The coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain isolated from an Iraqi patient was sequenced for the first-time using Illumina MiSeq technology. There was a D614G mutation in the spike protein-coding sequence. This report is valuable for better understanding the spread of the virus in Iraq.
ABSTRACT
The coding-complete genome sequence of a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) strain isolated from an Iraqi patient was sequenced for the first-time using Illumina MiSeq technology. There was a D614G mutation in the spike protein-coding sequence. This report is valuable for better understanding the spread of the virus in Iraq.
ANNOUNCEMENT
In December 2019, severe acute respiratorysyndrome coronavirus 2 (SARS-CoV-2) emerged in Wuhan, China. This virus belongs to the Betacoronavirus genus of the Coronaviridae family (1). RNA viruses are characterized by a high rate of genetic mutations, which enables them to escape host defenses and may also affect the discovery of an effective vaccine and the status of reverse transcription-quantitative PCR (RT-qPCR) coronavirus disease 2019 (COVID-19) detection (2, 3). Therefore, identifying the SARS-CoV-2 genome sequences and understanding their mutations are important. Here, we sequenced the coding-complete genome of a SARS-CoV-2 strain from an Iraqi patient.
A nasopharyngeal swab was collected from a patient with mild symptoms (26-year-old female infected in Samawah, Iraq) and added to Accuzol (Bioneer, Taejeon, South Korea) at a 1:3 (vol/vol) ratio. Viral RNA was extracted following the manufacturer’s protocol. Briefly, the pellet was resuspended in RNase-free water, and the RNA sample was concentrated using the RNA Clean and Concentrator kit (Zymo Research, CA, USA). The presence of SARS-CoV-2 RNA was confirmed using a Luna universal probe one-step RT-qPCR kit (NEB, MA, USA) and a primer/probe targeting the nucleocapsid gene (4), and viral RNA quantification was conducted using an in vitro-transcribed RNA standard (5). A Qubit 2.0 fluorometer (Thermo Fisher Scientific, MA, USA) and Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) were used to assess the RNA quantity and quality. From the total RNA, 100 ng was processed using a Swift Amplicon SARS-CoV-2 research panel (Swift Biosciences, USA). The library obtained passed a quality check and was quantified before being used at equimolar concentrations. High-throughput sequencing was conducted using an Illumina MiSeq sequencer following the standard procedure. The read length was 150 bp. This produced 1,222,270 reads. The raw sequence data were quality controlled using FastQC v0.11.9 (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Genome assembly was conducted using MEGAHIT v1.2.9 (6), and the genome size was 29,720 bp with a coverage depth of 5,340× (SAMtools v1.9) (7) and an overall GC content of 38.01%. This study was approved by the Research Ethics Committee at the University of Al-Muthanna (2529sci on 26 April 2020). Mapping of the obtained strain, hCoV-19/Iraq/ICGEB-5T (GISAID accession number EPI_ISL_582030), was accomplished using CoVsurver in GISAID (8), and the Wuhan strain (BetaCoV/Wuhan/WIV04/2019) was used as a reference. Analysis of this strain revealed eight mutations. These mutations were accompanied by four amino acid changes (Table 1). Among these changes, the following two infrequent amino acid changes were observed: NSP8-S76F, which has already been identified in three countries, and N-G215S, which has been identified in 10 countries to date. To estimate the effect of these amino acid changes on the stability of NSP8 and N proteins, the online server DUET (9) was used; it was predicted that these proteins are destabilized by the reported amino acid changes. Default parameters were selected for all the software and tools used in this work.
TABLE 1.
Genome features of the SARS-CoV-2 isolate in Iraq
| Isolate name | Sequence length (bp) | No. of mutations | Mutations | GISAID clade | Nextstrain clade |
|---|---|---|---|---|---|
| hCoV-19/Iraq/ICGEB-5T | 29,720 | 8 | C241T, C3037T, C10078T, C12318T, C18877T, A23403G, G25563T, G28916A | GH | 20A |
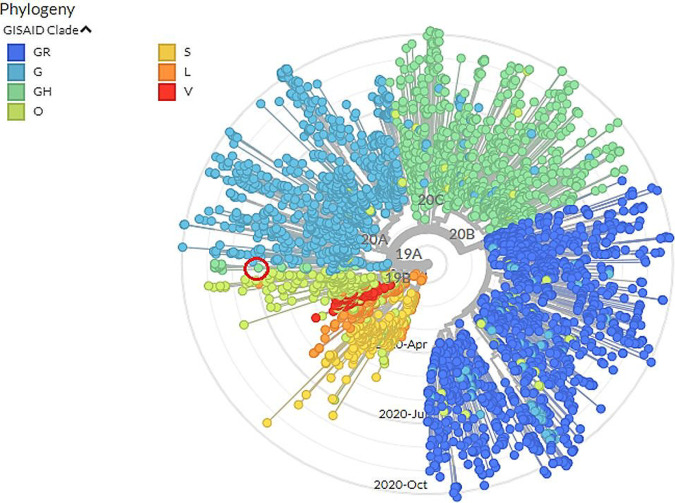
The Nextclade v0.8.1 Web tool (10) was employed to generate a phylogenetic tree and a clade assignment. The obtained phylogenetic tree was rooted with strains from Wuhan, China. The phylogenetic tree revealed that strain hCoV-19/Iraq/ICGEB-5T belongs to the Nextstrain clade 20A and GISAID clade GH (Fig. 1). However, the GH clade is much more prevalent in Europe and North America (11–13).
FIG 1.
Phylogenetic tree of SARS-CoV-2 strains, including an isolate from Iraq (June 2020) as depicted in the open red circle (GISAID accession number EPI_ISL_582030).
Data availability.
The coding-complete genome sequence of hCoV-19/Iraq/ICGEB-5T was deposited under GenBank accession number MW290973 and SRA accession number PRJNA689203 (BioProject). The GISAID accession number for hCoV-19/Iraq/ICGEB-5T is EPI_ISL_582030.
ACKNOWLEDGMENTS
We thank the ICGEB COVID-19 Resources Program (https://www.icgeb.org/covid19-resources/) and the fast-track sequencing program from the AREA Science Park of Trieste, Italy, for supporting this work. We thank the staff of the Public Health Laboratory in Samawa, Iraq. We also thank Nadia Al-Zahery (Department of Biology and Biotechnology, University of Pavia, Italy) for her assistance with and advice on accomplishing this work.
REFERENCES
- 1.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanjuán R, Domingo-Calap P. 2016. Mechanisms of viral mutation. Cell Mol Life Sci 73:4433–4448. doi: 10.1007/s00018-016-2299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruan Y, Wei CL, Ling AE, Vega VB, Thoreau H, Thoe SYS, Chia J-M, Ng P, Chiu KP, Lim L, Zhang T, Chan KP, Ean LOL, Ng ML, Leo SY, Ng LFP, Ren EC, Stanton LW, Long PM, Liu ET. 2003. Comparative full length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779–1785. doi: 10.1016/S0140-6736(03)13414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML, Mulders DGJC, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette J-L, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MPG, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Licastro D, Rajasekharan S, Dal Monego S, Segat L, D’Agaro P, Marcello A, The Regione FVG Laboratory Group on COVID-19 . 2020. Isolation and full-length genome characterization of SARS-CoV-2 from COVID-19 cases in northern Italy. J Virol 94:e00543-20. doi: 10.1128/JVI.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 7.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shu Y, McCauley J. 2017. GISAID: global initiative on sharing all influenza data—from vision to reality. Euro Surveill 22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pires DEV, Ascher DB, Blundell TL. 2014. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res 42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, Sagulenko P, Bedford T, Neher RA. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics 34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komissarov AB, Safina KR, Garushyants SK, Fadeev AV, Sergeeva MV, Ivanova AA, Danilenko DM, Lioznov D, Shneider OV, Shvyrev N, Spirin V, Glyzin D, Shchur V, Bazykin GA. 2020. Genomic epidemiology of the early stages of SARS-CoV-2 outbreak in Russia. medRxiv doi: 10.1101/2020.07.14.20150979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alm E, Broberg EK, Connor T, Hodcroft EB, Komissarov AB, Maurer-Stroh S, Melidou A, Neher RA, O’Toole A, Pereyaslov D, The WHO European Region sequencing laboratories and GISAID EpiCoV group . 2020. Geographical and temporal distribution of SARS-CoV-2 clades in the WHO European Region, January to June 2020. Euro Surveill 25:2001410. doi: 10.2807/1560-7917.ES.2020.25.32.2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mercatelli D, Giorgi FM. 2020. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol 11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The coding-complete genome sequence of hCoV-19/Iraq/ICGEB-5T was deposited under GenBank accession number MW290973 and SRA accession number PRJNA689203 (BioProject). The GISAID accession number for hCoV-19/Iraq/ICGEB-5T is EPI_ISL_582030.