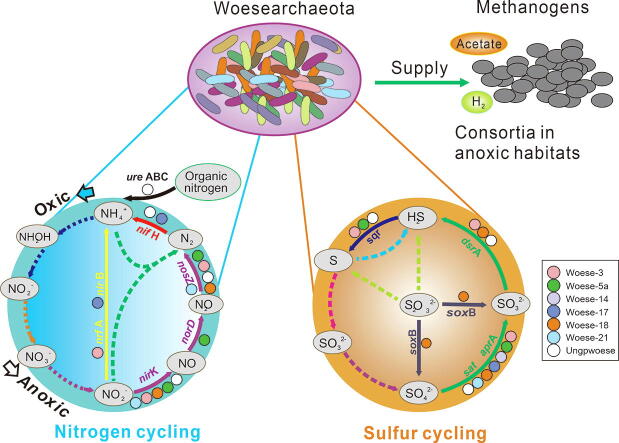
Graphical abstract
Keywords: Woesearchaeota, Ecological patterns, Anaerobic carbon cycling, Nitrogen cycling, Sulfur cycling
Highlights
-
•
Distribution of Woesearchaeota agrees with their potential biogeochemical roles.
-
•
Woesearchaeota might function with methanogens in anaerobic carbon cycling.
-
•
Woesearchaeota could participate in anaerobic nitrogen cycling.
-
•
Woesearchaeota may drive sulfur cycling in sulfuric or sulfidic-rich environments.
Abstract
Woesearchaeota as a newly established member of the superphylum DPANN (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaea) are surprisingly abundant and diverse in a wide variety of environments, including deep oil reservoir, sulfuric springs and anoxic aquifers, indicating a high diversity of their roles in global biogeochemical cycles. However, ecological functions of them remain elusive. To fill up this gap, we analyzed and compared the global distribution patterns of Woesearchaeota using the genomes available publicly. As a result, both ecological distribution patterns and metabolic predictions support a key role of woesearchaeotal lineages in cycling of carbon, nitrogen, and sulfur. Multivariate regression analysis reveals that Woesearchaeota might function in consortium with methanogens in the cycling of carbon in anaerobic environments, particularly in soils or sediments. Moreover, comparative genomic analysis and ecological distribution suggest the potential roles of Woesearchaeota in the processes of denitrification, nitrogen fixation, and dissimilatory nitrite reduction, especially in the wastewater treatment systems; and also uncovered the potential capability of sulfate reduction, sulfide oxidation and thiosulfate oxidation in sulfuric or sulfidic-rich environments. Our findings add more information into the ecological roles of archaea in the anoxic environment.
1. Introduction
Archaea are ubiquitous microbial members composed of more than 28 phyla and occupy one of the three domains in the tree of life [1], [2], [3]. Archaeal lineages have been detected in a wide variety of environments, ranging from extreme hydrothermal vents, hypersaline environments and polar permafrost soils to freshwater, wastewater treatment plants and coastal wetland [4], [5], [6], [7], [8]. Moreover, they are abundant but widely distributed in soils, marine or freshwater sediments and water columns, wetlands and estuaries [9], [10], [11], [12]. The rapid development of culture-independent sequencing technologies and bioinformatics tools have revolutionized our understanding of archaeal diversity and metabolic potentials in different ecosystems, which allows the discovery of the new archaeal lineages and their ecological roles [13]. Therefore, archaeal diversity, evolution and ecology have been of great interest in the field of microbial ecology.
The widely-distributed archaeal lineages play crucial roles in the global biogeochemical cycles [14]. For example, in global carbon cycling, many archaea have been implicated to participate in the methane metabolism that affects both the nutrient cycling and the global climate change [15]. In deep oceans, archaea with widespread occurrence are capable of scavenging of diverse organic substrates, such as carbohydrates, fatty acids and lipids as carbon and energy sources [16]. Asgard archaea can oxidize hydrocarbons, methane and butane in particular, driving anaerobic hydrocarbon cycling in the oceanic subsurface [17]. In nitrogen cycling, ammonia-oxidizing archaea (AOA) have been reported to play a crucial role in the oxidation of ammonia in a wide variety of environments [18], [19], [20], such as wastewater treatment systems [21], [22], sediments [23], stone cultural heritage [24], [25], [26], [27]. Moreover, genes involved in nitrogen metabolism have also been detected to be abundant in temperate marine sediments [28], hyperthermophilic environments [29], brine or salted pools [30], etc. In sulfur cycling, varieties of archaeal lineages are capable of using elemental sulfur and reduced inorganic sulfur species as energy source or electron donor in many sulfuric-rich environments [31], [32], [33], such as volcanic hot springs [34], peatlands [35] and hydrothermal vents [36]. Importantly, elemental sulfur can be used as not only the electron donor by aerobic archaea (e.g., Acidianus and Sulfolobus) but also the electron acceptor by anaerobic chemolithotrophic archaea (e.g., Acidianus and Pyrodictium) [37]. Therefore, archaea become one of essential participants in the global biogeochemical cycling of major elements. Exploration of novel metabolic potential of archaea will help understand their ecological roles in the global biogeochemical cycles.
Woesearchaeota as one of the members of the archaeal superphylum DPANN are widely distributed and abundant in soils, freshwater, sediments, wetlands, estuaries and hydrothermal vents [11], [38], [39], [40]. High diversity of woesearchaeotal lineages yields more than 26 subgroups at the class level based on the phylogenetic analysis [39]. High diversity and wide ecological distribution imply the important roles of woesearchaeotal lineages in the global biogeochemical cycles [41]. However, current studies have only revealed the roles in anaerobic carbon cycling [38], [39], and thus the major functions of Woesearchaeota remain still unknown. Identification of novel metabolic potential of woesearchaeotal lineages has great importance to unveil their ecological roles in the global biogeochemical cycles of major elements.
At present, a great number of woesearchaeotal 16S rRNA gene sequences (ca. 5000) in public databases are originated from a wide range of environments. This dataset provides large information available currently to explore the global ecological patterns and to imply more on the relationship between the distribution patterns and the ecological functions/roles of Woesearchaeota in the global biogeochemical cycles. Here, we retrieved the public databases of woesearchaeotal lineages and meta-genomes to (i) characterize the global distribution of woesearchaeotal communities and unveil the driving environmental factors that affect the woesearchaeotal distribution among diverse habitats, (ii) identify the metabolic potential on the basis of comparative genomic analysis of the woesearchaeotal genomes, and (iii) link the ecological distribution patterns to their potential ecological roles in the cycling of carbon, nitrogen, and sulfur. We expect to expand our understanding of the ecological roles of archaea in the global biogeochemical cycles.
2. Materials and methods
2.1. Construction of cultivation-independent libraries
We recovered both the published literature and the GenBank database for the construction of woesearchaeotal 16S rRNA clone libraries. The detailed steps involved are available elsewhere [39]. Specifically, the Esearch Utility was employed to capture archaeal 16S rRNA gene sequences from the database as per the following string ‘16S AND 600:2000 [Sequence Length] AND archaea [Organism] AND rrna [Feature key] AND isolation_source [All fields] NOT genome OR chromosome OR plasmid’. To make sure the availability of further analysis, sequences with isolation source tags missing were removed. To guarantee only studies or libraries that include woesearchaeotal sequences were retained, the remaining 122,559 archaeal sequences from 217 libraries were BLASTed against the reference database SILVA SSU 128 [42], [43]. Libraries or studies with the number of 16S rRNA gene sequences <10 were discarded. Finally, we accomplished the screening with 133 studies or libraries, which contain 15,012 archaeal sequences, including 3584 woesearchaeotal sequences. Such studies or libraries were grouped into seven distinct habitats as follows: 32 libraries from freshwater sediment (Fsed), 30 from freshwater (Fwc), 22 from soil (S), 19 from marine sediment (Msed), 12 from hydrothermal vent (Hdv), 11 from hypersaline environment (Hsal), and 7 from marine water column (Mwc) (Table S1). Furthermore, a semi-quantitative environmental matrix was constructed according to the range of environmental gradients among these habitats: temperature (cold, temperate, and hot), life style (water, sediment, and soil), salinity (non-saline, saline, and hypersaline), oxygen concentrations (oxic and anoxic), and trophic state (oligotrophic, mesotrophic, and hypertrophic) (Table S1).
2.2. Taxon-based analysis
The total 15,012 archaeal sequences formed 11,323 operational taxonomic units (OTUs) at an identity of 97%, including 2180 OTUs that belonged to Woesearchaeota (Table S1). Next, these OTU sequences were compared with the reference database SILVA SSU 128 and assigned to the corresponding taxon once one OTU matches with a reference or identified sequence with an identity higher than 97%. If an archaeal OTU does not match with any reference sequence, this OTU will be named ‘unknown archaea’. After the taxonomy assignment, each named OTU was dated back to the specific libraries that were associated with a given habitat or environmental matrix for further analyses, such as relative abundance and occurrence frequency.
2.3. Multivariate regression analysis
A multivariate regression tree (MRT) was determined and generated by the R package mvpart to illustrate the relationship between the environmental matrix (Table S1) and the table of lineage relative abundances [44]. Meanwhile, we introduced the indicator value (IndVal) index, which combines relative occurrence frequency and relative abundance to identify archaeal lineages, like the concept of ‘indicator species’, among the habitats. The indicator species represents the predominance of a species in both relative abundance and relative occurrence frequency in a given habitat, indicating the important ecological niche of a species in such habitat.
2.4. Comparative genomic analysis
Genomes of Woesearchaeota were retrieved from the NCBI Genome Database and further confirmed with the authors/owners. The quality of the retrieved 19 woesearchaeotal genomes was estimated by CheckM and the parameters, such as completeness (%), sequencing depth (x), relative GC content (%), and number of protein coding genes were listed in Table S2. To infer the metabolic functions of specific subgroups of Woesearchaeota, only genomes that have a 16S rRNA gene sequence were used for analysis. Genomic BLAST analysis for all the genomes was performed to provide the reciprocal best BLAST hits (rBBHs). The MCL76 algorithm was then introduced to cluster rBBHs into protein clusters. Finally, the proteins were ready for further annotation and identification.
2.5. Annotation and identification of genes
Open reading frames (ORFs) were predicted on their genomic scaffolds using the metagenome mode of Prodigal and were annotated by the NCBI Prokaryotic Genome Annotation Pipeline (Table S3) and by the comparison with KEGG PATHWAY Database (https://www.genome.jp/kegg/pathway.html). Genome-based functional gene annotation was determined by searching all predicted ORFs in a genome with the presence of the corresponding enzymes. If a metabolic process is dependent upon more than one enzyme, it was confirmed based on the presence of all the necessary enzymes. To find out the metabolic potential of the specific woesearchaeotal subgroups, we assigned the genomes to the corresponding subgroups on the basis of their 16S rRNA gene sequences if available [39] (Table S2).
3. Results
3.1. Ecological distribution patterns of Woesearchaeota
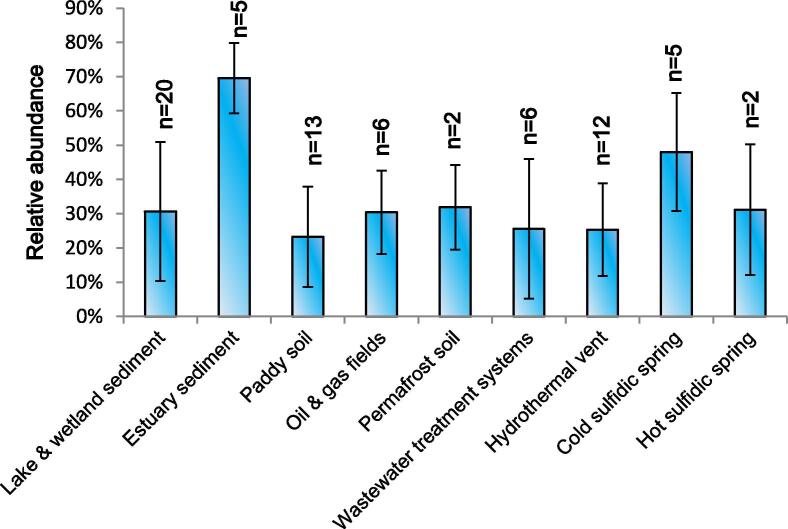
Woesearchaeota over the retrieved 133 archaeal libraries had a relative abundance of 23% and dominated differentially in the anoxic habitats, particularly in estuary sediments, lake and wetland sediments, and cold sulfuric springs (Fig. 1 and Table S1). For example, the abundance of Woesearchaeota throughout the 5 libraries of estuary sediments reached up to 70%, whereas the abundance over the 5 libraries of cold sulfuric springs was more than 50%. They tended to occur more frequently in paddy soils, lake and wetland sediments, and hydrothermal vents, where oxygen availability is largely limited or none existence.
Fig. 1.
Relative abundance of woesearchaeotal lineages (based on sequence abundance) within anoxic habitats. The number of libraries used for each kind of habitat is given above the bar. Error bars represent the standard deviation.
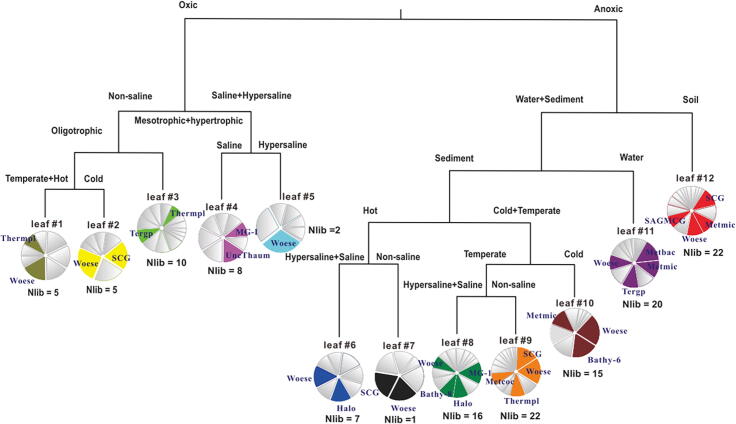
To link the abundance of the archaeal lineages to environmental factors, MRT was constructed (Fig. 2). The MRT analysis generated a twelve-leaf tree ordination based on oxic status for the first two branches, totally explaining 51.1% of the phylogenetic variance. Anoxia tended to combine S (soil with a depth of more than 10 cm) and anoxic water–sediment, Fsed and Fwc (sediments and water column from anoxic freshwaters or springs), as well as Hdv and Msed (hot- and cold-temperate anoxic marine sites), which agrees with previous study [4]. Woesearchaeota are the indicator lineage over the 7 leaves of the anoxic branch of the tree, covering 103 libraries. However, Woesearchaeota are detected only in 30 libraries on the oxic branch of 5 leaves, which represents only a small portion of the ecological distribution. Therefore, both distribution and abundance patterns indicate the predominance of Woesearchaeota in anoxic environments.
Fig. 2.
MRT of the interaction between archaeal lineage abundance and environmental factors. The model explained 51.1% of the variance of the whole data set. Libraries clustered in the tree leaves according to their sources (Table S1). Pie charts under each leaf indicate how the relative abundance of archaeal lineages contributed to the structure of the leaves. The IndVal index for each leaf (data not shown) revealed that most core lineages are still indicator lineages (p < 0.01), which are labeled on each pie. Abbreviation of taxonomy: Woese, Woesearchaeota; Thermpl, Thermoplasmata; SCG, Soil Crenarchaeotic Group; Tergp, Terrestrial group; MG-I, Marine Group I; UncThaum, Uncultured Thaumarchaeota; Halo, Halobacteria; Bathy-8/-6, Bathyarchaeota subgroup-8/-6; Metcoc, Methanococci; Metmic, Methanomicrobia; Metbac, Methanobacteria; SAGMCG, South African Gold Mine Group 1.
Notably, the MRT analysis showed that Woesearchaeota are the only lineage to be indicator for all anoxic leaves and also the co-indicator lineage of Methanomicrobia in soil, anoxic water and sediments (Fig. 2), such as paddy soil, permafrost soil, oil and gas fields, and freshwater sediments (Fig. 1). Moreover, Woesearchaeota are a co-indicator lineage of Methanococci in temperate fresh sediments and Methanobacteria in anoxic water. Such findings might further provide ecological evidence for Woesearchaeota as a potential partner of methanogens in methanogenesis, which takes place in the anaerobic environments lacking sulfate.
3.2. Potential functions in nitrogen cycling
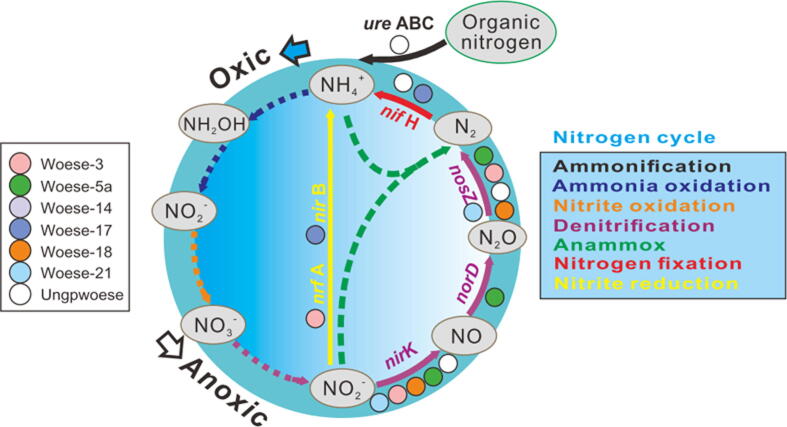
We identified some genes involved in the processes of denitrification (e.g., nirK, norD, and nosZ), nitrogen fixation (e.g., nifH), and dissimilatory nitrate reduction to ammonium (e.g., nrfA and nirB) as well as the genes relevant to the conversion of organic nitrogen (e.g., ure ABC) (Fig. 3). However, genes involved in the processes of anammox, aerobic ammonia oxidation, nitrate reduction, and nitrite oxidation are missing in Woesearchaeota. Such functional properties further agree with the distribution patterns that Woesearchaeota dominate in anoxic environments, particularly in the wastewater treatment systems (Fig. 1), where nitrogen is normally very rich and anaerobic nitrogen cycling processes are very active.
Fig. 3.
Metabolic potential of Woesearchaeota in nitrogen cycling. Colorful nodes indicate the corresponding woesearchaeotal subgroups that might hold the metabolic function of the corresponding pathways. Only genes detected are listed for the corresponding pathways. Dash lines indicate the pathways that are not detected in any woesearchaeotal subgroups based on the current genomic information available, whereas the solid lines indicate the pathways that might occur. The two arrows outside the chart indicate the start of the pathways that take place under anoxic or oxic condition.
3.3. Potential roles in sulfur cycling
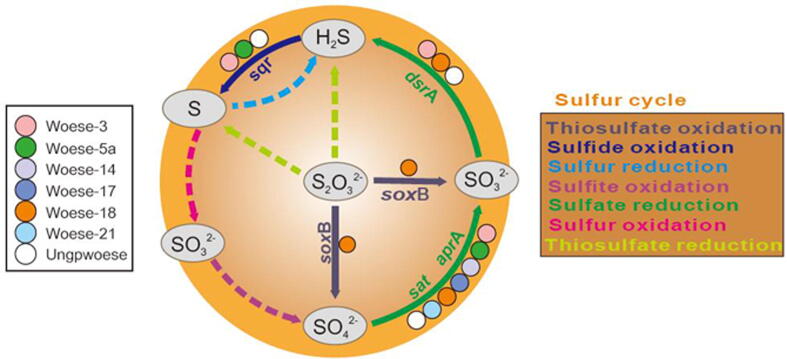
Comparative genomic analysis also revealed the potential roles of Woesearchaeota in sulfur cycling, particular for the reduction of sulfate (e.g., the genes sat, aprA, and dsrA) and thiosulfate oxidation (e.g., soxB) as well as sulfide oxidation (e.g., sqr) (Fig. 4). All of the genomes contained the genes sat and/or aprA that encode the enzymes that catalyze the reduction of sulfate to sulfite. Members of Woese-3, Woese-18 or the other ungrouped might complete the sulfate reduction to sulfide. Furthermore, members of Woese-3, Woese-5a or the other ungrouped might oxidize sulfide to sulfur. Only the members of Woese-18 show the potential to perform the thiosulfate oxidation. However, genes involved in the processes of thiosulfate reduction, sulfur oxidation, sulfur reduction or sulfite oxidation are not detected in the genomes of Woesearchaeota in this study. These functions in combination with the distribution patterns support the potential ecological role of Woesearchaeota in the sulfuric or sulfidic environments, particularly for hydrothermal vent and cold or hot sulfuric or sulfidic springs (Fig. 1), where sulfur is typically abundant and the element cycling is actively carried out.
Fig. 4.
Metabolic potential of Woesearchaeota in sulfur cycling. Colorful nodes indicate the corresponding woesearchaeotal subgroups that might hold the metabolic function of the corresponding pathways. Only genes detected are listed for the corresponding pathways. Dash lines indicate the pathways that are not detected in any woesearchaeotal subgroups based on the current genomic information, whereas the solid lines indicate the pathways that might occur.
4. Discussion
4.1. Dominance of Woesearchaeota in anoxic environments
The ecological distribution patterns indicated that Woesearchaeota dominate in anoxic environments, such as soil, sediments and other anoxic habitats. Meanwhile, high biodiversity of Woesearchaeota in such habitats has also been observed in our previous work [39]. These results reflect the important ecological niche specificity of Woesearchaeota to such environments, because niche differences over space allow the formation of biogeographic patterns of species [45]. The properties of the environments would reveal the ecological functions of woesearchaeotal communities in anaerobic biogeochemical cycles of elements [38]. For example, paddy soils [12], oil or gas fields [46], [47], [48], [49], wetlands or sediments [10], [50], [51], [52] are the typical habitats of methane-producing organisms, which contribute greatly to the global biological methane emission. In wastewater treatment systems, nitrogen cycling is actively driven by nitrifiers or denitrifiers [22], [53], [54], [55]. In sulfur-rich hydrothermal vents and cold or hot springs, sulfur-cycling archaea and bacteria manipulate the reduction and oxidation of elemental sulphur [36], [56], [57], [58]. These ecological implications reveal the potential roles of woesearchaeotal lineages in the specific biotopes. However, the frequency of the habitats may not represent the real ecological distribution of Woesearcheota, given the small sample sizes or the choice of habitats depent upon scientists’ interest in the studies.
4.2. Potential partner of methanogens in anaerobic carbon cycling
According to the distribution patterns and metabolic modeling, it seems that Woesearchaeota tends to co-occur with methanogens by a mean of consortium or syntrophy [39]. The MRT analysis showed that Woesearchaeota are the co-indicator lineage of methanogens in many anoxic environments, which further supports the hypothesis that Woesearchaeota might function as a partner with methanogens biochemically in anaerobic carbon cycling. Although the direct evidence has not yet been obtained or confirmed, many indirect findings support this hypothesis. Woesearchaeota have been found to perform the fermentation lifestyle and convert organic substrates (e.g., starch) into acetic acid or hydrogen [38], which might favor the growth of acetotrophic or hydrogenotrophic methanogens [39]. The retroelement-guided protein diversification mechanisms widely-existed in DPANN super-phylum, including Woesearchaeota, have revealed the presence of a versatile tool that could help them adapt to a dynamic, host-dependent existence [59]. As methanogenesis has a great impact on the global carbon cycle and climate change, further confirmation of the participation by Woesearchaeota is extremely important. Further work should be focused on the two directions to determine the effects. On the one hand, given the fact that the identified number of woesearchaeotal genomes is still very small, it is necessary to expand the genomic information of woesearchaeotal communities to support the direct evidence by exploring the methyl-coenzyme M reductase (Mcr) complex, which is the key enzyme that catalyses the final step in methanogenesis and the first step in methanotrophy [15]. Meanwhile, the analysis using more undiscovered genomes from both archaea groups for metabolic models’ reconstruction may provide further evidence to reveal their partnership in C cycling in anaerobic environments. On the other hand, co-culture of Woesearchaeota and methanogens or other highly co-occurring dominant microbial members based on metabolic complementation could also help prove the hypothesis of the partnership or syntrophic lifestyle.
4.3. Participant in nitrogen cycling
Woesearchaeota are also found to be abundant in the libraries of wastewater treatment systems, such as anaerobic wastewater bioreactor, activated sludge, and anaerobic digester plan. Such environments are characterized by the active nitrogen cycling driven by nitrifiers or denitrifiers, which include archaea, bacteria and fungi [21], [22], [60], [61], [62]. A few genes, nirK and nosZ in particular, involved in nitrogen cycling are detected in Woesearchaeota. The genes are mainly responsible for biochemical processes of nitrogen cycle under anoxic conditions, such as denitrification from nitrite to dinitrogen (N2), N2 fixation, dissimilatory nitrite reduction to ammonium as well as the ammonification of organic nitrogen. Thus, both ecological distributions and metabolic predictions revealed the potential roles of Woesearchaeota as a participant in nitrogen cycling in anoxic environments. However, it is also possible that the denitrification process from nitrite to N2 is a typical mechanism of detoxification in Woesearchaeota [38]. If this hypothesis is true, it is difficult to explain the dominance of Woesearchaeota in such nitrogen-rich environments because they may need more energy to cope with the damage from toxic nitrite or other products generated in the nitrogen cycle [63], [64]. Therefore, further experimental tests are still required to elucidate the functions of Woesearchaeota in the nitrogen cycle. Isotopic labeling test with specific substrates would be an effective way to confirm the actual expression of the genes in woesearchaeotal genomes.
4.4. Potential roles in sulfur cycling
In addition, the comparative genomic analysis indicated some potential roles of Woesearchaeota in sulfur cycling, such as sulfate reduction to sulfide, sulfide oxidation to sulfur, and thiosulfate oxidation. Importantly, such functions again agree with the high abundance of woesearchaeotal lineages in sulfuric or sulfidic-rich environments, such as hydrothermal vents and cold or hot springs [58], [65], [66], [67]. Therefore, these findings would well support the potential roles of Woesearchaeota in sulfur cycling. If it is true, this information will greatly advance our understanding of archaea in the global biogeochemical cycling of sulfur. Investigation into sulfur metabolism in archaea could help explore many novel enzymes and pathways of the archaea, particularly from extreme environments. For example, two anaerobic chemolithotrophic archaeal genera, Acidianus and Pyrodictium, have been found to be able to use elemental sulfur as the electron acceptor in a short electron transport chain based on a sulfur reductase and a membrane-bound hydrogenase in hyperthermal habitats [37]. In contrast, the relevant mechanisms remain unclear in heterotrophic anaerobic archaea, such as Pyrococcus and Thermococcus, though they are also found to use element sulfur as the electron acceptor. In a low-salt, sulfide- and sulfur-rich spring, Halobacteriales may adopt a novel sulfur-metabolic mechanism different from that in their typical high-salt habitats [68]. Such metabolic mechanisms and pathways would evolve diversely in the specific environments and, thus, could be one reason to explain archaeal distribution and diversity with ecological function on Earth.
Further studies for functional confirmation are still required. On the one hand, cultivation of the samples in these environments or in situ experiments might help us to confirm the roles of Woesearchaeota. On the other hand, molecular test based on synthetic biology is an effective approach to verify the function of the specific gene. Moreover, a comparative analysis of metabolic potential of Woesearchaeota from different environments will provide more insightful details on their ecological roles and functions in the biogeochemical cycles, given the fact that more genomes are discovered from different environments.
5. Conclusions
In this study, we investigated the ecological distribution patterns of woesearchaeotal lineages and linked the habitat properties to the predicted functions. The agreement between ecological distribution patterns and metabolic functions reveals the potential roles of Woesearchaeota in anaerobic biogeochemical cycling of carbon, nitrogen and sulfur. These findings will advance our understanding the ecological functions of the new archaea in the global biogeochemical cycles. Since thre is no pure culture of Woesearchaeota available now, information derived for this group is based entirely on 16S rRNA genes and Metagenome Assembled Genomes of this group, and further experimental demonstration is still required to confirm their ecophysiology in the ecosystems.
CRediT authorship contribution statement
Xiaobo Liu: Conceptualization, Methodology, Software, Data curation, Writing - original draft. Ji-Dong Gu: Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We appreciate the reviewers for their constructive comments and suggestions to improve an early version of this paper. This research is supported partially by National Natural Science Foundation of China (Grant No. 92051103). X.L. acknowledges the financial support of a research fellow from GTIIT.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.01.013.
Contributor Information
Xiaobo Liu, Email: xiaobo.liu@gtiit.edu.cn.
Ji-Dong Gu, Email: jidong.gu@gtiit.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hug L.A., Baker B.J., Anantharaman K., Brown C.T., Probst A.J., Castelle C.J. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 2.Woese C.R., Fox G.E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. PNAS. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auguet J.C., Barberan A., Casamayor E.O. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- 5.Oren A. The ecology of the extremely halophilic archaea. FEMS Microbiol Rev. 1994;13:415–439. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 6.Takai K., Horikoshi K. Genetic diversity of archaea in deep-sea hydrothermal vent environments. Genetics. 1999;152:1285–1297. doi: 10.1093/genetics/152.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takai K., Nakamura K. Archaeal diversity and community development in deep-sea hydrothermal vents. Curr Opin Microbiol. 2011;14:282–291. doi: 10.1016/j.mib.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Xue Y., Jonassen I., Øvreås L., Taş N. Bacterial and Archaeal Metagenome-Assembled Genome Sequences from Svalbard Permafrost. Microbiol Resour Announc. 2019;8:e00516–e519. doi: 10.1128/MRA.00516-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubo K., Lloyd K.G., Jennifer F.B., Amann R., Teske A., Knittel K. Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J. 2012;6:1949–1965. doi: 10.1038/ismej.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laskar F., Das Purkayastha S., Sen A., Bhattacharya M.K., Misra B.B. Diversity of methanogenic archaea in freshwater sediments of lacustrine ecosystems. J Basic Microbiol. 2018;58:101–119. doi: 10.1002/jobm.201700341. [DOI] [PubMed] [Google Scholar]
- 11.Liu X., Pan J., Liu Y., Li M., Gu J.-D. Diversity and distribution of Archaea in global estuarine ecosystems. Sci Total Environ. 2018;637–638:349–358. doi: 10.1016/j.scitotenv.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Pump J., Pratscher J., Conrad R. Colonization of rice roots with methanogenic archaea controls photosynthesis-derived methane emission. Environ Microbiol. 2015;17:2254–2260. doi: 10.1111/1462-2920.12675. [DOI] [PubMed] [Google Scholar]
- 13.Adam P.S., Borrel G., Brochier-Armanet C., Gribaldo S. The growing tree of Archaea: new perspectives on their diversity, evolution and ecology. ISME J. 2017;11:2407–2425. doi: 10.1038/ismej.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offre P., Spang A., Schleper C. Archaea in biogeochemical cycles. Annu Rev Microbiol. 2013;67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 15.Evans P.N., Boyd J.A., Leu A.O., Woodcroft B.J., Parks D.H., Hugenholtz P. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17:219–232. doi: 10.1038/s41579-018-0136-7. [DOI] [PubMed] [Google Scholar]
- 16.Li M., Baker B.J., Anantharaman K., Jain S., Breier J.A., Dick G.J. Genomic and transcriptomic evidence for scavenging of diverse organic compounds by widespread deep-sea archaea. Nat Commun. 2015;6:8933. doi: 10.1038/ncomms9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seitz K.W., Dombrowski N., Eme L., Spang A., Lombard J., Sieber J.R. Asgard archaea capable of anaerobic hydrocarbon cycling. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-09364-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao H., Auguet J.-C., Gu J.-D. Global ecological pattern of ammonia-oxidizing archaea. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0052853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erguder T.H., Boon N., Wittebolle L., Marzorati M., Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- 20.Könneke M., Bernhard A.E., José R., Walker C.B., Waterbury J.B., Stahl D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 21.Roy D., McEvoy J., Blonigen M., Amundson M., Khan E. Seasonal variation and ex-situ nitrification activity of ammonia oxidizing archaea in biofilm based wastewater treatment processes. Bioresour Technol. 2017;244:850–859. doi: 10.1016/j.biortech.2017.08.060. [DOI] [PubMed] [Google Scholar]
- 22.Yang Y., Daims H., Liu Y., Herbold C.W., Pjevac P., Lin J.-G. Activity and metabolic versatility of complete ammonia oxidizers in full-scale wastewater treatment systems. mBio. 2020;11:e03175–e3219. doi: 10.1128/mBio.03175-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J., Kan J., Zhang X., Xia Z., Zhang X., Qian G. Archaea dominate the ammonia-oxidizing community in deep-sea sediments of the Eastern Indian Ocean—from the equator to the bay of Bengal. Front Microbiol. 2017;8:415. doi: 10.3389/fmicb.2017.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X., Koestler R.J., Warscheid T., Katayama Y., Gu J.-D. Microbial deterioration and sustainable conservation of stone monuments and buildings. Nat Sustain. 2020;3:991–1004. [Google Scholar]
- 25.Liu X., Meng H., Wang Y., Katayama Y., Gu J.-D. Water is a critical factor in evaluating and assessing microbial colonization and destruction of Angkor sandstone monuments. Int Biodeterior Biodegrad. 2018;133:9–16. [Google Scholar]
- 26.Meng H., Katayama Y., Gu J.-D. More wide occurrence and dominance of ammonia-oxidizing archaea than bacteria at three Angkor sandstone temples of Bayon, Phnom Krom and Wat Athvea in Cambodia. Int Biodeterior Biodegrad. 2017;117:78–88. [Google Scholar]
- 27.Meng H., Luo L., Chan H.W., Katayama Y., Gu J.-D. Higher diversity and abundance of ammonia-oxidizing archaea than bacteria detected at the Bayon Temple of Angkor Thom in Cambodia. Int Biodeterior Biodegrad. 2016;115:234–243. [Google Scholar]
- 28.Reyes C., Schneider D., Lipka M., Thürmer A., Böttcher M.E., Friedrich M.W. Nitrogen metabolism genes from temperate marine sediments. Mar Biotechnol. 2017;19:175–190. doi: 10.1007/s10126-017-9741-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mehta M.P., Baross J.A. Nitrogen fixation at 92 °C by a hydrothermal vent archaeon. Science. 2006;314:1783–1786. doi: 10.1126/science.1134772. [DOI] [PubMed] [Google Scholar]
- 30.Mwirichia R., Alam I., Rashid M., Vinu M., Ba-Alawi W., Anthony Kamau A. Metabolic traits of an uncultured archaeal lineage -MSBL1- from brine pools of the Red Sea. Sci Rep. 2016;6:19181. doi: 10.1038/srep19181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anantharaman K., Hausmann B., Jungbluth S.P., Kantor R.S., Lavy A., Warren L.A. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018;12:1715–1728. doi: 10.1038/s41396-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Shi L., Gu J.-D. Microbial electrocatalysis: Redox mediators responsible for extracellular electron transfer. Biotechnol Adv. 2018;36:1815–1827. doi: 10.1016/j.biotechadv.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y., Beer L.L., Whitman W.B. Sulfur metabolism in archaea reveals novel processes. Environ Microbiol. 2012;14:2632–2644. doi: 10.1111/j.1462-2920.2012.02783.x. [DOI] [PubMed] [Google Scholar]
- 34.Elshahed M.S., Najar F.Z., Roe B.A., Oren A., Dewers T.A., Krumholz L.R. Survey of archaeal diversity reveals an abundance of halophilic Archaea in a low-salt, sulfide-and sulfur-rich spring. Appl Environ Microbiol. 2004;70:2230–2239. doi: 10.1128/AEM.70.4.2230-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hausmann B., Pelikan C., Herbold C.W., Köstlbacher S., Albertsen M., Eichorst S.A. Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 2018;12:1729–1742. doi: 10.1038/s41396-018-0077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sievert S.M., Hügler M., Taylor C.D., Wirsen C.O. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2008. Sulfur oxidation at deep-sea hydrothermal vents; pp. 238–258. [Google Scholar]
- 37.Kletzin A., Urich T., Müller F., Bandeiras T.M., Gomes C.M. Dissimilatory oxidation and reduction of elemental sulfur in thermophilic archaea. J Bioenerg Biomembr. 2004;36:77–91. doi: 10.1023/b:jobb.0000019600.36757.8c. [DOI] [PubMed] [Google Scholar]
- 38.Castelle C.J., Wrighton K.C., Thomas B.C., Hug L.A., Brown C.T., Wilkins M.J. Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol. 2015;25:690–701. doi: 10.1016/j.cub.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 39.Liu X., Li M., Castelle C.J., Probst A.J., Zhou Z., Pan J. Insights into the ecology, evolution, and metabolism of the widespread Woesearchaeotal lineages. Microbiome. 2018;6:102. doi: 10.1186/s40168-018-0488-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz-Alvarez R., Casamayor E.O. High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes. Environ Microbiol Rep. 2016;8:210–217. doi: 10.1111/1758-2229.12370. [DOI] [PubMed] [Google Scholar]
- 41.Castelle C.J., Brown C.T., Anantharaman K., Probst A.J., Huang R.H., Banfield J.F. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat Rev Microbiol. 2018;16:629–645. doi: 10.1038/s41579-018-0076-2. [DOI] [PubMed] [Google Scholar]
- 42.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 43.Glöckner F.O., Yilmaz P., Quast C., Gerken J., Beccati A., Ciuprina A. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J Biotechnol. 2017;261:169–176. doi: 10.1016/j.jbiotec.2017.06.1198. [DOI] [PubMed] [Google Scholar]
- 44.De'Ath G. Multivariate regression trees: a new technique for modeling species–environment relationships. Ecology. 2002;83:1105–1117. [Google Scholar]
- 45.Wiens J.J. The niche, biogeography and species interactions. Philos Trans R Soc B: Biol Sci. 2011;366:2336–2350. doi: 10.1098/rstb.2011.0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jeanthon C., Nercessian O., Corre E., Grabowski-Lux A. Hyperthermophilic and methanogenic archaea in oil fields. Petrol Microbiol Am Soc Microbiol. 2005:55–69. [Google Scholar]
- 47.Mochimaru H., Yoshioka H., Tamaki H., Nakamura K., Kaneko N., Sakata S. Microbial diversity and methanogenic potential in a high temperature natural gas field in Japan. Extremophiles. 2007;11:453–461. doi: 10.1007/s00792-006-0056-8. [DOI] [PubMed] [Google Scholar]
- 48.Zhou L., Wang D.-W., Zhang S.-L., Tang E.-G., Lu Y.-W., Jing Y.-F. Functional microorganisms involved in the sulfur and nitrogen metabolism in production water from a high-temperature offshore petroleum reservoir. Int Biodeterior Biodegrad. 2020;154 [Google Scholar]
- 49.Zhou L., Zhou Z., Lu Y.-W., Ma L., Bai Y., Li X.-X. The newly proposed TACK and DPANN archaea detected in the production waters from a high-temperature petroleum reservoir. Int Biodeterior Biodegrad. 2019;143 [Google Scholar]
- 50.Bukin S., Pavlova O., Kalmychkov G., Ivanov V., Pogodaeva T., Galach’yants YP Substrate specificity of methanogenic communities from Lake Baikal bottom sediments associated with hydrocarbon gas discharge. Microbiology. 2018;87:549–558. [Google Scholar]
- 51.Carr S.A., Schubotz F., Dunbar R.B., Mills C.T., Dias R., Summons R.E. Acetoclastic Methanosaeta are dominant methanogens in organic-rich Antarctic marine sediments. ISME J. 2018;12:330–342. doi: 10.1038/ismej.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang J., Jiao S., Lu Y. Biogeographic distribution of bacterial, archaeal and methanogenic communities and their associations with methanogenic capacity in Chinese wetlands. Sci Total Environ. 2018;622:664–675. doi: 10.1016/j.scitotenv.2017.11.279. [DOI] [PubMed] [Google Scholar]
- 53.Chai H., Xiang Y., Chen R., Shao Z., Gu L., Li L. Enhanced simultaneous nitrification and denitrification in treating low carbon-to-nitrogen ratio wastewater: Treatment performance and nitrogen removal pathway. Bioresour Technol. 2019;280:51–58. doi: 10.1016/j.biortech.2019.02.022. [DOI] [PubMed] [Google Scholar]
- 54.Chao Y., Mao Y., Yu K., Zhang T. Novel nitrifiers and comammox in a full-scale hybrid biofilm and activated sludge reactor revealed by metagenomic approach. Appl Microbiol Biotechnol. 2016;100:8225–8237. doi: 10.1007/s00253-016-7655-9. [DOI] [PubMed] [Google Scholar]
- 55.Shu D., He Y., Yue H., Wang Q. Metagenomic and quantitative insights into microbial communities and functional genes of nitrogen and iron cycling in twelve wastewater treatment systems. Chem Eng J. 2016;290:21–30. [Google Scholar]
- 56.Kato S., Itoh T., Yuki M., Nagamori M., Ohnishi M., Uematsu K. Isolation and characterization of a thermophilic sulfur-and iron-reducing thaumarchaeote from a terrestrial acidic hot spring. ISME J. 2019;13:2465–2474. doi: 10.1038/s41396-019-0447-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McKay L.J., Dlakić M., Fields M.W., Delmont T.O., Eren A.M., Jay Z.J. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat Microbiol. 2019;4:614–622. doi: 10.1038/s41564-019-0362-4. [DOI] [PubMed] [Google Scholar]
- 58.Meier D.V., Pjevac P., Bach W., Hourdez S., Girguis P.R., Vidoudez C. Niche partitioning of diverse sulfur-oxidizing bacteria at hydrothermal vents. ISME J. 2017;11:1545–1558. doi: 10.1038/ismej.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paul B.G., Burstein D., Castelle C.J., Handa S., Arambula D., Czornyj E. Retroelement-guided protein diversification abounds in vast lineages of Bacteria and Archaea. Nat Microbiol. 2017;2:17045. doi: 10.1038/nmicrobiol.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez-Martinez A., Sihvonen M., Muñoz-Palazon B., Rodriguez-Sanchez A., Mikola A., Vahala R. Microbial ecology of full-scale wastewater treatment systems in the Polar Arctic Circle: Archaea, bacteria and fungi. Sci Rep. 2018;8:1–11. doi: 10.1038/s41598-018-20633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pornkulwat P., Kurisu F., Soonglerdsongpha S., Banjongproo P., Srithep P., Limpiyakorn T. Incorporation of 13C-HCO3− by ammonia-oxidizing archaea and bacteria during ammonia oxidation of sludge from a municipal wastewater treatment plant. Appl Microbiol Biotechnol. 2018;102:10767–10777. doi: 10.1007/s00253-018-9436-0. [DOI] [PubMed] [Google Scholar]
- 62.Yang Y., Herbold C.W., Jung M.-Y., Qin W., Cai M., Du H. Survival strategies of ammonia-oxidizing archaea (AOA) in a full-scale WWTP treating mixed landfill leachate containing copper ions and operating at low-intensity of aeration. Water Res. 2020 doi: 10.1016/j.watres.2020.116798. [DOI] [PubMed] [Google Scholar]
- 63.Kuypers M.M.M., Marchant H.K., Kartal B. The microbial nitrogen-cycling network. Nat Rev Microbiol. 2018;16:263–276. doi: 10.1038/nrmicro.2018.9. [DOI] [PubMed] [Google Scholar]
- 64.Munzi S., Sheppard L.J., Leith I.D., Cruz C., Branquinho C., Bini L. The cost of surviving nitrogen excess: energy and protein demand in the lichen Cladonia portentosa as revealed by proteomic analysis. Planta. 2017;245:819–833. doi: 10.1007/s00425-017-2647-2. [DOI] [PubMed] [Google Scholar]
- 65.Gulecal-Pektas Y., Temel M. A Window to the subsurface: Microbial diversity in hot springs of a sulfidic cave (Kaklik, Turkey) Geomicrobiol J. 2017;34:374–384. [Google Scholar]
- 66.Wang L., Cheung M.K., Liu R., Wong C.K., Kwan H.S., Hwang J.-S. Diversity of total bacterial communities and chemoautotrophic populations in sulfur-rich sediments of shallow-water hydrothermal vents off Kueishan Island, Taiwan. Microb Ecol. 2017;73:571–582. doi: 10.1007/s00248-016-0898-2. [DOI] [PubMed] [Google Scholar]
- 67.Youssef N.H., Farag I.F., Hahn C.R., Premathilake H., Fry E., Hart M. Candidatus Krumholzibacterium zodletonense gen. nov., sp nov, the first representative of the candidate phylum Krumholzibacteriota phyl. nov. recovered from an anoxic sulfidic spring using genome resolved metagenomics. Syst Appl Microbiol. 2019;42:85–93. doi: 10.1016/j.syapm.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Elshahed M.S., Najar F.Z., Roe B.A., Oren A., Dewers T.A., Krumholz L.R. Survey of archaeal diversity reveals an abundance of halophilic archaea in a low-salt, sulfide- and sulfur-rich spring. Appl Environ Microbiol. 2004;70:2230–2239. doi: 10.1128/AEM.70.4.2230-2239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.