Highlights
-
•
To our knowledge, this is the first study to identify a peptide (named as “TAP1”) that specifically binds with PSA−/lo prostate cancer cells.
-
•
TAP1 inhibited PCa growth both in vitro and in vivo.
-
•
TAP1 also improved the anti-tumor effect of the anti-androgens and chemotherapeutic agents in vitro.
-
•
The effects of TAP1 might at least in part by shortening the lengths of telomeres and decreasing the expression of HOXB9 and TGF-β2.
-
•
Our results indicated that therapeutic peptides that specifically target prostate cancer stem cell might be a very valuable and promising approach to overcome chemoresistance and prevent recurrence in patients with PCa.
Keywords: Prostate cancer, Androgen deprivation therapy, Phage display, Peptide, Cancer stem cell
Abstract
Patients with prostate cancer (PCa) will eventually progress to castrate-resistant prostate cancer (CRPC) after androgen deprivation therapy (ADT) treatment. Prostate-specific antigen (PSA)−/lo cells which harbor self-renewing long-term tumor-propagating cells that can be enriched using ALDH+CD44+α2β1+ and can initiate tumor development may represent a critical source of CRPC cells. Our purpose was to find a peptide that specifically targets PSA−/lo PCa cells to retard the development of CRPC. PSA+ and PSA−/lo cells were successfully separated from LNCaP xenograft tumors after prostate- PSAP-GFP vector infection and FACS. A variety of PSA−/lo cells specifically targeting peptide (named as “TAP1” targeted affinity peptide 1) was identified by using phage display library screening. The highest binding rate in TAP1 binding cell subpopulations are identified to be among ALDH+CD44+CXCR4+CD24+ cells. TAP1 significantly inhibited PCa growth both in vitro and in vivo. TAP1 significantly improved the anti-proliferation effect of the anti-androgens (Charcoal dextran-stripped serum (CDSS)+Bicalutamide, Enzalutamide) and chemotherapeutic agents (Abiraterone, Docetaxel, Etoposide) in vitro. TAP1 treatment shortens the length of telomeres in ALDH+CD44+CXCR4+CD24+ cells and significantly reduces the expression of Homeobox B9 (HOXB9) and TGF-β2. In conclusion, PSA−/lo PCa cell-specific targeting peptide (TAP1) that suppressed PCa cell growth both in vitro and in vivo and improved the drug sensitivities of anti-androgens and chemotherapeutic agents at least through shortening the length of telomere and reducing the expression of HOXB9 and TGF-β2. Therapeutic peptides that specifically target prostate cancer stem cell might be a very valuable and promising approach to overcome chemoresistance and prevent recurrence in patients with PCa.
Introduction
Prostate cancer (PCa) is the second most frequently diagnosed cancer after lung cancer in men worldwide in 2018 [1]. Race, age, and a positive family history of PCa are the three confirmed risk factors for PCa [2]. A high incidence of PCa was shown in western countries. Oceania, followed by Northern America, Western Europe, Northern Europe, and the Caribbean have among the highest PCa incidence rates in the world [3]. The incidence of PCa in Asia is rising but is still significantly lower than that in western countries [4]. PCa incidence strongly increases with age. PCa risk begins to rise sharply after age 55 years and peaks at age 70–74, declining slightly thereafter [5]. Recent study showed that PCa incidence has increased in both older adolescents and young adult men. This may be caused by trends in obesity, physical inactivity, HPV infection, substance exposure, and environmental carcinogens [6]. A study that examined the current trends of metastatic PCa from 2004 to 2014 and forecast the annual burden and incidence rates by age and race for 2015–2025 showed that the annual burden is expected to increase by 42% by 2025, with 10,615 cases expected in 2015 and 15,097 in 2025 in the US[7]. There is an urgent need for determining the effective treatment options for PCa.
PCa is androgen dependent. Standard treatments for PCa include surgical removal, radiation, and hormone therapy, and in recent years, immunotherapy as an alternative method has gained increasing interest [8]. Androgen deprivation therapy (ADT), using surgical or chemical castration that block the synthesis of testosterone or block the androgen receptor, is a standard treatment used in all stages of recurrent PCa [9,10]. Management of metastatic hormone-sensitive prostate cancer (mHSPC) changed dramatically in the past several years. Data supported a survival benefit with the addition of four different agents (docetaxel, abiraterone acetate, enzalutamide, and apalutamide) to androgen deprivation among men with mHSPC [11]. Both chemotherapy and androgen-receptor signaling inhibitors (ARSi) demonstrated a significant survival benefit when combined with ADT compared to ADT alone for patients with mHSPC [12]. Nearly all patients eventually progress to castrate-resistant prostate cancer (CRPC) after about 18 months of ADT [13]. The mechanism of CRPC remains obscure. Chromosomal rearrangements and copy number gains/losses and gene alteration in the androgen axis and the kinase-dependent signal pathways are thought to be important mechanisms of CRPC genesis and development [14,15]. Accumulated evidences have indicated that tumor immunity, DNA repairing disorder, and other mechanisms might also participate in the progress of CRPC [16]. Prostate cancer stem cells (PCSC) are a small population of cells in PCa that display unlimited self-renewal potential and tumor-initiating capacities [17], [18], [19]. Studies showed that ADT resulted in a time-dependent increase in PCSC population numbers [20] that and PCSC is linked with high-risk disease, development of metastasis, biochemical failure, and poorer progression free survival [21]. PCSC can survive from chemotherapy or radiotherapy and are suggested to be responsible for the development of CRPC [22,23]. The expression of prostate-specific antigen (PSA), as a differentiation marker, in PCa positively correlates with its overall degree of differentiation [24,25]. PCa contains both differentiated (PSA+) and undifferentiated (PSA−/lo) tumor cells. It has been reported that PSA−/lo cells preferentially express stem cell genes, can initiate robust tumor development, possess long-term tumor-propagating capacity, and harbor highly tumorigenic castration-resistant PCa cells that can be prospectively enriched using the ALDH+CD44+α2β1+ phenotype [26]. Conversely, PSA+PCa cells possess a higher limited tumor-propagating capacity, undergo symmetric division, and are sensitive to castration. Thus, PSA−/lo cells may represent a critical source of CRPC cells. Novel therapeutic strategies targeting PSA−/lo cells may overcome and prevent PCa recurrence.
Therapeutic peptide is a promising and novel approach to treat many diseases, including cancer. It has several unique advantages over proteins or antibodies, such as ease of synthesis, high target specificity and selectivity, and low toxicity in normal tissues[27]. Phage display is a powerful technology for screening and isolating target specific peptides [28]. In this study, using phage display library (PDL) screening, we identified a PSA−/lo PCa cell-specific targeting peptide that can suppress PCa cell growth both in vitro and in vivo. Targeting PSA−/lo PCa cells with peptide might represent a novel therapeutic strategy for PCa treatment.
Materials and methods
Cell culture
Human prostate cancer cell line LNCaP was purchased from American Type Culture Collection (ATCC) (Manassas, VA, US). Cells were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, US) with 8% fetal bovine serum (FBS) (Sigma). Cells were maintained in a 37 °C humidified incubator with 5% CO2.
Reagents and chemotherapeutic agents
Dimethyl Sulfoxide (DMSO), hydrogen peroxide solution (H2O2), bicalutamide, abiraterone, docetaxel, and etoposide were obtained from Sigma. Charcoal dextran-stripped serum (CDSS) was obtained from Gemini (West Sacramento, CA, US). Enzalutamide was obtained from Selleck Chemicals (Houston, TX, US).
Animals
NOD-SCID mice (six weeks old, male) were purchased from Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China). The handling of the mice and all experimental procedures were carried out in strict accordance with Fudan University Guidelines for the Care and Use of Laboratory Animals. All animal studies were approved by the Ethics Committee of Fudan University Pudong Medical Center.
Separate PSA−/lo and PSA+ cells from lncap xenograft tumor
A measure of 1 × 106 LNCaP cells in 50 μL medium was mixed with 50 μL Matrigel (BD biosciences, San Jose, CA, US) and was injected into the subcutaneous area of NOD-SCID mice limbs. Mice were checked daily. When LNCaP xenograft tumors volume reached 1.5 cm3, tumors were harvested, minced into 1 mm3 pieces, and then digested with Accumax (Sigma) for 30 min at 37 °C. Digests were poured through a 70 μm cell strainer (Corning, NY, US). Cells were maintained in RPMI-1640 medium with 8% FBS. To separate PSA−/lo and PSA+ cells, as previously described[26], briefly, cells were infected with PSA promoter (PSAP)-GFP vector (which drives eGFP expression) for 72 h. Next, fluorescence-activated cell sorting (FACS) was performed to purify the top 10% GFP-bright (GFP+) and bottom 2%−6% GFP-negative/GFP-dim (GFP−/lo) cells. Through this separating, most purified GFP+cells were PSA+ cells and GFP− cells were PSA−/lo cells.
Phage display library screening
CX8C phage display peptide library was constructed using the fUSE55 vector which displays 1 × 109 CX8C (8 amino acid peptides in random sequences between 2 cysteine residues) unique peptide sequences. GFP+ (PSA+) and GFP− (PSA−/lo) cells were mixed in equal proportions and incubated with CX8C phage display library. A measure of 1 × 109 of the phage display peptide library in 5 ml of 1× phosphate buffered saline (PBS) was added to the confluent cell monolayer and incubated for 2 h on a rocker platform at 4 °C. The cells were washed four times with PBS containing 0.2% Tween 20. Cells were subjected to fluorescence-activated cell sorting (FACS) for GFP+ and GFP− cells. Sorted cells were lysed to release phage particles that were used to infect K91 bacteria. Individual bacterial colonies that emerged on the Tet/Kanamycin plates were isolated, amplified, and phage particles re-isolated and used in 2–3 more similar cycles. Peptides that specifically bind to GFP+ (PSA+) cells and GFP− (PSA−/lo) cells were identified and sequenced.
Synthetic peptides
Peptide TAP1 (TEWDYLTV), control peptide (TDEWLTYV), and biotinylated peptide TAP1 (TEWDYLTV) and biotinylated control peptide (TDEWLTYV) were synthesized and purified by Chinese peptide company (Hang Zhou, China). All peptides were dissolved and diluted in 1 × PBS.
Fluorescence-Activated cell sorting (FACS)
Cells from LNCaP xenograft tumors were incubated with a FcR blocking agent (Miltenyi Biotec, San Diego, CA, US) for 15 min at 4 °C and were then stained with CXCR4 antibody (BD, Biosciences, San Jose, CA, US) for 30 min on ice, followed by staining with APC-conjugated goat anti-mouse IgG (BD Biosciences) for 15 min on ice. Cells were then washed thrice and stained with PE conjugated anti-CD44 antibody (BD Bioscience) and PE-cy7 conjugated anti-CD24 (BD Biosciences) for 20 min. After washing with PBS, cells were incubated in a solution containing 1% bovine serum albumin (BSA) (Sigma-Aldrich) and 2.5 μg/ml insulin (Sigma-Aldrich). Then, the cells were suspended in an ALDEFLUOR assay buffer containing ALDH substrate (STEMCELL Technologies China Co., Ltd., Shanghai, China). After incubating at 37 °C for 40 min, cells were sorted by fluorescence-activated cell sorting (FACS).
Staining of biotinylated peptides
PSA+and PSA−/lo cells were seeded in a 6-well plate. The next day, cells were incubated with biotinylated peptide TAP1 (5 μM) or biotinylated control peptide (TDEWLTYV) (5 μM) for 72 h. Then cells were washed with 1 × PBS twice and fixed with methanol for 10 min and blocked with 5% FBS (in 1 × PBS) for 30 min. Cells were washed with 1 × PBS and stained with streptavidin-Alexafluor 594 (green) (Invitrogen, Carlsbad, CA, US). Cells nuclei were stained with Propidium Iodide (PI) (red).
TAP1 binding positivity calculating
Cell subpopulations (PSA+ and PSA−/lo cells, CD44+ and CD44− cells, ALDH+CD44+α2β1+ and ALDH−CD44−α2β1− cells, ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells) were seeded in a 6-well plate. The next day, cells were incubated with biotinylated peptide TAP1 (5 μM) or biotinylated control peptide (TDEWLTYV) (5 μM) for 72 h. Then cells were stained with streptavidin-Alexafluor 594 (green) (Invitrogen, Carlsbad, CA, US). Cells nuclei were stained with Propidium Iodide (PI) (red). The percentage of biotinylated TAP1 or biotinylated control peptide binding positivity was quantified at 10 randomly selected fields under an inverted light microscope (Olympus, Tokyo, Japan).
Cell proliferation assay
Proliferation assays were conducted using WST-1 assay (Beyotime, Shanghai, China). ALDH+CD44+CXCR4+CD24+ cells were isolated and purified from LNCaP xenograft tumor and seeded in 96-well plates and treated with peptide TAP1 or control peptide at 1, 2, 5, 10, 20, and 40 μM for 72 h. ALDH+CD44+CXCR4+CD24+ cells were incubated with peptide TAP1 (20 μM) or control peptide (20 μM) for 24, 48, and 72 h. ALDH+CD44+CXCR4+CD24+ cells and ALDH−CD44−CXCR4−CD24− cells were incubated with anti-androgens or chemotherapeutic agents at an indicated concentration and with peptide TAP1 (5 μM) or control peptide (5 μM) for 72 h. Culture medium was removed, and 100 μl fresh medium containing 10 μl of WST-1 reagents was added into the wells. After 1–2 h, the absorbance was measured at 450 nm using an ELISA Microplate Reader (Biocompare, San Francisco, CA).
Transwell assay
Cell migration and invasion abilities were evaluated by the transwell assay with Corning BioCoat Matrigel Invasion Chamber (Tewksbury, MA, US). According to the manufacturer's instructions, 500 μL cells suspensions (2.5 × 104 cells) were loaded to the 24-well chambers and then incubated for 22 h in a 37 °C humidified incubator with 5% CO2. After incubation, the non-invading cells were removed from the upper surface of the membrane by scrubbing. The invading cells on the lower surface of the membrane were fixed and stained using crystal violet. The stained cells were counted in 10 randomly selected fields under an inverted light microscope (Olympus, Tokyo, Japan).
ALDH+CD44+CXCR4+CD24+ xenograft model and treatment of TAP1 by intratumoral injection
ALDH+CD44+CXCR4+CD24+ cells were isolated and purified from LNCaP xenograft tumors. A measure of 1 × 106 ALDH+CD44+CXCR4+CD24+ cells was injected subcutaneously into the NOD-SCID mice (n = 12). After 7 days of tumor cell implantation, mice were randomly divided into two groups (n = 6 per group) and treated with 10 μL (20 μM) peptide TAP1 or 10 μL (20 μM) control peptide every other day by intratumoral injection. Tumor volume was measured every week. Tumor diameters are measured with digital calipers, and the tumor volume in mm3 is calculated by the formula: Volume = (width)[2] x length/2. At 28 days of tumor cell implantation, tumors were harvested and weighed.
ALDH−CD44−CXCR4−CD24− xenograft model and western blot analysis
A measure of 1 × 106 ALDH−CD44−CXCR4−CD24− cells were injected subcutaneously into the NOD-SCID mice. Five weeks after tumor cell implantation, mice were castrated and received Bicalutamide (150 mg/kg body weight/day) by subcutaneous injection. After 2 weeks of castration and Bicalutamide treatment, tumors received 10 μL (5 μM) biotinylated peptide TAP1 by intratumoral injection. Forty-eight hours after intratumoral injection, tumors were excised and we performed fluorescein-5-isothiocyanate (FITC) staining to identify TAP1 binding.
Using laser capture microdissection (LCM) to obtain biotinylated TAP1 targeted tumor tissue (Zone1) and non-biotinylated TAP1 targeted tumor tissue (Zone2). Total protein was extracted using a protein extraction kit (Beyotime, Shanghai, China). Protein concentration was measured using the BCA Protein Assay Kit (Beyotime, Shanghai, China). Protein fractions were separated on sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis (Bio-Rad, Hercules, CA, US) and transferred to a nitrocellulose membrane (Bio-Rad). After blocking in 5% skimmed milk in PBS for 1 h at room temperature, the membranes were incubated overnight at 4 °C with primary antibodies. Primary antibodies against PSA, PSMA, ALDH, CD44, CXCR4, CD24, HOXB9, TGF-β1, TGF-β2, and GAPDH were obtained from Cell Signaling Technology (Beverly, MA, US). Secondary antibodies IRDye800CW Goat anti-Mouse IgG and IRDye800CW Goat anti-Rabbit IgG were obtained from LI-COR (LI-COR Biotechnology, Lincoln, NE, US). Western bolt images were detecting using a Li-COR Odyssey 9120 Imaging System (LI-COR Biotechnology).
Flow-fluorescence in situ hybridization (flow-FISH)
Flow-FISH was conducted to measure the telomere lengths of the cells. Flow cytometer calibration, cell fixation, staining protocol, and normalization were conducted using mouse lymphoma cells with known telomere lengths. A measure of 5 × 105 ALDH+CD44+CXCR4+CD24+ cells, ALDH−CD44−CXCR4−CD24− cells, or mouse lymphoma cells were washed in hybridization buffer and resuspended in hybridization solution containing formamide and 0.3 μg/ml FITC-conjugated pentose nucleic acid (PNA) probe. Control samples were incubated in hybridization solution without a PNA probe. Lymphoma cells were distinguished from cell derivatives by immunostaining with CD45 antibody (Sigma). The DNA content was quantified using propidium iodide staining. Cells were gated at G0/G1 based on DNA content, and the telomere fluorescence intensity was calculated. Detections were conducted on an FACSCanto flow cytometer (Becton Dickinson; Franklin Lakes, NJ, US).
Statistical analysis
Data represent the mean ± SD. Statistical analysis was performed using GraphPad Prism (version 5.0, GraphPad Software, La Jolla, CA). One-way analysis of variance along with Bonferroni adjustment and student's t-test were used to evaluate the differences between groups. A p value < 0.05 was considered statistically significant.
Results
Identification of a peptide that specifically binds with PSA−/lo PCa cells
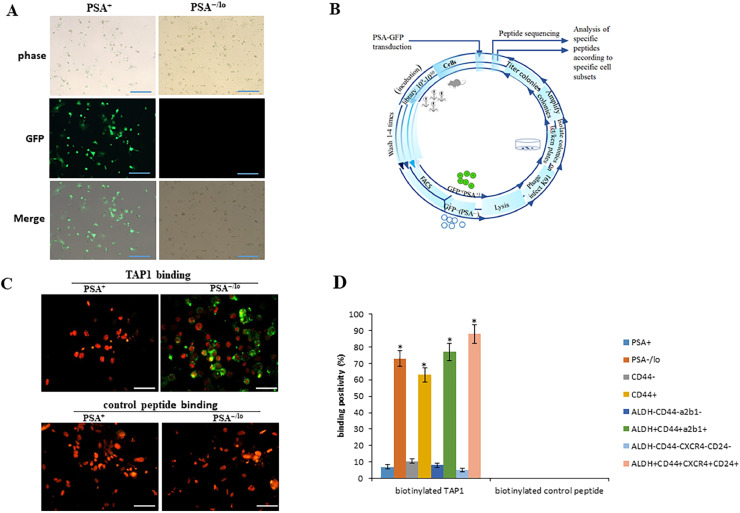
It has been reported that PSA−/lo cells may represent a critical cell source of CRPC[26]. Our goal was to find a therapeutic peptide that specifically targets PSA−/lo PCa cells. To separate PSA+ and PSA−/lo PCa cells, LNCaP xenograft tumors were established and then xenograft tumors were harvested, minced, and digested and infected with PSAP-GFP lentiviral reporter (which drives eGFP expression) for 72 h. Perform FACS to purify out the top 10% GFP-bright (GFP+) and bottom 2%−6% GFP−/lo cells (supplementary Figure S1). As shown in Fig. 1A, we successfully separated PSA+ and PSA−/lo PCa cells from a LNCaP xenograft tumor. Most GFP+cells were PSA+ cells, and GFP− cells were PSA−/lo cells. Next, PSA+and PSA−/lo cells were mixed in equal proportions and incubated with CX8C phage display library. Through the phage display assay (schematic in Fig. 1B), after 4 rounds, we sequenced 272 and 257 specific peptides that bound with PSA+ and PSA−/lo cell clones, respectively. The majority of (about 72%) the sequences were shared between PSA+ and PSA−/lo clones. Clones specific to PSA−/lo cells were identified, among them the most abundant was a peptide with a sequence of TEWDYLTV, which we named as “targeted affinity peptide 1 (TAP1)”.
Fig. 1.
Identification of a peptide that specifically binds with PSA−/lo PCa cells. (A) Cells from LNCaP xenograft tumors were infected with PSAP-GFP lentiviral reporter for 72 h. Perform FACS to purify the top 10% GFP-bright (GFP+) and bottom 2%−6% GFP−/lo cells. Representative images of GFP+ (PSA+) and GFP− (PSA−/lo) cells. Scale bars 25 μm. (B) Schematic diagram of phage display library screening. (C) Purified PSA−/lo and PSA+ cells were incubated with biotinylated peptide TAP1 (5 μM) or biotinylated control peptide (5 μM) for 72 h. Biotinylated peptide TAP1 was stained with streptavidin-Alexafluor 594 (green) and cells nuclei were stained with Propidium Iodide (PI) (red).Scale bars 10 μm. (D) Cell subpopulations (PSA+ and PSA−/lo cells, CD44+ and CD44− cells, ALDH+CD44+α2β1+ and ALDH−CD44−α2β1− cells, ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells) were incubated with biotinylated peptide TAP1 (5 μM) or biotinylated control peptide (5 μM) for 72 h. Biotinylated peptide TAP1 was stained with streptavidin-Alexafluor 594 (green) and cells nuclei were stained with Propidium Iodide (PI) (red). The percentage of peptide binding positivity was quantified in 10 randomly selected fields under an inverted light microscope. Data are presented as the mean ± SD. *p < 0.05. Experiments were performed in triplicate.
To further confirm that TAP1 specifically binds with PSA−/lo cells, purified PSA−/lo and PSA+cells were incubated with biotinylated TAP1 (5 μM) or biotinylated control peptide (5 μM) for 72 h followed by streptavidin-Alexafluor 594, as shown in Fig. 1C, biotinylated TAP1 specifically binds to PSA−/lo cells, no TAP1 binding was observed in PSA+ cells. Biotinylated control peptide did not bind to PSA+or PSA−/lo cells. A study showed that PSA−/lo cells harbor self-renewing cancer stem cells that can be prospectively enriched using ALDH+CD44+α2β1+ phenotype [26]. Aldehyde dehydrogenase (ALDH) [29], CD44 [30], α2β1 [31], CXCR4 [32], and CD24 [33] are well established cancer stem cell markers. To determine the binding affinity of TAP1 in PSA−/lo cells and PCSC, cells were incubated with biotinylated peptide TAP1 (5 μM) or biotinylated control peptide (5 μM) for 72 h followed with streptavidin-Alexafluor 594. The percentage of biotinylated TAP1 and biotinylated control peptide binding positivity was calculated. As shown in Fig. 1D, biotinylated TAP1 showed highest affinity and specificity in ALDH+CD44+CXCR4+CD24+ cells (almost 90% positivity). Moreover, biotinylated control peptide (TDEWLTYV) did not significantly bind to either PSA+ or PSA−/lo cells (Fig. 1D). Taken together, these findings indicated that peptide TAP1 specifically binds with PSA−/lo cells in vitro. ALDH+CD44+CXCR4+CD24+ cell subpopulation has the highest TAP1 binding affinity. Our preliminary data showed that ALDH+CD44+CXCR4+CD24+ cells are the most resistant cells to CRPC among these cell subpopulations. In the following experiments, we used ALDH+CD44+CXCR4+CD24+ cells.
TAP1 inhibits PCa cell growth and invasion in vitro
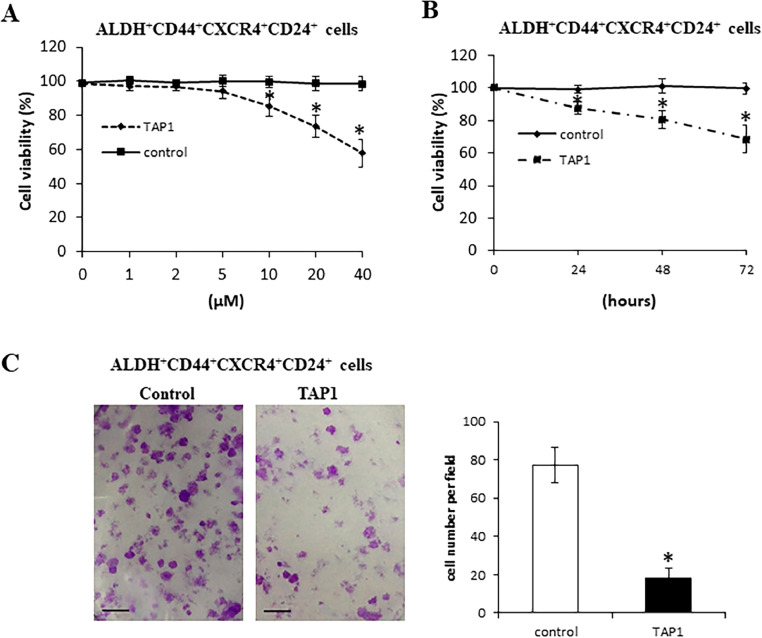
To investigate the role of TAP1 in ALDH+CD44+CXCR4+CD24+ cells progression, ALDH+CD44+CXCR4+CD24+ were incubated with TAP1 or control peptide at various concentrations for 72 h. Cell proliferation was evaluated by WST-1. As shown in Fig. 2A, TAP1 did not affect cell viability at 5 μM; however, TAP1 significantly decreased cell viability starting at 10 μM and caused a dose-dependent decrease in cell viability over a 72 h incubation period compared with control peptide. ALDH+CD44+CXCR4+CD24+ cells were incubated with TAP1 (20 μM) or control peptide (20 μM) for 24, 48, and 72 h. As shown in Fig. 2B, TAP1 significantly inhibited PCa cell growth with increasing time compared with control peptide. Control peptide has no effect on the cell viability. The effect of TAP1 on ALDH+CD44+CXCR4+CD24 cell migration and invasion capacity was analyzed using transwell assay. As shown in Fig. 2C, TAP1 significantly inhibited ALDH+CD44+CXCR4+CD24 cell migration and invasion compared with the control peptide. These findings indicated that TAP1 significantly inhibited ALDH+CD44+CXCR4+CD24+ cell proliferation, migration, and invasion in vitro.
Fig. 2.
TAP1 inhibits PCa growth in vitro. (A) ALDH+CD44+CXCR4+CD24+ cells were incubated with TAP1 or control peptide at 1, 2, 5, 10, 20, and 40 μM for 72 h. Cell viability was measured by WST-1. (B) ALDH+CD44+CXCR4+CD24+ cells were incubated with TAP1 or control peptide at 20 μM for 24, 48, and 72 h. Cell viability was measured by WST-1. (C) Transwell migration assay of ALDH+CD44+CXCR4+CD24+ cells incubated with TAP1 or control peptide at 20 μM. Left panel: representative images (scale bars = 50 μM). Right panel: quantification. Data are presented as the mean ± SD. *p < 0.05. All experiments were performed in triplicate.
TAP1 inhibits PCa growth in vivo
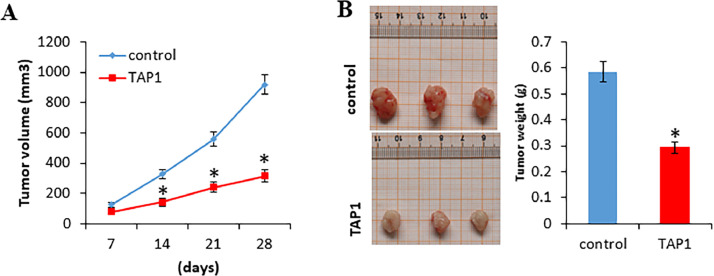
To examine whether TAP1 inhibits tumor growth in vivo, 1 × 106 ALDH+CD44+CXCR4+CD24+ cells were injected subcutaneously into the NOD-SCID mice. After 7 days of tumor cell implantation, mice were treated with 10 μL (20 μM) TAP1 or 10 μL (20 μM) control peptide every other day by intratumoral injection. As shown in Fig. 3A and 3B, TAP1 treatment significantly decreased tumor volume and tumor weight compared with the control peptide. These results suggested that TAP1 inhibited PCa growth in vivo.
Fig. 3.
TAP1 inhibits PCa growth in vivo. (A) 1 × 106 ALDH+CD44+CXCR4+CD24+ cells were injected subcutaneously into the NOD-SCID mice, n = 12. After 7 days of tumor cell implantation, mice were randomly divided into two groups (n = 6 per group) and treated with 10 μL (20 μM) peptide TAP1 or 10 μL (20 μM) control peptide every other day by intratumoral injection. Tumor vol`ume was measured every week. Volume = (width)[2] × length/2. (B) At 28 days of tumor cell implantation, tumors were harvested, photographed, and weighed. Data are presented as the mean ± SD. *p < 0.05.
TAP1 enhances the anti-proliferation effect of anti-androgens and chemotherapeutic agents in vitro
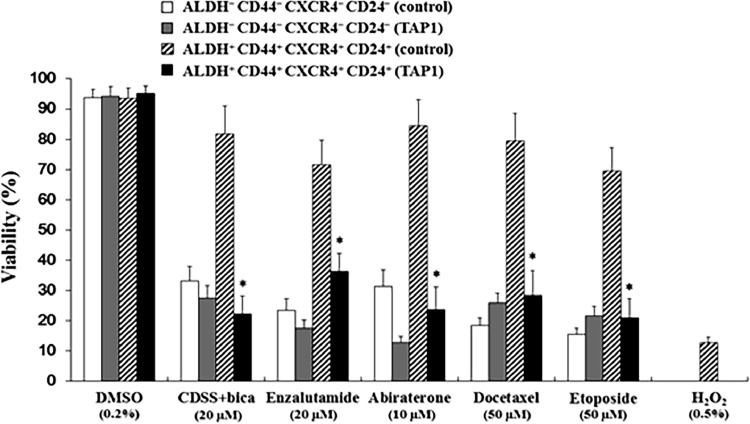
We tested whether TAP1 affects the drug sensitivities of anti-androgens and chemotherapeutic agents in vitro. ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells were incubated with 5 μM TAP1 (at this concentration, TAP1 did not affect cell viability) or 5 μM control peptide with anti-androgens (CDSS+ Bicalutamide, Enzalutamide), chemotherapeutic agents (Abiraterone, Docetaxel, Etoposide), and H2O2 therapy at indicated concentration for 72 h. Cell viability was assessed by WST-1. As shown in Fig. 4, when anti-androgens or chemotherapeutic agents were co-treated with 5 μM TAP1, the anti-proliferation of anti-androgens and chemotherapeutic agents in ALDH+CD44+CXCR4+CD24+ cells, but not in ALDH−CD44−CXCR4−CD24− cells, was significantly improved compared to control peptide co-treatment. In addition, our result showed that ALDH+CD44+CXCR4+CD24+ cells were resistant to H2O2 and that TAP1 improved the sensitivity of ALDH+CD44+CXCR4+CD24+ cells to H2O2. These findings indicated that TAP1 specifically enhanced the anti-proliferation efficacy of anti-androgens, chemotherapeutic agents, and H2O2 in ALDH+CD44+CXCR4+CD24+ cells. Our findings suggested that TAP1 is not only a single therapeutic peptide but might be considered as a good, combined administration for PCa therapy.
Fig. 4.
TAP1 enhances the anti-proliferation effect of anti-androgens and chemotherapeutic agents in vitro. ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells were incubated with anti-androgens (CDSS+ Bicalutamide, Enzalutamide), chemotherapeutic agents (Abiraterone, Docetaxel, Etoposide), and H2O2 at indicated concentration with TAP1 (5 μM) or control peptide (5 μM) for 72 h. DMSO was used as control. Cell viability was assessed by WST-1. Data are presented as the mean ± SD. *p < 0.05. Experiments were performed in triplicate.
TAP1 specifically binds with PSA−/lo cells in vivo
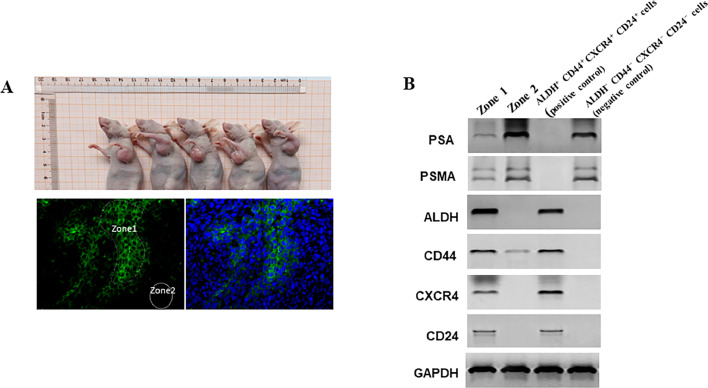
To investigate the targeting cells of TAP1 in vivo, ALDH−CD44−CXCR4−CD24− cells were injected subcutaneously into the NOD-SCID mice. Five weeks after tumor cell implantation, mice were castrated and received Bicalutamide (an antiandrogen medication that is used to treat PCa) (150 mg/kg body weight/day) by subcutaneous injection. It has been reported that ADT resulted in a time dependent increase in PCSC population numbers [20]. After 2 weeks of castration and Bicalutamide treatment, tumors received 10 μL (5 μM) biotinylated peptide TAP1 by intratumoral injection. Forty-eight hours after intratumoral injection, tumors were harvested. The in vivo binding of biotinylated-TAP1 was demonstrated by fluorescence staining of tumor tissue. As shown in Fig. 5A, the Zone1 area showed strong binding to the biotinylated-TAP1, whereas there was no apparent accumulation in Zone2. Using laser capture microdissection, we obtained Zone1 and Zone2 tumor tissue. Prostate-specific membrane antigen (PSMA) is a receptor on the surface of PCa cells which is expressed at low levels in normal human prostate epithelium and is highly expressed in PCa [34]. Western blot analysis of protein lysates from Zone1 and Zone2 showed that the expressions of PAS and PMSA in Zone1 are significantly lower than Zone2 area (Fig. 5B). The expressions of cancer stem cell markers, ALDH, CD44, CXCR4, and CD44 were significantly higher in Zone1 compared with Zone2 area. The protein levels in ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells were used as positive and negative controls, respectively. These findings indicated that ALDH+CD44+CXCR4+CD24+ cells were generated in ALDH−CD44−CXCR4−CD24− xenograft tumors and that TAP1 specifically binds with ALDH+CD44+CXCR4+CD24+ cells in vivo.
Fig. 5.
TAP1 specifically binds with PSA−/lo cells in vivo. (A) 1 × 106 ALDH−CD44−CXCR4−CD24− cells were injected subcutaneously into the NOD-SCID mice. Five weeks after tumor cell implantation, mice were castrated and received Bicalutamide (150 mg/kg body weight/day) by subcutaneous injection. After 2 weeks of castration and Bicalutamide treatment, tumors received 10 μL (5 μM) biotinylated peptide TAP1 by intratumoral injection. Forty-eight hours after intratumoral injection, tumors were excised and we performed fluorescein-5-isothiocyanate (FITC) staining. Zone1 represented biotinylated peptide TAP1 targeted tumor tissue and Zone2 represented no biotinylated peptide targeted tumor tissue (Lower and left panel). FITC and DAPI staining were shown in the lower and right panel. Scale bars 20 μm. (B) Using laser capture microdissection (LCM) to obtain Zone1 and Zone2 tumor tissue. Western blot analysis of protein expression in Zone1 and Zone2. Protein levels in ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells are as positive and negative control, respectively. GAPDH is used as a loading control.
TAP1 shortens the length of telomere and targets HOXB9 and TGF-β2
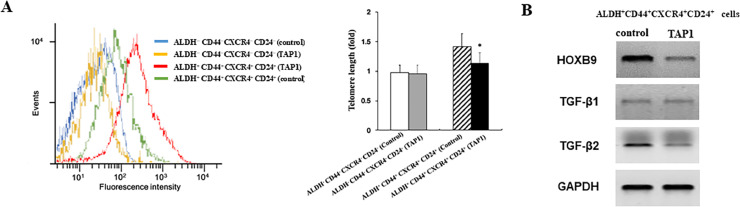
Cancer stem cells (CSCs) have the capacity to self-renew, differentiate, and give rise to entire new tumors [35]. Telomeres are protective structures of chromosome ends, are essential for the maintenance of chromosomal integrity, and are gradually shortened by each cell division [36]. Functional telomeres are essential for the developmental pluripotency of embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), and CSCs [37,38]. ESCs with long telomeres exhibit authentic developmental pluripotency, whereas, short telomeres decrease the proliferative rate or capacity of ESCs and disrupt germ cell differentiation [37]. iPSCs with longer telomeres generate chimeras with higher efficiency than those with short telomeres[37]. We examined the effect of TAP1 on the length of telomere through flow-FISH. As shown in Fig. 6A, TAP1 treatment significantly decreased the lengths of telomeres in ALDH+CD44+CXCR4+CD24+ cells compared with control peptide. There was no significant effect of TAP1 on the lengths of telomeres in ALDH−CD44−CXCR4−CD24− cells. These finding suggested that TAP1 led to telomere shortening which may cause the loss of pluripotency in ALDH+CD44+CXCR4+CD24+ cells and inhibit cell proliferation.
Fig. 6.
TAP1 shortens the length of telomere and targets HOXB9 and TGF-β2. (A) ALDH+CD44+CXCR4+CD24+ and ALDH−CD44−CXCR4−CD24− cells were incubated in TAP1 (20 μM) for 72 h followed by flow-FISH analysis to determine telomere lengths. Representative images (left panel). Summary data (right panel). (B) ALDH+CD44+CXCR4+CD24+ were incubated with TAP1 (20 μM) or control peptide (20 μM) for 72 h. Western blot analysis of the protein expression. GAPDH is used as a loading control. Data are presented as the mean±SD. *p < 0.05.
Homeobox B9 (HOXB9) is a member of the homeobox family of transcription factors. Silencing of HOXB9 suppresses PCa cell proliferation and invasion in vitro [39]. HOXB9 activates transforming growth factors beta (TGF-β) pathway and promotes tumorigenicity and lung metastasis in breast cancer [40]. TGF-β subfamily contains five members named TGF-β1 through β5. TGF-β1, -β2, and -β3 have been identified in mammals [41]. TGF-β cytokines regulate multiple cellular functions, and depending on the cellular context, different TGF-βs exhibit significant pleiotropic effects on cells [42]. A study showed that repression of TGF-β signaling suppresses CRPS progression [43]. Our Western blotting assay showed that TAP1 treatment significantly decreased the expression of HOXB9 and TGF-β2 but not TGF-β1 in ALDH+CD44+CXCR4+CD24+ cells (Fig. 6B). Taken together, our finding demonstrated that TAP1 inhibits ALDH+CD44+CXCR4+CD24+ cell growth at least through telomere shortening and the inhibition of HOXB9 and TGF-β2.
Discussion
Despite robust progress in CRPC therapeutics development, CRPC remains a lethal malignancy [44]. Although the cell origin of CRPC remains controversial, several studies clearly indicate the presence of PCSC in CRPC [45]. PCSC plays a major role in PCa initiation, progression, and metastasis and is considered to be a driving force of cancer evolution and resistance to therapy [45], [46]. Most therapeutic regimens only target the proliferative tumor cells. Therefore, targeting PCSC to decrease the expansion of CSCs population may lead to new strategies to overcome chemoresistance. Therapeutic peptide has been of great interest due to advantages such as a low molecular weight, the ability to specifically target tumor cells, and low toxicity in normal tissues [47]. Peptide-based therapies have shown promising results in both in vitro and in vivo studies. For example, a bi-functional peptide designed by coupling the cancer recognition peptide to the toxic peptide showed toxicity to breast cancer, PCa, and neuroblastoma cell lines [48]. Han et al. developed a small peptide specific to Extradomain-B fibronectin (EDB-FN) for PCa targeting and imaging [49]. Wada et al. identified peptides that specifically target xenografted PCa cells and suppressed cell growth [50]. In this study, by using phage display library screening, we identified a peptide TAP1 that specifically targets PSA−/lo PCa cells and inhibited cell growth both in vitro and in vivo. In addition, we observed that the anti-tumor effects of the anti-androgens, chemotherapeutic agents, and H2O2 in ALDH+CD44+CXCR4+CD24+ cells were significantly improved when the anti-androgens and chemotherapeutic agents were co-treated with TAP1. Our finding indicated that this selective targeting peptide may be a very promising agent which avoids adverse effects on non-target cells and can be used either alone or in combination with standard therapies.
Telomere length maintenance is critical for the unlimited self-renewal and pluripotency of a stem cell. Pluripotent stem cells need to preserve their telomere length and homeostasis to maintain their self-renewal ability and pluripotency [51], [52], [53]. We found that ALDH+CD44+CXCR4+CD24+ cells exhibited shorter-length telomeres after TAP1 treatment. These findings indicated that TAP1 might prohibit telomere elongation, disrupt telomere, inhibit the pluripotency of PCSC, and eventually cause the retardation of cell proliferation and tumor initiation. Studies suggest that CSC self-renewal promotes tumor recurrence and differentiation therapy may constitute a strategy to sensitize CSCs to chemotherapy. It is also possible that TAP1 increases sensitivity of ALDH+CD44+CXCR4+CD24+ cells to chemotherapeutic agents through inducing CSC differentiation. HOXB9 is highly expressed in many types of cancers and promotes invasion and metastasis of PCa [39]. TGF-β signaling has divergent roles in regulating tumor proliferation. TGF-β1 in early-stage lesions suppresses cellular proliferation, whereas it promotes tumor progression in late stages [54]. High level of TGF-β1 was associated with poor prognosis in patients with PCa [55]. Targeting the TGF-β2-associated mechanism could provide novel opportunities to prevent lethal PCa metastasis [56]. Aberrant TGF-β signaling can drive CRPC [57]. In this study, we observed that the expression of HOXB9 and TGF-β2, but not TGF-β1, was significantly decreased in ALDH+CD44+CXCR4+CD24+ cells after TAP1 treatment. This may be due to the presence of HOX-binding sites in the promoter regions of TGF-β2 but not in TGF-β140. Therefore, TAP1 reduced the expression of TGF-β2 possibly through inhibiting HOXB9.
In summary, our findings indicated that TAP1 specifically targets PSA−/lo/ALDH+CD44+CXCR4+CD24+ cells, inhibits PCa growth both in vitro and in vivo, and enhances the anti-tumor effect of the anti-androgens and chemotherapeutic agents in vitro. TAP1 suppressed PCa cell growth and invasion and improved the drug sensitivities of anti-androgens and chemotherapeutic agents maybe at least through telomeres shortening and decreased the expression of HOXB9 and TGF-β2. Our findings provided a support for the strategy that therapeutic peptides might be used either alone or combined with other standard therapies for the treatment of PCa. Targeted therapies based on small peptides may be a very valuable and promising approach to overcome therapeutic resistance and achieve a significant therapeutic response to increase the therapeutic efficacy.
CRediT author statement
Yi Sui: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data Curation, Writing - Original Draft, Funding acquisition. Rujian Zhu: Conceptualization, Methodology, Investigation, Resources, Data Curation, Writing - Original Draft. Wei Hu: Conceptualization, Methodology, Investigation, Resources, Data Curation. Wei Zhang: Investigation, Resources, Data Curation. Hongbo Zhu: Investigation, Resources, Data Curation. Min Gong: Investigation, Data Curation. Lili Gao: Investigation, Data Curation. Ting Cao: Investigation, Data Curation. Tao Tang: Investigation, Data Curation. Bo Yu: Data Curation, Formal analysis. Tao Yang: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The work was financially supported by the National Natural Science Foundation of China (Grant no. 81572518 & 81372750 to T.Y., Grant no. 81660150 to Y.S.) and Academic Leaders Training Program of Pudong Health Bureau of Shanghai (Grant no. PWRd2018–07) to T.Y. Clinical Plateau Discipline Project of Pudong Health Bureau of Shanghai (Grant no. PWYgy2018-08) to B.Y., and Science and Technology Development Fund of Shanghai Pudong New Area (Grant no. PKJ2020-Y47) to T.Y.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101020.
Contributor Information
Bo Yu, Email: yubo120@hotmail.com.
Tao Yang, Email: taoyang@tongji.edu.cn.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Leitzmann M.F., Rohrmann S. Risk factors for the onset of prostatic cancer: age, location, and behavioral correlates. Clin. Epidemiol. 2012;4:1–11. doi: 10.2147/CLEP.S16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawla P. Epidemiology of Prostate Cancer. World J Oncol. 2019;10:63–89. doi: 10.14740/wjon1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ha Chung B., Horie S., Chiong E. The incidence, mortality, and risk factors of prostate cancer in Asian men. Prostate Int. 2019;7:1–8. doi: 10.1016/j.prnil.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gann P.H. Risk factors for prostate cancer. Rev. Urol. 2002;4(5):S3–S10. Suppl. [PMC free article] [PubMed] [Google Scholar]
- 6.Bleyer A., Spreafico F., Barr R. Prostate cancer in young men: an emerging young adult and older adolescent challenge. Cancer. 2020;126:46–57. doi: 10.1002/cncr.32498. [DOI] [PubMed] [Google Scholar]
- 7.Kelly S.P., Anderson W.F., Rosenberg P.S., Cook M.B. Past, current, and future incidence rates and burden of metastatic prostate cancer in the United States. Eur. Urol. Focus. 2018;4:121–127. doi: 10.1016/j.euf.2017.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janiczek M., Szylberg L., Kasperska A., Kowalewski A., Parol M., Antosik P., Radecka B., Marszalek A. Immunotherapy as a promising treatment for prostate cancer: a systematic review. J. Immunol. Res. 2017;2017 doi: 10.1155/2017/4861570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamat M., McNeel D.G. Androgen deprivation and immunotherapy for the treatment of prostate cancer. Endocr. Relat. Cancer. 2017;24:T297–T310. doi: 10.1530/ERC-17-0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans A.J. Treatment effects in prostate cancer. Mod. Pathol. 2018;31:S110–S121. doi: 10.1038/modpathol.2017.158. [DOI] [PubMed] [Google Scholar]
- 11.Hall M.E., Huelster H.L., Luckenbaugh A.N., Laviana A.A., Keegan K.A., Klaassen Z., Moses K.A., Wallis C.J.D. Metastatic hormone-sensitive prostate cancer: current perspective on the evolving therapeutic landscape. Onco Targets Ther. 2020;13:3571–3581. doi: 10.2147/OTT.S228355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattrini C., Castro E., Lozano R., Zanardi E., Rubagotti A., Boccardo F., Olmos D. Current treatment options for metastatic hormone-sensitive prostate cancer. Cancers. 2019;11(9):1355. doi: 10.3390/cancers11091355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karantanos T., Corn P.G., Thompson T.C. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: new prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grasso C.S., Wu Y.M., Robinson D.R., Cao X., Dhanasekaran S.M., Khan A.P., Quist M.J., Jing X., Lonigro R.J., Brenner J.C., Asangani I.A., Ateeq B., Chun S.Y., Siddiqui J., Sam L., Anstett M., Mehra R., Prensner J.R., Palanisamy N., Ryslik G.A., Vandin F., Raphael B.J., Kunju L.P., Rhodes D.R., Pienta K.J., Chinnaiyan A.M., Tomlins S.A. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K., Ruan H., Xu T., Liu L., Liu D., Yang H., Zhang X., Chen K. Recent advances on the progressive mechanism and therapy in castration-resistant prostate cancer. Onco Targets Ther. 2018;11:3167–3178. doi: 10.2147/OTT.S159777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maitland N.J., Bryce S.D., Stower M.J., Collins A.T. Prostate cancer stem cells: a target for new therapies. Ernst Schering Found Symp. Proc. 2006;5:155–179. doi: 10.1007/2789_2007_050. [DOI] [PubMed] [Google Scholar]
- 18.Visvader J.E., Lindeman G.J. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat. Rev. Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 19.Patrawala L., Calhoun T., Schneider-Broussard R., Li H., Bhatia B., Tang S., Reilly J.G., Chandra D., Zhou J., Claypool K., Coghlan L., Tang D.G. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 20.Lee S.O., Ma Z., Yeh C.R., Luo J., Lin T.H., Lai K.P., Yamashita S., Liang L., Tian J., Li L., Jiang Q., Huang C.K., Niu Y., Yeh S., Chang C. New therapy targeting differential androgen receptor signaling in prostate cancer stem/progenitor vs. non-stem/progenitor cells. J. Mol. Cell Biol. 2013;5:14–26. doi: 10.1093/jmcb/mjs042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Reilly D., Johnson P., Buchanan P.J. Hypoxia induced cancer stem cell enrichment promotes resistance to androgen deprivation therapy in prostate cancer. Steroids. 2019;152 doi: 10.1016/j.steroids.2019.108497. [DOI] [PubMed] [Google Scholar]
- 22.Hurt E.M., Kawasaki B.T., Klarmann G.J., Thomas S.B., Farrar W.L. CD44+ CD24(-) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br. J. Cancer. 2008;98:756–765. doi: 10.1038/sj.bjc.6604242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tu S.M., Lin S.H. Prostate cancer stem cells. Clin. Genitourin Cancer. 2012;10:69–76. doi: 10.1016/j.clgc.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feiner H.D., Gonzalez R. Carcinoma of the prostate with atypical immunohistological features. Clinical and histologic correlates. Am. J. Surg. Pathol. 1986;10:765–770. doi: 10.1097/00000478-198611000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Gallee M.P., Visser-de Jong E., van der Korput J.A., van der Kwast T.H., ten Kate F.J., Schroeder F.H., Trapman J. Variation of prostate-specific antigen expression in different tumour growth patterns present in prostatectomy specimens. Urol. Res. 1990;18:181–187. doi: 10.1007/BF00295844. [DOI] [PubMed] [Google Scholar]
- 26.Qin J., Liu X., Laffin B., Chen X., Choy G., Jeter C.R., Calhoun-Davis T., Li H., Palapattu G.S., Pang S., Lin K., Huang J., Ivanov I., Li W., Suraneni M.V., Tang D.G. The PSA(-/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marqus S., Pirogova E., Piva T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017;24:21. doi: 10.1186/s12929-017-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saw P.E., Song E.W. Phage display screening of therapeutic peptide for cancer targeting and therapy. Protein Cell. 2019;10:787–807. doi: 10.1007/s13238-019-0639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raha D., Wilson T.R., Peng J., Peterson D., Yue P., Evangelista M., Wilson C., Merchant M., Settleman J. The cancer stem cell marker aldehyde dehydrogenase is required to maintain a drug-tolerant tumor cell subpopulation. Cancer Res. 2014;74:3579–3590. doi: 10.1158/0008-5472.CAN-13-3456. [DOI] [PubMed] [Google Scholar]
- 30.Korski K., Malicka-Durczak A., Breborowicz J. Expression of stem cell marker CD44 in prostate cancer biopsies predicts cancer grade in radical prostatectomy specimens. Pol. J. Pathol. 2014;65:291–295. doi: 10.5114/pjp.2014.48190. [DOI] [PubMed] [Google Scholar]
- 31.Collins A.T., Berry P.A., Hyde C., Stower M.J., Maitland N.J. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 32.Miki J., Furusato B., Li H., Gu Y., Takahashi H., Egawa S., Sesterhenn I.A., McLeod D.G., Srivastava S., Rhim J.S. Identification of putative stem cell markers, CD133 and CXCR4, in hTERT-immortalized primary nonmalignant and malignant tumor-derived human prostate epithelial cell lines and in prostate cancer specimens. Cancer Res. 2007;67:3153–3161. doi: 10.1158/0008-5472.CAN-06-4429. [DOI] [PubMed] [Google Scholar]
- 33.Shen Y.A., Wang C.Y., Chuang H.Y., Hwang J.J., Chi W.H., Shu C.H., Ho C.Y., Li W.Y., Chen Y.J. CD44 and CD24 coordinate the reprogramming of nasopharyngeal carcinoma cells towards a cancer stem cell phenotype through STAT3 activation. Oncotarget. 2016;7:58351–58366. doi: 10.18632/oncotarget.11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmett L., Willowson K., Violet J., Shin J., Blanksby A., Lutetium Lee J: PSMA radionuclide therapy for men with prostate cancer: a review of the current literature and discussion of practical aspects of therapy. J. Med. Radiat. Sci. 2017;64(177):52–60. doi: 10.1002/jmrs.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Y., He K., Goldkorn A. Telomerase targeted therapy in cancer and cancer stem cells. Clin. Adv. Hematol. Oncol. 2011;9:442–455. [PubMed] [Google Scholar]
- 36.Stewart J.A., Chaiken M.F., Wang F., Price C.M. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutat. Res. 2012;730:12–19. doi: 10.1016/j.mrfmmm.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang J., Wang F., Okuka M., Liu N., Ji G., Ye X., Zuo B., Li M., Liang P., Ge W.W., Tsibris J.C., Keefe D.L., Liu L. Association of telomere length with authentic pluripotency of ES/iPS cells. Cell Res. 2011;21:779–792. doi: 10.1038/cr.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kong F., Zheng C., Xu D. Telomerase as a "stemness" enzyme. Sci. China Life Sci. 2014;57:564–570. doi: 10.1007/s11427-014-4666-6. [DOI] [PubMed] [Google Scholar]
- 39.Xu H., Wu S., Shen X., Wu D., Qin Z., Wang H., Chen X., Sun X. Silencing of HOXB9 suppresses cellular proliferation, angiogenesis, migration and invasion of prostate cancer cells. J. Biosci. 2020;45:40. [PubMed] [Google Scholar]
- 40.Hayashida T., Takahashi F., Chiba N., Brachtel E., Takahashi M., Godin-Heymann N., Gross K.W., Vivanco M., Wijendran V., Shioda T., Sgroi D., Donahoe P.K., Maheswaran S. HOXB9, a gene overexpressed in breast cancer, promotes tumorigenicity and lung metastasis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:1100–1105. doi: 10.1073/pnas.0912710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Itoh N., Patel U., Cupp A.S., Skinner M.K. Developmental and hormonal regulation of transforming growth factor-beta1 (TGFbeta1), -2, and -3 gene expression in isolated prostatic epithelial and stromal cells: epidermal growth factor and TGFbeta interactions. Endocrinology. 1998;139:1378–1388. doi: 10.1210/endo.139.3.5787. [DOI] [PubMed] [Google Scholar]
- 42.Ahel J., Hudorovic N., Vicic-Hudorovic V., Nikles H. Tgf-Beta in the natural history of prostate cancer. Acta Clin. Croat. 2019;58:128–138. doi: 10.20471/acc.2019.58.01.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song B., Park S.H., Zhao J.C., Fong K.W., Li S., Lee Y., Yang Y.A., Sridhar S., Lu X., Abdulkadir S.A., Vessella R.L., Morrissey C., Kuzel T.M., Catalona W., Yang X., Yu J. Targeting FOXA1-mediated repression of TGF-beta signaling suppresses castration-resistant prostate cancer progression. J. Clin. Invest. 2019;129:569–582. doi: 10.1172/JCI122367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong L., Zieren R.C., Xue W., de Reijke T.M., Pienta K.J. Metastatic prostate cancer remains incurable, why? Asian J Urol. 2019;6:26–41. doi: 10.1016/j.ajur.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vander Griend D.J., Karthaus W.L., Dalrymple S., Meeker A., DeMarzo A.M., Isaacs J.T. The role of CD133 in normal human prostate stem cells and malignant cancer-initiating cells. Cancer Res. 2008;68:9703–9711. doi: 10.1158/0008-5472.CAN-08-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mei W., Lin X., Kapoor A., Gu Y., Zhao K., Tang D. The contributions of prostate cancer stem cells in prostate cancer initiation and metastasis. Cancers. 2019;11(4):434. doi: 10.3390/cancers11040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao Y.F., Jie M.M., Li B.S., Hu C.J., Xie R., Tang B., Yang S.M. Peptide-based treatment: a promising cancer therapy. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/761820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meschenmoser K., Kim Y., Franken S., Nowak M., Feldmann G., Bendas G., Wolfgarten M., Messmer D., Schmidt-Wolf I.G. Targeting cancer with a bi-functional peptide: in vitro and in vivo results. In vivo. 2013;27:431–442. [PubMed] [Google Scholar]
- 49.Han Z., Zhou Z., Shi X., Wang J., Wu X., Sun D., Chen Y., Zhu H., Magi-Galluzzi C., Lu Z.R. EDB fibronectin specific peptide for prostate cancer targeting. Bioconjug. Chem. 2015;26:830–838. doi: 10.1021/acs.bioconjchem.5b00178. [DOI] [PubMed] [Google Scholar]
- 50.Wada A., Terashima T., Kageyama S., Yoshida T., Narita M., Kawauchi A., Kojima H. Efficient prostate cancer therapy with tissue-specific homing peptides identified by advanced phage display technology. Mol Ther Oncolytics. 2019;12:138–146. doi: 10.1016/j.omto.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhao S., Wang F., Liu L. Alternative Lengthening of Telomeres (ALT) in tumors and pluripotent stem cells. Genes. 2019;10(12):1030. doi: 10.3390/genes10121030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li F., Ge Y., Liu D., Songyang Z. The role of telomere-binding modulators in pluripotent stem cells. Protein Cell. 2020;11:60–70. doi: 10.1007/s13238-019-0651-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu L. Linking Telomere Regulation to Stem Cell Pluripotency. Trends Genet. 2017;33:16–33. doi: 10.1016/j.tig.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Yumoto K., Eber M.R., Wang J., Cackowski F.C., Decker A.M., Lee E., Nobre A.R., Aguirre-Ghiso J.A., Jung Y., Taichman R.S. Axl is required for TGF-beta2-induced dormancy of prostate cancer cells in the bone marrow. Sci. Rep. 2016;6:36520. doi: 10.1038/srep36520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reis S.T., Pontes-Junior J., Antunes A.A., Sousa-Canavez J.M., Abe D.K., Cruz J.A., Dall'oglio M.F., Crippa A., Passerotti C.C., Ribeiro-Filho L.A., Viana N.I., Srougi M., Leite K.R. Tgf-beta1 expression as a biomarker of poor prognosis in prostate cancer. Clinics. 2011;66:1143–1147. doi: 10.1590/S1807-59322011000700004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ruppender N., Larson S., Lakely B., Kollath L., Brown L., Coleman I., Coleman R., Nguyen H., Nelson P.S., Corey E., Snyder L.A., Vessella R.L., Morrissey C., Lam H.M. Cellular adhesion promotes prostate cancer cells escape from dormancy. PLoS One. 2015;10 doi: 10.1371/journal.pone.0130565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pu H., Begemann D.E., Kyprianou N. Aberrant TGF-beta signaling drives castration-resistant prostate cancer in a male mouse model of prostate tumorigenesis. Endocrinology. 2017;158:1612–1622. doi: 10.1210/en.2017-00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.