Abstract
Purpose
Successful oral feeding and speech emergence are dependent upon the coordination of shared oral muscles and facial nerves. We aimed to determine if the speech-associated genes, forkhead box P2 (FOXP2), contactin-associated protein-like 2 (CNTNAP2), glutamate receptor, ionotropic, N-methyl D-aspartate 2A (GRIN2A), and neurexin 1, were detectable in neonatal saliva and could predict feeding outcomes in premature newborns.
Method
In this prospective, observational, preliminary study, saliva collected from 51 premature infants (gestational ages: 30–34 6/7 weeks) at different stages of oral feeding development underwent gene expression analysis. Binary (+/–) expression profiles were explored and examined in relation to days to achieve full oral feeds.
Results
GRIN2A and neurexin 1 rarely amplified in neonatal saliva and were not informative. Infants who amplified FOXP2 but not CNTNAP2 at the start of oral feeds achieved oral feeding success 3.20 (95% CI [−2.5, 8.9]) days sooner than other gene combinations.
Conclusions
FOXP2 and CNTNAP2 may be informative in predicting oral feeding outcomes in newborns. Salivary analysis at the start of oral feeding trials may inform feeding outcomes in this population and warrants further investigation.
Successful oral feeding and speech emergence are dependent upon pathways involved in oral motor coordination, planning, and execution and use shared muscles and cranial nerve innervations (Matsuo & Palmer, 2008; McFarland & Tremblay, 2006). Infants born prematurely are at risk for disrupted and/or delayed maturation of these developmental pathways often resulting in poor oral feeding skills (Lau, Alagugurusamy, Schanler, Smith, & Shulman, 2000), impaired growth and nutrition (Johnson, Wootton, Leaf, & Jackson, 2012), and language delays (Stephens & Vohr, 2009; van Noort-van der Spek, Franken, & Weisglas-Kuperus, 2012). Early identification of infants at risk for these developmental impairments may allow for timely and targeted interventions that could significantly improve care and outcomes for this vulnerable population.
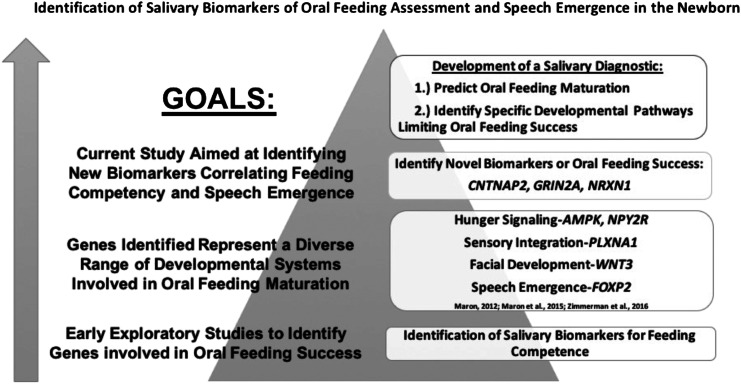
In recent years, our understanding of the molecular and genetic pathways regulating the complex and coordinated interplay of muscles and nerves required for speech emergence and oral feeding has expanded (Khanna, Maron, & Walt, 2017; Maron, 2012; McFarland &Tremblay, 2006). Specifically, advances in molecular techniques have allowed caregivers to analyze single drops of neonatal saliva to understand real-time development as it relates to oral feeding maturation in the preterm infant. By convention, genes (ribonucleic acid [RNA]) are transcribed from the genetic code (deoxyribonucleic acid [DNA]) and translated into proteins to perform the biological mechanisms of cells. Through a series of experiments and published articles, the Maron Laboratory has demonstrated that neonatal saliva serves as a rich source of both gene (RNA) and protein expression (Khanna, Johnson, et al., 2017; Khanna, Maron, et al., 2017; Maron et al., 2015). By performing comparative analyses of saliva obtained from successful and unsuccessful neonatal oral feeders, Dr. Maron's lab has identified salivary biomarkers related to oral feeding maturation (see Figure 1). These biomarkers represent a diverse group of developmental systems related to successful oral feeding including facial development, hunger signaling, and sensory integration (see Figure 1). Some biomarkers increase their gene expression (up-regulation) during feeding maturation; other biomarkers decrease their gene expression (down-regulation) during feeding maturation (Maron et al., 2015). Given the shared pathways involved in both oral feeding and speech emergence, the work further extended into exploring the diagnostic accuracy of neonatal forkhead box P2 (FOXP2) expression to predict feeding maturation in the newborn.
Figure 1.
Neonatal salivary biomarker discovery and validation to assess oral feeding in the premature newborn.
FOXP2 was first described in the multigenerational family, “KE,” who exhibited language impairment when the gene was mutated (Hurst, Baraitser, Auger, Graham, & Norell, 1990). Heterozygous mutations of FOXP2 in humans cause severe speech-language delays (Fisher & Scharff, 2009; Lai, Fisher, Hurst, Vargha-Khadem, & Monaco, 2001; Lai, Gerrelli, Monaco, Fisher, & Copp, 2003; Liégeois et al., 2003; MacDermot et al., 2005), specifically verbal apraxia—a speech disorder of motor planning. Bowers, Perez-Pouchoulen, Edwards, and McCarthy (2013) examined quantitative levels of FOXP2 protein in the left cortical hemisphere in children and found that 4-year-old boys had significantly lower FOXP2 expression levels than aged-matched girls (Bowers et al., 2013). This research suggests that FOXP2 can vary based on sex. Zimmerman, Maki, and Maron (2016) demonstrated a relation between quantitative expression levels of salivary FOXP2 and a shorter duration of time to achieve full oral feeds in the preterm neonate and also reported the first case study linking a deletion in the gene to impaired oral feeding skills in the newborn (Zimmerman & Maron, 2016). Taken together, these data suggest that FOXP2 may play a critical role in both oral feeding and speech competency.
Beyond FOXP2, there are emerging data identifying other genes that play a key role in speech development. One gene, contactin-associated protein-like 2 (CNTNAP2), is located on chromosome 7 downstream from its regulator FOXP2. Patients with CNTNAP2 mutations have been found to share core phenotypes, including language problems characterized as speech difficulty, delayed language development, or absent language (Rodenas-Cuadrado, Ho, & Vernes, 2014). Pitt-Hopkins–like syndrome 1 is associated with deletions in CNTNAP2. Symptoms of Pitt-Hopkins–like syndrome include ataxia, spasticity, and generalized and focal seizures with absent speech (Poot, 2017). Thus, CNTNAP2 may be an additional gene that can provide insight into the complex molecular mechanism of oral feeding and speech development.
Another gene, glutamate receptor, ionotropic, N-methyl D-aspartate 2A (GRIN2A), located on chromosome 16, encodes the NR2A subunit of the glutamate N-methyl-D-aspartate receptor. GRIN2A mutations, both inherited and de novo, have been found in patients with epilepsy–aphasia syndrome (Carvill et al., 2013). Interestingly, Turner et al. (2015) have identified family members of epilepsy–aphasia syndrome patients with different GRIN2A mutations who have speech impairments without a seizure disorder. This finding supports GRIN2A and its association with speech production. Finally, neurexin 1 (NRXN1), located on chromosome 2, encodes a membrane protein in the neurexin family. Deletions in NRXN1 features developmental delays, most commonly speech delays (Dabell et al., 2013). NRXN1 deletion syndrome is associated with heterozygous deletions of NRXN1 and includes autism spectrum disorder, attention-deficit/hyperactivity disorder, intellectual disability, seizures, schizophrenia, mood disorders, and congenital malformations (Al Shehhi et al., 2019). Although it is clear that these genes play a role in speech development, it remains unclear if they are related to oral feeding development.
Oral feeding competency is a complex developmental task requiring the integration of the oromotor, sensory, neurodevelopmental, and gastrointestinal systems, along with a mature gut–brain axis. Although oromotor maturation represents only one developmental system required for oral feeding success, identifying disruption in this biological network may be informative of both oral feeding difficulties and delayed speech maturation. The aim of this study was to determine if expression of speech genes, FOXP2, CNTNAP2, GRIN2A, and NRXN1, could be amplified in neonatal saliva and predict feeding outcomes in premature newborns. Specifically, we hypothesized that genes involved in speech development could predict oral feeding readiness in the preterm newborn and aimed to determine if expression of these genes, alone or in combination, could serve as noninvasive, objective biomarkers of oral feeding success in this vulnerable population.
Materials and Method
This study was approved by the Tufts Medical Center Institutional Review Board. With parental consent, premature infants born between 30 and 35 weeks' gestational age (GA) were recruited from the neonatal intensive care unit (NICU) at Tufts Medical Center (Boston, MA) between December 2015 and March 2017. The Tufts Medical Center NICU is a 42-bed Level III NICU with a large geographical referral area across Eastern Massachusetts. Tufts Medical Center practices a distributive care model whereby newborns with a stable respiratory status who are > 32 weeks' postconceptional age (PCA) and weigh > 1,500 g are often transferred to Level II NICUs within the health system to be closer to their homes for convalescent care. Tufts Medical Center has five regional Level II NICU hospitals within their own network, but will transfer infants to the nearest Level II home hospital, even if outside of their own network, in order to provide optimal family-centered care. Exclusion criteria for this study included major congenital anomalies; intraventricular hemorrhage > Grade II; necrotizing enterocolitis; and prolonged respiratory support, defined as ventilator support, noninvasive positive pressure support, or continuous positive airway pressure.
Oral Feeding Outcomes
Tufts Medical Center and its affiliated hospitals utilize the infant-driven, cue-based feeding protocol of Ludwig and Waitzman (2007), which allows for standardization of feeding practices across network sites. Per the protocol, nurses begin to assess infants for oral feeding readiness beginning at 32 weeks' PCA. Ludwig and Waitzman developed a 5-point infant feeding assessment scale and a 5-point quality of nippling scale. The feeding assessment scale considers an infant's alertness, tone, respiratory status, and feeding cues (i.e., rooting, sucking on pacifier) to determine if an infant should be allowed an oral feeding trial. Infants are observed at each feed and scored from a high likelihood feeding score of 1 (alert, good tone, rooting) to a low likelihood feeding score of 5 (sleepy, tachypneic, needs oxygen). An infant must score no less than four 1s or 2s on the feeding scale in a 24-hr period before being allowed to orally feed. During the feeding, the nippling quality evaluates coordination of suck, swallow, and breathing and number of events during feeding such as apnea, bradycardia, and desaturations. Through the process of learning to feed, therapists and nurses provide feeding therapy utilizing positioning, pacing, and bottle system changes, as needed.
Oral feeding percentages were calculated using the nursing clinical bedside flow sheet and were defined as the amount of volume taken by mouth (termed per os, or PO) divided by the total fluid intake either provided via intravenous fluids or via an indwelling nasogastric tube (Barlow et al., 2017; Poore, Zimmerman, Barlow, Wang, & Gu, 2008; Zimmerman et al., 2016). Full oral feeding was achieved when an infant was able to take their minimum required volume orally without the use of a nasogastric tube for supplementation for greater than 24 hr. The number of days required to achieve oral feeding was calculated from the start of oral feeding attempts until the nasogastric tube was removed.
Salivary Collection, RNA Extraction, and Reverse Transcription–Quantitative Polymerase Chain Reaction Analysis
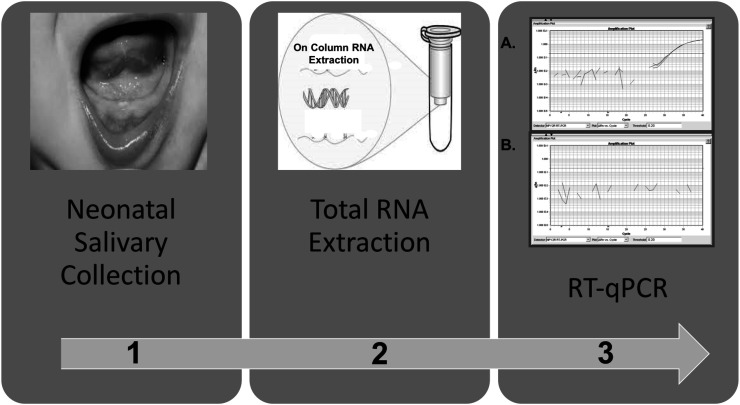
Saliva samples were collected from each participant at three time points: at the start of oral feeding, during the learning process of oral feeding (15%–75% PO), and at 100% oral feeding. All salivary samples were collected prior to a feed to limit contamination from breast milk or formula. Saliva was collected with the use of a 1-ml syringe, plunger removed, and attached to low wall suction at the bedside. The infant's mouth was gently suctioned for approximately 10–20 s. Salivary volume need only to be visualized in the tip of the syringe. Once collected, the syringe was detached from suction, the plunger was replaced, and the saliva was flushed into 500 μL of RNAProtect Saliva stabilizing agent (QIAGEN) in a 2-ml Eppendorf tube directly at the bedside. The sample was immediately placed on ice and stored at 4 °C for a minimum of 48 hr but not longer than 28 days before total RNA extraction using the RNeasy Protect Saliva Mini Kit (QIAGEN) per manufacturer's instructions (Maron et al., 2010). To minimize DNA contamination and improve RNA purification, on-column DNase digestion was performed on all samples. Extracted total RNA was stored at −80 °C pending analysis (see Figure 2).
Figure 2.
Salivary processing methods: (1) Saliva is collected at the bedside via gentle, low wall suction. Saliva tends to pool under tongue and in gingival crevices; (2) saliva undergoes total ribonucleic acid (RNA) extraction with the Qiagen RNeasy Protect Saliva Mini Kit; (3) total RNA undergoes reverse transcription–quantitative polymerase chain reaction (RT-qPCR) application. (3A) Gene targets either amplify (+ gene expression) or (3B) fail to amplify (− gene expression).
For quality assurance measures of reverse transcription–quantitative polymerase chain reaction (RT-qPCR) data, minimum information for quantitative real-time PCR experiments guidelines were followed (Bustin et al., 2009). Extracted saliva samples were analyzed for gene expression using a two-step RT-qPCR method. The first step generated first-strand complementary DNA (cDNA) using SuperScript VILO cDNA Synthesis kit per manufacturer's instructions (Applied Biosystems, Thermo Fisher Scientific) in the Eppendorf Mastercycler pro S thermal cycler (Eppendorf). Temperatures during incubation were as follows: 25 °C for 10 min, 42 °C for 60 min, and 85 °C for 5 min. cDNA samples were stored at −20 °C until preamplification was performed.
Due to the anticipated low starting quantities of total RNA extracted from neonatal saliva, target genes (FOXP2, CNTNAP2, GRIN2A, and NRXN1), along with the reference genes (glyceraldehyde 3-phosphate dehydrogenase [GAPDH], hypoxanthine phosphoribosyltransferase 1 [HPRT1], 14-3-3 protein zeta/delta [YWHAZ]) were preamplified using Taqman Preamp Master Mix kit (Applied Biosystems, Thermo Fisher Scientific) per manufacturer's instructions. This targeted preamplification was utilized to limit amplification bias inherent in a whole transcriptomic amplification approach. The preamplification reaction was run on the Eppendorf Mastercycler pro S thermal cycler at 95 °C for 10 min for enzyme activation, followed by 14 cycles of amplification at 95 °C for 15 s, and 60 °C for 4 min. Preamplified samples were stored at −20 °C until qPCR amplification.
The preamplified product was diluted 1:5 with nuclease-free water prior to qPCR amplification. A nonpreamplified control sample was run as a comparison to ensure uniform amplification across all gene targets. Diluted samples were mixed with Taqman Fast Advanced Master Mix (2×) and nuclease-free water. Sample mix was plated on Taqman Array 96-Well Plate Custom Format 8 (Applied Biosystems, Thermo Fisher Scientific). The 96-Well Plate was customized for singleplex PCR for expression of four target genes and three reference genes; Applied Biosystems includes ribosomal 18s in all premade plates as an additional control gene. Expression of this gene was not used in our analysis.
qPCR occurred on the Applied Biosystems QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Each sample was run in triplicate, and each plate was run with a positive control (infant whose saliva was not included in analysis) to assess for plate-to-plate variability. A negative control of nuclease-free water was also analyzed to detect primer–primer amplification. The qPCR cycle profile was as follows: 50 °C for 2 min, 95 °C for 20 s, and 40 fast run cycles at 95 °C for 1 s for denaturing and at 60 °C for 20 s for annealing and extension. Only samples that amplified all three reference genes (GAPDH, HPRT1, and YWAHZ) were included in the analysis. These three reference genes have been previously shown by the Maron Laboratory to maintain stable gene expression in saliva between the sexes and across the premature GA (Khanna, Johnson, & Maron, 2017).
Sample Size and Statistical Analyses
Zimmerman et al. (2016) completed a study on 21 infants that showed a significant association between oral feeding and FOXP2 expression levels with an effect size of 1.79. For this study, we conservatively expect an effect size that is less than that for this preliminary study of 0.82, which yielded a sample size of n = 45. Therefore, we planned to enroll 50 infants for this study. We used SAS (Version 9.4_M3) with SAS Enterprise Guide (Version 7.15HF3, SAS Institute Inc.) for the data analysis. RT-qPCR data were analyzed in binary (+/−) gene expression manner. Descriptive analyses were completed examining the relation between gene expression and oral feeding emergence.
Results
Salivary Samples
Fifty-four infants were initially recruited for this study. However, three infants were excluded due to prolonged respiratory support or death, resulting in a sample size of 51 babies. From 51 babies, 78 of the samples met quality control standards and inclusion criteria for analysis (63%). On average, enrolled infants were born at 33.20 weeks' GA (SD = 1.30 days) with a birth weight of 1,959 g (SD = 427 g).
PCR Quality Control Metrics
Amplification results of the reference genes in our positive control analyzed on each premade plate to assess plate-to-plate variability and performance were as followed: GAPDH: mean cycle threshold (Ct) = 22.07, SD = 0.39, coefficient of variation (CV) = 1.75; YWHAZ: mean Ct = 20.02, SD = 0.18, CV = 0.89; and HPRT1: mean Ct = 30.40, SD = 0.42, CV = 1.37. For our negative control, three of 39 wells amplified for GAPDH and two of 39 wells amplified for YWHAZ. Four of the five negative control wells amplified at Ct > 37. Conversely, subject samples on these plates amplified each of these reference genes at Ct < 33; therefore, the samples were not impacted by the primer to primer amplification. One of the five negative control wells was considered a contaminant with amplification at Ct = 31. There was equal amplification across gene targets in the nonamplified-to-amplified sample comparisons: nonamplified samples Ct = 33.98 (GAPDH) and Ct = 31.49 (YWHAZ); amplified samples at Ct = 21.27 (GAPDH) and Ct = 20.06 (YWHAZ).
Gene Binary Analysis (Expressed vs. not Expressed)
Expression of all target genes were analyzed in a binary fashion (e.g., the genes were either expressed or not expressed) and reported as a percentage. Table 1 provides expression data of FOXP2, CNTNAP2, GRIN2A, and NRXN1 independently for each infant during the three time points. For each collection time point during the learning process of oral feeding, samples were grouped as having either (+/−) expression of each gene. Mean PCA and days to full feed were described for each group. Due to the low expression levels of GRIN2A and NRXN1, these genes were not used in our oral feeding analysis. FOXP2 and CNTNAP2 were considered individually and in combination. Table 2 summarizes the gene models for each infant at each time point.
Table 1.
The expression of each gene was evaluated at all three time points.
| Gene | Time point | Expression | n | PCA | Days to full feed | % expressed | Effect size (Cohen's d) |
|---|---|---|---|---|---|---|---|
| FOXP2 | Start of PO feeding | + | 28 | 34.11 (0.62) | 10.50 (6.61) | 93 | 0.34 |
| – | 2 | 34.29 (0.61) | 14.50 (14.85) | ||||
| Learning PO feeding | + | 16 | 35.06 (0.82) | 15.75 (6.07) | 76 | 1.06 | |
| – | 5 | 34.51 (0.65) | 10.60 (3.21) | ||||
| 100% PO feeding | + | 19 | 35.84 (1.00) | 10.74 (8.29) | 70 | 0.72 | |
| – | 8 | 35.14 (0.51) | 6.13 (3.52) | ||||
| CNTNAP2 | Start of PO feeding | + | 7 | 34.06 (0.75) | 12.57 (7.04) | 23 | 0.34 |
| – | 23 | 34.14 (0.58) | 10.22 (7.08) | ||||
| Learning PO feeding | + | 4 | 34.79 (0.55) | 15.25 ( 4.27) | 19 | 0.16 | |
| – | 17 | 34.97 (0.87) | 14.35 (6.31) | ||||
| 100% PO feeding | + | 8 | 35.66 (1.16) | 7.38 (6.67) | 30 | 0.38 | |
| – | 19 | 35.62 (0.86) | 10.21 (7.76) | ||||
| GRIN2A | Start of PO feeding | + | 5 | 34.37 (0.56) | 11.00 (6.04) | 17 | 0.04 |
| – | 25 | 34.07 (0.61) | 10.72 (7.17) | ||||
| Learning PO feeding | + | 0 | 0 | ||||
| – | 21 | 34.96 (0.84) | 14.52 (5.90) | ||||
| 100% PO feeding | + | 1 | 34.57 | 3.00 | 3.7 | ||
| – | 26 | 35.68 (0.93) | 9.62 (7.48) | ||||
| NRXN1 | Start of PO feeding | + | 4 | 34.50 (0.38) | 7.00 ( 7.16) | 13 | 0.64 |
| – | 26 | 34.06 (0.62) | 11.35 (7.08) | ||||
| Learning PO feeding | + | 2 | 34.21 (0.51) | 13.00 ( 4.24) | 9.5 | 0.32 | |
| – | 19 | 35.02 (0.80) | 14.68 ( 6.11) | ||||
| 100% PO feeding | + | 2 | 34.93 (0.51) | 2.50 (0.71) | 7.4 | 1.38 | |
| – | 25 | 35.69 ( 0.94) | 9.92 (7.47) |
Note. Postconceptional age (PCA) and Days to full feed are mean (SD). n = no. of infants; FOXP2 = forkhead box P2; PO = per os; (+) = gene expressed; (−) = gene not expressed; CNTNAP2 = contactin-associated protein-like 2; GRIN2A = glutamate receptor, ionotropic, N-methyl D-aspartate 2A; NRXN1 = neurexin 1.
Table 2.
Mean days to full feed for gene models at different time points.
| Time point | Gene model | n | PCA | Days to full feed |
|---|---|---|---|---|
| Start of PO feeding | FOXP2+ CNTNAP2+ | 7 | 34.06 (0.75) | 12.57 (7.04) |
| FOXP2+ CNTNAP2− | 21 | 34.12 (0.59) | 9.81 (6.49) | |
| FOXP2− CNTNAP2− | 2 | 34.29 (0.61) | 14.50 (14.85) | |
| FOXP2− CNTNAP2+ | 0 | |||
| Learning PO feeding | FOXP2+ CNTNAP2+ | 4 | 34.78 (0.55) | 15.25 (4.27) |
| FOXP2+ CNTNAP2− | 12 | 35.15 (0.90) | 15.92 (6.72) | |
| FOXP2− CNTNAP2− | 5 | 34.51 (0.65) | 10.60 (3.21) | |
| FOXP2− CNTNAP2+ | 0 | |||
| 100% PO feeding | FOXP2+ CNTNAP2+ | 6 | 35.74 (1.18) | 7.40 (7.02) |
| FOXP2+ CNTNAP2− | 13 | 35.79 (0.96) | 11.62 (8.87) | |
| FOXP2− CNTNAP2− | 6 | 35.51 (0.77) | 8.43 (4.578) | |
| FOXP2− CNTNAP2+ | 2 | 34.78 (0.71) | 3.00 (1.41) |
Note. Postconceptional age (PCA) and Days to full feed are mean (SD). n = no. of infants; PO = per os; FOXP2 = forkhead box P2; CNTNAP2 = contactin-associated protein-like 2.
Infants who expressed FOXP2 (FOXP2+) at the start of PO feeding (n = 28) achieved full oral feeding 4 days sooner than infants who did not express FOXP2 (FOXP2–; n = 2). However, during the learning process and at full oral feeding, FOXP2+ infants (n = 16, n = 19, respectively) took an average of 5.20 and 4.60 days longer to achieve full oral feeding than FOXP2– infants (n = 5, n = 8, respectively) during the same time point. Conversely, CNTNAP2+ infants at the start of PO feeding (n = 7) and during the learning process (n = 4) learned to fully feed orally 2.40 and 0.90 days longer, respectively, than CNTNAP2– infants at the same time point. At 100% PO feeding, CNTNAP2+ infants (n = 8) had 2.83 days shorter to full oral feeding.
Infants with the gene model FOXP2+ CNTNAP2– at the initiation of oral feeding trials had a mean of 3.20 fewer days to full oral feeding in comparison to all other infants (p = .26; 95% CI [−2.50, 8.90]). However, during the learning process, infants with the gene model FOXP2+ CNTNAP2– took a mean of 3.30 days longer to full oral feeding in comparison to other gene models (p = .22; 95% CI [−2.10, 8.60]). At 100% oral feeding, infants with the gene model FOXP2+ CNTNAP2– took a mean of 4.30 days longer to achieve full oral feeding in comparison to other gene models (p = .13; 95% CI [−1.40, 10.10]).
Discussion
This study examined if the speech associated genes, FOXP2, CNTNAP2, GRIN2A, and NRXN1, were detectable in neonatal saliva and if these gene expression levels related to oral feeding outcomes in premature newborns. Despite exploring these four speech-language genes of interest, only FOXP2 and CNTNAP2 were detected consistently in neonatal saliva. NRXN1 and GRIN2A were rarely amplified in our cohort, and therefore, we were not able to examine these genes in relation to oral feeding development. However, it is possible that in this small pilot study, we were unable to attain a diverse enough group of subjects to account for biological variability in expression of all biomarkers and that in larger, multicenter trials, each may provide insight into oral feeding readiness in this population. It is important to note that, to our knowledge, this is the first time that CNTNAP2, GRIN2, and NRXN1 have been examined in preterm infant saliva.
Next, we examined how the genes of interest related to oral feeding. We found that infants who expressed FOXP2 at the start of PO feeding learned to orally feed 4 days sooner than infants who did not express the gene (95% CI [−7, 14]) with effect sizes ranging from 0.30 to 1.06 indicating that with a larger sample size, these trends may reach significance. Conversely for CNTNAP2, we found that infants who expressed this gene at the start of oral feeding fed 2 days later (95% CI [−4, 9]). Because CNTNAP2 is regulated by FOXP2, binary expression was also examined in combination. Infants with the gene model FOXP2+ CNTNAP2− at the start of feeding achieved full oral feeds 3 days faster than the other models (95% CI [−3, 9]). Results from this study are similar to previous research showing that increased expression of salivary FOXP2 is associated with a shorter time to full oral feeding (Zimmerman et al., 2016). The relation between FOXP2 and oral feeding is worth further investigation when considering the financial cost on the health care system and the neurodevelopmental impact that prolonged hospitalization may have on the infant. NICU costs are approximately $4,000/day, length of stay is highly correlated to feeding success (Niknajad, Ghojazadeh, Sattarzadeh, Bashar Hashemi, & Dezham Khoy Shahgholi, 2012) and can have negative impacts on parent–child attachment (Flacking et al., 2012).
Interestingly, FOXP2 and CNTNAP2 expression neither steadily increased nor decreased during the learning process of oral feeding. Although the biological significance of this finding remains unknown, to our knowledge, this is the first research note to report expression levels over time. Previous studies explored feeding outcomes based upon salivary gene expression profiles from a single time point (Maron et al., 2015; Zimmerman et al., 2016). We believe it is unlikely these changes are due to maturing circadian rhythms or hunger signaling, as all samples were collected during the day and prior to a feed. Rather, these data suggest that biological variability (i.e., age) over time may impact expression profiles and/or changing levels of these genes play an essential regulatory mechanism in neonatal feeding development. Of note, interpretation of these findings is limited by the paucity of infants in this pilot study who had complete salivary acquisition data analysis throughout the learning process of oral feeding. Infant transfer likely impacted our observation that expression profiles at the initiation of feeds proved most informative of long-term feeding outcomes. Only large scale studies will be able to appropriately explore these variations across GAs and sex to determine which time point(s) best predict feeding outcomes.
Oral feeding is a complex developmental task. Oral feeding development requires proper structural development, physiologic stability, tone, and coordination (Crowe, Chang, & Wallace, 2016; McGrath & Braescu, 2004; Mizuno & Ueda, 2003). In utero, the development of feeding occurs with fetal swallowing noted as early as 12 weeks' GA, and by 34 weeks' GA, the fetus is known to have a coordinated suck and swallow reflex (Humphrey, 1970). However, for preterm infants, this typical developmental process is disrupted and improper timing of feeding may have negative results such as aspiration, hypoxia, bradycardia, fatigue, and oral aversion. Another key factor in oral feeding success for preterm infants is their birth GA. Infants who are born at higher birth GA are able to learn to feed faster (Davis, Liu, & Rhein, 2013; Mizuno & Ueda, 2003). This finding is likely due to the fact that these infants are older, have a more mature neurodevelopmental system, and have had more practice sucking and swallowing in the womb. Given the complexity of feeding and the risk for early aversions and negative clinical outcomes, more objective assessment tools are needed to predict oral feeding readiness.
Current feeding practices in the NICU are largely limited to subjective assessment of feeding cues by neonatal caregivers to determine oral feeding readiness. While these approaches may identify which babies are mature enough to attempt oral feeding trials, they are incapable of assessing developmental delays limiting oral feeding success. Our goal is to develop an objective assay that moves beyond feeding readiness assessment to one that holds the potential of identifying specific developmental pathways (i.e., oromotor, sensory, hunger signaling) that are impacting an infant's ability to feed (see Figure 1). We hypothesize that this individualized approach may allow caregivers to develop personalized care plans to improve oral feeding skills, as well as alert them to the potential of additional developmental delays, including delays in speech emergence. In turn, this approach may reduce feeding-associated morbidities and decrease hospitalization length with associated health care costs.
This study was limited by a small sample size and the fact that infants were transferred to multiple Level II nurseries in our community, reducing the number of salivary samples collected for analysis. In addition, infants who were transferred outside the Tufts Medical Center neonatal network may have been exposed to varying feeding practices, which in turn may have impacted our primary outcome. Nevertheless, these data support our previous study that demonstrated levels of FOXP2 at the initiation of oral feeding in the preterm infant result in a shorter time to achieve full oral feeds. In addition, we have identified at least one subsequent speech-language gene, CNTNAP2, that may inform feeding outcomes. Thus, there is a strong need for large, prospective, multicenter research studies in order to better understand gene ontogeny, specifically as it relates to feeding outcomes. Incorporating speech follow-up on these infants may provide insight into how these genes relate to future development.
Conclusion
FOXP2 and CNTNAP2 are readily detectable in neonatal saliva and may prove informative, alone or in combination, to help predict oral feeding outcomes in the preterm infant. NRXN1 and GRIN2A were often not amplified and were noninformative in relation to feeding progression in this pilot study. Further studies incorporating larger sample sizes are needed to more fully understand the expression patterns of speech-language genes to objectively and accurately predict oral feeding readiness. Importantly, long-term developmental follow-up of these infants is warranted to determine if aberrant expression patterns of speech-language genes in the neonatal period predict delayed speech emergence in infancy and toddlerhood.
Acknowledgments
This study was funded by the Tufts CTSI Pilot Award (UL1TR001054, awarded to PIs: Emily Zimmerman and Jill Maron). We would like to thank the families of those who participated in the study and Robin Ruthazer for her assistance with statistical analyses.
Funding Statement
This study was funded by the Tufts CTSI Pilot Award (UL1TR001054, awarded to PIs: Emily Zimmerman and Jill Maron).
References
- Al Shehhi M., Forman E. B., Fitzgerald J. E., McInerney V., Krawczyk J., Shen S., … Lynch S. A. (2019). NRNX1 deletion syndrome: Phenotypic penetrance data from 34 families. European Journal of Medical Genetics, 62(3), 204–209. [DOI] [PubMed] [Google Scholar]
- Barlow S. M., Maron J. L., Alterovitz G., Song D., Wilson B. J., Jegatheesan P., … Rosner A. O. (2017). Somatosensory modulation of salivary gene expression and oral feeding in preterm infants: Randomized controlled trial. JMIR Research Protocols, 6(6), e113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J. M., Perez-Pouchoulen M., Edwards N. S., & McCarthy M. M. (2013). Foxp2 mediates sex differences in ultrasonic vocalization by rat pups and directs order of maternal retrieval. The Journal of Neuroscience, 33(8), 3276–3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., … Wittwer C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622. [DOI] [PubMed] [Google Scholar]
- Carvill G. L., Regan B. M., Yendle S. C., O'Roak B. J., Lozovaya N., Bruneau N., … Mefford H. C. (2013). GRIN2A mutations cause epilepsy-aphasia spectrum disorders. Nature Genetics, 45(9), 1073–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe L., Chang A., & Wallace K. (2016). Instruments for assessing readiness to commence suck feeds in preterm infants: Effects on time to establish full oral feeding and duration of hospitalization. Cochrane Database Systematic Review, 23(8), CDD005586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabell M. P., Rosenfeld J. A., Bader P., Escobar L. F., El-Khechen D., Vallee S. E., … Shaffer L. G. (2013). Investigation of NRXN1 deletions: Clinical and molecular characterization. American Journal of Medical Genetics, 161A(4), 717–731. [DOI] [PubMed] [Google Scholar]
- Davis N. L., Liu A., & Rhein L. (2013). Feeding immaturity in preterm neonates: Risk factors for oropharyngeal aspiration and timing of maturation. Journal of Pediatric Gastroenterology and Nutrition, 57(6), 735–740. [DOI] [PubMed] [Google Scholar]
- Fisher S. E., & Scharff C. (2009). FOXP2 as a molecular window into speech and language. Trends in Genetics, 25(4), 166–177. [DOI] [PubMed] [Google Scholar]
- Flacking R., Lehtonen L., Thomson G., Axelin A., Ahlqvist S., Moran V. H., …. Separation and Closeness Experiences in the Neonatal Environment Group. (2012). Closeness and separation in neonatal intensive care. Acta Paediatrica, 101(10), 1032–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey T. (1970). The development of human fetal activity and its relation to postnatal behavior. Advances in Childhood Development and Behavior, 5, 1–57. [DOI] [PubMed] [Google Scholar]
- Hurst J. A., Baraitser M., Auger E., Graham F., & Norell S. (1990). An extended family with a dominantly inherited speech disorder. Developmental Medicine and Child Neurology, 32(4), 352–355. [DOI] [PubMed] [Google Scholar]
- Johnson M. J., Wootton S. A., Leaf A. A., & Jackson A. A. (2012). Preterm birth and body composition at term equivalent age: A systematic review and meta-analysis. Pediatrics, 130(3), e640–e649. [DOI] [PubMed] [Google Scholar]
- Khanna P., Johnson K. L., & Maron J. L. (2017). Optimal reference genes for RT-qPCR normalizatioon in the newborn. Biotechnic & Hisotchemistry, 92(7), 459–466. [DOI] [PubMed] [Google Scholar]
- Khanna P., Maron J. L., & Walt D. R. (2017). Development of a rapid salivary proteomic platform for oral feeding readiness in the preterm newborn. Frontiers in Pediatrics, 5, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C. S., Fisher S. E., Hurst J. A., Vargha-Khadem F., & Monaco A. P. (2001). A forkhead-domain gene is mutated in a severe speech and language disorder. Nature, 413(6855), 519–523. [DOI] [PubMed] [Google Scholar]
- Lai C. S., Gerrelli D., Monaco A. P., Fisher S. E., & Copp A. J. (2003). FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain, 126(Pt. 11), 2455–2462. [DOI] [PubMed] [Google Scholar]
- Lau C., Alagugurusamy R., Schanler R. J., Smith E. O., & Shulman R. J. (2000). Characterization of the developmental stages of sucking in preterm infants during bottle feeding. Acta Paediatrica, 89(7), 846–852. [PubMed] [Google Scholar]
- Liégeois F., Baldeweg T., Connelly A., Gadian D. G., Mishkin M., & Vargha-Khadem F. (2003). Language fMRI abnormalities associated with FOXP2 gene mutation. Nature Neuroscience, 6(11), 1230–1237. [DOI] [PubMed] [Google Scholar]
- Ludwig S. M., & Waitzman K. A. (2007). Changing feeding outcomes to reflect infant-driven feeding practice. Newborn and Infant Nursing Reviews, 7(3), 155–160. [Google Scholar]
- MacDermot K. D., Bonora E., Sykes N., Coupe A. M., Lai C. S., Vernes S. C., … Fisher S. E. (2005). Identification of FOXP2 truncation as a novel cause of developmental speech and language deficits. American Journal of Human Genetics, 76(6), 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron J. L. (2012). Insights into neonatal oral feeding through the salivary transcriptome. International Journal of Pediatrics, 2012, 195153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron J. L., Hwang J. S., Pathak S., Ruthazer R., Russell R. L., & Alterovitz G. (2015). Computational gene expression modeling identifies salivary biomarker analysis that predict oral feeding readiness in the newborn. The Journal of Pediatrrics, 166(2), 282.e5–288.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron J. L., Johnson K. L., Rocke D. M., Cohen M. G., Liley A. J., & Bianchi D. W. (2010). Neonatal salivary analysis reveals global developmental gene expression changes in the premature infant. Clinical Chemistry, 56(3), 409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo K., & Palmer J. B. (2008). Anatomy and physiology of feeding and swallowing: Normal and abnormal. Physical Medicine and Rehabilitation Clinics of North America, 19(4), 691–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland D. H., & Tremblay P. (2006). Clinical implications of cross-system interactions. Seminars in Speech and Language, 27(4), 300–309. [DOI] [PubMed] [Google Scholar]
- McGrath J. M., & Braescu A. V. (2004). State of the science: Feeding readiness in the preterm infant. Journal of Perinatal and Neonatal Nursing, 18(4), 353–370. [DOI] [PubMed] [Google Scholar]
- Mizuno K., & Ueda A. (2003). The maturation and coordination of sucking, swallowing, and respiration in preterm infants. Journal of Pediatrics, 142(1), 36–40. [DOI] [PubMed] [Google Scholar]
- Niknajad A., Ghojazadeh M., Sattarzadeh N., Bashar Hashemi F., & Dezham Khoy Shahgholi F. (2012). Factors affecting the neonatal intensive care unit stay duration in very low birth weight premature infants. Journal of Caring Science, 1(2), 85–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore M., Zimmerman E., Barlow S. M., Wang J., & Gu F. (2008). Patterned orocutaneous therapy improves sucking and oral feeding in preterm infants. Acta Paediatrica, 97(7), 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M. (2017). Intragenic CNTNAP2 deletions: A bridge too far? Molecular Syndromology, 8(3), 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodenas-Cuadrado P., Ho J., & Vernes S. C. (2014). Shining a light on CNTNAP2: Complex functions to complex disorders. European Journal of Human Genetics, 22(2), 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens B. E., & Vohr B. R. (2009). Neurodevelopmental outcome of the premature infant. Pediatric Clinics of North America, 56(3), 631–646. [DOI] [PubMed] [Google Scholar]
- Turner S. J., Mayes A. K., Verhoeven A., Mandelstam S. A., Morgan A. T., & Scheffer I. E. (2015). GRIN2A: An aptly named gene for speech dysfunction. Neurology, 84(6), 586–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noort-van der Spek I. L., Franken M. C., & Weisglas-Kuperus N. (2012). Language functions in preterm-born children: A systematic review and meta-analysis. Pediatrics, 129(4), 745–754. [DOI] [PubMed] [Google Scholar]
- Zimmerman E., Maki M., & Maron J. (2016). Salivary FOXP2 expression and oral feeding success in premature infants. Cold Spring Harbor Molecular Case Studies, 2(1), a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E., & Maron J. L. (2016). FOXP2 gene deletion and infant feeding difficulties: A case report. Cold Spring Harbor Molecular Case Studies, 2(1), a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]