Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infects the nasopharynx and lungs and causes coronavirus disease-2019 (COVID-19). It may impact the heart, brain, kidney, and liver.1 Although functional impairment of the liver has been correlated with worse clinical outcomes, little is known about the pathophysiology of hepatic injury and repair in COVID-19.2,3 Histologic evaluation has been limited to small numbers of COVID-19 cases with no control subjects2,4 and demonstrated largely heterogeneous patterns of pathology.2,3
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infects the nasopharynx and lungs and causes coronavirus disease-2019 (COVID-19). It may impact the heart, brain, kidney, and liver.1 Although functional impairment of the liver has been correlated with worse clinical outcomes, little is known about the pathophysiology of hepatic injury and repair in COVID-19.2 , 3 Histologic evaluation has been limited to small numbers of COVID-19 cases with no control subjects2 , 4 and demonstrated largely heterogeneous patterns of pathology.2 , 3
Methods
Liver tissues of 60 patients who died of COVID-19 pneumonia were obtained from complete autopsies performed in Hamburg between March and June 2020. A cohort of 13 patients with fatal pneumonia in the absence of SARS-CoV-2 infection served as control subjects.
Hematoxylin and eosin staining and immunohistochemistry were applied to comprehensively evaluate the pathophysiology and regeneration aspects at the level of the hepatic microarchitecture. Label-free coherent Raman scattering and second harmonic generation imaging visualized major morphologic changes, revealing steatosis and dilatation of sinusoids.
Results
There were no significant differences in the patient characteristics except for preexisting neurologic conditions (P = .04) (Supplementary Table 1). Only 5 patients with COVID-19 and 1 control subject had history of liver disease. A minority of patients in the COVID-19 group (17%) received intensive care at the time of death with a trend within that group toward home or nursing home care, when compared with control subjects (Supplementary Table 1).
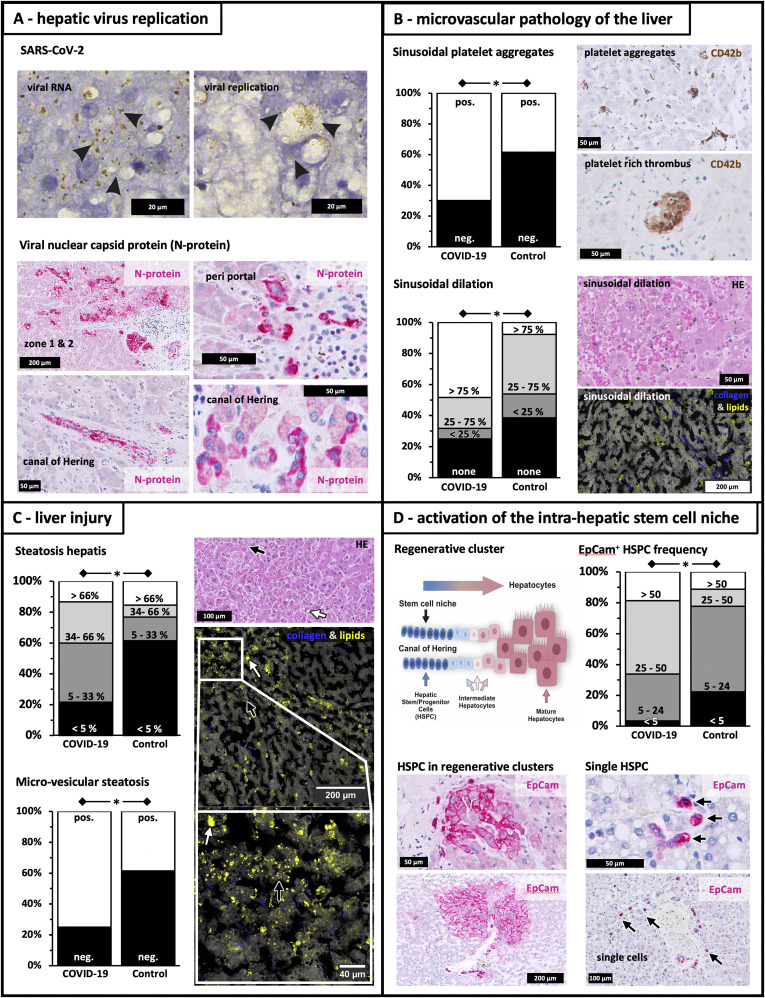
Quantitative real-time polymerase chain reaction and in situ hybridization revealed viral RNA and replicative intermediates in liver tissues. Viral nucleocapsid protein was detected in hepatic stem/progenitor cells (HSPC), cholangiocytes, and hepatocytes (Figure 1 A). SARS-CoV-2 was detected at the RNA and/or protein level in 25% of livers in COVID-19 patients (Supplementary Table 1). Quantitative real-time polymerase chain reaction demonstrated 2 out of 5 bile samples positive for SARS-CoV-2 (data not shown).
Figure 1.
Microvascular pathology and regenerative responses in livers of COVID-19 patients. SARS-CoV-2 replication, histopathology, and regeneration responses of the liver in fatal COVID-19. (A) Viral replication. SARS-CoV-2 viral RNA (“viral RNA”; +strand, brown dots indicated by black arrows) detected by in situ hybridization and replicative viral intermediates (“viral replication”; -RNA, brown dots indicated by black arrows). Immunohistochemistry shows immunoreactivity against viral nucleocapsid protein (red) in hepatocytes within acinar zones I and II, periportal HSPC, cholangiocytes of portal bile ducts, and premature hepatocytes along the canal of Hering. (B) Microvascular pathology. Sinusoidal platelet aggregates and thrombi (anti-CD42b staining), frequent in COVID-19 patients. Sinusoidal dilatation is noted in hematoxylin and eosin (HE) staining. Coherent Raman scattering microscopy provided label-free contrast for tissue structure (grey) and lipid droplet identification (yellow), whereas second harmonic generation provided contrast for collagen fibers (blue). (C) Liver injury. High levels of hepatocyte steatosis in COVID-19 patients, in comparison with control subjects, as detected by HE staining and coherent Raman scattering/second harmonic generation (lipid droplets in yellow and collagen in blue). White arrows indicate macrovesicular steatosis, whereas black arrows show microvesicular steatosis. (D) Intrahepatic stem cell niche. Drawing of the bipotent intrahepatic stem cell compartment, which is located in the canal of Hering. On severe hepatic injury HSPC emerge and expand, contributing to liver repair. HSPC are detected as single cells (indicated by black arrows) or in regenerative clusters comprised of EpCAM+ HSPC, hepatobiliary intermediate cells, and premature hepatocytes, which are adjacent to the canal of Hering. Stem cell marker EpCam (red) is also expressed by numerous HSPC and intermediates of hepatocytes/cholangiocytes and regenerative clusters of periportal hepatocytes of irregular size and shape. HSPC frequency in column diagram represents numbers of HSPC per 10 field views. ∗P < .05.
Sinusoidal platelet-aggregates were predominantly observed in the hepatic microvasculature of COVID-19 patients when compared with control subjects (70% vs 30%; P = .032) (Figure 1 B). Likewise, sinusoidal (P = .024) (Figure 1 B) and portal dilatation were observed (P = .002) (Supplementary Table 1). Hepatic microvascular thrombosis in COVID-19 patients was predominantly observed in nonhospitalized patients (32%; not receiving anticoagulant therapy), when compared with general in-patients (3%) and those requiring intensive care (10%; P = .016; data not shown).
COVID-19 patients demonstrated more hepatic steatosis (P = .046), mainly the microvesicular variant (P = .01), when compared with SARS-CoV-2-negative control subjects (Figure 1 C). Severe intrahepatic injury was associated with the activation of the intrahepatic stem cell niche along the canal of Hering (Figure 1 D), resulting in regenerative clusters of EpCAM+HSPC, hepatobiliary intermediate cells, and premature hepatocytes (Figure 1 D). Increased numbers of EpCAM+HSPC were observed alone, and in regenerative clusters, in patients with COVID-19, when compared with control livers (>25 HSPC/10 field views: 66% vs 22%; P = .020) (Figure 1 D).
Discussion
We show here secretion of virus into bile and that replication of SARS-CoV-2 occurs in liver tissues during COVID-19. Viral infection of HSPC was observed, comparable with that noted in lung alveolocyte progenitors in the first variant of SARS-CoV infections.5
The major pathologic finding is that microvascular changes and platelet-rich thromboembolic phenomena reflect disordered thromboregulation and vascular insults as described previously.4 , 6 As noted with other prothrombotic features of SARS-CoV-2 infections,4 , 7 our observation of microthrombotic pathology predominantly occurring at time of death in nonhospitalized patients suggests that early initiation of anticoagulant therapy in those hospitalized may attenuate microvascular disease of the liver.
Platelet activation, sinusoidal injury, and parenchymal damage result in necrosis, which may be combined with predominantly microvesicular hepatocyte steatosis. Previous reports on postmortem evaluations have demonstrated hepatic steatosis to variable degrees in COVID-19 and seem to demonstrate macrovesicular over microvesicular forms of steatosis.3 , 4
We demonstrate that vascular injury is accompanied by activation of the intrahepatic stem cell compartment showing features of aberrant regeneration. Similar scenarios of HSPC activation, correlating with the extent of liver cell necrosis, as noted here, have been also shown in hepatitis B–mediated liver injury.8
We conclude that SARS-CoV-2 infects the liver as part of systemic illness and results in substantial microvascular thrombotic disease, organ injury, and attempts at regeneration.
Acknowledgments
Barbara Kaltschmidt, Antonia D.E. Fitzek, Julia Schaedler, and Christine Förster contributed equally.
Martin Krüger, Simon C. Robson, Ludwig Wilkens, and Jan Schulte am Esch contributed equally.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Beatrice A. Windmöller is funded by a grant of the Bethel foundation. Christian Pilger and Cihang Kong were supported by the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie Grant (Agreement No. 766181, project DeLIVER). Simon C. Robson acknowledges support from Department of Defense Award W81XWH-16-0464.
Note: To access the supplementary material accompanying this article, visit the online version of Clinical Gastroenterology and Hepatology at www.cghjournal.org, and at http://doi.org/10.1016/j.cgh.2021.01.044.
Supplementary Methods
Autopsies and Liver Sampling
We analyzed liver samples from a cohort of 60 consecutive patients deceased with COVID-19 including cases of none to moderate levels of autolysis. We excluded samples with a high degree of autolysis. Autopsies were carried out by order of the Hamburg health authorities in accordance to the German Infection Protection Act §25 para. 4. The Ethics Committee of the Hamburg Medical Association agreed in principle to this procedure in a vote on April 22, 2020 (file number PV7311, “SARS-CoV-2: Post-mortem description of COVID-19-associated deaths”). The study complied with the tenets of the Declaration of Helsinki. During the autopsies, liver tissue samples were taken and fixed in buffered 4% formaldehyde and virologic swabs were retained. All deceased patients in the COVID-19 group were positive for SARS-CoV-2 viral RNA using a throat smear followed by immediate quantitative real-time polymerase chain reaction (qRT-PCR). A control group was constituted of 13 patients with fatal pneumonia, but absence of SARS-CoV-2 infection.
In Situ Hybridization
Viral RNA and viral replication of SARS-CoV-2 were analyzed in liver sections of patients who died of COVID-19, using RNAscope (ACDbio, Newark, CA). Microphotographs were taken using bright field microscopy.
Quantitative Real-Time Polymerase Chain Reaction
RNA extracted from paraffin sections using the Maxwell RSC (Promega, Madison, WI) was used for RT-PCR analysis (RealStar SARS-CoV-2 RT-PCR Kit 1.0, Altona Diagnostics, Hamburg, Germany).
Histopathology
Standard staining
One tissue block was taken from a representative sampling area of each patient and embedded in paraffin. Three micrometer paraffin sections were stained with hematoxylin and eosin, Elastica van Gieson, periodic acid–Schiff, reticulin stain (Gomori), and iron stain (Perls Prussian blue).
Immunohistochemical staining
Staining was done using Dako Omnis (DAKO, Agilent, Santa Clara, CA) with the following antibodies: anti-CK7, anti-EpCAM, anti-CD31, anti-CD34, anti-CD42b, and anti-SARS-CoV nucleocapsid. Detection was either with DAB or alkaline phosphatase/red. Microphotographs were taken using bright field microscopy.
Nonlinear Optical Microscopy
A custom-built coherent Raman scattering microscope was used to image fixed liver sections. Coherent Raman scattering visualizes lipids in the sample by probing the 2845 cm-1 molecular resonance. Second harmonic generation imaging was performed using a home-built fiber-based femtosecond laser. The overlay of coherent Raman scattering and second harmonic generation channels in different look-up tables were done using the open-source software FIJI.
Statistical Analyses
Dichometric and nominal variables were analyzed applying Pearson chi-square test. Ordinal variables were subjected to Wilcoxon rank sum test (Mann-Whitney) analyses. Statistical significance was taken as P values < .05. Stata/SE version 15.1 for Mac software (StataCorp, College Station, TX) was used for statistical analyses.
Supplementary Table 1.
Clinical Characteristics and Autopsy Findings in COVID-19 Pneumonia and the Control Patient Group of Fatal Cases of Pneumonia Unrelated to SARS-CoV-2
| Classification |
COVID-19 negative |
COVID-19 positive |
P value |
|
|---|---|---|---|---|
| n = 13 | n = 60 | |||
| n = 13 | n = 60 | |||
| Gender | .347 | |||
| Female | 31 (4) | 45 (27) | ||
| Male | 69 (9) | 55 (33) | ||
| Age, y | .422 | |||
| ≤80 | 54 (7) | 42 (25) | ||
| >80 | 46 (6) | 58 (35) | ||
| Liver disease in medical history | 8 (1) | 8 (5) | .939 | |
| Heart disease in medical history | 85 (11) | 93 (56) | .299 | |
| Lung disease in medical history | 77 (10) | 63 (38) | .349 | |
| Neurologic disease in medical history | 15 (2) | 47 (28) | .038 | |
| Renal disease in medical history | 46 (6) | 33 (20) | .381 | |
| Oncologic disease in medical history | 8 (1) | 22 (13) | .246 | |
| Level of care at time of death | .096 | |||
| Home/nursing home | 23 (3) | 32 (19) | ||
| Hospital, low care | 31 (4) | 52 (31) | ||
| Intensive care unit | 46 (6) | 17 (10) | ||
| Autolysis of hepatic tissue | .171 | |||
| None | 31 (4) | 45 (27) | ||
| Little | 31 (4) | 37 (22) | ||
| Moderate | 39 (5) | 18 (11) | ||
| Liver tissue: SARS-CoV-2 and/or nucleocapsid-protein | Positive | 0 (0) | 22 (13) | .043 |
| Sinusoidal platelet aggregates | Present | 39 (5) | 70 (42) | .031 |
| Sinusoidal ectasia | .024 | |||
| None | 39 (5) | 25 (15) | ||
| Focal (<25%) | 15 (2) | 7 (4) | ||
| Multifocal (25%–75%) | 39 (5) | 20 (12) | ||
| Diffuse (>75%) | 8 (1) | 48 (29) | ||
| Portal ectasia | .002 | |||
| None | 8 (1) | 3 (2) | ||
| Focal (<25%) | 46 (6) | 13 (8) | ||
| Multifocal (25%–75%) | 31 (4) | 25 (15) | ||
| Diffuse (>75%) | 15 (2) | 5 (35) | ||
| Portal fibrosis | .293 | |||
| Widen, no septae | 53 (7) | 72 (43) | ||
| Widen with septae | 15 (2) | 5 (3) | ||
| Incomplete cirrhosis | 31 (4) | 22 (13) | ||
| Cirrhosis | 0 (0) | 2 (1) | ||
| Confluent necrosis | .175 | |||
| None | 85 (11) | 66 (38) | ||
| Focal | 8 (1) | 10 (6) | ||
| Moderate | 0 (0) | 5 (3) | ||
| Frequent | 8 (1) | 19 (11) | ||
| Apoptosis (cell count per field view) | .838 | |||
| 0–1 | 77 (10) | 81 (47) | ||
| 2–4 | 23 (3) | 12 (7) | ||
| >5 | 0 (0) | 7 (4) | ||
| Hepatic steatosis | .046 | |||
| <5% | 62 (8) | 22 (13) | ||
| 5%–33% | 15 (2) | 38 (23) | ||
| 34%–66% | 8 (1) | 27 (16) | ||
| >66% | 15 (2) | 13 (8) | ||
| Macrovesicular steatosis | Positive | 39 (5) | 67 (40) | .058 |
| Microvesicular steatosis | Positive | 39 (5) | 75 (45) | .010 |
| Kupffer cell siderosis | .771 | |||
| None | 85 (11) | 88 (53) | ||
| Little | 15 (2) | 7 (4) | ||
| Moderate | 0 (0) | 3 (2) | ||
| Severe | 0 (0) | 2 (1) | ||
| Hepatocyte siderosis | .720 | |||
| None | 92 (12) | 88 (53) | ||
| Little | 0.00 (0) | 8 (5) | ||
| Moderate | 8 (1) | 2 (1) | ||
| Severe | 0 (0) | 2 (1) | ||
| Ductular reaction (CK7) | Positive | 77 (10) | 87 (52) | .348 |
| Cholestasis (histologically) | Positive | 0 (0) | 3 (2) | .793 |
| EpCam+ regenerative cluster | Positive | 11 (1) | 29 (17) | .262 |
| EpCam+ hepatic stem/progenitor cells (count per 10 field views) | .020 | |||
| <5 | 22 (2) | 3 (2) | ||
| 5–24 | 56 (5) | 31 (18) | ||
| 25–50 | 11 (1) | 48 (28) | ||
| >50 | 11 (1) | 19 (11) |
NOTE. Data given are % (n). P value: bold < .05.
References
- 1.Puelles V.G., Lutgehetmann M., Lindenmeyer M.T., et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020;383:590–592. doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lagana S.M., Kudose S., Luga A.C., et al. Hepatic pathology in patients dying of COVID-19: a series of 40 cases including clinical, histologic, and virologic data. Mod Pathol. 2020;33:2147–2155. doi: 10.1038/s41379-020-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saviano A., Wrensch F., Ghany M.G., et al. Liver disease and COVID-19: from pathogenesis to clinical care. Hepatology. 2020 doi: 10.1002/hep.31684. online ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapkiewicz A.V., Mai X., Carsons S.E., et al. Megakaryocytes and platelet-fibrin thrombi characterize multi-organ thrombosis at autopsy in COVID-19: A case series. EClinicalMedicine. 2020;24:100434. doi: 10.1016/j.eclinm.2020.100434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y., Chen V.S., Zheng B., et al. A novel subset of putative stem/progenitor CD34+Oct-4+ cells is the major target for SARS coronavirus in human lung. J Exp Med. 2007;204:2529–2536. doi: 10.1084/jem.20070462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aid M., Busman-Sahay K., Vidal S.J., et al. Vascular disease and thrombosis in SARS-CoV-2-infected rhesus macaques. Cell. 2020;183:1354–1366. doi: 10.1016/j.cell.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ionescu F., Jaiyesimi I., Petrescu I., et al. Association of anticoagulation dose and survival in hospitalized COVID-19 patients: A retrospective propensity score weighted analysis. Eur J Haematol. 2021;106:165–174. doi: 10.1111/ejh.13533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissim O., Melis M., Diaz G., et al. Liver regeneration signature in hepatitis B virus (HBV)-associated acute liver failure identified by gene expression profiling. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049611. [DOI] [PMC free article] [PubMed] [Google Scholar]