n = 33) conditions. Rats were trained to self-administer oxycodone-HCL (NIDA Pharmacy, Baltimore, MD) for one 3 h daily session for the short-access (ShA) condition (n = 10) or one to three 3 h sessions for long-access (LgA) condition (n = 23) (Fig. 1A). For the LgA group, the 3 h sessions were separated by 30 min intervals from day 6 to day 20 (Fig. 1A). Lever presses were reinforced using a fixed ratio-1 with a 20-s timeout accompanied by a 5-s compound tone-light cue. We used a scheduling pattern of 5 days of drug SA and 2 days off to control for weight loss, a common side effect of oxycodone intake in laboratory animals25. Rats self-administered oxycodone at a dose of 0.1 mg/kg per infusion over 3.5-s (0.1 ml per infusion). The house light was turned off, and the active lever retracted at the end of the 3 h session.
Figure 1.
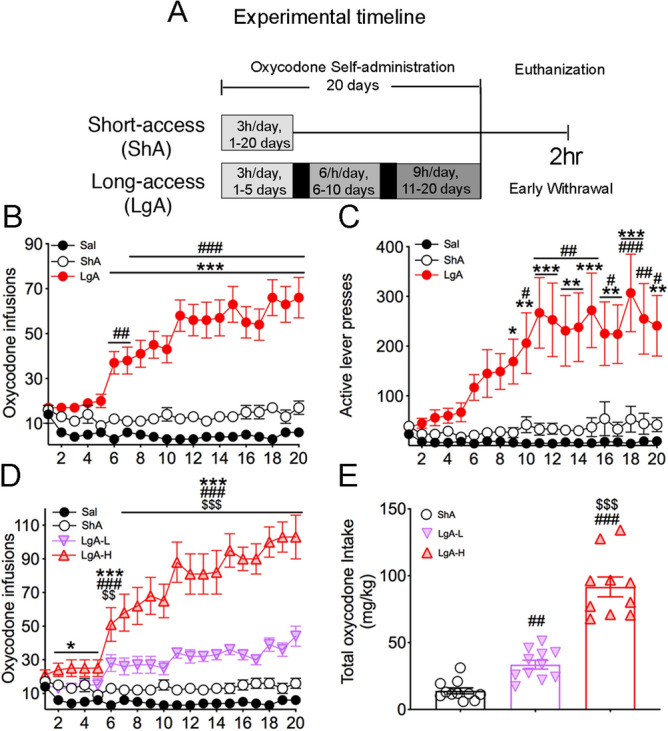
Rats exposed to long-access, but not short-access, oxycodone SA escalate their drug intake. (A) Experimental timeline of oxycodone self-administration (SA) training. Rats self-administered oxycodone using either short-access (ShA) (n = 10) (trained for 3 h for 20 days) or long-access (LgA) (n = 21–23) (trained for 3 h for 1–5 days, 6 h for 6–10 days, then 9 h for 11–20 days) paradigms. (B) LgA rats escalate their intake of oxycodone after the first 5 days of SA training. (C) LgA rats show significant increases in active lever presses during SA training. (D) LgA-H rats show two distinct intake phenotypes, high (LgA-H) (n = 10) and low (LgA-L) (n = 13) oxycodone takers, during the escalation phase. (E) LgA-H rats took substantially more oxycodone than LgA-L and ShA rats. Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to Sal rats; #, ##, ### = p < 0.05, 0.01, 0.001, respectively, in comparison to SHA rats; $$, $$$ = p < 0.01, 0.001 in comparison to LgA-L rats. Stats were performed by either one-way or two-way ANOVA followed by Bonferroni or Fisher’s PLSD post hoc test.
Tissue collection
Rats were euthanized during early withdrawal, which is defined as the 2 h time point after cessation of drug self-administration. Dorsal striata tissue was dissected as previously described26. In brief, we used stereotaxic coordinates (A/P + 2 to -2 mm bregma, M/L ± 2 to 5 mm, D/V − 3 to − 6 mm) according to the rat atlas27 and we used the position of anatomical structures (corpus callosum and lateral ventricles) for further accuracy. In brief, the dorsal striata was removed from the skulls and snap frozen on dry ice. Tissue was later used for western blotting and quantitative RT-PCR experiments.
Western blotting
Western blotting was conducted as previously described19. Ten—twenty µg of lysate was prepared in solutions that contained 1 × NuPage LDS Sample Buffer (ThermoFisher Scientific, Waltham, MA), and 1% β-Mercaptoethanol. Protein samples were heated to 70 °C and loaded on 3–8% Tris-Acetate Protein Gels (ThermoFisher Scientific, Waltham, MA) or NuPAGE 4–12% Bis–Tris Protein Gels (ThermoFisher Scientific, Waltham, MA). Proteins were electrophoretically transferred on the Trans-Blot Turbo System (Bio-Rad, Hercules, CA). Membrane blocking, antibody incubations, and chemiluminescence reactions were performed according to the manufacturer’s instructions. Primary and secondary antibodies are listed in Supplemental Table S1. Supplemental Table S1 also includes Research Resource Identifiers (RRIDs) where antibodies were previously validated. All antibodies ran at approximate predicted sizes according to manufacturer’s instructions. Cyclophilin B or alpha-tubulin was used as loading controls. Following secondary antibody incubation, ECL clarity (Bio-Rad, Hercules, CA) was used to visualize gel bands on ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA), and intensities were quantified with Image Lab version 6.0 (Bio-Rad, Hercules, CA) software.
Quantitative PCR
Total RNA was collected as previously described19. PCR experiments were performed using the LightCycler 480 II (Roche Diagnostics, Indianapolis, IN) with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). Primers were purchased from Johns Hopkins University (Baltimore, MD) Synthesis and Sequence Facility. Primer sequences are listed in Supplementary Table S2. The data was normalized to Qaz1 or B2m reference genes. The standard curve method was used to analyze data and the results are reported as fold change relative to Sal.
Statistical analyses
Behavioral data were analyzed using either one-way or two-way analysis of variance (ANOVA) as previously described19. In brief, dependent variables were the number of oxycodone infusions on training days. Independent variables were between-subject factor reward types (Sal, ShA, LgA-L, LgA-H), within-subject factor SA day (training days 1–20), and their interactions. If the main effects were significant (p < 0.05), Bonferroni post hoc tests were used to compare reward types on each training day. Biochemical data were analyzed using one-way ANOVA followed by the Fisher’s PLSD post hoc test. Regression analyses were performed using the correlation function in Prism version 8.3.0 (GraphPad Software, San Diego, CA). Statistical significance for all hypothesis tests was set at p < 0.05. Behavioral and biochemical data were analyzed with Prism version 8.3.0 (GraphPad Software, San Diego, CA).
Results
Long-access self-administration leads to escalated oxycodone intake in rats
Figure 1 shows the experimental timeline and behavioral results for oxycodone SA. As described in details under methods, rats were given short-access (ShA) or long-access (LgA) to oxycodone during the experiment19. The repeated-measures ANOVA for reward earned included the between-subject factor, groups (Saline, ShA, LgA), the within-subject factor of SA days (training days 1–20), and the group day interaction. This analysis showed statistically significant effects of group [F(2, 890) = 307.5, p < 0.001], day [F(19, 890) = 3.016, p < 0.0001], and significant group day interaction [F(38, 890) = 3.958, p < 0.0001]. A comparison of the LgA rats to Saline rats showed that the LgA rats increased their oxycodone intake substantially after training day 5 compared to Saline rats [F(1, 507) = 35, p < 0.0001; Fig. 1B], with there being significant increases in the number of active lever presses [F(1, 507) = 87, p < 0.0001; Fig. 1C] by LgA rats during the drug SA experiments. As previously reported19, LgA rats could be further divided into two SA phenotypes depending on whether they took high (LgA-H) and lower (LgA-L) amounts of oxycodone (Fig. 1D). Figure 1E shows that the LgA-H rats consumed significantly more oxycodone than the ShA and LgA-L rats [F(2, 29) = 85.00, p < 0.0001].
Effects of early withdrawal and oxycodone SA on the activation of PKC
Rats were euthanized 2 h after cessation of oxycodone SA and their dorsal striata were used in Western Blot analyses of several phospho-proteins involved in the MAPK/MSK signaling pathway. The kinase, PKC, is known to be involved in the MAPK signaling cascade stimulated by opioid receptors28–30. Supplementary Figure S1 shows the effects of oxycodone SA on PKC and pPKC protein expression in the dorsal striata of rats euthanized at 2 h after cessation of drug SA. There were no significant changes in striatal PKC protein levels [F(3, 18) = 0.73, p = 0.540; Supplementary Fig. S1]. However, changes in phosphorylated PKC (pPKC) abundance trended towards significance [F(3, 18) = 2.82, p = 0.068] in LgA-H rats, with planned tests showing small increases in LgA-H in comparison to Saline and ShA rats (Supplementary Fig. S1). Importantly, pPKC/PKC ratios were significantly increased [F(3, 18) = 4.45, p = 0.0165] in the LgA-H group compared to the other groups (Supplementary Fig. S1). Regression analysis revealed a significant positive correlation between pPKC/PKC ratios and the amount of total oxycodone taken during the SA experiment (Supplementary Fig. S1). By contrast, there were no changes in protein kinase A (PKA) protein expression [F(3, 20) = 1.98, p = 0.1498] or pPKA abundance [F(3, 20) = 2.22, p = 0.1178] (Supplementary Fig. S2), suggesting that changes in the PKA signaling pathway were not involved in oxycodone SA after 20 days of drug exposure in a manner consistent with a previous report that cAMP/PKA cascade might not be involved in the rewarding properties of morphine31.
Withdrawal from oxycodone SA increases ERK phosphorylation in LgA rats
ERK1/2 are members of MAPK kinases that are regulated by opioid drugs32 and are also activated by PKC33. We thus measured their expression in oxycodone-exposed rats and found no significant changes [F(3, 20) = 2.18, p = 0.1224] in striatal ERK1/2 protein expression (Fig. 2A,B). However, there were increases [F(3, 20) = 6.41, p = 0.0032] in the abundance of pERK1/2 in the LgA-H group in comparison to Saline and ShA groups (Fig. 2A,C). pERK/ERK ratios were also significantly increased [F(3, 20) = 7.10, p = 0.0020] in the LgA-H group in comparison to the other 3 groups (Fig. 2D). Regression analysis showed oxycodone amount-dependent increases in pERK/ERK ratios (Fig. 2E).
Figure 2.
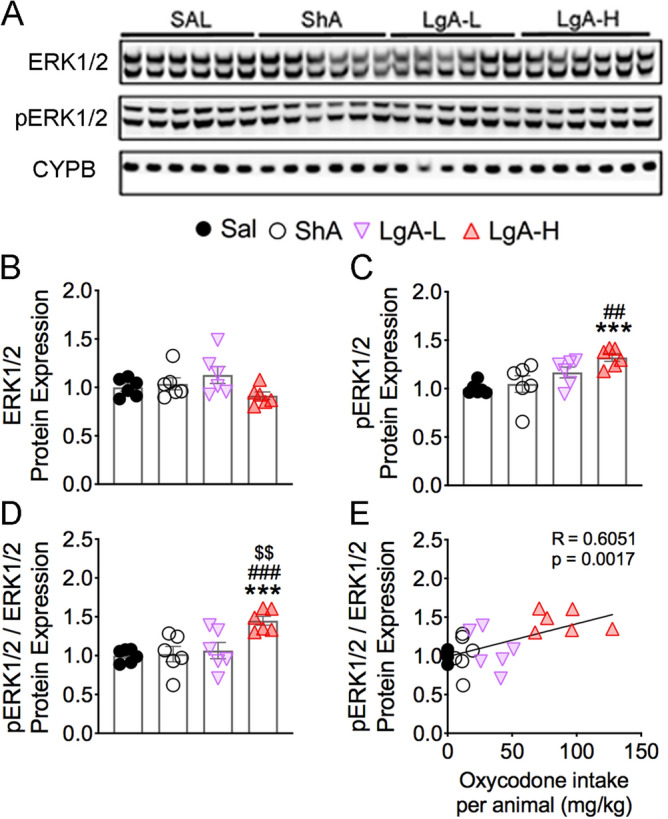
Effects of oxycodone SA on ERK1/2 phosphorylation. (A) Images of western blot and (B, C) quantification of ERK1/2, and pERK1/2. (B) ERK1/2 protein levels were not significantly impacted by oxycodone. (C) pERK1/2 abundance was increased in only LgA-H rats. (D) pERK1/2/ERK1/2 ratios are substantially increased in the LgA-H rats. (E) pERK1/2/ERK1/2 ratios correlate with amount of oxycodone taken. (n = 6 Sal; n = 6 ShA; n = 6 LgA-L; n = 6 LgA-H). Full-length blots are presented in Supplementary Fig. S5. Key to statistics: *** = p < 0.001 in comparison to Sal rats; ##, ### = p < 0.01, 0.001, respectively, in comparison to ShA rats; $$ = p < 0.01, in comparison to LgA-L rats. Statistical analyses are as described in Fig. 2.
Effects of oxycodone and early withdrawal on MSK1 and MSK2 proteins
ERK1/2 kinases phosphorylate MSK1 and MSK2 proteins in the MAPK/MSK cascade34,35. MSK1 is also activated in neurons in response to stress and neurotrophins36. We therefore tested the possibility that MSK1 and MSK2 phosphorylation might be affected in oxycodone SA animals. Figure 3 shows the effects of oxycodone SA on MSK1, pMSK1, MSK2, and pMSK2 levels. MSK1 protein expression was significantly decreased [F(3, 20) = 6.18, p = 0.0038] in both LgA-L and LgA-H rats in comparison to the Saline group (Fig. 3A,B). However, pMSK1 abundance was substantially increased [F(3, 20) = 4.53, p = 0.0140] in the LgA-H group in comparison to Sal and ShA groups (Fig. 3A,C). In addition, pMSK1/MSK ratios were increased [F(3, 20) = 16.3, p < 0.0001] in both LgA-L and LgA-H rats in comparison to Sal and ShA animals (Fig. 3D). Regression analysis revealed significant correlation between pMSK1/MSK ratios and amount of oxycodone self-administered (Fig. 3E).
Figure 3.
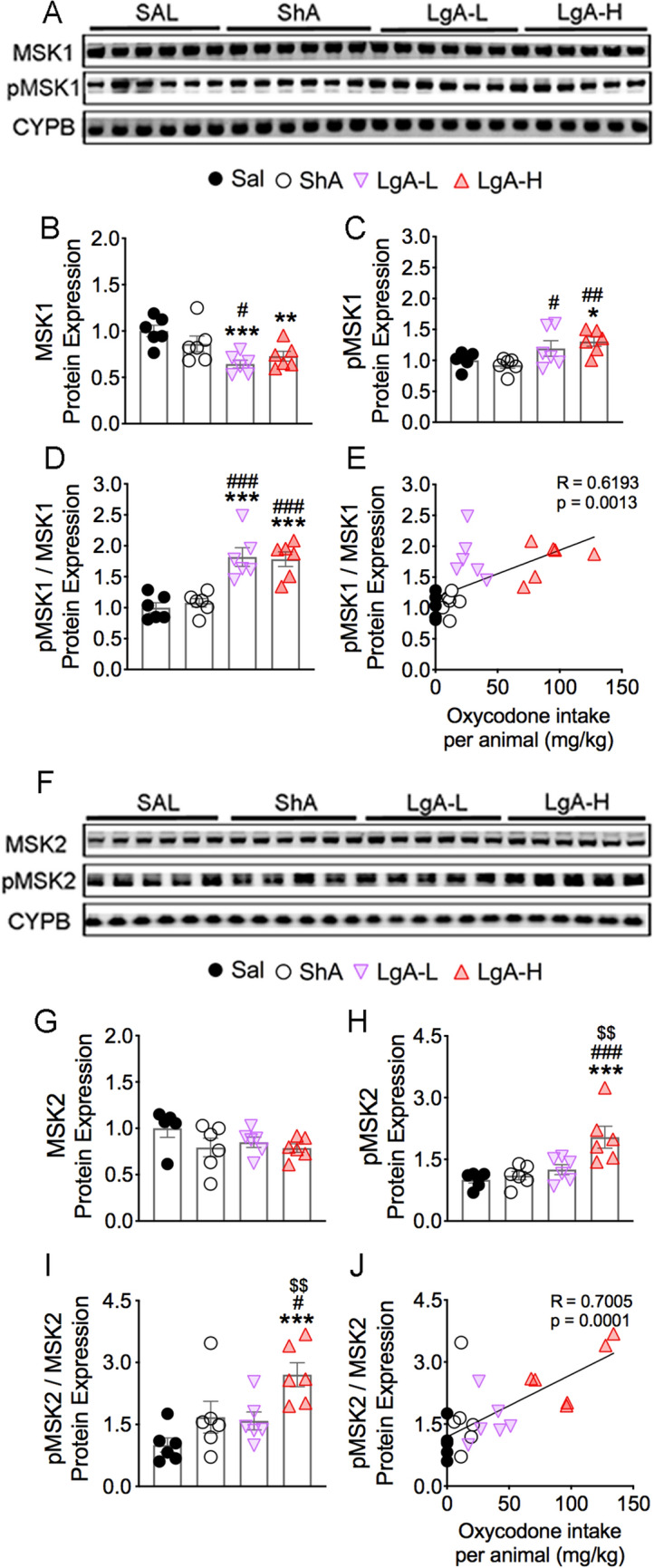
Increased MSK1 and MSK2 protein phosphorylation in LgA-H rats. (A,F) Images of western blot and (B,C,G,H) quantification of MSK1, pMSK1, MSK2, pMSK2. (B) MSK1 protein levels are decreased in LgA-L and LgA-H rats. (C) pMSK1 protein abundance is upregulated in LgA-L and LgA-H rats. (D) pMSK1/MSK1 ratios are increased in LgA rats; (E) Changes in pMSK1/MSK1 ratios are dependent on oxycodone intake. (G) MSK2 protein levels were not significantly affected by oxycodone. (H) pMSK2 abundance is increased in LgA-H rats. (I) pMSK2/MSK2 ratios are increased in LgA-H rats and (J) correlated with amount of oxycodone. (n = 5–6 Sal; n = 6 ShA; n = 6 LgA-L; n = 6 LgA-H). Full-length blots are presented in Supplementary Figure S5. Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to Sal rats; #, ##, ### = p < 0.05, 0.01, 0.001, respectively, in comparison to SHA rats; $$ = p < 0.01 in comparison to LgA-L rats. Statistical analyses are as described in Fig. 2.
There were no significant changes [F(3, 19) = 1.52, p = 0.2413] in MSK2 protein expression (Fig. 3F,G). There were, however, significant increases [F(3, 20) = 8.73, p = 0.0007] in pMSK2 abundance only in LgA-H rats in comparison to other groups (Fig. 3F,H). pMSK2/MSK2 ratios were also significantly increased [F(3, 20) = 6.50, p = 0.0030] in only LgA-H rats (Fig. 3I), with significant positive correlation observed between these ratios and amount of oxycodone taken during the SA experiment (Fig. 3J).
Increased pCREB in LgA-H rats
Because CREB phosphorylation can be mediated by several upstream kinases that include PKC, ERK1/2 and MSKs37, we examined the possibility that activation of these kinases might have led to increased pCREB after oxycodone SA. There were no significant changes [F(3, 18) = 2.97, p = 0.0592] in CREB protein expression (Fig. 4A,B). However, the abundance of pCREB was significantly increased [F(3, 20) = 36.0, p < 0.001] in the LgA-H groups in comparison to the Saline group (Fig. 4A,C). Furthermore, pCREB/CREB ratios were also substantially increased [F(3, 18) = 24.8, p < 0.001] in the LgA-H group compared to other groups (Fig. 4D), with there being a significant positive correlation between these ratios and amount of oxycodone taken (Fig. 4E).
Figure 4.
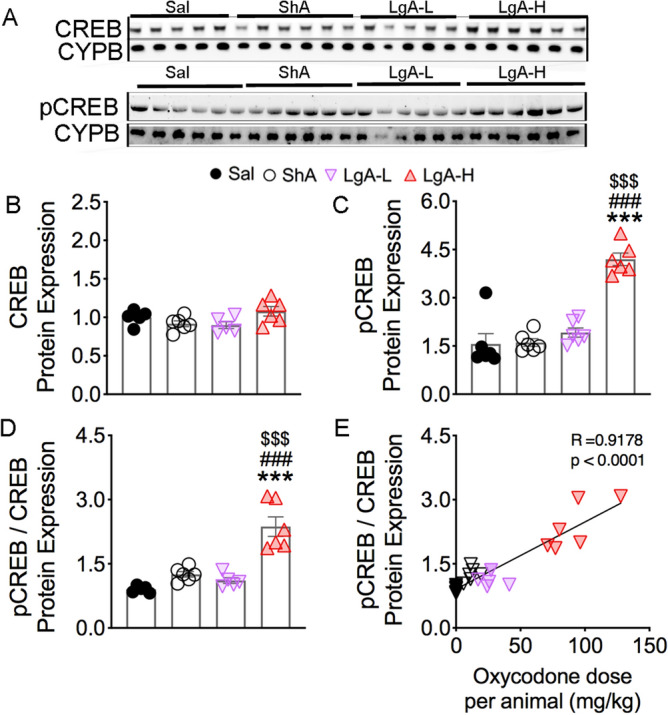
Increased phosphorylation of CREB protein levels in the LgA-H rats. (A) Images of western blot and (B, C) quantification of CREB and pCREB. (B) CREB protein levels show no significant changes. (C) pCREB is significantly increases in LgA-L and LgA-H rats. (D) pCREB/CREB ratios are increased in the LgA-H rats and (E) correlated with amount of oxycodone. (n = 5–6 Sal; n = 6 ShA; n = 5–6 LgA-L; n = 6 LgA-H). Full-length blots are presented in Supplementary Figure S5. Key to statistics: *, ** = p < 0.05, 0.01, respectively, in comparison to Sal rats; #, ## = p < 0.05, 0.01, respectively, in comparison to SHA rats; $$ = p < 0.01 in comparison to LgA-L rats. Statistical analyses are as described in Fig. 2.
Oxycodone SA induces increased phosphoacetylation of histone H3 in LgA rats
In addition to CREB phosphorylation, activated MSK1 and MSK2 participate in the phosphorylation of histone H3 at serine residue S10 and can cause increases in histone H3 phosphoacetylation of H3S10pK14Ac34,37,38. We therefore sought to determine the effects of oxycodone SA on the abundance of this histone marker. Figure 5 shows the results for histone H3 and H3S10pK14Ac. Unexpectedly, we found significant decreases [F(3, 20) = 5.02, p = 0.0094] in histone H3 protein levels in the ShA group in comparison with Sal and the LgA-H groups (Fig. 5A,B). In contrast, H3S10pK14Ac abundance was significantly increased [F(3, 20) = 20.0, p < 0.0001] in the LgA-L and LgA-H groups compared with Saline rats. Moreover, H3S10pK14Ac in the LgA-H group was substantially increased compared to the other 3 groups (Fig. 5A,C). H3S10pK14Ac/H3 ratios were also increased [F(3, 20) = 20.0, p < 0.0001] in the LgA-L and LgA-H groups compared to the Saline and ShA groups (Fig. 5D). Regression analyses revealed that both H3S10pK14Ac abundance and H3S10pK14Ac/H3 ratios positively correlated with the amount of total oxycodone taken during the experiment (Fig. 5E, F).
Figure 5.
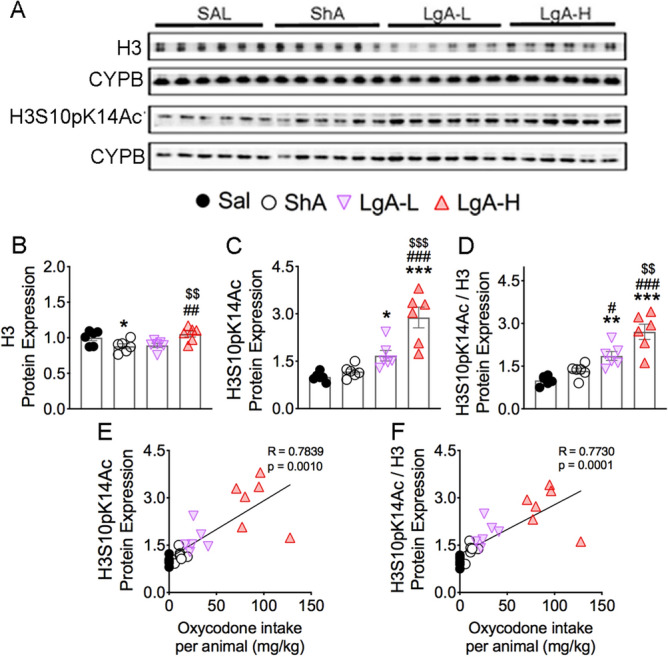
Effects of oxycodone SA and early withdrawal on histone H3 and H3S10pK14Ac. (A) Images of western blot and quantification of (B) histone H3 and (C) H3S10pK14Ac protein levels. (B) Protein levels of histone H3 display decrease in the ShA group. (C) Protein expression of H3S10pK14Ac shows significant increases in LgA-L and LgA-H groups. (D) Ratio of H3S10pK14Ac/H3 displays significant increases in the LgA-L and LgA-H groups. (E) Protein expression of H3S10pK14Ac and (F) H3S10pK14Ac/H3 positively correlated with doses of oxycodone taken during the experiment (n = 5–6 Sal; n = 6 ShA; n = 6 LgA-L; n = 6 LgA-H). Key to statistics: *, **, *** = p < 0.05, 0.01, 0.001, respectively, in comparison to Sal rats; #, ##, ### = p < 0.05, 0.01, 0.001, respectively, in comparison to SHA rats; $$, $$$ = p < 0.01, 0.001 in comparison to LgA-L rats. Statistical analyses are as described in Fig. 2.
Differential protein expression of CBP and H3K27Ac in oxycodone exposed rats
Phosphorylated CREB recruits CBP, under certain circumstances, to promote changes in gene expression39,40. We thus tested the possibility that oxycodone SA might have influenced striatal CBP protein expression and found that CBP protein expression was significantly increased [F(3, 17) = 6.82, p = 0.0032] in the LgA-L and LgA-H rats compared with the Saline and ShA groups (Fig. 6A,B). Increased CBP expression correlated with the amount of oxycodone consumed by the rats (Fig. 6C).
Figure 6.
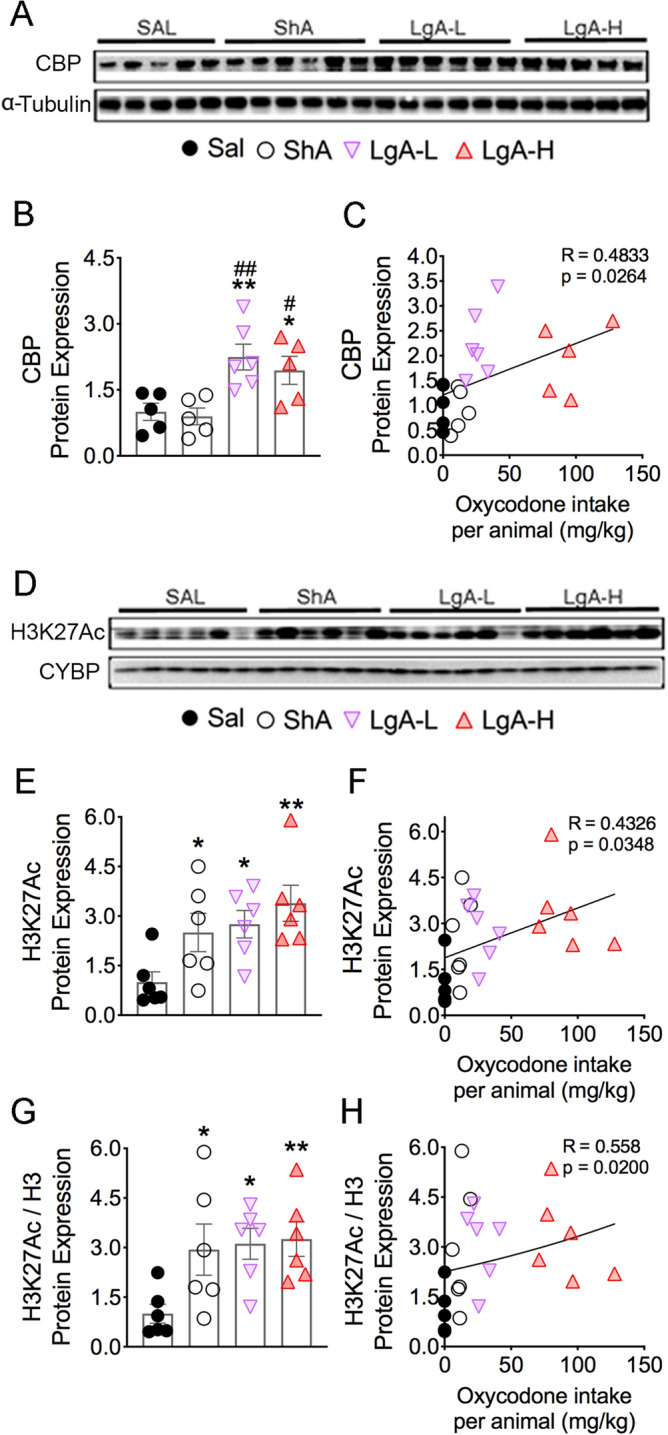
Differential effects on the protein expression of CBP and H3K27Ac after oxycodone SA. (A,D) Images of western blot and quantification of (B) CBP and (E) H3K27Ac protein levels. (B) CBP protein levels display significant increases in LgA groups. The protein levels of (E) H3K27Ac and (G) H3K27Ac/H3 protein were significantly increased in all drug groups. The regression analyses of (C) CBP, (F) H3K27Ac, and (H) H3K27Ac/H3 correlates with the amount of oxycodone taken (n = 5–6 Sal; n = 5–6 ShA; n = 6 LgA-L; n = 5–6 LgA-H). Key to statistics: *, ** = p < 0.05, 0.01, respectively, in comparison to Sal rats; #, ## = p < 0.05, 0.01, respectively, in comparison to ShA rats. Statistical analyses are as described in Fig. 2.
In addition to CBP’s transcriptional co-activity, it functions as a histone acetyltransferase41,42 that mediates acetylation of H3K2743–45, a marker of active enhancers46–48 that is involved in regulating neuronal gene expression. We therefore measured the abundance of H3K27Ac, which had been previously shown to be impacted in the brains of heroin addicts49. We found significant increases [F(3, 20) = 4.53, p = 0.0140] in H3K27Ac abundance in all oxycodone groups, including ShA rats that did not escalate their intake (Fig. 6D,E), suggesting that oxycodone exposure is enough to increase striatal H3K27Ac abundance. We found that H3K27Ac abundance positively correlated with the amount of oxycodone self-administered by rats (Fig. 6F). These increases confirm the data in heroin-using individuals49. H3K27Ac/H3 ratios were also significantly increased [F(3, 20) = 3.82, p = 0.0259] in the 3 oxycodone groups (Fig. 6G) and correlated with the amount of oxycodone taken (Fig. 6H).
Because CBP expression was only increased in the two LgA groups while H3K27Ac was increased in the 3 oxycodone groups, we sought to determine if the expression of another histone H3 acetyltransferase, Tip60, with putative activity towards the lysine 27 residue50 was affected in the 3 oxycodone groups. Indeed, Tip60 protein expression was significantly increased [F(3, 20) = 3.63, p = 0.0307] in all three oxycodone groups (Supplementary Fig. S3).
Oxycodone SA increases GluA2 and GluA3 glutamate receptor mRNA levels
Changes in gene expression in response to exogenous stimuli include many target genes that are expressed with different time courses of induction. Because respective changes in histone phosphorylation and acetylation generated by MSKs and CBP are known regulators of gene expression 51–54, we tested the idea that some of their target genes might be affected in the striata of oxycodone-exposed rats. We also measured the mRNA levels of some glutamatergic genes whose expression was altered in the brains of heroin users based on a previous microarray study49. Figure 7 shows the effects of oxycodone SA on the mRNA expression of GluA1, GluA2, GluA3, and GluA4 subunits of AMPA receptors55. We found no substantial changes in GluA1 [F(3, 29) = 2.03, p = 0.1322; Fig. 7A] and GluA4 [F(3, 34) = 1.60, p = 0.2084; Fig. 7G] mRNA levels, with no relationships between their levels and the amount of oxycodone consumed during the experiment (Fig. 7B,H). In contrast, striatal GluA2 [F(3, 27) = 3.49, p = 0.0291; Fig. 7C] and GluA3 [F(3, 31) = 15.7, p < 0.0001; Fig. 7E] were increased in the LgA-H group compared to other groups. Regression analyses revealed significant oxycodone amount-dependent changes in their mRNA levels (Fig. 7D,F). Moreover, the changes in GluA2 and GluA3 mRNAs correlated with changes in pMSK1 (Fig. 8A,B). However, only changes in GluA3, but not in GluA2, mRNA levels correlated with changes in pMSK2, H3S10pK14Ac, and CBP protein expression (Fig. 8C–H).
Figure 7.
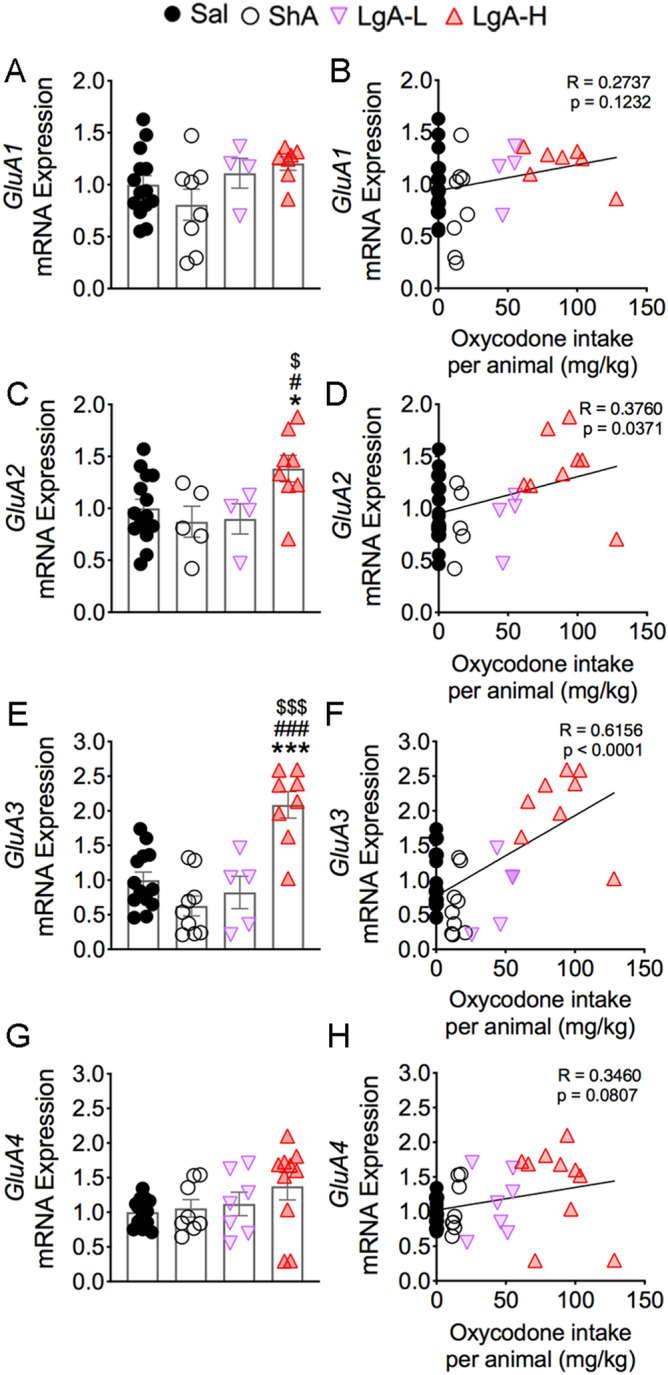
Consequences of oxycodone SA and early withdrawal on mRNA on GluA1, GluA2, GluA3, and GluA4 mRNA levels. mRNA levels of (A) GluA1 and (G) GluA4 were not significantly affected. There were no correlations between (B) GluA1 and (H) GluA4 mRNA levels and the consumption of oxycodone. However, (C) GluA2 and (E) GluA3 mRNA levels were significantly increased in the LgA-H rats, with significant positive correlations between mRNA expression of (D) GluA2 and (F) GluA3 with the amount of oxycodone taken. Key to statistics: *, *** = p < 0.05, 0.001, respectively, in comparison to Sal rats; #, ### = p < 0.05, 0.001, respectively, in comparison to SHA rats; $, $$$ = p < 0.05, 0.001 in comparison to LgA-L rats (n = 10–11 Sal; n = 5–9 ShA; n = 4–7 LgA-L; n = 7–10 LgA-H). Statistical analyses are as described in Fig. 2.
Figure 8.
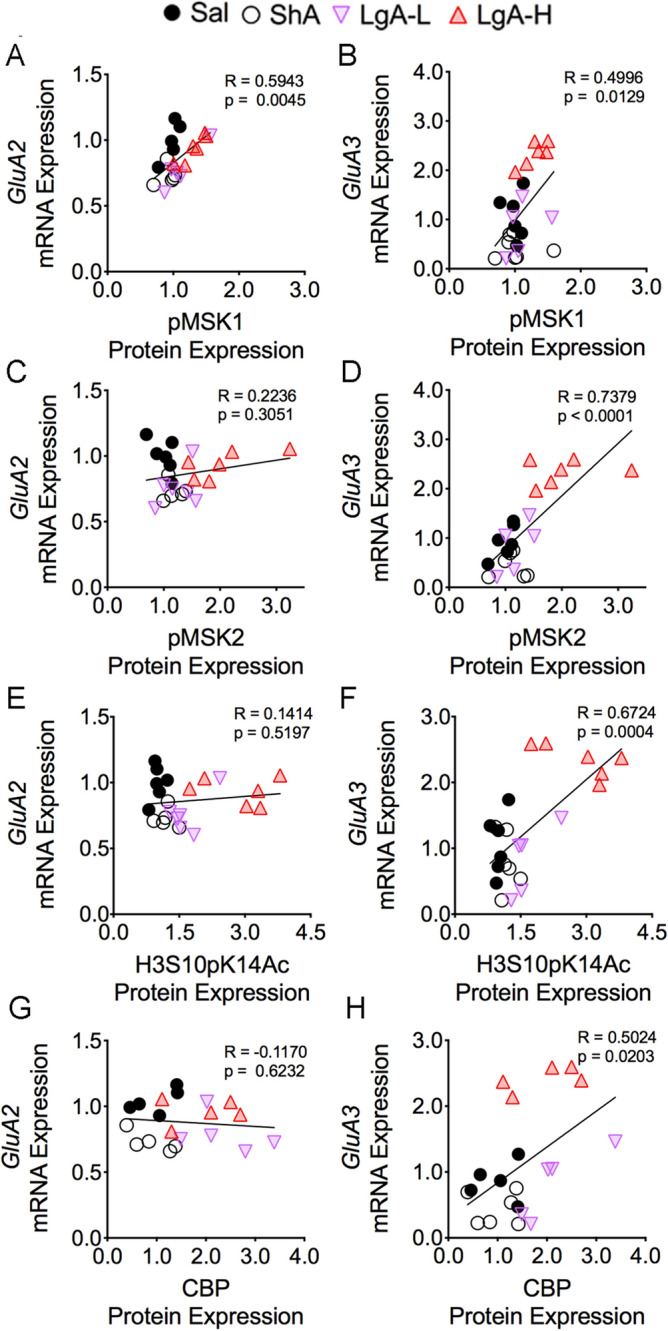
GluA3 mRNA correlated with pMSK1, pMSK2, H3S10pK14Ac, and CBP protein expression. The mRNA expression of GluA2 shows significant linear relationship with (A) pMSK1, but not with (C) pMSK2, (E) H3S10pK14Ac, and (G) CBP protein expression. However, the mRNA levels of GluA3 display a positive correlation with the protein abundance of (B) pMSK1, (D) pMSK2, (F) H3S10pK14Ac, and (H) CBP (n = 5–6 Sal; n = 5–6 ShA; n = 5–6 LgA-L; n = 5–6 LgA-H). The correlation coefficients and p values are shown on the graph.
mRNA levels of acetyltransferases, Ncoa1-3, are increased in LgA oxycodone rats
Egervari et al. (2017) had recently reported that the expression of the acetyltransferase, nuclear receptor coactivator 1 (Ncoa1), was significantly increased in the ventral striatum of heroin users. We therefore measured the expression of Ncoa1-3 in our experiment. Supplemental Fig. 4 shows significant increases in the mRNA levels of Ncoa1 [F(3, 35) = 12.6; p < 0.0001], Ncoa2 [F(3, 31) = 4.55; p = 0.0094], and Ncoa3 [F(3, 28) = 12.5; p < 0.0001] in the LgA-H group compared to the other groups. Furthermore, there were significant correlations between their expression and changes in pMSK1, pMSK2, and H3S10pK14Ac protein abundance (Supplementary Fig. S4). However, we observed no significant correlations between Ncoa1, Ncoa2, and Ncoa3 mRNA levels and CBP suggesting that histone phosphorylation might play a more important role in regulating their expression than histone acetylation (Supplementary Fig. S4).
Discussion
In the present study, we show that long-access to oxycodone SA over a period of 20 days leads to activation of several kinases involved in the MAPK/MSK signaling pathway with consequent CREB and histone H3 phosphorylation in the rat dorsal striatum. These results are consistent, in part, with previous evidence of the involvement of the MAPK in the biochemical effects of morphine56,57. We also found significant increases in CBP and H3K27 acetylation in oxycodone-exposed rats. These findings are consistent with observations that epigenetic mechanisms are involved in models of opioid abuse58. The changes in signaling pathways are accompanied by oxycodone-induced increased gene expression of GluA2 and GluA3 subunits of AMPA receptors and of the acetyltransferases, Ncoa1-3. Our results provide novel insights into the role of H3S10pK14Ac in oxycodone-induced gene expression and hint to a model whereby this histone marker is involved in the regulation of genes that might be responsible for some long-term molecular adaptations that drive compulsive oxycodone intake.
The dorsal striatum is a brain region that is integral to various behavioral changes consequent to drug taking behaviors including habit forming and drug seeking during periods of opioid withdrawal19,22,23,59. Similar to other observations with cocaine, methamphetamine, and other drugs58,60–66, we found increased phosphorylation of PKC, ERK1/2, MSK1/2, and pCREB in the rat dorsal striatum after repeated exposure to oxycodone SA. Increased phosphorylation of H3S10pK14Ac, a marker that is downstream of these kinases34,37,67 is of interest because these findings suggest that repeated exposure to long-access oxycodone self-administration might engender a permissive molecular state characterized by increased histone H3 phosphoacetylation and a more open chromatin structure. The hypothesized permissive state might also facilitate pCREB binding at the cAMP-Response Element (CRE) on the promoters of genes that have been implicated in the regulation of synaptic plasticity51,68,69. CREB activation is also known to enhance the recruitment of co-activators39,40 such as CBP, an acetyltransferase that acetylates H3K2743–45,70 to increase the transcription of downstream genes in diverse cell populations39,71,72. This suggestion is further supported by observations of increased expression of co-activators for the steroid hormone receptor family, Ncoa173, Ncoa274, and Ncoa375, that can enhance transcription, in part, via histone acetylation76,77 and recruitment of CBP75,78,79, whose expression is also increased in rats exposed to relatively large quantities of oxycodone. Our proposal of an oxycodone-induced permissive state in the dorsal striatum is consistent with observations that histone H3 phosphorylation and acetylation can work in concert to regulate gene expression80. This discussion is supported by the observations that histone H3 phosphoacetylation also participates in heroin-induced conditioned place preference81, thus indicating a role of phosphoacetylation in the effects of opioids in general. His discussion notwithstanding, given the diversity of neurons within the dorsal striata82, follow-up studies are needed to identify which specific neuronal subtypes might exhibit the oxycodone-associated changes in that structure.
Our observations of increased histone H3 phosphorylation and acetylation led us to test the possibility that some genes downstream of these molecular events might show differential expression in the brains of oxycodone-exposed rats. Indeed, we found increased expression of several genes in the LgA-H rats that showed increased abundance of striatal pMSK1, pMSK2, and histone H3S10pK14Ac. Of interest among those are the changes in AMPA receptor subunits, GluA2 and GluA3, in LgA-H rats in an oxycodone amount- and pMSK1-dependent fashion. MSK1 and MSK2 are known to play substantial roles in a number of biological events including synaptic plasticity83. The altered expression of GluA2 and GluA3 is of singular interest because Egervari et al. (2017) had reported that their microarray analyses, using tissues from the ventral striatum of heroin users, had detected changes in the expression of several genes, including GluA3, which are involved in glutamate neurotransmission. Increases in the expression of GluA2 and GluA3 receptor subunits in our study are consistent with the proposed roles of glutamate receptors in models of substance use disorders including cocaine84, methamphetamine85, and opioids86,87. For example, chronic cocaine increases GluA2 expression in the nucleus accumbens and increased expression of GluA2 via viral injections enhanced the sensitivity of mice to the behavioral effects of cocaine84, thus suggesting that increased GluA2 expression in the present study might have served to facilitate escalation of oxycodone intake in the LgA-H rats. A similar argument could be made for our novel findings of increased GluA3 expression after oxycodone SA. GluA3-containing AMPA receptors are located in various brain regions88,89. Because GluA3 exists in GluA2A3 combinations or GluA3 monomers or dimers90, it is possible that increased expression of both GluA2 and GluA3 might potentiate AMPAR-mediated changes in synaptic plasticity during repeated oxycodone exposure. Alternatively, GluA3 alone may regulate oxycodone intake because GluA3 knockout mice show decreased alcohol intake91. Thus, elucidation of the specific roles that GluA3 alone or in combination with GluA2 play in oxycodone SA will await future genetic and pharmacological studies.
In conclusion, we have demonstrated that rats that self-administer large quantities of oxycodone showed increased histone and CREB phosphorylation via activation of the MAPK/MSK phosphorylation signaling pathway in the rat dorsal striatum. Rats exposed to large quantities of oxycodone also showed increased striatal CBP and histone acetylation in oxycodone-exposed rats. Changes in histone modifications are proposed to lead to more permissive chromatin states that promoted changes in the expression in a diversity of classes of genes as exemplified by increased mRNA levels of acetyltransferases, Ncoa1-3, and of AMPA receptor subunits, GluA2 and GluA3, in an oxycodone amount-dependent fashion. Importantly, changes in the expression of both GluA2 and GluA3 mRNA levels correlated with altered abundance of pMSK1, a kinase that is involved in the regulation of synaptic plasticity92. These observations are illustrated schematically in Fig. 9. Furthermore, additional experiments that include genetic manipulations and pharmacological interventions that impact these pathways are necessary to identify the specific role that these transcriptional and post-transcriptional changes might play in promoting oxycodone self-administration or cessation of drug intake. Finally, confirmation of the involvement of MAPK/MSK/histone phosphorylation in oxycodone SA may serve as a stimulus to develop potential pharmacological agents against oxycodone use disorder.
Figure 9.
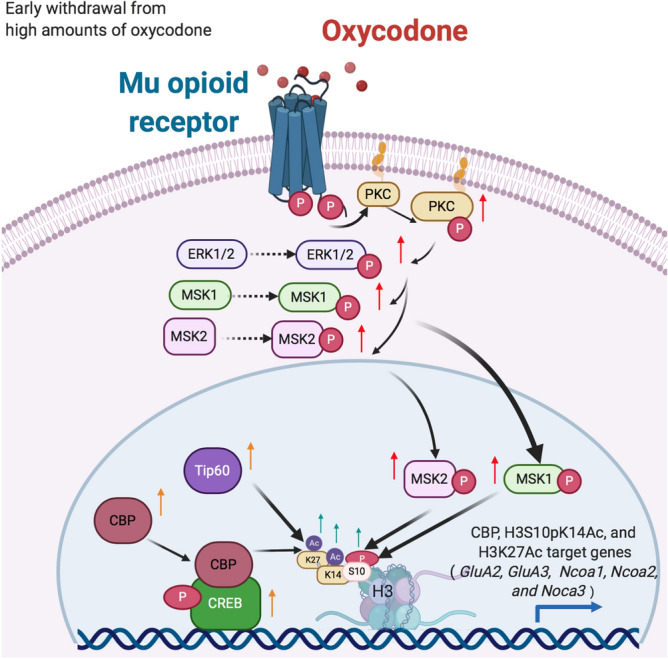
Illustration of the activation of MAPK/MSK signaling pathway in LgA-H rats. Intake of large amount of oxycodone during long-access over 20 days caused increased phosphorylation of PKC, ERK1/2, MSK1, and MSK2. Increased MSK phosphorylation is accompanied by increased histone H3 phosphoacetylation and CREB phosphorylation. In addition, there were increases in the protein expression of two acetyltransferases, CBP and Tip60 that acetylate H3K27. Recruitment of CBP by CREB and histone modifications serve to create a permissible transcriptional environment that led to increased mRNA levels of GluA2 and GluA3 in a MSK1-dependent fashion. This kinase-histone modification cascade may serve as targets for therapeutic interventions against oxycodone use disorder.
Supplementary Information
Author contributions
C.A.B, M.T.M, and B.L performed self-administration, western blot and RT-qPCR experiments. C.A.B and J.L.C prepared manuscript. J.L.C supervised the overall project.
Funding
Open Access funding provided by the National Institutes of Health (NIH). This work was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82206-3.
References
- 1.Cicero TJ, Inciardi JA, Munoz A. Trends in abuse of Oxycontin and other opioid analgesics in the United States: 2002–2004. J. Pain. 2005;6:662–672. doi: 10.1016/j.jpain.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Skolnick P. The opioid epidemic: crisis and solutions. Annu. Rev. Pharmacol. Toxicol. 2017 doi: 10.1146/annurev-pharmtox-010617-052534. [DOI] [PubMed] [Google Scholar]
- 3.Balyan R, Hahn D, Huang H, Chidambaran V. Pharmacokinetic and pharmacodynamic considerations in developing a response to the opioid epidemic. Expert Opin. Drug Metab. Toxicol. 2020;16:125–141. doi: 10.1080/17425255.2020.1721458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaskell H, Derry S, Stannard C, Moore RA. Oxycodone for neuropathic pain in adults. Cochrane Database Syst. Rev. 2016;7:CD010692. doi: 10.1002/14651858.CD010692.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt-Hansen M, Bennett MI, Arnold S, Bromham N, Hilgart JS. Oxycodone for cancer-related pain. Cochrane Database Syst. Rev. 2017;8:CD003870. doi: 10.1002/14651858.CD003870.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Zee A. The promotion and marketing of oxycontin: commercial triumph, public health tragedy. Am. J. Public Health. 2009;99:221–227. doi: 10.2105/AJPH.2007.131714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rudd RA, Seth P, David F, Scholl L. Increases in drug and opioid-involved overdose deaths—United States, 2010–2015. MMWR Morb. Mortal. Wkly. Rep. 2016;65:1445–1452. doi: 10.15585/mmwr.mm655051e1. [DOI] [PubMed] [Google Scholar]
- 8.Seth P, Scholl L, Rudd RA, Bacon S. Overdose deaths involving opioids, cocaine, and psychostimulants—United States, 2015–2016. MMWR Morb. Mortal. Wkly. Rep. 2018;67:349–358. doi: 10.15585/mmwr.mm6712a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schuckit MA. Treatment of opioid-use disorders. N. Engl. J. Med. 2016;375:357–368. doi: 10.1056/NEJMra1604339. [DOI] [PubMed] [Google Scholar]
- 10.Cadet JL, Bisagno V. Neuropsychological consequences of chronic drug use: relevance to treatment approaches. Front. Psychiatry. 2015;6:189. doi: 10.3389/fpsyt.2015.00189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cadet JL, Bisagno V, Milroy CM. Neuropathology of substance use disorders. Acta Neuropathol. 2014;127:91–107. doi: 10.1007/s00401-013-1221-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noble F, Lenoir M, Marie N. The opioid receptors as targets for drug abuse medication. Br. J. Pharmacol. 2015;172:3964–3979. doi: 10.1111/bph.13190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein C. Opioid receptors. Annu. Rev. Med. 2016;67:433–451. doi: 10.1146/annurev-med-062613-093100. [DOI] [PubMed] [Google Scholar]
- 14.Ehrich JM, et al. Kappa opioid receptor-induced aversion requires p38 MAPK activation in VTA dopamine neurons. J. Neurosci. 2015;35:12917–12931. doi: 10.1523/JNEUROSCI.2444-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz R, Eisinger DA, Wehmeyer A. Opioid control of MAP kinase cascade. Eur. J. Pharmacol. 2004;500:487–497. doi: 10.1016/j.ejphar.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 16.Wagley Y, Law PY, Wei LN, Loh HH. Epigenetic activation of mu-opioid receptor gene via increased expression and function of mitogen- and stress-activated protein kinase 1. Mol. Pharmacol. 2017;91:357–372. doi: 10.1124/mol.116.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Hasani R, Bruchas MR. Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology. 2011;115:1363–1381. doi: 10.1097/ALN.0b013e318238bba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruchas MR, Roth BL. New technologies for elucidating opioid receptor function. Trends Pharmacol. Sci. 2016;37:279–289. doi: 10.1016/j.tips.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackwood CA, et al. Molecular adaptations in the rat dorsal striatum and hippocampus following abstinence-induced incubation of drug seeking after escalated oxycodone self-administration. Mol. Neurobiol. 2019;56:3603–3615. doi: 10.1007/s12035-018-1318-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blackwood CA, Leary M, Salisbury A, McCoy MT, Cadet JL. Escalated oxycodone self-administration causes differential striatal mRNA expression of FGFs and IEGs following abstinence-associated incubation of oxycodone craving. Neuroscience. 2019;415:173–183. doi: 10.1016/j.neuroscience.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belin D, Everitt BJ. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron. 2008;57:432–441. doi: 10.1016/j.neuron.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 22.Everitt BJ, Robbins TW. Drug addiction: updating actions to habits to compulsions ten years on. Annu. Rev. Psychol. 2016;67:23–50. doi: 10.1146/annurev-psych-122414-033457. [DOI] [PubMed] [Google Scholar]
- 23.Hodebourg R, et al. Heroin seeking becomes dependent on dorsal striatal dopaminergic mechanisms and can be decreased by N-acetylcysteine. Eur. J. Neurosci. 2018 doi: 10.1111/ejn.13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cadet JL, et al. Genome-wide DNA hydroxymethylation identifies potassium channels in the nucleus accumbens as discriminators of methamphetamine addiction and abstinence. Mol. Psychiatry. 2017;22:1196–1204. doi: 10.1038/mp.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade CL, Vendruscolo LF, Schlosburg JE, Hernandez DO, Koob GF. Compulsive-like responding for opioid analgesics in rats with extended access. Neuropsychopharmacology. 2015;40:421–428. doi: 10.1038/npp.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blackwood CA, McCoy MT, Ladenheim B, Cadet JL. Escalated oxycodone self-administration and punishment: differential expression of opioid receptors and immediate early genes in the rat dorsal striatum and prefrontal cortex. Front. Neurosci. 2019;13:1392. doi: 10.3389/fnins.2019.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paxinos GAWC. The Rat Brain in Stereotaxic Coordinates. 6. Burlington, MA: Academic Press; 1998. [Google Scholar]
- 28.Kramer HK, Simon EJ. Role of protein kinase C (PKC) in agonist-induced mu-opioid receptor down-regulation: I. PKC translocation to the membrane of SH-SY5Y neuroblastoma cells is induced by mu-opioid agonists. J. Neurochem. 1999;72:585–593. doi: 10.1046/j.1471-4159.1999.0720585.x. [DOI] [PubMed] [Google Scholar]
- 29.Kramer HK, Simon EJ. Role of protein kinase C (PKC) in agonist-induced mu-opioid receptor down-regulation: II. Activation and involvement of the alpha, epsilon, and zeta isoforms of PKC. J. Neurochem. 1999;72:594–604. doi: 10.1046/j.1471-4159.1999.0720594.x. [DOI] [PubMed] [Google Scholar]
- 30.Williams JT, et al. Regulation of mu-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol. Rev. 2013;65:223–254. doi: 10.1124/pr.112.005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borgkvist A, Usiello A, Greengard P, Fisone G. Activation of the cAMP/PKA/DARPP-32 signaling pathway is required for morphine psychomotor stimulation but not for morphine reward. Neuropsychopharmacology. 2007;32:1995–2003. doi: 10.1038/sj.npp.1301321. [DOI] [PubMed] [Google Scholar]
- 32.Ortiz J, et al. Extracellular signal-regulated protein kinases (ERKs) and ERK kinase (MEK) in brain: regional distribution and regulation by chronic morphine. J. Neurosci. 1995;15:1285–1297. doi: 10.1523/JNEUROSCI.15-02-01285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang JM, et al. Acetylcholine induces mesenchymal stem cell migration via Ca2+ /PKC/ERK1/2 signal pathway. J. Cell. Biochem. 2012;113:2704–2713. doi: 10.1002/jcb.24148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adewumi I, Lopez C, Davie JR. Mitogen and stress- activated protein kinase regulated gene expression in cancer cells. Adv. Biol. Regul. 2019;71:147–155. doi: 10.1016/j.jbior.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Wiggin GR, et al. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 2002;22:2871–2881. doi: 10.1128/mcb.22.8.2871-2881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arthur JS, et al. Mitogen- and stress-activated protein kinase 1 mediates cAMP response element-binding protein phosphorylation and activation by neurotrophins. J. Neurosci. 2004;24:4324–4332. doi: 10.1523/JNEUROSCI.5227-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deak M, Clifton AD, Lucocq LM, Alessi DR. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 1998;17:4426–4441. doi: 10.1093/emboj/17.15.4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyson MH, et al. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J. Cell Sci. 2005;118:2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- 39.Cardinaux JR, et al. Recruitment of CREB binding protein is sufficient for CREB-mediated gene activation. Mol. Cell. Biol. 2000;20:1546–1552. doi: 10.1128/mcb.20.5.1546-1552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwok RP, et al. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 41.Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 42.Weinert BT, et al. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell. 2018;174:231–244 e212. doi: 10.1016/j.cell.2018.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin Q, et al. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249–262. doi: 10.1038/emboj.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raisner R, et al. Enhancer activity requires CBP/P300 bromodomain-dependent histone H3K27 acetylation. Cell Rep. 2018;24:1722–1729. doi: 10.1016/j.celrep.2018.07.041. [DOI] [PubMed] [Google Scholar]
- 45.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nat. Neurosci. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egervari G, et al. Striatal H3K27 acetylation linked to glutamatergic gene dysregulation in human heroin abusers holds promise as therapeutic target. Biol. Psychiatry. 2017;81:585–594. doi: 10.1016/j.biopsych.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hsu CC, et al. Recognition of histone acetylation by the GAS41 YEATS domain promotes H2A.Z deposition in non-small cell lung cancer. Genes Dev. 2018;32:58–69. doi: 10.1101/gad.303784.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Impey S, et al. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 52.Kasper LH, Qu C, Obenauer JC, McGoldrick DJ, Brindle PK. Genome-wide and single-cell analyses reveal a context dependent relationship between CBP recruitment and gene expression. Nucleic Acids Res. 2014;42:11363–11382. doi: 10.1093/nar/gku827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramos YF, et al. Genome-wide assessment of differential roles for p300 and CBP in transcription regulation. Nucleic Acids Res. 2010;38:5396–5408. doi: 10.1093/nar/gkq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wiersma M, et al. Protein kinase Msk1 physically and functionally interacts with the KMT2A/MLL1 methyltransferase complex and contributes to the regulation of multiple target genes. Epigenetics Chromatin. 2016;9:52. doi: 10.1186/s13072-016-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol. Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Duraffourd C, Kumala E, Anselmi L, Brecha NC, Sternini C. Opioid-induced mitogen-activated protein kinase signaling in rat enteric neurons following chronic morphine treatment. PLoS ONE. 2014;9:e110230. doi: 10.1371/journal.pone.0110230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jia W, et al. Differential regulation of MAPK phosphorylation in the dorsal hippocampus in response to prolonged morphine withdrawal-induced depressive-like symptoms in mice. PLoS ONE. 2013;8:e66111. doi: 10.1371/journal.pone.0066111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Browne CJ, Godino A, Salery M, Nestler EJ. Epigenetic mechanisms of opioid addiction. Biol. Psychiatry. 2020;87:22–33. doi: 10.1016/j.biopsych.2019.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bossert JM, et al. Role of mu, but not delta or kappa, opioid receptors in context-induced reinstatement of oxycodone seeking. Eur. J. Neurosci. 2018 doi: 10.1111/ejn.13955. [DOI] [PubMed] [Google Scholar]
- 60.Brami-Cherrier K, Roze E, Girault JA, Betuing S, Caboche J. Role of the ERK/MSK1 signalling pathway in chromatin remodelling and brain responses to drugs of abuse. J. Neurochem. 2009;108:1323–1335. doi: 10.1111/j.1471-4159.2009.05879.x. [DOI] [PubMed] [Google Scholar]
- 61.Garcia-Pardo MP, Roger-Sanchez C, Rodriguez-Arias M, Minarro J, Aguilar MA. Pharmacological modulation of protein kinases as a new approach to treat addiction to cocaine and opiates. Eur. J. Pharmacol. 2016;781:10–24. doi: 10.1016/j.ejphar.2016.03.065. [DOI] [PubMed] [Google Scholar]
- 62.Krasnova IN, et al. CREB phosphorylation regulates striatal transcriptional responses in the self-administration model of methamphetamine addiction in the rat. Neurobiol. Dis. 2013;58:132–143. doi: 10.1016/j.nbd.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mattson BJ, et al. Cocaine-induced CREB phosphorylation in nucleus accumbens of cocaine-sensitized rats is enabled by enhanced activation of extracellular signal-related kinase, but not protein kinase A. J. Neurochem. 2005;95:1481–1494. doi: 10.1111/j.1471-4159.2005.03500.x. [DOI] [PubMed] [Google Scholar]
- 64.Miller BW, et al. Cocaine craving during protracted withdrawal requires PKCepsilon priming within vmPFC. Addict. Biol. 2017;22:629–639. doi: 10.1111/adb.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shin EJ, et al. Significance of protein kinase C in the neuropsychotoxicity induced by methamphetamine-like psychostimulants. Neurochem. Int. 2019;124:162–170. doi: 10.1016/j.neuint.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Torres OV, Jayanthi S, McCoy MT, Cadet JL. Selective activation of striatal NGF-TrkA/p75NTR/MAPK intracellular signaling in rats that show suppression of methamphetamine intake 30 days following drug abstinence. Int. J. Neuropsychopharmacol. 2018;21:281–290. doi: 10.1093/ijnp/pyx105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chung YW, Kim HK, Kim IY, Yim MB, Chock PB. Dual function of protein kinase C (PKC) in 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced manganese superoxide dismutase (MnSOD) expression: activation of CREB and FOXO3a by PKC-alpha phosphorylation and by PKC-mediated inactivation of Akt, respectively. J. Biol. Chem. 2011;286:29681–29690. doi: 10.1074/jbc.M111.264945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kandel ER. The molecular biology of memory: cAMP, PKA, CRE, CREB-1, CREB-2, and CPEB. Mol. Brain. 2012;5:14. doi: 10.1186/1756-6606-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lakhina V, et al. Genome-wide functional analysis of CREB/long-term memory-dependent transcription reveals distinct basal and memory gene expression programs. Neuron. 2015;85:330–345. doi: 10.1016/j.neuron.2014.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. U. S. A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Y, et al. Transcriptional landscape of the human cell cycle. Proc. Natl. Acad. Sci. U. S. A. 2017;114:3473–3478. doi: 10.1073/pnas.1617636114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paauw ND, et al. H3K27 acetylation and gene expression analysis reveals differences in placental chromatin activity in fetal growth restriction. Clin. Epigenetics. 2018;10:85. doi: 10.1186/s13148-018-0508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Onate SA, Tsai SY, Tsai MJ, O'Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 74.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. doi: 10.1002/j.1460-2075.1996.tb00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Torchia J, et al. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 76.Chen H, et al. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 77.Spencer TE, et al. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 78.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 79.Olivares AM, Moreno-Ramos OA, Haider NB. Role of nuclear receptors in central nervous system development and associated diseases. J. Exp. Neurosci. 2015;9:93–121. doi: 10.4137/JEN.S25480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheung P, et al. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- 81.Sheng J, Lv Z, Wang L, Zhou Y, Hui B. Histone H3 phosphoacetylation is critical for heroin-induced place preference. NeuroReport. 2011;22:575–580. doi: 10.1097/WNR.0b013e328348e6aa. [DOI] [PubMed] [Google Scholar]
- 82.Le Moine C, Bloch B. D1 and D2 dopamine receptor gene expression in the rat striatum: sensitive cRNA probes demonstrate prominent segregation of D1 and D2 mRNAs in distinct neuronal populations of the dorsal and ventral striatum. J. Comp. Neurol. 1995;355:418–426. doi: 10.1002/cne.903550308. [DOI] [PubMed] [Google Scholar]
- 83.Reyskens KM, Arthur JS. Emerging roles of the mitogen and stress activated kinases MSK1 and MSK2. Front. Cell Dev. Biol. 2016;4:56. doi: 10.3389/fcell.2016.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kelz MB, et al. Expression of the transcription factor deltaFosB in the brain controls sensitivity to cocaine. Nature. 1999;401:272–276. doi: 10.1038/45790. [DOI] [PubMed] [Google Scholar]
- 85.Jayanthi S, et al. Methamphetamine downregulates striatal glutamate receptors via diverse epigenetic mechanisms. Biol. Psychiatry. 2014;76:47–56. doi: 10.1016/j.biopsych.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Billa SK, et al. Increased insertion of glutamate receptor 2-lacking alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors at hippocampal synapses upon repeated morphine administration. Mol. Pharmacol. 2010;77:874–883. doi: 10.1124/mol.109.060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang Y, et al. Chronic oxycodone self-administration altered reward-related genes in the ventral and dorsal striatum of C57BL/6J mice: an RNA-seq analysis. Neuroscience. 2018;393:333–349. doi: 10.1016/j.neuroscience.2018.07.032. [DOI] [PubMed] [Google Scholar]
- 88.Renner MC, et al. Synaptic plasticity through activation of GluA3-containing AMPA-receptors. Elife. 2017 doi: 10.7554/eLife.25462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schwenk J, et al. Regional diversity and developmental dynamics of the AMPA-receptor proteome in the mammalian brain. Neuron. 2014;84:41–54. doi: 10.1016/j.neuron.2014.08.044. [DOI] [PubMed] [Google Scholar]
- 90.Reimers JM, Milovanovic M, Wolf ME. Quantitative analysis of AMPA receptor subunit composition in addiction-related brain regions. Brain Res. 2011;1367:223–233. doi: 10.1016/j.brainres.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sanchis-Segura C, et al. Involvement of the AMPA receptor GluR-C subunit in alcohol-seeking behavior and relapse. J. Neurosci. 2006;26:1231–1238. doi: 10.1523/JNEUROSCI.4237-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Correa SA, et al. MSK1 regulates homeostatic and experience-dependent synaptic plasticity. J. Neurosci. 2012;32:13039–13051. doi: 10.1523/JNEUROSCI.0930-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


