Abstract
Golden Rice with β-carotene in the grain helps to address the problem of vitamin A deficiency. Prior to commercialize Golden Rice, several performance and regulatory checkpoints must be achieved. We report results of marker assisted backcross breeding of the GR2E trait into three popular rice varieties followed by a series of confined field tests of event GR2E introgression lines to assess their agronomic performance and carotenoid expression. Results from confined tests in the Philippines and Bangladesh have shown that GR2E introgression lines matched the performance of the recurrent parents for agronomic and yield performance, and the key components of grain quality. Moreover, no differences were observed in terms of pest and disease reaction. The best performing lines identified in each genetic background had significant amounts of carotenoids in the milled grains. These lines can supply 30–50% of the estimated average requirements of vitamin A.
Subject terms: Biotechnology, Genetics, Plant sciences
Introduction
Rice (Oryza sativa) is the major source of energy and nutrition for more than half the world’s population1. However, rice supplies minimal micronutrients in its milled form and completely lacks β-carotene which is the precursor for vitamin A. Thus, resource-poor people primarily dependent on rice with little access to diverse diets suffer from micronutrient deficiencies, also termed hidden hunger2,3. Even though efforts are being made to address micronutrient deficiencies by supplementation, fortification, and dietary diversification, the problem still persists globally. Biofortification of major staple crops has been recognized as one of the sustainable means to tackle micronutrient deficiencies especially in the vulnerable target groups in rural areas4.
Vitamin A is essential for various functions in the human body such as development and functioning of the visual system, differentiation and maintenance of cells, epithelial membrane integrity, and production of red blood cells, immune system, reproduction, and iron metabolism5,6. An estimated 190 million children and 19 million pregnant women have vitamin A deficiency (VAD), and almost a million children go blind every year7. In the Philippines, VAD ranges between 19.6 to 27.9% in infants and preschool children8, while in Bangladesh, over half of the preschool age (56.3%) and school age children (53.3%) at the national level were found to exhibit at least a mild grade of VAD9.
Several crops such as maize, cassava, and sweet potato have been successfully biofortified with elevated levels of provitamin A10,11. However, there is no naturally-occurring variation for provitamin A in grains in rice germplasm, so this has been achieved by using genetic engineering approaches. The genetic modification was made by the addition of two genes, phytoene synthase (Zmpsy1) from Zea mays and carotene desaturase (crtI) gene from the common soil bacterium, Pantoea ananatis (syn. Erwinia uredovora) into a temperate japonica rice variety, Kaybonnet, from the USA. This completed the carotenoid pathway in the grain and resulted in the accumulation of β-carotene in the endosperm12,13. However, the transfer of this golden rice trait from Kaybonnet into additional locally-adapted and widely-grown rice varieties is required for the successful release and adoption of golden rice in Asia.
Among the six second-generation Golden Rice (GR2) events received by the International Rice Research Institute (IRRI), event GR2E was found to contain a single intact copy of the inserted DNA integrated at a single site within the rice genome, giving rise to agronomically desirable progeny with suitable grain carotenoid content. This event has been transferred into Asian rice varieties through marker-assisted backcrossing (MABC). MABC has been successfully used to transfer high value genes/QTLs for disease resistance, submergence and drought tolerance traits into popular rice varieties without altering their desirable traits14–16.
Development of stable golden rice breeding lines with nutritionally relevant levels of provitamin A and without trait-associated yield, grain quality, or disease resistance penalties relative to the recipient parental varieties is essential for the successful adoption of golden rice. Introgression of the GR2E locus from GR2E Kaybonnet into PSBRc82, IR64, and BRRI dhan29 (BR29) was performed at IRRI through MABC along with selections for desirable agronomic and grain quality traits. The phenotypic evaluation was conducted under screen house conditions. Selection of homozygous plants and lines were carried out under field conditions at IRRI. Agronomic evaluations of selected lines were carried out under field conditions in a series of confined field tests (CTs) at IRRI, PhilRice and BRRI.
The main objectives of the present work were to: develop agronomically desirable lines of provitamin A enriched GR2E golden rice in the genetic backgrounds of popular rice varieties from Asia; to understand the effects of genetic background and environment on carotenoid expression, and to identify stable and productive lines of GR2E golden rice for varietal evaluation.
Results
Introgression of event GR2E into multiple genetic backgrounds
A series of five backcrosses of event GR2E Kaybonnet into three widely-grown rice varieties, IR64, PSBRc82, and BR29, resulted in the identification of introgression lines that were agronomically similar to their respective recipient parents. The stability and inheritance of the GR2E locus was confirmed using event-specific PCR in every generation, where it was found to segregate without distortion in a typical 1:1 Mendelian ratio in all the backcross generations (BC1 to BC5) and genetic backgrounds. All seeds containing the GR2E event showed the typical golden yellow color, indicating the expression of the provitamin A trait in the endosperm. Hemizygous (It is a condition in a diploid organism, where only one copy of the locus is present) plants phenotypically similar to their respective recipient parents were identified, backcrossed and advanced up to BC5F1, and with each successive backcross there was a progressive increase in similarity of the progenies to their respective recurrent (recipient) parents (Fig. 1). A total of 400, 190, and 94 BC5F1 plants of IR64, PSBRc82, and BR29, respectively, were phenotyped and genotyped by event-specific PCR. Yellow BC5F2 seeds were selected and analyzed for total carotenoid content, which ranged from 3.6–6.2 ppm in IR64, 3.1–6.4 ppm in PSBRc82, and 3.2–8.0 ppm in BR29. The BC5F2 plants were closer to respective recipient parents for key agronomic traits with average days to flowering (DTF), plant height (PH) and number of panicles (NP) of the selected BC5 progenies were 71.5 days, 108 cm and 15 for IR64, 82.5 days, 122.3 cm and 15.4 for PSBRc82 and 83 days, 117 cm and 17 for BR29 respectively. The final set of BC5F3 selected lines had background recovery of more than 98%. Agro-morphological traits, panicle characteristics, and grain parameters were similar to the recipient parents and no unintended, unexpected, effects due to the presence of the GR2E event were observed throughout the backcross breeding program. Based on the overall agronomic performance, carotenoid levels, and genetic background recovery, 40 BC5F1 plants in the IR64 background, and 20 BC5F1 plants in each of the PSBRc82 and BR29 backgrounds were selected. The BC5F2 seeds produced by each of these plants were further evaluated under field conditions in confined tests and plants homozygous for the GR2E locus were selected.
Figure 1.

(a–c) GR2E introgression lines.
Selection of homozygous and agronomically acceptable GR2E lines
The first confined field test of GR2E breeding lines was carried out during the 2015WS at IRRI to make individual homozygous plant selections. From among 8000 BC5F2 plants tested, a total of 602, 439, and 471 plants homozygous for the GR2E locus were identified in IR64, PSBRc82, and BR29, respectively (Fig S1). Efforts were focused on the lines homozygous for GR2E; however, hemizygous and null plants were also phenotyped to determine the impact of the presence of the GR2E locus on agronomic traits. The pair-wise t-tests were conducted between families derived from single BC5F1 plants within each of the three genetic backgrounds. Significant differences between families for total carotenoids were noted in a number of the possible pair-wise comparisons (data not shown). The mean comparisons between homozygous, hemizygous and null GR2E plants within each of the three populations did not show any abnormal deviations for key agronomic traits (Fig S2). The mean PH of lines carrying GR2E were marginally shorter than the respective recipient parent. For the remaining traits there were no clear differences between plants carrying GR2E and the respective parent variety. A total of 70 BC5F3 ILs similar to their respective parents and having higher levels of carotenoids were selected for IR64 and PSBRc82 genetic backgrounds.
Evaluation of GR2E introgression lines in multi-location replicated confined tests
Agronomic performance of GR2E Introgression Lines (ILs) and their respective control varieties were assessed in a series of CTs at IRRI (2015WS, 2016DS and 2016WS), PhilRice (2015WS and 2016DS) and BRRI in Bangladesh (2016 Boro). A total of 70 ILs similar to their respective parents in agronomic performance and having the greatest levels of carotenoids were selected from each of IR64 and PSBRc82 backgrounds. A total of 14 agronomic, yield and yield-related traits and carotenoid content were measured from the different confined tests. Among the 70 ILs tested during the 2015WS at IRRI, PSBRc82 GR2E ILs showed small but statistically significant differences from non-transgenic PSBRc82 for eight traits including days to flowering (DTF), plant height (PH), Flag leaf length (FL), flag leaf width (FW), filled spikelets (FS), total number of spikelets per plant (TSP), grain length (GL) and hundred seed weight (HSW) (Table 1). However, in successive CTs conducted using 32 GR2E PSBRc82 ILs at IRRI and PhilRice, only FL, GL and HSW (2016DS), and GL, HSW and plot yield (PY) (2016WS; IRRI) showed significant differences. On the other hand, no significant differences were observed during the 2016DS and only GL and HSW showed significant differences at PhilRice in 2016WS (Table 2). Similarly GR2E IR64 ILs showed small but significant differences to the recipient parent for FL, TSP, GL, GW and HSW in 2015DS and for FW, FS, spikelet fertility (SF) and PY in 2016DS, while only GL showed significant difference in 2016WS. For the CT conducted with GR2E BR29 ILs in Bangladesh in the 2016 Boro season there were no significant differences from BR29 for all the traits measured (Table 3). Significant variations in total carotenoids among different families were observed in all backgrounds. The highest concentration of total carotenoids was observed in the BR29 background, followed by the PSBRc82 background, while the IR64 background had the lowest concentration of total caroteneoids (Tables 1, 2, 3). The grain samples of GR2E ILs along with recipient parents are shown in Fig. 2. Grain quality traits amylose content (AC), gel consistency (GC) and alkali spreading value (ASV) were measured for PSBRc82, IR64 and BR29 (Tables 1, 2, 3). There were no significant differences for AC between GR2E PSBRc82 ILs and PSBRc82 in all the trials. There were no significant differences in ASV and AC between GR2E IR64 ILs and the IR64 parent, while for BR29 there were no differences between the transgenic and the control except for AC. The background recovery of final set of selected BC5F3 ILs showed more than 98% recipient genome in all the three genetic backgrounds (Fig S3–S5). There was no significant difference in AC except in BR29, similarly for GC some minor significant differences were observed in PSBRc82 and IR64 in some seasons.
Table 1.
Agronomic and grain quality performance of GR2EPSBRc82 at IRRI.
| Traits | 2015WS-IRRI | 2016DS-IRRI | 2016WS-IRRI | ||||||
|---|---|---|---|---|---|---|---|---|---|
| GR2EPSBRc82 | PSBRc82 | p value | GR2EPSBRc82 | PSBRc82 | p value | GR2EPSBRc82 | PSBRc82 | p value | |
| DTF | 83.2 ± 0.1 | 86.7 ± 0.3 | 0.000 | 79.2 ± 0.1 | 78.3 ± 0.8 | 0.325 | 89.4 ± 0.3 | 87.0 ± 1.7 | 0.179 |
| PH (cm) | 126.2 ± 0.4 | 131.1 ± 1.0 | 0.012 | 95.5 ± 0.4 | 98.5 ± 2.0 | 0.145 | 132.0 ± 0.5 | 136.2 ± 2.7 | 0.128 |
| TN | 13.7 ± 0.2 | 13.1 ± 0.5 | 0.952 | 15.1 ± 0.2 | 17.2 ± 1.1 | 0.068 | 14.0 ± 0.2 | 13.2 ± 1.1 | 0.474 |
| PN | 13.0 ± 0.2 | 12.2 ± 04 | 0.809 | 13.8 ± 0.2 | 15. 7 ± 1.1 | 0.065 | 13.6 ± 0.2 | 12.9 ± 1.1 | 0.512 |
| PL (cm) | 25.5 ± 0.1 | 25.5 ± 0.2 | 0.663 | 23.8 ± 0.1 | 23.5 ± 0.6 | 0.699 | 26. 5 ± 0.2 | 27.7 ± 1.1 | 0.299 |
| FL (cm) | 36.5 ± 0.4 | 32.5 ± 0.9 | 0.024 | 40.4 ± 0.5 | 33.7 ± 2.7 | 0.015 | 42.0 ± 0.5 | 38.6 ± 3.0 | 0.282 |
| FW(cm) | 1.6 ± 0.01 | 1.5 ± 0.02 | 0.028 | 1.3 ± 0.03 | 1.6 ± 0.2 | 0.245 | 1.5 ± 0.008 | 1.5 ± 0.05 | 0.283 |
| FS | 1298.6 ± 19.3 | 1099.9 ± 44.9 | 0.029 | 1538.5 ± 22.5 | 1477.9 ± 127.0 | 0.640 | 1176. 1 ± 21.4 | 1155.8 ± 120.9 | 0.869 |
| TSP | 1806.3 ± 24.7 | 1485.0 ± 57.7 | 0.002 | 1892.7 ± 29.0 | 1852.1 ± 164.1 | 0.808 | 1725.1 ± 25.5 | 1501.4 ± 144.1 | 0.130 |
| SF (%) | 72.0 ± 0.6 | 74.1 ± 1.5 | 0.131 | 81.5 ± 0.5 | 80.4 ± 2.7 | 0.677 | 68.5 ± 1.0 | 77.1 ± 5.6 | 0.135 |
| GL(mm) | 9.2 ± 0.02 | 9.8 ± 0.06 | 0.000 | 9.0 ± 0.03 | 9.6 ± 0.2 | 0.001 | 9.2 ± 0.03 | 9.7 ± 0.2 | 0.003 |
| GW (mm) | 2.5 ± 0.005 | 2.5 ± 0.01 | 0.179 | 2.4 ± 0.005 | 2.4 ± 0.02 | 0.353 | 2.5 ± 0.008 | 2.4 ± 0.04 | 0.042 |
| HSW (g) | 2.6 ± 0.01 | 2.5 ± 0.02 | 0.000 | 2.5 ± 0.01 | 2.6 ± 0.07 | 0.014 | 2.5 ± 0.02 | 2.8 ± 0.09 | 0.012 |
| PY (g) | 337.9 ± 4.9 | 327.5 ± 11.4 | 0.121 | 807.5 ± 6.3 | 821.0 ± 35.8 | 0.711 | 705.6 ± 12.0 | 878.1 ± 68.0 | 0.014 |
| TC (ppm) | 4.4 ± 0.06 | – | 0.000 | 4.7 ± 0.06 | – | 0.000 | 5.0 ± 0.1 | – | 0.000 |
| AC (%) | 20.6 ± 0.2 | 19.2 ± 1.1 | 0.248 | 22.8 ± 0.2 | 22.9 ± 0.9 | 0.909 | 23.1 ± 0.3 | 22.7 ± 1.6 | 0.666 |
| GC | 59.7 ± 1.6 | 62.7 ± 9.1 | 0.750 | 82.4 ± 1.5 | 84.3 ± 8.7 | 0.830 | 68.4 ± 1.6 | 77.7 ± 9.2 | 0.325 |
| ASV | HI/I | HI/I | – | HI/I | HI/I | – | HI/I | HI/I | – |
DTF days to fifty percent flowering, PH plant height, TN Tiller number, PN Panicle number, PL Panicle length, FL flag leaf length, FW flag leaf width, FS number of filled spikelets, TSP total number of spikelets per plant, SF spikelet fertility, GL grain length, GW grain width, HSW hundred seed weight, PY plot yield, TC total carotenoids, AC amylose content, GC gel consistency, ASV alkali spreading value.
Table 2.
Agronomic and grain quality performance of GR2EPSBRc82 at PhilRice.
| Traits | 2016DS-PhilRice | 2016WS-PhilRice | ||||
|---|---|---|---|---|---|---|
| GR2E PSBRc82 | PSBRc82 | p value | GR2E PSBRc82 | PSBRc82 | p value | |
| DTF | 86.0 ± 0.2 | 85.8 ± 0.6 | 0.231 | 81.5 ± 0.3 | 81.3 ± 1.4 | 0.985 |
| PH (cm) | 130.5 ± 0.4 | 129.2 ± 1.5 | 0.276 | 96.1 ± 0.4 | 98.3 ± 2.1 | 0.288 |
| TN | 9.9 ± 0.1 | 9.4 ± 0.4 | 0.357 | 10.8 ± 0.1 | 10.7 ± 0.8 | 0.867 |
| PN | 9.7 ± 0.1 | 9.4 ± 0.4 | 0.361 | 10.8 ± 0.1 | 10.7 ± 0.8 | 0.875 |
| PL (cm) | 26.7 ± 0.1 | 26.6 ± 0.5 | 0.093 | 24.1 ± 0.2 | 23.7 ± 1.4 | 0.792 |
| FL (cm) | 31.9 ± 0.4 | 31.6 ± 1.2 | 0.687 | 27.9 ± 0.3 | 25.8 ± 1.7 | 0.236 |
| FW(cm) | 1.3 ± 0.01 | 1.3 ± 0.03 | 0.643 | 1.2 ± 0.007 | 1.1 ± 0.05 | 0.205 |
| FS | 1098. 2 ± 20.1 | 1145.8 ± 69.8 | 0.781 | 1274.6 ± 27.8 | 1007.0 ± 158.0 | 0.085 |
| TSP | 1262. 9 ± 21.3 | 1329.0 ± 74.0 | 0.754 | 1410.1 ± 30.7 | 1089.2 ± 174.5 | 0.063 |
| SF (%) | 86.9 ± 0.5 | 86.3 ± 1.8 | 0.060 | 90.4 ± 0.4 | 92.6 ± 2.3 | 0.376 |
| GL(mm) | 9.7 ± 0.4 | 9.3 ± 1.5 | 0.149 | 8.9 ± 0.02 | 9.5 ± 0.1 | 0.000 |
| GW (mm) | 2.4 ± 0.01 | 2.5 ± 0.03 | 0.233 | 2.6 ± 0.01 | 2.5 ± 0.08 | 0.555 |
| HSW (g) | 2.6 ± 0.1 | 2.5 ± 0.4 | 0.951 | 2.4 ± 0.01 | 2.6 ± 0.07 | 0.002 |
| PY (g) | 376. 8 ± 5.0 | 404. 9 ± 17.4 | 0.583 | 654.7 ± 9.8 | 681.3 ± 55.5 | 0.658 |
| TC (ppm) | 4.8 ± 0.06 | – | 0.000 | 4.1 ± 0.05 | – | 0.000 |
| AC (%) | 20.0 ± 0.2 | 19.3 ± 1.4 | 0.644 | 20.6 ± 0.2 | 19.2 ± 1.1 | 0.248 |
| GC | 65.3 ± 1.3 | 80.3 ± 7.4 | 0.048* | 59.7 ± 1.6 | 62.7 ± 9.1 | 0.750 |
| ASV | HI/I | HI/I | – | HI/I | HI/I | – |
DTF days to fifty percent flowering, PH plant height, TN Tiller number, PN Panicle number, PL Panicle length, FL flag leaf length, FW flag leaf width, FS number of filled spikelets, TSP total number of spikelets per plant, SF spikelet fertility, GL grain length, GW grain width, HSW hundred seed weight, PY plot yield, TC total carotenoids, AC amylose content, GC gel consistency, ASV alkali spreading value.
Table 3.
Agronomic performance of GR2EIR64 at IRRI and GREBR29 at BRRI.
| Traits | 2015WS IRRI | 2016DS IRRI | 2016WS IRRI | 2016 Boro | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GR2E IR64 | IR64 | p value | GR2E IR64 | IR64 | p value | GR2E IR64 | IR64 | p value | GR2E BR29 | BR29 | p value | |
| DTF | 78.1 ± 0.3 | 81.0 ± 0.9 | 0.073 | 79.8 ± 0.2 | 80.0 ± 1.2 | 0.825 | 80.4 ± 0.2 | 82.3 ± 1.2 | 0.377 | 122.38 ± 0.2 | 124.3 ± 0.3 | 0.090 |
| PH (cm) | 118.6 ± 0.4 | 116.4 ± 1.2 | 0.285 | 96.4 ± 0.4 | 94.8 ± 2.5 | 0.535 | 115. 5 ± 0.5 | 120.5 ± 2.8 | 0.078 | 106.76 ± 0.4 | 107.9 ± 2.7 | 0.594 |
| TN | 13.8 ± 0.2 | 13.9 ± 0.5 | 0.525 | 14.3 ± 0.2 | 14.6 ± 1.2 | 0.825 | 14.1 ± 0.2 | 15.1 ± 1.2 | 0.377 | - | - | - |
| PN | 12.4 ± 0.2 | 12.3 ± 0.5 | 0.622 | 13.2 ± 0.2 | 13.7 ± 1.1 | 0.653 | 13.7 ± 0.08 | 13.5 ± 0.5 | 0.610 | 13.58 ± 1.0 | 12.93 ± 0.3 | 0.336 |
| PL (cm) | 27.0 ± 0.3 | 27.1 ± 0.1 | 0.460 | 24.7 ± 0.1 | 25.6 ± 0.7 | 0.232 | 27.3 ± 0.2 | 28.6 ± 1.0 | 0.215 | 26.59 ± 0.1 | 27.4 ± 0.9 | 0.210 |
| FL (cm) | 38.2 ± 0.3 | 34.2 ± 0.9 | 0.000 | 34.0 ± 0.4 | 33.8 ± 2.1 | 0.924 | 41.7 ± 0.4 | 40.9 ± 2.4 | 0.753 | – | – | – |
| FW(cm) | 1.7 ± 0.009 | 1.73 ± 0.02 | 0.389 | 1.5 ± 0.009 | 1.7 ± 0.05 | 0.001 | 1.6 ± 0.009 | 1.7 ± 0.05 | 0.233 | – | – | – |
| FS | 1045.1 ± 15.0 | 943.1 ± 46.2 | 0.068 | 1337.7 ± 22.8 | 898.4 ± 128.9 | 0.001 | 1110.0 ± 23.4 | 1045.0 ± 132.2 | 0.630 | 2147.61 ± 37.5 | 2022.15 ± 49.7 | 0.491 |
| TSP | 1751.4 ± 22.5 | 1580.9 ± 69.1 | 0.043 | 1679.2 ± 26.4 | 1510.9 ± 149.1 | 0.269 | 1593.9 ± 24.0 | 1596.9 ± 135.6 | 0.983 | 2603.29 ± 36.7 | 2574.99 ± 61.2 | 0.873 |
| SF (%) | 59.8 ± 0.5 | 59.6 ± 1.6 | 0.995 | 79.8 ± 0.7 | 59.5 ± 4.0 | 0.000 | 69.3 ± 0.9 | 65.1 ± 5.2 | 0.433 | 17.7 ± 0.5 | 21.47 ± 0.4 | 0.140 |
| GL (mm) | 9.6 ± 0.0.02 | 9.8 ± 0.04 | 0.015 | 9.7 ± 0.03 | 10.1 ± 0.2 | 0.069 | 10.0 ± 0.03 | 10.4 ± 0.2 | 0.027 | 8.43 ± 0.02 | 8.58 ± 0.09 | 0.134 |
| GW (mm) | 2.5 ± 0.007 | 2.4 ± 0.02 | 0.002 | 2.4 ± 0.008 | 2.4 ± 0.04 | 0.616 | 2.4 ± 0.007 | 2.4 ± 0.04 | 0.766 | 1.8 ± 0.005 | 1.81 ± 0.03 | 0.880 |
| HSW (g) | 2.5 ± 0.009 | 2.7 ± 0.03 | 0.003 | 2.6 ± 0.02 | 2.4 ± 0.09 | 0.069 | 2.6 ± 0.02 | 2.7 ± 0.09 | 0.323 | 2.81 ± 0.01 | 2.14 ± 0.05 | 0.289 |
| PY (g) | 297.0 ± 5.3 | 227.8 ± 16.4 | 0.062 | 757.6 ± 10.3 | 421.6 ± 58.3 | 0.000 | 638.1 ± 15.7 | 656.7 ± 88.8 | 0.837 | 3479.39 ± 0.03 | 3511.26 ± 0.01 | 0.864 |
| TC (ppm) | 4.4 ± 0.05 | – | 0.000 | 3.9 ± 0.04 | – | 0.000 | 4.4 ± 0.05 | – | 0.000 | 10.99 ± 0.55 | – | – |
| AC (%) | 19.4 ± 0.2 | 19.7 ± 0.9 | 0.785 | 22.1 ± 0.1 | 22.4 ± 0.9 | 0.528 | 19.5 ± 0.2 | 19.6 ± 1.4 | 0.956 | 24.0 | 27.0 | 0.004 |
| GC | 56.2 ± 1.1 | 74.7 ± 6.0 | 0.004 | 76.6 ± 1.1 | 69.7 ± 9.1 | 0.475 | 52.7 ± 1.1 | 68.3 ± 6.1 | 0.012 | – | – | – |
| ASV | I | I | I | I | – | I | I | – | 4.5 | 4.0 | 0.669 | |
DTF days to fifty percent flowering, PH plant height, TN Tiller number, PN Panicle number, PL Panicle length, FL flag leaf length, FW flag leaf width, FS number of filled spikelets, TSP total number of spikelets per plant, SF spikelet fertility, GL grain length, GW grain width, HSW hundred seed weight, PY plot yield, TC total carotenoids, AC amylose content, GC gel consistency, ASV alkali spreading value.
Figure 2.
Grain samples of GR2E golden rice and respective recipient parents.
Correlation between yield, yield related traits and carotenoid content
The correlation among yield and yield related traits; and with total carotenoid content is presented in the Figs S6–S8. Over all there was no specific trend in correlations among different yield and yield related traits. Except in one environment carotenoid content was negatively but non-significantly associated with PY in all the three genetic backgrounds. The correlation analysis of carotenoid content between different seasons showed highly significant correlation in all the three genetic backgrounds.
Effect of genetic background and environment on expression of carotenoids
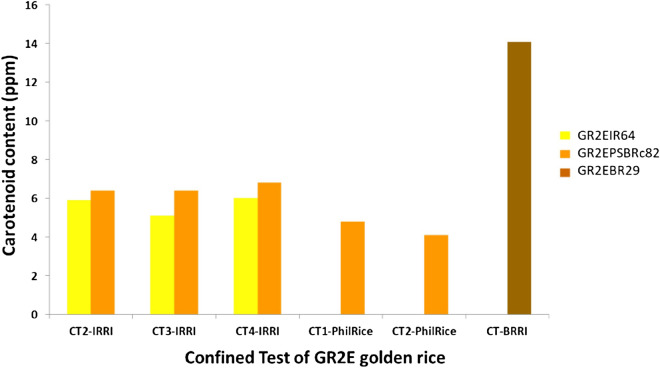
The combined analysis of variance for carotenoid content at two months after harvest showed that there were significant genotypic, seasonal and location effects on the expression of carotenoid content. However, there were no significant genotype and environmental interactions (G × E) for carotenoid content except CT2 PR vs CT4 (Table 4). However, among the three genetic backgrounds, expression of carotenoids was higher in GR2E BR29 ILs followed by PSBRc82 and lowest in GR2EIR64 ILs (Fig. 3, Fig S9). There were very highly positive significant correlations for carotenoid content estimated in different locations both within and between seasons (Figs S10–S12). In general carotenoids expression was bit higher in WS than in DS, but also among most of the CTs no significant G × E interaction was observed (Table 4).
Table 4.
Analysis of variance and G × E for carotenoid content.
| Trials | Genotype | Season or Location | Genotype × season or genotype × location | |||
|---|---|---|---|---|---|---|
| F value | p value | χ2 value | p value | χ2 value | p value | |
| CT3 vs CT4 IRRI | 9.526 | 0.001 | 2.788 | 0.095 | 2.271 | 0.131 |
| CT2 PR vs CT3 IRRI | 10.138 | 0.001 | 7.901 | 0.005 | 0.134 | 0.714 |
| CT2 PR vs CT4 IRRI | 6.289 | 0.001 | 7.298 | 0.007 | 10.308 | 0.001 |
| Pooled (location effect) | 41.228 | 0.001 | 9.315 | 0.002 | 0.182 | 0.669 |
| Pooled (environment effect) | 37.570 | 0.001 | 10.370 | 0.001 | 0.000 | 0.999 |
CT confined test, IRRI International Rice Research Institute, PR Philrice.
Figure 3.
Carotenoid levels in different genetic backgrounds.
Identification of superior GR2E NILs for multi-location evaluation
We selected five GR2E introgression lines each for PSBRc82 and IR64, for BR29 eight lines were selected from the CTs. These lines will be further evaluated in multi-location field testing in the Philippines and Bangladesh respectively. The list of selected lines and their corresponding agronomic performance is provided in Table 5. The ILs were similar to the respective recipient parents in all the agronomic, yield and yield traits measured, and the total carotenoids ranged from 3.8 to 5.5 ppm in the DS and 4.1 to 6.1 in the WS. Among the eight selected GR2E BR29 ILs no significant variation was observed in any trait except yield, with an advantage of 12.8% over BR29.
Table 5.
Agronomic performance of GR2E lines selected for multi-location evaluation.
| Designation | DTF | PH (cm) | GL (mm) | GW (mm) | HSW (g) | GY (kg/ha) | TC (ppm) | AC (%) | ASV | GC | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | DS | WS | ||||
| IRRI | |||||||||||||||||
| IR 112017 GR 2-E:23-30-98-B | 78.3 | 92.0 | 94.6 | 134.5 | 8.9 | 8.9 | 2.4 | 2.5 | 2.4 | 2.4 | 6718.8 | 5749.8 | 4.7 | 5.2 | 19.8 | HI | 67 |
| IR 112017 GR 2-E:23-40-51-B | 80.0 | 88.7 | 96.3 | 136.6 | 9.1 | 9.4 | 2.5 | 2.5 | 2.5 | 2.6 | 5958.5 | 6280.6 | 4.8 | 5.0 | 22.0 | I | 40 |
| IR 112019 GR 2-E:38-4-13-B | 80.0 | 86.7 | 91.6 | 124.7 | 8.6 | 8.6 | 2.4 | 2.5 | 2.5 | 2.4 | 6305.0 | 5327.7 | 5.3 | 5.8 | 21.3 | I | 31 |
| IR 112019 GR 2-E:38-4-27-B | 79.7 | 87.0 | 91.1 | 120.8 | 8.8 | 9.0 | 2.4 | 2.4 | 2.4 | 2.4 | 6221.5 | 5110.6 | 5.5 | 6.0 | 17.9 | HI/I | 71 |
| IR 112020 GR 2-E:57-18-75-B | 79.3 | 93.3 | 94.9 | 133.5 | 9.1 | 9.3 | 2.4 | 2.5 | 2.3 | 2.5 | 6040.2 | 5900.3 | 5.1 | 4.9 | 19.2 | HI | 63 |
| PSBRc82 | 78.3 | 87.0 | 98.5 | 136.2 | 9.6 | 9.6 | 2.4 | 2.5 | 2.6 | 2.5 | 6414.3 | 6859.9 | 0.6 | 0.5 | 17.4 | HI/I | 62 |
| PhilRice | |||||||||||||||||
| IR 112017 GR 2-E:23-30-98-B | 80.7 | 86.3 | 95.4 | 127.5 | 8.9 | 9.2 | 2.5 | 2.4 | 2.3 | 2.3 | 9060.7 | 4737.4 | 3.9 | 5.1 | 17.4 | I | 71 |
| IR 112017 GR 2-E:23-40-51-B | 80.7 | 84.8 | 98.3 | 126.7 | 8.9 | 8.9 | 2.6 | 2.6 | 2.3 | 2.4 | 8484.4 | 5027.0 | 3.8 | 4.6 | 18.6 | HI | 63 |
| IR 112019 GR 2-E:38-4-13-B | 80.0 | 84.3 | 92.5 | 127.3 | 8.6 | 9.1 | 2.5 | 2.5 | 2.3 | 2.4 | 8541.5 | 4414.4 | 4.1 | 4.9 | 23.2 | I | 30 |
| IR 112019 GR 2-E:38-4-27-B | 81.3 | 86.4 | 90.7 | 127.8 | 8.7 | 9.1 | 2.5 | 2.4 | 2.1 | 2.4 | 8327.5 | 4276.4 | 5.1 | 6.1 | 23.0 | HI/I | 47 |
| IR 112020 GR 2-E:57-18-75-B | 80.7 | 85.8 | 98.6 | 133.1 | 9.2 | 9.1 | 2.5 | 2.5 | 2.3 | 2.5 | 8726.6 | 5737.3 | 4.2 | 4.1 | 18.1 | HI | 81 |
| PSBRc82 | 81.3 | 85.9 | 98.3 | 129.2 | 9.5 | 9.3 | 2.5 | 2.5 | 2.6 | 2.5 | 8527.6 | 5061.2 | 0.6 | 0.3 | 19.3 | HI | 80 |
| BRRI | |||||||||||||||||
| IR 112060 GR2E:2-9-89-16-55 | 127.8 | 106.6 | 8.3 | 2.1 | 1.9 | 5700 | 13.8 | 25 | – | – | |||||||
| IR 112060 GR2E:2-7-63-1-60 | 126.3 | 108.4 | 8.4 | 2.1 | 2.0 | 5950 | 12.3 | 23 | – | – | |||||||
| IR 112060 GR2E:2-7-63-2-96 | 126.8 | 108.4 | 8.4 | 2.1 | 2.0 | 6373 | 12.1 | 24 | 4.5 | – | |||||||
| IR 112060 GR2E:2-17-36-10-71 | 126.1 | 106.5 | 8.4 | 2.2 | 1.9 | 5751 | 13.3 | 23 | – | – | |||||||
| IR 112062 GR2E:14-40-7-16-22 | 126.9 | 108.3 | 8.4 | 2.1 | 1.9 | 6287 | 13.2 | 23 | – | – | |||||||
| IR 112062 GR2E:14-40-7-21-4 | 127.3 | 107.9 | 8.4 | 2.1 | 1.9 | 6178 | 12.8 | 25 | – | – | |||||||
| IR 112062 GR2E:14-40-7-8-64 | 125.2 | 108.1 | 8.4 | 2.1 | 2.0 | 5865 | 12.0 | 23 | – | – | |||||||
| IR 112062 GR2E:14-40-7-23-8 | 126.8 | 108.6 | 8.5 | 2.1 | 2.0 | 5670 | 12.7 | 24 | – | – | |||||||
| BR29 | 125.5 | 109.0 | 8.5 | 2.2 | 2.0 | 5649 | 0.0 | 26 | 4.0 | – | |||||||
DTF days to fifty percent flowering, PH plant height, TN Tiller number, PN Panicle number, PL Panicle length, FL flag leaf length, FW flag leaf width, FS number of filled spikelets, TSP total number of spikelets per plant, SF spikelet fertility, GL grain length, GW grain width, HSW hundred seed weight, PY plot yield, TC total carotenoids, AC amylose content, GC gel consistency, ASV alkali spreading value, DS dry season, WS wet season.
Discussion
Most of the dietary vitamin A is of plant origin in the form of provitamin A that is converted to vitamin A in the body17. VAD is persistent in most of the rice eating countries in Asia, Africa and Latin America18,19. Therefore, enriching rice with provitamin A through biofortification is a viable and complementary intervention to tackle the VAD. The provitamin A trait was introduced into the rice variety Kaybonnet through genetic engineering13, which has a temperate japonica genetic background and is not well adapted to the tropical conditions in most rice growing Asian countries. We developed GR2E event introgressed golden rice ILs in the genetic backgrounds of IR64, PSBRc82 and BR29.
Introgression of the GR2E produced agronomically superior plants
Golden rice GR2E is genetically stable and molecularly clean event useful for breeding (https://www.dropbox.com/sh/qpiz0cftefcaceq/AAByIpj_HED3zgqH7ufW7A-ta?dl=0; https://www.foodstandards.gov.au/code/applications/Documents/A1138%20Application_Redacted.pdf). The breeding process to develop GR2E introgression lines did not show any abnormal plant phenotypes both in homozygous and hemizygous conditions indicating the genetic stability of the GR2E gene and trait expression. Both the phenotypic and genotypic based segregation analysis showed typical Mendelian segregation ratio in different segregating generations. GR2E advance backcross progenies were phenotypically very similar to their respective recipient parents. Transgenic events with single copy, clean integration and showing normal Mendelian segregation are considered ideal for research and breeding purposes, as they do not alter the host plant genome20–22.
Agronomic performance at field level and G × E studies showed that the GR2E gene did not alter any of the traits of the recipient parents in all its zygosity conditions. Overall plant performance was better during DS and among the genetic backgrounds the GR2EPSBRc82 lines performed better than the GR2EIR64 lines. Morphological traits such as panicle type, panicle exertion, grain shape, flag leaf length and width were similar for the GR2E ILs. Many lines performed equally similar to the respective recurrent parents, allowing the selection of advanced lines in all backgrounds for further testing in multi-location trials. The results showed that back cross process recovered almost all the desirable agronomic, yield and grain quality traits of the respective parents with significant expression of vitamin A. Despite many typhoons, heavy rains and high winds during the trials. There were no severe lodging incidences observed. Insects and diseases incidences were monitored during the two growing seasons at two different plant growth stages: maximum tillering stage (vegetative stage) and 50% flowering. Generally, crop stand was good with manageable level of insect pests and diseases during the growing seasons. Insects observed (both pest and beneficial insects) were found to be present in both test materials. We did not notice any difference between GR2E introgression lines and their respective recipient parents for the pest or diseases pressure on the crop across the confined field tests.
Woodfield and White23, and Badenhorst et al.24 opined that development of transgenic product is not limited only to transformation, but also includes breeding through further backcrossing of transgenes with recipient parents and selection for desired traits of interest, in order to expedite commercial product development. For commercial deployment of any new variety with one or more introduced new trait(s) of a staple crop, in parallel to yield and other key agronomic traits, the newly developed variety should have essentially similar or better performance against biotic and abiotic stresses and grain quality traits compared to recipient variety; the introduced trait(s) should not alter these traits of the recipient variety25,26.
Grain quality and proximate composition of GR is similar to recipient rice varieties
Furthermore, different cooking and eating quality traits like, AC and ASV did not show any significant difference between the ILs and their respective recipient parents in any CTs. The golden rice breeding lines with significant amount of provitamin A accumulated in the grains helps to tackle VAD in high risk countries such as Bangladesh and the Philippines. However, it is a requirement to assess the composition of genetically modified crops to see if any significant changes in grain quality, nutrients and anti-nutrients contents in comparison to traditional counterpart and to assess the safety of the intended or unintended changes27,28. The compositional analysis of golden rice showed that all the compounds measured are within the biologically acceptable range and does not pose any risk to human health29. Earlier reports on transgenic products for insect and herbicide tolerance have also shown that little biologically meaningful changes in grain quality, nutrient and anti-nutrient composition30. There was a clear environmental effect, even though total carotenoids varied with environments, the genotypes with high carotenoids were always the best in all the locations. Such variations in trait expression due to environmental and agronomic factors and genetic basis have been well explained31,32.
Genetic background and environment influences carotenoid expression
Stable trait expression and minimal G × E for any trait of importance, especially for grain micronutrients and vitamins is essential for varietal release as well as for their successful adoption4,33,34. Total carotenoids were well correlated across the sites and generations; and expressed stably across the environments but there is a genetic background effect. Carotenoids expression varied even within segregating lines of different generations in each of the genetic backgrounds. So targeted breeding and careful selection of progenies with carotenoids test in each generation is necessary for advancing the lines. Mapping background QTLs and genes and using them in MAB can provide opportunity for precise development of GR lines with highest expression. The carotenoid levels were found to vary across the genetic backgrounds, locations and seasons but there were no significant G × E interactions. The highest expression of carotenoids was observed in BR29 background and the lowest in IR64 background. Several earlier attempts to develop golden rice events and introgression lines had to face the genetic background effects. Transgenic events developed in the indica backgrounds of IR64 and BR29 reported lower expression of GR genes in IR64 and higher expression in BR29 transformants, even ILs developed in IR64 showed lesser expression35. Moreover, ILs did not show any significant difference in yield when expressing the genes in the carotenoid pathway36. In our study also lowest expression was noticed in IR64. Simultaneously efforts are being made to develop next generation golden rice events with elevated levels of carotenoids with longer stability37–39. However, a genetic background effect is still a major bottle neck for introgression of carotenoid trait. Background effect on the expression of introduced traits was reported in rice for submergence tolerance, yield and related traits, disease resistance and drought tolerance15,16,40,41.
The variation in carotenoid concentration in grains might be due to variations in sunlight exposure and intensity across the locations and seasons42. Differential accumulation of β-carotene due to variation in exposure period and intensity of sunlight was also observed in algae, carrots, pumpkin and maize43–46. Moreover, like other carotenoids containing crops the carotenoid concentration in the grains of golden rice degrades over time after harvest. The degradation rate is very high at first few weeks after harvest and it becomes very slow after 6–8 weeks (data not shown). The carotenoids degradation rate is highly influenced by the storage temperature, moisture and exposure to light of the storage environment22,47. So, development of golden rice varieties with stable carotenoids expression is essential to achieve the impact37. However, there might be genotypic effect on the retention ability for carotenoids in rice grain. Understanding background effect and standardization of post-harvest handling is needed to achieve desired level of carotenoids in the introgression lines of multiple backgrounds.
Superior introgression lines were identified for multi-location trials
The five back crosses of GR2E gene into three genetic backgrounds resulted in identification of ILs similar to respective recipient parents. Adoption by the farmers and preference by the consumers for a specific crop variety particularly rice introduced with a new trait largely depends on its yield, grain quality and eating quality parameters. The introduced trait should be stable over locations and seasons to expedite the adoption level. Considering the present levels of carotenoids and per capita consumption in these target countries, the resulting ILs would be able to supply 30–50% of the EAR for vitamin A for the high risk population group if GR2E rice is consumed regularly.
Materials and methods
Development of GR2E near isogenic lines
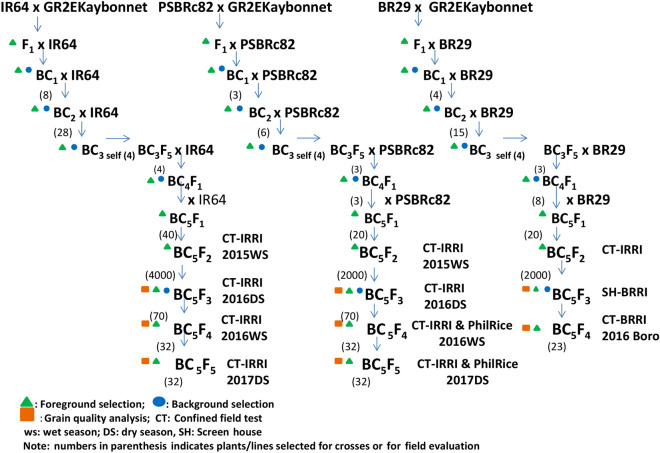
Kaybonnet is a high yielding japonica rice variety with blast resistance and excellent milling quality commercially cultivated in the USA. The genetic modification was made by the addition of two genes, phytoene synthase (Zmpsy1) from Zea mays and carotene desaturase (crtI) gene from the common soil bacterium, Pantoea ananatis (syn. Erwinia uredovora). The GR2E Kaybonnet was crossed with the popular high yielding and adopted rice varieties such as IR64, PSBRc82, and BR29. IR64 is popular in most of the Asian countries, PSBRc82 in the Philippines, and BR29 in Bangladesh. In each generation, segregating materials were genotyped using GR2E event specific molecular marker. Plants containing the GR2E event and phenotypically similar to respective recipients were selected and backcrossed in each backcross generation to advance the materials to BC5F2. Background selections were performed using 100 randomly selected SSR markers in BC1 and BC2, while selected plants from BC3, BC4 and BC5 were genotyped using the 6 K SNPs set at Genotyping Service Laboratory, IRRI. Only yellow-colored BC5F2 seeds were separated and analyzed for total carotenoid content. A total of 40 BC5F2 families for IR64 and 20 families each for PSBRc82 and BR29 were selected for evaluation in the confined test at IRRI. We have provided details of MAB scheme and evaluation of introgression lines in the Fig. 4.
Figure 4.
Development and evaluation of GR2E introgression lines.
Experimental materials used in the confined tests
A total of 8000 individual plants comprised of 4000 BC5F2 plants from GR2E IR64, 2000 plants each from GR2E PSBRc82 and GR2E BR29 were included in a CT in the dry season of 2015 (2015DS). Plants were genotyped using GR2E specific markers and homozygous plants were selected. Selected BC5F3 homozygous plants from each genetic background along with the respective recipient and donor parents were evaluated in a series of CTs at IRRI and PhilRice in the Philippines and at BRRI in Bangladesh. The list of GR2E materials evaluated and the details of the CTs is provided in the Supplementary Table S1. Three CTs were conducted for GR2E IR64 and GR2E PSBRc82 at IRRI, while the selected lines of GR2E PSBRc82 were evaluated for two seasons at PhilRice. Further, BC5F3 seeds of GR2E BR29 were sent to Bangladesh, multiplied in the screen house, and further evaluated in a CT at BRRI, Gazipur, for one season in 2016.
Crop management and observations
Seeds of the selected plants of GR2E introgression lines, recipient and donor parents were seeded in trays. Seedlings were transplanted at 21 days after sowing with a standard spacing of 20 × 20 cm. Details of the experimental design and layout are provided in Tables S1 and S2. Standard agronomic practices were followed to raise a good crop, including the application of need-based plant protection measures to protect the crop from diseases and insect pests. Data were gathered on key agronomic, yield and yield-related traits; and total carotenoid content was measured two months after harvest. Grain quality data were generated from the selected lines of CT2 and from all lines included in CT3 and CT4. Insect pest infestations and disease incidences were recorded at maximum tillering and at 50% flowering. Agronomic traits were measured on five random plants from each entry. Days to 50% flowering was recorded on a whole plot basis. At maturity, five selected plants were harvested from individual plots and the remaining inner plants were harvested in bulk. Final plot yield was adjusted to a uniform grain moisture content of 14%.
Genotyping
DNA was extracted using fresh leaf samples and following a modified cetyl trimethylammonium bromide (CTAB) protocol48. Nanopore was used to check the quality and quantity of the DNA extracted. The DNA samples were diluted with distilled water into an equal concentration of 25 ng/µl. Amplification of event specific markers using polymerase chain reaction (PCR) was carried out with a 10 µl reaction mixture that contained 1.5 µl of DNA template, 1.0 µl of 10 × PCR buffer with MgCl2, 0.5 µl each of forward and reverse primers, 0.2 µl of 1 mM dNTP and 0.1 µl of Taq DNA polymerase and 5.7 µl distilled water. The amplification reaction was carried out in a 96-well PCR plate in a thermocycler using the following temperature profile: denaturation, 95 °C for 5 min; 35 cycles of denaturation at 95 °C for 45 s, annealing at 55 °C for 45 s and extension at 72 °C for 45 s; and final extension at 72 °C for 8 min and long-term storage at 10 °C. Amplification products were separated by gel electrophoresis on 1.2% agarose (0.5 × TBE; 160 V for 45 min) and visualized using SYBR Safe DNA stain and imaging using an AlphaImager HP (Protein Simple, San Jose, CA) gel documentation system. The GR2E specific primer sequences as follows.
ZD-E1-P1 5′-GCTTAAACCGGGTGAATCAGCGTTT-3′
ZD-E1-P2 5′-CGAGAGGAAGGGAAGAGAGGCCACCAA-3′
ZD-E1-P3 5′-CTCCCTCACTGGATTCCTGCTACCCATAGTAT-3′
Grain quality analysis
Grain quality analysis was carried out at the Analytical Service Laboratory (ASL) of IRRI. We measured/analyzed grain length and width, amylose content, alkali spreading value and gel consistency, using standard protocols49. Similar analyses were performed at BRRI on grain samples of GR2E BR29.
Amylose content
Amylose content (AC) was determined on milled rice extracts using a segmented flow analyzer. Rice samples were ground to a fine powder using a cyclone mill. Sodium Hydroxide and Ethanol were added to a test portion of the sample and heated in a boiling bath for 10 min. Acetic acid and Iodine solution was mixed with the aliquot of the test solution to form a blue starch iodine complex and its absorbance was measured at 620 nm using a colorimeter49. The result of the analysis was reported as apparent amylose to take into account the contribution of amylopectin present in the rice, which also forms a blue color starch iodine complex.
Gelatinization temperature
Rice starch gelatinization temperature (GT) was estimated by determining the alkali spreading value (ASV) of milled rice grains in potassium hydroxide solution. Six kernels of whole milled rice were incubated with 10 ml of 1.7% KOH for 23 h at ambient temperature (25 °C). The appearance and disintegration of the endosperm was visually rated depending on the intensity of spreading and swelling. ASV of 1–2 was classified as high GT, 3 for intermediate to high GT, 4–5 for intermediate GT and 6–7 for low GT.
Gel consistency
Samples of milled rice were ground to a fine powder, placed in a culture tube and suspended in a mixture of ethanol and 0.2 N KOH containing thymol blue and incubated in a boiling water bath for 15 min, followed by cooling to room temperature (15 min) and placing in an ice bath (20 min). Gel consistency of the rice paste (4.4% w/v) was determined by measuring the length of the cold gel in the culture tube after placing horizontally for 1 h. Rice was differentiated into three consistency types—soft (61 to 100 mm), medium (41 to 60 mm) and hard (27 to 40 mm).
Carotenoid concentrations
Total carotenoid concentration was estimated following the protocol developed by Gemmecker et al.50. Dehulled and polished rice seeds were ground to a fine powder using a modified paint shaker and accurately weighed amounts (ca. 500 mg) were dispensed into 15-ml Falcon tubes, mixed by sonication with 2 ml distilled water and incubated for 10 min at 60 °C. Cooled samples were centrifuged (3000g, 5 min) and the supernatant fractions were transferred to new 15-ml tubes. Acetone (2 ml) and 100 μl of the lipophilic metallo organic dye, VIS682A (20 μg/ml; QCR Solutions Corp.), as an internal standard were added to each sample followed by mixing with short pulses of sonication and centrifugation (3000g, 5 min). Supernatants were transferred to 15-ml tubes and the pellets were re-extracted twice more with 2-ml volumes of acetone and the resulting supernatant fractions were combined. Two ml petroleum ether (PE): di-ethyl ether (DE) (2:1 v/v) was added to each combined supernatant fraction (ca. 8 ml) and volumes were adjusted to 14 ml with distilled water. After vortexing, phase separation was achieved by centrifugation (3000g, 5 min). The organic phase was recovered by pipetting out and transferred into a 2 ml graduated Eppendorf tube and the remaining aqueous phase was re-extracted with another 2 ml PE:DE (2:1 v/v), followed by centrifugation (3000g, 5 min). The combined organic phases were dried using a vacuum-concentrator (Eppendorf concentrator 5301) and re-dissolved in 1 ml acetone. Maximum absorbance of sample extract at 450 nm and maximum absorbance of internal standard at 680 nm was determined using DU730 Beckman Coulter UV/VIS spectrophotometer. Concentrations of total carotenoids were determined from A450 nm assuming an average E450 nm = 142, 180 l mol−1 cm−1 in acetone using the Beer-Lambert law corrected for sample dilution and normalized to the internal standard.
Statistical analysis
All statistical analyses were performed as a linear mixed model using R51 and PB Tools v1.052.
Mixed model for single site analysis:
where µi denotes the mean of the ith entry (fixed effect), bj denotes the effect of the jth block, and eij denotes the residual error.
Mixed model for multiple site analysis:
where µi denotes the mean of the ith entry (fixed effect), lk denotes the effect of the kth site, bj(k) denotes the effect of the jth block within the kth site, (µl)ik denotes the interaction between the entries and sites (random effect), and eijk denotes the residual error.
Mean comparison and correlation analysis
The differences in least square (LS)-mean values between GR2E rice and the control rice were tested at first step followed by significant difference (p < 0.05) was identified in the multi-year combined-sites analysis53. Correlation among different traits from all the replicated trials was carried out using R Program51.
Supplementary Information
Acknowledgements
This work was made possible through support of grants provided from Bill and Melinda Gates Foundation and US Agency for International Development.
Author contributions
B.P.M.S. designed the study, conducted experiments, analyzed data, prepared manuscript, S.M. conducted experiments, data analysis, draft preparation, M.S. involved in genotyping, data collection, carotenoid analysis, R.L.O., D.B.R., R.M., R.R.S., A.A., A.T.R., M.A.T., A.A.A., P.S.B. and M.A.K. conducted field experiments, reviewed manuscript; R.F.R., R.B. and D.J.M. involved in experimental design and edited the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-82001-0.
References
- 1.Muthayya S, Sugimoto JD, Montgomery S, Maberly GF. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014;1324:7–14. doi: 10.1111/nyas.12540. [DOI] [PubMed] [Google Scholar]
- 2.Kennedy E. Healthy lifestyles, healthy people-the mega country health promotion network. Asian Pac. J. Clin. Nutr. 2003;11:S738–S739. doi: 10.1046/j.1440-6047.11.s8.17.x. [DOI] [Google Scholar]
- 3.Alavi S, Bugusu B, Cramer G, et al. Rice fortification in developing countries: A critical review of the technical and economic feasibility. Washington DC: A2Z Project/Academy for Educational Development; 2008. [Google Scholar]
- 4.Bouis HE, Welch RM. Biofortification-A sustainable agricultural strategy for reducing micronutrient malnutrition in the global South. Crop Sci. 2010;50:20–32. doi: 10.2135/cropsci2009.09.0531. [DOI] [Google Scholar]
- 5.Bates CV. Vitamin A. Lancet. 1995;345:31–35. doi: 10.1016/S0140-6736(95)91157-X. [DOI] [PubMed] [Google Scholar]
- 6.De Pee S. Proposing nutrients and nutrient levels for rice fortification. Ann. N. Y. Acad. Sci. 2014;1324:55–66. doi: 10.1111/nyas.12478. [DOI] [PubMed] [Google Scholar]
- 7.WHO . Global Prevalence of Vitamin A Deficiency in Populations at Risk 1995–2005. WHO Global Database on Vitamin A Deficiency. Geneva: World Health Organization; 2009. [Google Scholar]
- 8.FNRI. Vitamin A deficiency (VAD) among Filipino preschool children, pregnant, and lactating women: 1993–2013. In 42nd FNRI Seminar Series, First 100 Days, 1–12, Los Banos (2016).
- 9.ICDDRB. National micronutrients status survey 2011–2012. Technical report January. Centre for Nutrition and Food Security, icddr,b; UNICEF; GAIN; IPHN, Dhaka (2013).
- 10.La Frano MR, Woodhouse LR, Burnett DJ, Burri BJ. Biofortified cassava increases β-carotene and vitamin A concentrations in the TAG-rich plasma layer of American women. Br. J. Nutr. 2013;110:310–320. doi: 10.1017/S0007114512005004. [DOI] [PubMed] [Google Scholar]
- 11.De Moura FF, et al. Are biofortified staple food crops improving vitamin A and iron status in women and children? New evidence from efficacy trials. Adv. Nutr. 2014;5:568–570. doi: 10.3945/an.114.006627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye X, et al. Engineering the provitamin A (β-Carotene) biosynthetic pathway into (carotenoids free) rice endosperm. Science. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 13.Paine JA, et al. Improving the nutritional value of golden rice through increased provitamin A content. Nat. Biotechnol. 2005;23:482–487. doi: 10.1038/nbt1082. [DOI] [PubMed] [Google Scholar]
- 14.Das G, Rao GJN. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015 doi: 10.3389/fpls.2015.00698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Septiningsih EM, et al. Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann. Bot. 2009;103:151–160. doi: 10.1093/aob/mcn206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swamy BPM, et al. Genetic, physiological, and gene expression analyses reveal multiple QTLs that enhance yield of rice mega-variety IR64 under drought. PLoS ONE. 2013;8:e62795. doi: 10.1371/journal.pone.0062795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivakumar B. Current controversies in carotene nutrition. Ind. J. Med. Res. 1998;108:157–166. [PubMed] [Google Scholar]
- 18.Sommers A. New imperatives for an old vitamin (A) J. Nutr. 1988;119:96–100. doi: 10.1093/jn/119.1.96. [DOI] [PubMed] [Google Scholar]
- 19.Gillespie, S. & Haddad, L. Attacking the double burden of malnutrition in Asia and the Pacific. ADB Nutrition and Development Series 4 (2001).
- 20.Yin Z, Plader W, Malepszy S. Transgenic inheritance in plants. J. Appl. Genet. 2004;45:127–144. [PubMed] [Google Scholar]
- 21.Trijatmiko KR, et al. Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 2016;6:19792. doi: 10.1038/srep19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bollinedi HS, et al. Molecular and functional characterization of GR2-R1 event based backcross derived lines of Golden Rice in the genetic background of a mega rice variety Swarna. PLoS ONE. 2017;12:e0169600. doi: 10.1371/journal.pone.0169600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodfield DR, White DWR. Breeding strategies for developing transgenic white clover cultivars. Agron. Soc. 1996;11:125–130. [Google Scholar]
- 24.Badenhorst PE, Smith KF, Spangenberg G. Development of a molecular breeding strategy for the integration of transgenic traits in outcrossing perennial grasses. Agronomy. 2016;6:56. doi: 10.3390/agronomy6040056. [DOI] [Google Scholar]
- 25.Collard BCY, Mackill DJ. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 2008;363:557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasan MM, et al. Marker-assisted backcrossing: a useful method for rice improvement. Biotechnol. Biotechnol. Equip. 2015;29:237–254. doi: 10.1080/13102818.2014.995920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codex. Guideline for the conduct of food safety assessment of foods derived from recombinant-DNA plants CAC/GL 45-2003 (2003).
- 28.Jiao Z, Si XX, Li GK, Zhang ZM, Xu XP. Unintended compositional changes in transgenic rice seeds (Oryzasativa L.) studied by spectral and chromatographic analysis coupled with chemometrics methods. J. Agric. Food Chem. 2010;58:1746–1754. doi: 10.1021/jf902676y. [DOI] [PubMed] [Google Scholar]
- 29.Swamy BPM, et al. Compositional analysis of genetically engineered GR2E “Golden Rice” compared to conventional rice. J. Agric. Food Chem. 2019;67:7986–7994. doi: 10.1021/acs.jafc.9b01524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herman RA, Price WD. Unintended compositional changes in genetically modified (GM) crops: 20 years of research. J. Agric. Food Chem. 2013;61:11695–70110. doi: 10.1021/jf400135r. [DOI] [PubMed] [Google Scholar]
- 31.Harrigan GG, et al. Impact of genetics and environment on nutritional and metabolite components of maize grain. J. Agric. Food Chem. 2007;55:6177–6185. doi: 10.1021/jf070494k. [DOI] [PubMed] [Google Scholar]
- 32.Ricroch AE. Assessment of GE food safety using ‘-omics’ techniques and long-term animal feeding studies. New Biotechnol. 2013;30:349–354. doi: 10.1016/j.nbt.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Bishaw Z, van Gastel AJG. Variety release and policy options. In: Ceccarelli S, Guimaraes EP, Weltzien E, editors. Plant Breeding and Farmer Participation. Rome: FAO; 2009. [Google Scholar]
- 34.Inabangan-Asilo MA, et al. Stability and G × E analysis of zinc-biofortified rice genotypes evaluated in diverse environments. Euphytica. 2019;215:61. doi: 10.1007/s10681-019-2384-7. [DOI] [Google Scholar]
- 35.Datta SK, et al. Golden rice: Introgression, breeding, and field evaluation. Euphytica. 2007;154:271–278. doi: 10.1007/s10681-006-9311-4. [DOI] [Google Scholar]
- 36.Baisakh N, et al. Marker-free transgenic MFT near-isogenic introgression lines NIILs of ‘golden’ indica rice cv IR64 with accumulation of provitamin A in the endosperm tissue. Plant Biotechnol. J. 2006;4:467–475. doi: 10.1111/j.1467-7652.2006.00196.x. [DOI] [PubMed] [Google Scholar]
- 37.Datta K, Sahoo G, Krishnan S, Ganguly M, Datta SK. Genetic stability developed for β-carotene synthesis in BR29 rice line using dihaploid homozygosity. PLoS ONE. 2014;9:e100212. doi: 10.1371/journal.pone.0100212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian YS, et al. Enhancing carotenoid biosynthesis in rice endosperm by metabolic engineering. Plant Biotechnol. J. 2019;17:849. doi: 10.1111/pbi.13059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bai C, et al. Bottlenecks in carotenoid biosynthesis and accumulation in rice endosperm are influenced by the precursor–product balance. Plant Biotechnol. J. 2016;14:195–205. doi: 10.1111/pbi.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie XW, et al. Genetic background and environmental effects on QTLs for sheath blight resistance revealed by reciprocal introgression lines in rice. Acta Agron. Sin. 2008;34:1885–1893. doi: 10.1016/S1875-2780(09)60013-4. [DOI] [Google Scholar]
- 41.Wang X, et al. Genetic background effects on QTL and QTL × environment interaction for yield and its component traits as revealed by reciprocal introgression lines in rice. Crop J. 2014;2:345–357. doi: 10.1016/j.cj.2014.06.004. [DOI] [Google Scholar]
- 42.Jaswir I, et al. Effects of season and storage period on accumulation of individual carotenoids in pumpkin flesh (Cucurbita moschata) J. Oleo Sci. 2014;63:761–767. doi: 10.5650/jos.ess13186. [DOI] [PubMed] [Google Scholar]
- 43.Horváth G, Kissimon J, Faludi-Dániel Á. Effect of light intensity on the formation of carotenoids in normal and mutant maize leaves. Phytochemistry. 1972;11:183–187. doi: 10.1016/S0031-9422(00)89987-2. [DOI] [Google Scholar]
- 44.Ben-Amotz A, Avron M. On the factors which determine massive beta-carotene accumulation in the halotolerant alga Dunaliella bardawil. Plant Physiol. 1983;72:593–597. doi: 10.1104/pp.72.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuentes P, et al. Light-dependent changes in plastid differentiation influence carotenoid gene expression and accumulation in carrot roots. Plant Mol. Biol. 2012;79:47–59. doi: 10.1007/s11103-012-9893-2. [DOI] [PubMed] [Google Scholar]
- 46.Norshazila S, Othman R, Jaswir I, Yumi Zuhanis HH. Effect of abiotic stress on carotenoids accumulation in pumpkin plants under light and dark conditions. Int. Food Res. J. 2017;24:S387–S394. [Google Scholar]
- 47.Bechoff A, et al. Effect of drying and storage on the degradation of total carotenoids in orange-fleshed sweet potato cultivars. J. Sci. Food Agric. 2010;90:622–629. doi: 10.1002/jsfa.3859. [DOI] [PubMed] [Google Scholar]
- 48.Murray MG, Thompson WF. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juliano B. Physicochemical Properties of Starch and Protein in Relation to Grain Quality and Nutritional Value of Rice. Los Baños: International Rice Research Institute. Rice Breeding; 1972. pp. 389–405. [Google Scholar]
- 50.Gemmecker S, et al. Phytoene Desaturase from Oryza sativa: Oligomeric assembly, membrane association and preliminary 3D-analysis. PLoS ONE. 2015;10:e0131717. doi: 10.1371/journal.pone.0131717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Team . A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2012. [Google Scholar]
- 52.IRRI 2014. Plant Breeding Tools (PB Tools), Version: 1.4, International Rice Research Institute, Los Banos.
- 53.Lenth, R. lsmeans: Least-squares means. R package version 1.06-05. R Core Team (2015). R: A language and environment for statistical computing. Technical report (R Foundation for Statistical Computing, Vienna).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.