Abstract
The nuclease MRE11A is often included in genetic test panels for hereditary breast and ovarian cancer (HBOC) due to its BRCA1-related molecular function in the DNA repair pathway. However, whether MRE11A is a true predisposition gene for HBOC is still questionable. We determined to investigate this notion by dissecting the molecular genetics of the c.1516G > T;p.E506* truncating MRE11A variant, that we pinpointed in two unrelated French-Canadian (FC) HBOC patients. We performed a case–control study for the variant in ~ 2500 breast, ovarian, and endometrial cancer patients from the founder FC population of Quebec. Furthermore, we looked for the presence of second somatic alterations in the MRE11A gene in the tumors of the carriers. In summary, these investigations suggested that the identified variant is not associated with an increased risk of developing breast or ovarian cancer. We finally performed a systematic review for all the previously reported MRE11A variants in breast and ovarian cancer. We found that MRE11A germline variants annotated as pathogenic on ClinVar often lacked evidence for such classification, hence misleading the clinical management for affected patients. In summary, our report suggests the lack of clinical utility of MRE11A testing in HBOC, at least in the White/Caucasian populations.
Subject terms: Breast cancer, Gynaecological cancer, Clinical genetics
Introduction
Ten to twenty percent of breast and ovarian cancer cases, respectively, are hereditary1,2. About 25% of hereditary breast and ovarian cancer (HBOC) cases are attributed to pathogenic variants in the highly penetrant BRCA1/2 genes3. This fueled the hunt for other potential predisposition genes. As BRCA1/2 and PALB2, the most established HBOC predisposing genes, act mainly to orchestrate the dynamics of DNA repair, it has been postulated that genes feeding into the same functional circuit are potential breast and ovarian cancer predisposing candidate genes4. One of such proposed candidate genes is MRE11A. Biallelic pathogenic MRE11A germline variants cause the autosomal recessive ataxia-telangiectasia-like disorder (ATLD; OMIM #604391) which presents with a milder course than ataxia-telangiectasia. Together with NBS1/NBN and RAD50, the nuclease MRE11A forms the MRN complex that binds to BRCA1 upon DNA damage to maintain genomic integrity. This notion strengthened MRE11A’s candidacy as an HBOC risk gene, prompting its integration in HBOC screening panels. For instance, Castera et al. screened 708 HBOC patients with an NGS focused panel (27 genes including MRE11A) and defined 3 truncating variants in MRE11A5. In another report, Couch et al. screened around 2000 breast cancer patients for germline variants using a 17-gene sequencing panel and defined two truncating variants in MRE11A6, as well. However, whether MRE11A is a true cancer susceptibility gene remains unclear for two main reasons. First, there is no established cancer risk in the MRE11A-deficient ATLD patients. So far, only two siblings with ATLD were diagnosed with lung adenocarcinoma at the ages of 9 and 15, without a family history of cancer7. Second, the lack of comprehensive analysis of individual MRE11A variants such as nonsense and rare missense variants through population studies back-to-back with molecular analysis. Indeed, the latter could complement the holistic gene-level analyses, such as the one reported in the recent study by LaDuca et al. who demonstrated the lack of association between the MRE11A predicted pathogenic variants and increased risk for different cancer types, including approximately 89,000 breast cancer patients8.
In this study, we identified the truncating c.1516G > T;p.E506* MRE11A variant (rs587781384) in two unrelated French-Canadian women with breast or ovarian cancer following whole-exome sequencing and a broad clinical HBOC panel, respectively. Since, studying founder populations can help in risk estimation and as we have been studying French Canadians (FCs) for some years9,10, we thought we could tackle the questionable MRE11A HBOC risk candidacy by investigating the molecular genetics of this single truncating variant, which appears to be overrepresented in the FC population of Quebec compared with other populations studied thus far.
Results
Identifying the truncating c.1516G > T;p.E506* MRE11A variant in two unrelated HBOC patients
A 36 year-old woman was referred to the cancer genetics clinic with a diagnosis of invasive breast ductal carcinoma. Her clinical test for germline pathogenic mutations in BRCA1/2 and PALB2 resulted negative. We then performed whole exome sequencing (WES) in DNA from her peripheral blood mononuclear cells (pbmc) and identified a germline truncating variant c.1516G > T;p.E506* in the MRE11A gene (Methodology, and Table S1). A few months later, a 55 years-old woman diagnosed with ovarian high-grade serous carcinoma tested negative for BRCA1/2 and PALB2 in a broad HBOC clinical panel (Ref.11 for the panel), however the same truncating variant c.1516G > T;p.E506* in MRE11A was identified (Fig. 1a,b). Both probands were of a French-Canadian ancestry. The c.1516G > T;p.E506* variant was previously reported in three BRCA1/2 negative breast and/or ovarian cancer patients6,12,13 and twice in ATLD cases on ClinVar. The variant also has a gnomAD frequency of 3.19E-05 and 4E–05 in the UK biobank European-ancestry individuals14. Given the variant’s low frequency in public databases, we hypothesized that this rare MRE11A variant might be a candidate founder pathogenic variant (PV) for HBOC in FCs.
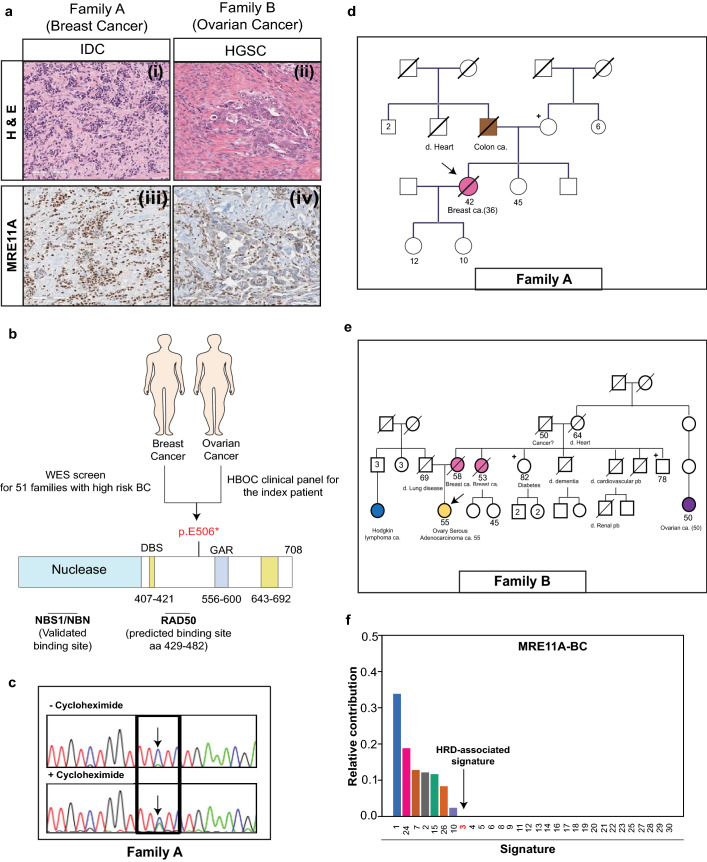
Figure 1.
Investigating the c.1516G > T;p.E506* MRE11A variant in two unrelated HBOC patients of French-Canadian origins. (a) (i) and (ii) H&E staining of the two probands’ breast (invasive ductal carcinoma; IDC) and ovarian tumors (high grade serous carcinoma; HGSC), respectively. (iii) and (iv) Immunohistochemistry staining demonstrating the MRE11A protein expression levels in the two probands’ tumors. (b) Schematic representation of the identified variant’s position within the MRE11A protein. (c) Cycloheximide chase assay showing that the identified variant undergoes nonsense-mediated mRNA decay, as denoted by the arrow, in the Family A proband-derived lymphoblastoid cell line. (d,e) Pedigrees of Family A and B, respectively. Carriers of c.1516G > T;p.E506* MRE11A are indicated by + and probands are indicated by arrows. (f) Contribution of the different COSMIC single base substitution (SBS) mutational signatures. The SBS3, the Homologous Repair Deficiency (HRD)-associated mutational signature, is indicated by an arrow. (Pedigrees were generated by the PhenoTips opensource https://phenotips.com/index.html).
Further genetic analysis of the c.1516G > T;p.E506* MRE11A variant
During the discovery phase of the project, a segregation analysis was done on the available members of the two families. In family A, the unaffected mother of the proband resulted to be a carrier of the variant. In family B, two healthy relatives of the proband (a maternal aunt and uncle) tested negative (Fig. 1d,e).
To address the possibility of this variant being a founder PV in FCs, we performed a case–control study including 1925 breast, 341 ovarian, and 367 endometrial cancer patients, in addition to 2,287 adult controls and 1932 newborns, all of FC origin (Table 1). The c.1516G > T;p.E506* variant was not observed in any of the cancer patients, but it was found in 4 control subjects (0.002%; 2 males and 2 females). This suggested that c.1516G > T;p.E506*, which is the most reported likely pathogenic variant in MRE11A on ClinVar, is overrepresented in the FC population, but is unlikely to be associated with a risk of breast or ovarian cancer.
Table 1.
Case–control study for the C.1516G > T;p.E506* MRE11A variant in French-Canadians.
| Cases | Controls | ||||||
|---|---|---|---|---|---|---|---|
| BC | OC | EC | Adult FC controls Unselected | Adult FC control Selected | FC Newborns | ||
| Number of persons studied | 1925 | 341 | 367 | 396 | 1891 (50% Females) | 1932 | |
|
< 70 y/o: 512 Medium Risk: 1201 High Risk: 212 | |||||||
| MRE11A p.E506* carriers | Male | 0/5 | NA | NA | 0 | 2/946 | |
| Female | 0/1920 | 0/341 | 0/341 | 0 | 2/945 | ||
| Total (%) | 0/1925 | 0/341 | 0/341 | 0/396 |
4/1891 (0.002%) |
1 (0.00052%) |
|
BC breast cancer, OC ovarian cancer, EC endometrial cancer, MR moderate risk, HR high risk.
Nonsense-mediated mRNA decay (NMD) of alleles harboring a truncating mutation has been postulated as a molecular mechanism by which tumor suppressor genes get inactivated15,16. According to the recently-established four rules of NMD, the c.1516G > T;p.E506* variant is predicted to trigger NMD15,16. Hence, we carried out a nonsense-mediated mRNA decay (NMD) assay, where a lymphoblastoid cell line derived from the proband of Family A was treated either with cycloheximide to inhibit mRNA degradation or DMSO as a control, as previously described17. This assay confirmed that the c.1516G > T;p.E506* variant can indeed trigger NMD as expected (Fig. 1c).
We next investigated the presence of second pathogenic alterations in the tumors of the two probands. No other somatic mutations in MRE11A was found by Sanger sequencing and none of the tumors showed any signs of loss-of-heterozygosity, an expected genetic event in the course of developing tumors through inactivating tumor suppressor genes. We then performed WES in the breast tumor and mutational signature analysis18 showed that the breast tumor lacked the characteristic Homologous Recombination Repair Deficiency (HRD) mutational signature, MutSign3, arguing against the involvement of c.1516G > T;p.E506* in breast cancer tumorigenesis (Fig. 1f). DNA quality of the ovarian tumor did not meet requirements for WES. At the tumor level, immunohistochemistry staining demonstrated that the breast tumor sample retained MRE11A protein expression (Fig. 1a). In contrast, despite the lack of additional somatic hits in the MRE11A gene, the ovarian tumor showed loss of MRE11A protein expression (Fig. 1a). However, the mechanism is uncertain given that the wild-type allele is retained at the DNA level. Altogether, these results indicated that the c.1516G > T;p.E506* variant does not increase the risk for breast and ovarian cancer in FCs.
Review of the reported MRE11A variants in breast and/or ovarian cancer
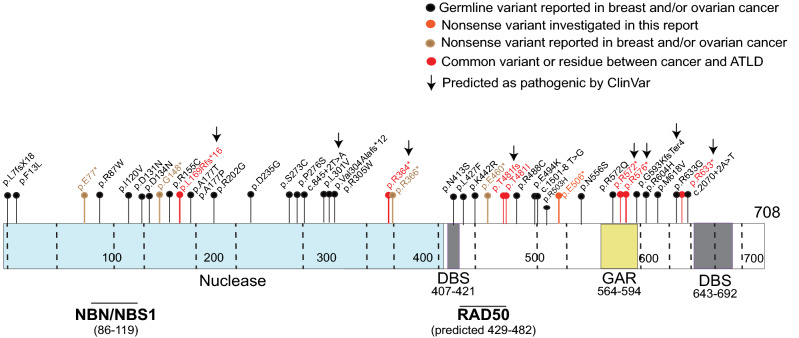
To further complement our investigation and to generally address the initial question of the debatable pathogenicity and clinical utility of MRE11A testing in HBOC, we performed a systematic review of all previously reported MRE11A germline variants in breast and ovarian cancer patients. A total of 43 distinct MRE11A germline variants have been reported in such patients over the past 17 years (Table S2). Eight were annotated as pathogenic variants in ClinVar. The main criteria for pathogenic annotation in ClinVar included being a truncating allele, undetectable/absent in control populations, and/or being reported in ATLD or in HBOC patients. Five of those variants had been published or reported in ClinVar as commonly altered among ATLD and cancer cases (Fig. 2 and Table S2).
Figure 2.
Schematic representation of the previously reported 43 MRE11A germline variants in breast and ovarian cancer. Eight variants are classified as pathogenic on ClinVar (denoted to by arrows). Five variants in addition to a residue were commonly reported between ATLD and cancer patients (red circles/sticks). GAR glycine and arginine rich motif, DBS DNA-binding site.
The c.1516G > T;p.E506* variant was classified as pathogenic in two ATLD cases. However, it had conflicting classifications in hereditary cancer cases. One submission annotated the variant as pathogenic and another as a variant of uncertain significance.
Altogether, these observations demonstrate a clear limitation in the pathogenicity classification of the MRE11A germline variants on ClinVar, particularly as applied to HBOC.
Discussion
Advancement in DNA sequencing technologies has enabled the parallel testing of up to 100 genes within multiple-gene sequencing panels19. However, whether all the tested genes are true risk/predisposition genes is questionable. Two main criteria were previously proposed to guide indication for clinical gene screening: clinical validity and utility20,21. Both require the candidate to possess genetic variants that are associated with a phenotype (i.e. increased risk of developing cancer) and to predict the clinical outcome and help guide patient management. One of the questionable genes is MRE11A that has been included in HBOC gene panels for over a decade mainly for its BRCA1-closely related DNA repair molecular actions.
In this report, using a well-studied founder population, we provide strong support for the notion that protein truncating germline variants in MRE11A are not associated with an increased risk of breast, ovarian, or likely, endometrial cancer. This is in accordance with a recently published retrospective study that analyzed the associated risk with predicted pathogenic variants of 32 cancer predisposition genes, including MRE11A, in 165,000 persons8. Seventy two percent (~ 120,000 persons) had cancer, of which ~ 89,000 were women with breast cancer, ~ 13,000 with ovarian cancer, in addition to ~ 5500 women with endometrial cancer. The team identified 140 predicted pathogenic MRE11A variants in total in different cancers (90 in breast cancer, 11 in ovarian cancer, and 5 in endometrial cancer). The authors did not identify a significant association between the MRE11A predicted pathogenic variants and any cancer phenotype, including breast, ovarian, and endometrial cancer8, suggesting a lack of MRE11A screening utility in HBOC and likely endometrial cancer patients. In another recent study with a similar objective 22, Lee et al. investigated the association between the frequently included genes in genetic screening panels, including MRE11A, and familial breast and ovarian cancer. The authors used a point-based curation system that takes into account genetic and experimental evidence to score the gene association with the tested cancer type. The categories included Definitive, Strong, Moderate, Limited, Refuted, Disputed, or No Reported Evidence for association. MRE11A association with familial breast and/or ovarian cancer was disputed, denoting to the conflicting data and views of its association with the cancer. Our findings, taken together with the recent report of LaDuca et al.8 call for re-evaluating the MRE11A classification in HBOC. Additional investigations targeting rare missense variants in MRE11A, such as those listed in Table S2 and shown in Fig. 2, and their possible association with HBOC are, however, needed to complete the picture for MRE11A and HBOC risk. Additionally, limited by the relatively low number of tested endometrial cancer patients in the current report, we cannot completely exclude a role for MRE11A truncating variants in the development of endometrial cancer. It is also worth noting in this context that most of the studies focused on Caucasian populations and rarely focused on Black populations. Importantly, Hart et al. recently reported threefold and twofold increase in MRE11A variants prevalence in ovarian and breast cancer, respectively, in Black population versus Non-Hispanic Whites23.
Our systematic review highlights some of the gaps in our current knowledge of the validity and utility of MRE11A testing. Classifying the MRE11A ATLD and truncated variants as pathogenic in HBOC should be supported, as previously recommended24, by further clinical evidence (e.g., population and/or segregation studies) and/or functional studies to assess the possible associated MRE11A function perturbation. Notably, the truncating c.1516G > T;p.E506* variant, investigated here, was not associated with HRD (Fig. 1f), an expected consequence of a disfunctional MRE11A.
MRE11A variants are, however, associated with a clinically important cancer-related phenotype: clonal hematopoiesis. Two independent studies have found that heterozygous rare variants in MRE11A are subject to copy-neutral loss of heterozygosity, which is strongly associated with clonal hematopoiesis25,26. These clones become much more evident with age, such that clonal hematopoiesis affects 40% of Japanese persons over age 90 years. Therefore, MRE11A variants deserve further attention as they are associated with adverse health outcomes.
In conclusion, we report the detailed investigation of a truncating variant in the candidate breast and ovarian cancer susceptibility gene, MRE11A. Our findings, taken together with other recently published studies, show that truncating variants in MRE11A are not a cause of breast or ovarian cancer. Based on the current evidence, testing for MRE11A variants should not be offered to women with, or at risk for, breast or ovarian cancer.
Patients and methods
Patients and samples
In total, 2633 cancer patients and a control group of 2287 adults and 1932 newborns of French-Canadian ancestry were included in this study, as explained in detail in Table 1 and the study population section.
Ethics statement
The initial two families and control subjects were recruited following approval of the Institutional Review Board of the Faculty of Medicine of McGill University (IRB 2019-5465) and the Jewish General Hospital (2016-397), respectively and written informed consent was obtained.
Newborn blood left-over after delivery was collected at the Hôpital Saint-François d’Assise maternity unit between 1996 and 2003. DNA was extracted and a biobank with over 7000 samples was constituted. These samples were made anonymous and unlinked. About 15% of the samples were randomly excluded from the collection in order to preserve even further the anonymity of the samples because any woman having given birth in this hospital during that period has a 15% chance of not being part of the studied samples. The Hôpital Saint-François d’Assise Clinical Research Ethics Committee approved the research project. For the present study a subset of n = 1932 samples from the collection was randomly selected based uniquely on amount of DNA available as it is not possible to identify the subset of samples included in the study, sex information is not available.
Samples from healthy controls (1891 individuals) were obtained through CARTaGENE biobank27 (https://www.cartagene.qc.ca/en/home). Ovarian and endometrial cancer patients in addition to the under 70 years old breast cancer patients’ group were obtained through the Banque de tissus et de données of the Réseau de recherche sur le cancer of the FRQS. All participants were recruited in compliance with the second edition of the Canadian Tri-Council Policy Statement of Ethical Conduct for Research Involving Humans and Eligible Persons or Designates and signed a consent form in accordance with the Institutional Review Board approvals. All methods were performed in accordance with the relevant guidelines and regulations.
Index patients and their families
The proband of family A was diagnosed at the age of 36 with invasive ductal carcinoma. This patient is part of a 51-high risk BRCA1/2 negative BC patients study previously carried out by our team28. The patient had been screened for germline variants by whole exome sequencing (WES) revealing the presence of a likely pathogenic variant in MRE11A (c.1516G > T;p.E506*) (Fig. 1a,b). Proband of Family B was a 55-year old woman diagnosed with a high-grade serous carcinoma (HGSC) that was referred to the cancer genetics clinic from the gynecology clinic at the Jewish General Hospital (Montreal, Canada) for genetic testing (Fig. 1a,b). The patient had a family history strongly suggestive of hereditary breast and ovarian cancer (HBOC) (mother and aunt diagnosed with breast cancer). A clinical HBOC screening panel identified the same nonsense MRE11A c.1516G > T;p.E506*variant independently found in the proband of family A. Both women were of French-Canadian ancestry.
Study population (case–control study)
During the validation phase for this study, 2633 cancer patients and a control group of 2287 adults and 1932 newborns of French-Canadian ancestry were studied (Table 1). In detail, 1925 BC including 212 (207 women, 5 men) high-risk cases (age, 19–73; average, 45) in addition to 1201 (all women) moderate risk BC cases (age, 25–68; average, 48) that we previously described in detail9 and a group of 512 BC diagnosed under 70 years of age29,30. Briefly, the risk classification of the BC patients depended mainly on the age of diagnosis and presence of family history (high risk = early onset and/or strong family history; moderate risk is defined as age at diagnosis < 50 years or ≥ 50 years but with affected first- or second-degree relatives). The study included 341 HGSC31 (age, 36–87; median age 61 years) and 367 endometrial cancer patients (age, 21–92; average 62) of French-Canadian origins9. In parallel, 1891 healthy adults with no personal history of cancer or diagnosed cancer in 1st degree relative were recruited as controls. Those controls were aged 40 to 70 that were ascertained between 2009 and 2015 through the CARTaGENE biobank27 (https://www.cartagene.qc.ca/en/home) living in the Greater Montreal area and their FC ancestry was defined as French as their first language with their four grandparents being of Canadian origin30. Additionally, 396 control subjects were recruited as unselected group9. Finally, to assess the prevalence of the identified variant in FCs, we screened 1932 newborns of French-Canadian ancestry28.
Whole exome sequencing (WES)
The proband A was among a 51-high risk BC patient series we previously screened for germline variants in genomic DNA extracted from peripheral blood leukocytes as previously outlined in detail28. Breast cancer from the proband studied here was subjected to WES that was performed at the McGill University and Genome Quebec Innovation Centre (MUGQIC). Breast cancer FFPE-derived DNA sample underwent exome capture (Nextera Rapid Capture Exome Kit), followed by 100 bp paired-end sequencing on Illumina HiSeq 2500 (details in Ref26). Bioinformatics analysis of exome sequencing data was performed using our WES pipeline as previously described9,32–34. In brief, alignment of sequenced reads to the reference genome (hg19) was performed using BWA35 (v. 0.5.9). Subsequently, the Genome Analysis Toolkit (GATK) was used to perform local realignment of reads around small insertions and deletions (indels) and to assess capture efficiency and coverage for all samples. The coverage statistics are listed in Table 2. Likely pathogenic variants in 152 known cancer susceptibility genes from Huang et al.36 and in 468 cancer panel gene list elaborated by the MSK-IMPACT were selected. Candidate somatic mutations were subjected to several filtering steps and eliminated if they fulfilled any one of the following criteria: (1) genomic position of variant covered by < 5-reads, (2) < 5 reads support the alternative variant, (3) variant has allelic ratio < 10% for SNVs or < 15% for indels, (4) variant has allele frequency > 0.001 in TCGA-ExAC databases (release 0.3 2016-01-13) or seen as homozygote in ExAC database (build). (5) missense variants that were not predicted to be disease causing by 3 out of 6 bioinformatic algorithms (SIFT, PolyPhen, MutationTaster, Revel, MCAP and CADD)37–42.
Table 2.
Exome sequencing coverage summary.
| Sample | Mean coverage | %CCDS bases ≥ 5× coverage | %CCDS bases ≥ 10× coverage | %CCDS bases ≥ 20× coverage | %CCDS bases ≥ 50× coverage |
|---|---|---|---|---|---|
| Blood-family A | 122 | 96 | 95 | 91 | 75 |
| Breast cancer-family A | 67 | 97 | 93 | 84 | 54 |
CCDS coverage of consensus coding sequence.
Mutational signature
SomaticSignatures, a Bioconductor package, was used to analyze all the filtered synonymous and nonsynonymous somatic mutations. Briefly, 96 trinucleotides mutational context [A|C|G|T] [C > A|C > G|C > T|T > A|T > C|T > G] [A|C|G|T] were extracted and compared with the current known set of COSMIC single base substitution signatures (SBS) listed by the Sanger Institute18 https://cancer.sanger.ac.uk/cosmic/signatures/SBS/index.tt. The Homologous Repair Deficiency (HRD) mutational signature corresponds to SBS3 as denoted in the text and Fig. 1f.
MRE11A c.1516G > T genotyping
Moderate-risk breast cancer samples were genotyped using a TaqMan SNP Genotyping assay. For the HGSC samples, DNA from peripheral blood lymphocytes (n = 305), ovarian tumours (n = 34) or ascites (n = 2) were genotyped using The Custom TaqMan SNP Genotyping Assay protocol performed as previously described43. Using a similar pipeline to our previous work28, we genotyped samples from the below 70 years old breast cancer group (n = 512), newborns of French Canadian ancestry (n = 1932) in addition to the healthy control adults (CARTaGENE biobank; n = 1891) for MRE11A c.1516G > T; p.E506* allele using the iPLEX MassARRAY platform (Sequenom) at the McGill University and Genome Quebec Innovation Centre (MUGQIC). A high-resolution melting (HRM) assay was used to test all the remaining patient and control samples included in this study as previously described9. For validations, Sanger sequencing was used, and chromatograms were aligned with the NM_005591.4 RefSeq transcript. Probes and primers sequences are available upon request.
Databases and software used in this report
gnomAD. We interrogated the gnomAD database44 to assess the frequency of the p.E506* MRE11A variant and its prevalence in the reported ethnicities. gnomAD v2.1.1 was last accessed on 30 April 2020 and the MRE11 p.E506* variant (variant ID:11-94189489-C-A; GRCh37) prevalence was detected as detailed in the text.
The PhenoTips software was used to generate pedigrees (https://phenotips.com/index.html). The ProteinPaint tool45 from https://proteinpaint.stjude.org/ was used for elaboration of Fig. 2.
Immunohistochemistry
Formalin-fixed, paraffin-embedded (FFPE) samples from the breast and ovarian tumors were studied by IHC. MRE11A (Abcam #ab214, 1:1500) was optimized in a Ventana machine following recommended protocols. MRE11A expression and localization was analyzed by a pathologist (OA) independently for diagnosis and mutational analysis. Aperio ImageScope software was used to obtain the images.
NMD assay
A lymphoblastoid cell line was established from a blood sample from the proband of Family A. The cells were cultured using RPMI medium. For assessing whether the mutant allele undergoes NMD, cells were plated in 6 well plates, and either treated with cycloheximide (28 ug/ml)17 or (DMSO) for 3 h. At the experimental endpoint, RNA was extracted from both the treated or untreated cells using Trizol and RT-PCR was performed to obtain cDNA. cDNA was then subjected to sanger sequencing.
MRE11A germline variants literature review
We did a literature review for the reported MRE11A germline variants as of May 13 2020. We found 21 studies reporting different variants of MRE11A in mainly breast and ovarian cancers over the past 17 years (from 2003 to 2020). One study was excluded as it did not focus on HBOC. All the related info and references are included in Table S2.
Supplementary Information
Acknowledgments
We would like to thank the Genome Quebec Innovation Centre for technical services. I.E.E. is a recipient of an FRQS Doctoral scholarship and was supported by an IRCM Foundation-TD scholarship. M.D. was supported by both the Mach-Gaensslen Foundation and Dr. Clarke K. McLeod research scholarships. B.R. holds a Junior Leader Fellowship (LCF/BQ/PI19/11690009) from La Caixa Foundation (ID100010434). J.F.C holds the Transat Chair in Breast Cancer Research. A.M-M.M. is a researcher of the CRCHUM/ICM, and P.N.T and W.D.F. are researchers of the RI MUHC, research institutes that both receive support from the Fonds de recherche du Québec—Santé (FRQS). Tissue banking was supported by the Banque de tissus et de données of the Réseau de recherche sur le cancer (RRCancer) of the FRQS affiliated with the Canadian Tumor Repository Network (CTRNet). This research was partially funded by the Cancer Research Society grant OG-24377 to B.R and the Quebec Breast Cancer Foundation (QBCF) to W.D.F, and the RRCancer of the FRQS to P.N.T.
Author contributions
I.E.E. and M.D.I. performed experiments and analysis; S.F. performed W.E.S. analysis; S.L.A., H.R.H., C.N., S.B., N.H., S.G., M.D.L., M.A. contributed to the experiments; O.A. provided pathology revision and IHC evaluation; W.H.G. performed clinical details and follow up of the patient and provided sample; J.F.C., F.R., P.T., D.P., M.A. and A.M.M. contributed to experimental supervision, sample set collections and data interpretation; B.R. and W.D.F. supervised the study; I.E.E., B.R. and W.D.F. co-wrote the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-81106-w.
References
- 1.Constantinou P, Tischkowitz M. Genetics of gynaecological cancers. Best Pract. Res. Clin. Obstet. Gynaecol. 2017;42:114–124. doi: 10.1016/j.bpobgyn.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Economopoulou P, Dimitriadis G, Psyrri A. Beyond BRCA: new hereditary breast cancer susceptibility genes. Cancer Treat. Rev. 2015;41:1–8. doi: 10.1016/j.ctrv.2014.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Kast K, et al. Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J. Med. Genet. 2016;53:465–471. doi: 10.1136/jmedgenet-2015-103672. [DOI] [PubMed] [Google Scholar]
- 4.Foulkes WD, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castera L, et al. Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur. J. Hum. Genet. 2014;22:1305–1313. doi: 10.1038/ejhg.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Couch FJ, et al. Inherited mutations in 17 breast cancer susceptibility genes among a large triple-negative breast cancer cohort unselected for family history of breast cancer. J. Clin. Oncol. 2015;33:304–311. doi: 10.1200/JCO.2014.57.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchisaka N, et al. Two brothers with ataxia-telangiectasia-like disorder with lung adenocarcinoma. J. Pediatr. 2009;155:435–438. doi: 10.1016/j.jpeds.2009.02.037. [DOI] [PubMed] [Google Scholar]
- 8.LaDuca H, et al. A clinical guide to hereditary cancer panel testing: evaluation of gene-specific cancer associations and sensitivity of genetic testing criteria in a cohort of 165,000 high-risk patients. Genet. Med. 2020;22:407–415. doi: 10.1038/s41436-019-0633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rivera B, et al. Functionally null RAD51D missense mutation associates strongly with ovarian carcinoma. Cancer Res. 2017;77:4517–4529. doi: 10.1158/0008-5472.CAN-17-0190. [DOI] [PubMed] [Google Scholar]
- 10.Tonin PN, et al. Founder BRCA1 and BRCA2 mutations in French Canadian breast and ovarian cancer families. Am. J. Hum. Genet. 1998;63:1341–1351. doi: 10.1086/302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh T, et al. Mutations in 12 genes for inherited ovarian, fallopian tube, and peritoneal carcinoma identified by massively parallel sequencing. Proc. Natl. Acad. Sci. U.S.A. 2011;108:18032–18037. doi: 10.1073/pnas.1115052108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaDuca H, et al. Utilization of multigene panels in hereditary cancer predisposition testing: analysis of more than 2000 patients. Genet. Med. 2014;16:830–837. doi: 10.1038/gim.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shirts BH, et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet. Med. 2016;18:974–981. doi: 10.1038/gim.2015.212. [DOI] [PubMed] [Google Scholar]
- 14.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindeboom RG, Supek F, Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nat. Genet. 2016;48:1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindeboom RGH, Vermeulen M, Lehner B, Supek F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019;51:1645–1651. doi: 10.1038/s41588-019-0517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rio Frio T, et al. Premature termination codons in PRPF31 cause retinitis pigmentosa via haploinsufficiency due to nonsense-mediated mRNA decay. J. Clin. Investig. 2008;118:1519–1531. doi: 10.1172/JCI34211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, Stratton MR. A mutational signature in gastric cancer suggests therapeutic strategies. Nat. Commun. 2015;6:8683. doi: 10.1038/ncomms9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurian AW, Ford JM. Multigene panel testing in oncology practice: How should we respond? JAMA Oncol. 2015;1:277–278. doi: 10.1001/jamaoncol.2015.28. [DOI] [PubMed] [Google Scholar]
- 20.Easton DF, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N. Engl. J. Med. 2015;372:2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phimister EG. Curating the way to better determinants of genetic risk. N. Engl. J. Med. 2015;372:2227–2228. doi: 10.1056/NEJMe1506276. [DOI] [PubMed] [Google Scholar]
- 22.Lee K, et al. Clinical validity assessment of genes frequently tested on hereditary breast and ovarian cancer susceptibility sequencing panels. Genet. Med. 2019;21:1497–1506. doi: 10.1038/s41436-018-0361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hart SN, et al. Mutation prevalence tables for hereditary cancer derived from multi-gene panel testing. Hum. Mutat. 2020 doi: 10.1002/humu.24053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terao C, et al. Chromosomal alterations among age-related haematopoietic clones in Japan. Nature. 2020;584:130–135. doi: 10.1038/s41586-020-2426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh PR, Genovese G, McCarroll SA. Monogenic and polygenic inheritance become instruments for clonal selection. Nature. 2020;584:136–141. doi: 10.1038/s41586-020-2430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Awadalla P, et al. Cohort profile of the CARTaGENE study: Quebec's population-based biobank for public health and personalized genomics. Int. J. Epidemiol. 2013;42:1285–1299. doi: 10.1093/ije/dys160. [DOI] [PubMed] [Google Scholar]
- 28.Cybulski C, et al. Germline RECQL mutations are associated with breast cancer susceptibility. Nat. Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- 29.Behl S, et al. Founder BRCA1/BRCA2/PALB2 pathogenic variants in French-Canadian breast cancer cases and controls. Sci. Rep. 2020;10:6491. doi: 10.1038/s41598-020-63100-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glentis S, et al. Exome sequencing in BRCA1- and BRCA2-negative Greek families identifies MDM1 and NBEAL1 as candidate risk genes for hereditary breast cancer. Front. Genet. 2019;10:1005. doi: 10.3389/fgene.2019.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belanger MH, et al. A targeted analysis identifies a high frequency of BRCA1 and BRCA2 mutation carriers in women with ovarian cancer from a founder population. J. Ovarian Res. 2015;8:1. doi: 10.1186/s13048-015-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fahiminiya S, et al. Molecular analyses reveal close similarities between small cell carcinoma of the ovary, hypercalcemic type and atypical teratoid/rhabdoid tumor. Oncotarget. 2016;7:1732–1740. doi: 10.18632/oncotarget.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang KL, et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–370. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jagadeesh KA, et al. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 39.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 41.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat. Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 43.Ancot F, Arcand SL, Mes-Masson AM, Provencher DM, Tonin PN. Double PALB2 and BRCA1/BRCA2 mutation carriers are rare in breast cancer and breast-ovarian cancer syndrome families from the French Canadian founder population. Oncol. Lett. 2015;9:2787–2790. doi: 10.3892/ol.2015.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Karczewski KJ, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, et al. Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 2016;48:4–6. doi: 10.1038/ng.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.