Graphical abstract
Keywords: Covid-19, Pangenomics, sRNA transcriptomics, Proteomics, Drug repurposing, Molecular dynamics
Abstract
The recent pandemic Coronavirus disease-19 outbreak had traumatized global countries since its origin in late December 2019. Though the virus originated in China, it has spread rapidly across the world due its firmly established community transmission. To successfully tackle the spread and further infection, there needs a clear multidimensional understanding of the molecular mechanisms. Henceforth, 942 viral genome sequences were analysed to predict the core genomes crucial in virus life cycle. Additionally, 35 small interfering RNA transcripts were predicted that can target specifically the viral core proteins and reduce pathogenesis. The crystal structure of Covid-19 main protease-6LU7 was chosen as an attractive target due to the factors that there were fewer mutations and whose structure had significant identity to the annotated protein sequence of the core genome. Drug repurposing of both recruiting and non recruiting drugs was carried out through molecular docking procedures to recognize bitolterol as a good inhibitor of Covid-19 protease. The study was extended further to screen antiviral phytocompounds through quantitative structure activity relationship and molecular docking to identify davidigenin, from licorice as the best novel lead with good interactions and binding energy. The docking of the best compounds in all three categories was validated with molecular dynamics simulations which implied stable binding of the drug and lead molecule. Though the studies need clinical evaluations, the results are suggestive of curbing the pandemic.
1. Introduction
Coronaviruses are giant viruses with positive single stranded RNA genomes with characteristic crown like structures called spikes [2]. These viruses are widespread in animals and bats, but very few are reported to affect humans. By the end of December 2019, there was a cluster of pneumonia-like severe respiratory illnesses in Wuhan, China, and it is reported that a novel coronavirus is responsible [9]. The recent emergence of a novel coronavirus is named as SARS-CoV2 due to its similarities with the past pandemics of coronaviruses, Severe Acute Respiratory Syndrome (2002) and Middle East Respiratory Syndrome (2012) [24]. On March 11th 2020, the World Health Organisation (WHO) has declared the coronavirus disease (CoViD-19), a pandemic and as per the situation reports on June 10th, 2020, there stands 7, 145, 539 confirmed cases and 408, 025 deaths across the world (www.who.int.) The virus is airborne and the symptoms include fever, cough, gastrointestinal disorders, causes cytokine storm, and ultimately death. Spread of the virus is due human to human transmission and thereby community transmission at an alarmingly increased rate [8], [6].
Research is growing in leaps and bounds across the world to investigate effective interventions to considerably stop the spread of contagious novel coronavirus [53]. Currently studies including reverse vaccinology [18], deep learning networks in the diagnosis [8], are proposed. There are 2,251 studies under evaluation by clinical trials (clinicaltrials.gov/) including dozens of antiviral drugs like ramdesvir, and favipiravir [16](www.covid19treatmentguidelines.nih.gov) mRNA therapies [63], and diagnostics for CoViD-19. To deal with the difficult situation advancements of computational biology techniques, offer fast paced diagnostics and solutions [30] whereas a combination of genomics, transcriptomics, proteomics, and metabolomics approaches will join the dots to arrive at the key to curb the viral pathogenesis from all possible directions [25], [59]. Pan-genome analysis is a method to enumerate from the entire repertoire of genomes or coding sequences, the core genes that are shared by all the strains and accessory genes that are shared in the subset of genes. The core genomes summarily contain the majority of genes involved in viral life cycle and multiplication whereas dispensable or accessory genes are minimally essential to the viral pathogenesis [69]. Small interfering RNAs are exogenous, which have an external origin and enter the cell during an RNA viral infection [15]. They are recognised by dicer-drosha complex systems as exogenous nucleic acids, which are processed by the argonaute protein and initiate cleavage of the exogenous RNAs such as the viral genome or viral mRNA [34]. Hence the viral amplification and pathogenesis are restricted. As we already know molecular docking, quantitative structure activity relationships and molecular dynamics altogether offer an excellent source of drug screening and development. The decisive aspect of the computational approaches is dependent on the analysis of large datasets and arriving at an accurate conclusion [29].
Viral pathogenesis is multifaceted and hence the investigation adhered in the study would be the need of the hour. This study aims to identify the promising core genome fragments from the complete genome sequences of SARS-CoV-2 and small RNA transcripts that can target the core genes. A comprehensive analysis of conserved genes across multiple isolates proposes to reveal potential protein targets. The research as well focuses on drug repurposing of already available drugs using high throughput molecular screening protocols, in addition we intend to identify potent natural compound inhibitors of the chosen target protein. The present study has extensively evaluated the SARS-CoV2 geneome to curb and evade viral pandemic.
2. Methodology
2.1. Pan genome analysis
A total of 942 sequences representing all the countries were retrieved and pan genome analysis was performed using the Nucmer algorithm of the Spine webserver, and predicted the conserved core genome fragments along with the accessory genome fragments [68]. The top ten core genome fragments were clustered using clustage tool [47]. Genes coded by both core and accessory fragments were predicted based on the ab initio gene finding program FGENESH [51]. The motifs and domains of the core fragments were also identified from the pfam [21] database.
2.2. Identification of siRNAs to target the core genomes
Small Interfering RNAs are significant key factors in eliminating viral infections. The siRNAs with ~ 20 base pairs are exogenous and are proven to suppress viral pathogenesis [40]. The preliminary step was to identify the siRNAs already available in the database VirSiRNA [62] with the core fragments being the query sequences due to the fact that off target effects are reduced and strong antiviral activity [20]. From the resultant 196 hits of siRNA predicted for SARS virus, mutually efficient against SARS COV-2, only those with high silencing efficiency above the threshold of 85% were filtered. The guide strands and the 3′ UTR matching seed regions were predicted with high scores and matches above 4, respectively. Few additional siRNAs were modeled from the core genomes using InvivoGen's siRNA wizard software. Understanding virus host interaction is the most significant feature in any viral pathogenesis [10] and so predicting the host genes targeted by the viral core genes. Host - SARS CoV2 viral interactions were identified from the literature [23] and a network plot of interacting siRNAs, viral targets, and human interactions were constructed using cytoscape.
2.3. Compounds selection and optimisation
A total of 18,000 recruiting, non-recruiting, and FDA approved drugs were retrieved from the website clinicaltrials.gov. One thousand (1,000) antiviral natural phytocompound libraries were identified and retreived from Traditional Knowledge Digital library (http://www.tkdl.res.in) and Indian Medicinal Plant Phtochemicals And Therapeutics database (https://cb.imsc.res.in/imppat/home). All the retrieved 3-D compounds were prepared and optimized with the ligprep module using OPLS 2005 force field. Stereoisomers were limited to 10 per ligand structure and only the ligand with the least energy was taken further [50], [17].
2.4. Prediction of ADMET properties
The Pharmacokinetic properties of the natural compound library including the Absorption, Distribution, Metabolism, Elimination, and Toxicity (ADMET) parameters were predicted only for the natural compounds using the qikprop module, Schrodinger [70]. Pharmacokinetic properties such as atom-based LogP, Molecular weight, membrane permeability, intestinal absorption, skin permeability levels, P-glycoprotein substrate or inhibitor are significant in determining the absorption levels of drugs. The distribution of drugs depend on the blood–brain barrier descriptors. Drugs acting as substrates or inhibitors of Cytochrome P450 enzymes represent their metabolism ability whereas toxicity can be predicted based on AMES toxicity, hERG inhibition and LD50 parameters [27]. These properties were predicted computationally to deduce the druggability of natural compounds.
2.5. Quantitative structure activity relationship (QSAR) using python
QSAR based virtual screening predicts a large set of active molecules with the desired activity. QSAR coupled with machine learning methods are the latest reliable strategies in screening the promising lead compounds to be used in drug development [31]. The pandas, numpy, scipy and sklearn modules of python are used to calculate simple linear regression of the independent descriptor set [36]. This method is more useful in understanding the structure activity relationships. Feature selection was done for the predicted variables by removing the null values and the insignificant variables less than the threshold of 0.1% [38]. The molecules that adhered to the Lipinski’s rule of five were filtered to create training and test sets. Actual and predicted values were computed and a plot of linear regression was drawn to filter out the best molecules [49], [65].
2.6. Target identification
The target proteins of Covid-19 were identified based on the ‘bottom up approach’ [64] and compared against the core translated proteins. Protein Data bank has an extensive collection of 3 dimensional protein structures and an appropriate Covid-19 target was chosen based on the criteria: 1) availability of X-ray crystal structure complexed with an inhibitor, 2) a significant match with the core translated protein fragment in pfam and 3) significant match with the Protein Data bank structures and 4) a good resolution of structure preferably less than 3.0 A0 [13]. The target that obeyed the above criteria was found to be the X-Ray crystallographic structure of main protease of SARS CoV-2 (PDB ID-6LU7) with 2.6 A0 resolution which had a complex inhibitor, and matched with 97% identity to translated core genome sequence [57]. This target is one of the vastly studied protein targets is the SARS-CoV2 main protease which is also called MPRO and 3CLPro which is essential for processing of polyproteins from viral mRNA [32], [19]. Though there are enormous silico studies performed with main protease as target, the same protein is again taken as target because it is the predominant core gene obtained from pangenomics and has lots of significance in the viral pathogenesiss [26], [22]. In the present study, we concentrated on extensive docking analysis of existing FDA drugs and carefully restricted our analysis to inhibitor compounds that were not reported yet.
2.7. Target preparation and binding site prediction
Protein crystal structure- the Main protease of covid-19 in complex with the peptide like inhibitor N3 (6LU7) was retrieved and viewed in the maestro window of the Glide module of Schrodinger drug design suite 2020–1 [1], [41]. Using the protein preparation wizard, water molecules of the protein were removed, hydrogen atoms were assigned, and the protein structure was energy minimized and optimised using OPLS (Optimised Potential for Liquid Simulations) force field [14]. Ramachandran plot was constructed for the minimized energy structure. The binding pockets of the proteins were predicted using the site map module and compared with the binding pocket of the original ligand. The receptor grid was generated for the active site pocket that was mutual to the sitemap prediction and crystallographic binding pocket to enable efficient docking of ligands and drugs [11].
2.8. Highthroughput virtual screening
The processed and optimised drugs and natural compounds were screened as separate groups through high throughput virtual screening (HTVS) against the active site of the prepared Covid main protease. The screened ligands in their groups were docked using the Standard Precision (SP) mode of induced fit docking against the target protein [52]. This step filtered out the ligands that had good interactions with the active site and were still in separate groups. Finally, the groups of ligands were docked using the eXtra Precision (XP) mode to narrow down the analysis to result in better lead compounds [7].
2.9. Molecular dynamics simulation of the target and lead molecules
Molecular dynamics simulations of the docked ligand and protein complex files were performed using Gromacs 2020. The top best XP docked complex output file from each group was prepared with the PDB reader and ligand reader and modeler module of CHARMM GUI server and the resultant trajectory and topology files were processed with gromacs [54], [58]. The parametrization was done with charmm-36 force fields and the processed complex files structure was solvated in a cubic box using the model TIPS3 (Transferable Intermolecular potential water molecules) [44]. Modified Berendsen thermostat and Parrinello-Rahman barostat were used for temperature and pressure coupling, respectively [35]. Steepest descent energy minimization was performed, followed by two steps of equilibration ensembles - NVT (Number of molecules, volume and temperature) and NPT (number of molecules, pressure and temperature). MD simulation was done at 100 ns at 5000 steps for each equilibration ensemble [37]. Long range forces and van der Waals interactions were treated with Particle Mesh Ewald summation and Lennard Jones potential with a cut-off distance of 10 A0 for non bonded interactions. A plot of RMSD per residue was constructed to confirm the binding of the ligands [55].
2.10. Binding free energy calculation with MMPBSA methods
The binding energy of the protein and ligand complexes were calculated by the molecular Mechanic/Poisson-Boltzmann Surface Area (MM-PBSA) method which constitutes the potential energy, polar and non polar solvation energies [45].
| ΔGbind,aq = ΔH − TΔS≈ΔEMM + ΔGbind,solv − TΔS, | (1) |
| ΔEMM = ΔEcovalent + ΔEelectrostatic + ΔEvdW, | (2) |
| ΔEcovalent = ΔEbond + ΔEangle + ΔEtorsion, | (3) |
| ΔGbind,solv = ΔGpolar + ΔGnon-polar, | (4) |
The changes, ΔEMM (the gas-phase molecular mechanical energy change), ΔGbind, solv, (the solvation free energy change) and – TΔS (conformational entropy change upon binding) are computed via ensemble averages [67]. ΔEMM is the combination of molecular mechanics (MM): the covalent energy change (ΔEcovalent), the electrostatic energy change (ΔEelectrostatic), and the van der Waals energy change (ΔEvdW). The bond terms (ΔEbond), the angle terms (ΔEangle), and the torsion terms (Δetorsion) are represented as ΔEcovalent. The solvation free energy change (ΔGbind, solv) includes polar and non-polar contributions (ΔGpolar and ΔGnon-polar). The most difficult parameter to compute is entropy term which approximated with a normal mode method using a few selected snapshots taken from MD simulations. The binding affinity calculation of all the ensemble averages in equations 1–4 were calculated using MMPBSA methods [60]. The MD scripts at last 10 ns were extracted and MM-PBSA binding energy was calculated using the g_mmpbsa module of GROMACS to estimate the interaction of the two best docked complexes with the target [61].
3. Results
3.1. Pan genome analysis of CoViD-19
Pangenome analysis of 942 complete genome sequences using the spine webserver revealed 35 genome fragments that were highly conserved throughout the isolates and around 5–10 accessory sequences specific to each sequence [12]. We filtered out 10 core genome fragments that had identifiable motifs and clustered them as a clustage plot of the mutual core fragments and represented in Fig. 1 . The coding sequences of core fragments were identified and translated into protein sequences. The motifs and domains of the protein sequences of each core were predicted and represented in Supplementary Table 1.
Fig. 1.
Core genome fragments clustered with clustage tool.
3.2. Identification of silencing RNA to target the core fragments
Small RNAs are of great help in specific gene silencing with the recognition of complementary DNA or RNA targets. The small interfering RNAs with high inhibition efficiency of >85% and an estimated binding energy of less than 34 Kcal/mol [5] were filtered from the ViSiRNA database and represented in Table 1 . The viral siRNAs, targeting the viral core genes which interact with the human proteins are plotted and mapped with cytoscape and are represented as an interaction network of siRNAs in Fig. 2 . In the figure the viral siRNAs mutual for SARS CoV and SARS CoV-2 are colored in pink whereas the predicted siRNAs of SARS CoV2 core genome are colored in orange. The viral targets are colored in blue and the interacting host genes in green. Few off-target sites of host genes like GPC5, PDE4D, ILIRAPL2, NSPAS3, Zinc finger, and FBOX were also predicted for the siRNA though are very negligible. The most dominant and significant interactions of the clusters were predicted using MCode module of cytoscape and is represented in Fig. 2B.
Table 1.
Silencing RNAs of CoViD-19.
| S. No | siRNA | Target | Sequence | Inhibition efficacy | GC% |
|---|---|---|---|---|---|
| 1 | virsi2257 | ORF5, M | AACCUAGUAAUAGGUUUCCUA | 91 | 33 |
| 2 | virsi1323 | N | GAACAAACCCAAGGAAAUU | 83 | 37 |
| 3 | virsi1304 | N | GAACAAACCCAAGGAAAUU | 94 | 37 |
| 4 | virsi1374 | N | UCAAGGAACAACAUUGCCAAA | 51 | 38 |
| 5 | virsi1382 | N | AAUAAUACUGCGUCUUGGUUC | 80 | 38 |
| 6 | virsi2267 | ORF9a, N-Protein | AAAUUGGCUACUACCGAAGAG | 91 | 43 |
| 7 | virsi1330 | Pol | UAUGACUAUGUCAUAUUCA | 80 | 26 |
| 8 | virsi2233 | ORF1b, NSP-13 | AAGGUGACUAUGGUGAUGCUG | 92 | 48 |
| 9 | virsi1351 | S | AUCUGUUCUCUAAACGAAC | 93.4 | 35 |
| 10 | virsi2241 | orf1, spike | AACCUUACAGAGUUGUAGUAC | 91 | 32 |
| 11 | VIRSI2237 | ORF2, Spike | AAGCUCCUAAUUACACUCAAC | 91.2 | 35 |
| 12 | VIRSI1584 | 3CL Protease | GGAUGAAGAAGAUGGCCAU | 91 | 40 |
| 13 | VIRSI1696 | E | CGGAGACAUCAGACAACUA | 90 | |
| 14 | VIRSI1611 | NS1 | GGAUGAAGAAGAUGGCCAU | 88 | 40 |
| 15 | SiCoV2-1 | Orf 1a | GUUUAUACCUUCCCAGGUA | 93.72 | 42.8 |
| 16 | SiCoV2-2 | Orf 1a | GCACUAGUACUGAUGUCGUAU | 93.36 | 52.38 |
| 17 | SiCoV2-3 | Orf 1a | GGACAACAGGGCAACCUUACA | 92.98 | 47.62 |
| 18 | SiCoV2-4 | N | GACAACAGGGCAACCUUACAA | 92.85 | 42.8 |
| 19 | SiCoV2-5 | S | AGGCUGUUGCUAAUGGUGAUU | 92.85 | 47.62 |
| 20 | SiCoV2-6 | E | GGCUGUUGCUAAUGGUGAUUC | 92.74 | 42.86 |
| 21 | SiCoV2-7 | Orf1a | AGAUGGCUGAUCAAGCUAUGA | 92.42 | 38.1 |
| 22 | SiCoV2-8 | Orf 1b | AGUUGGAUAAUGAUGCACUCA | 92.28 | 38.1 |
| 23 | SiCoV2-9 | Nsp5 | GUUGGAUAAUGAUGCACUCAA | 92.1 | 33.33 |
| 24 | SiCoV2-10 | E | GAACAUAAUACCUCUUACAAC | 91.63 | 38.1 |
| 25 | SiCoV2-11 | S | GUUGUCAUACCAGACUAUAAC | 91.62 | 42.86 |
| 26 | SiCoV2-12 | M | GUUGACAUACUAGGACCUCUU | 91.31 | 38.1 |
| 27 | SiCoV2-13 | M | GGUUCUGUUGCUUAUGAAAGU | 91.41 | 47.62 |
| 28 | SiCoV2-14 | Orf1a | GUGCUCAUGGAUGGCUCUAUU | 90.69 | 41.86 |
| 29 | SiCoV2-15 | Orf 1b | GAAGGUUCUGUUAGAGUGGUA | 90.63 | 52.38 |
| 30 | SiCoV2-16 | Orf 1b | GGCACGGCACUUGUGAAAGAU | 90.28 | 33.33 |
| 31 | SiCoV2-17 | Orf 1ab | GAUGGGUACUUAACAAUGAUU | 90.2 | 33.33 |
| 32 | SiCoV2-18 | Orf 1b | GACAUAUCAGCAUCUAUAGUA | 90.04 | 38.1 |
| 33 | SiCoV2-19 | N | GGUAUUGUAGCUAUCGUAGUA | 90.3 | 37 |
| 34 | SiCoV2-20 | M | GAGCAACAAGAGUCGAAUGUA | 90 | 42.86 |
Fig. 2.
A(Left) Complete interactions of siRNAs with the viral targets and the human guide strands. SiRNAs of SARS-CoV are represented in pink color and siRNAs of SARS-Cov2 are shown in Orange colour. The human targets are shown in green and the viral targets in blue. Fig. 2B (right) shows the significant interactions of the clusters with SiRNAs of SARS-CoV are represented in green color and siRNAs of SARS-Cov2 are shown in Orange colour with viral targets in cyan. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. ADMET properties
Two sets of compounds including 18,000 FDA approved drugs and 1,000 natural phytocompounds were downloaded from pubchem zinc, drugbank and were added hydrogens and energy minimised. The structures were optimised with OPLS 2005 force field and considered suitable for further investigations. The natural compounds were screened based on the Absorption Distribution Metabolism Excretion Toxicity (ADMET) properties calculated from the qikprop. ADMET properties of the selected phytocompounds are shown in table 2 . The compounds davidigenin, 7-hydroxy 2(4-hydroxyphenyl)chroman-4-one, agarol, Eugenin exhibited high gastro intestinal (GI) absorption, solubility and tested negative for Ames test for mutagenicity. Further these compounds ahered to the Lipinski’s rule of five and attributed to promising drug likliness.
Table 2.
ADMET properties of the selected natural phytocompounds.
| S.No | Compound | Molecular weight g/mol | Log P | HBond Donor | HBond acceptor | GI absorption | Solubility | LD 50 | Ames test for mutagenicity |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Davidigenin | 258.27 | 2.62 | 3 | 4 | high | soluble | 2.114 | negative |
| 2 | 7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | 256.25 | 2.48 | 2 | 4 | high | soluble | 2.213 | negative |
| 3 | Agarol | 248.32 | 2.98 | 1 | 3 | high | soluble | 2.536 | negative |
| 4 | Eugenin | 470 | 3.35 | 2 | 6 | high | soluble | 4.03 | negative |
3.4. Quantitative structure activity relationship
A dataset with properties was created using python 3.2, cleaned from all the null values. They were grouped into a test set and a training set predicted with 20% and 80% data respectively [48]. Fig. 3 A shows a scatter plot with molecular weight and partition coefficient (LogP) plotted with a sklearn module of python. The feature selection criteria called Lipinski’s rule of five was applied. The data which had a molecular weight less than 500 Da, with hydrogen bond donors less than 5, hydrogen bond acceptors less than 10 and partition coefficient (log P) less than 5 were filtered for further analysis. A regression fit and coefficient of regression were computed for the filtered compounds and a plot representing predicted vs actual values is shown in the Fig. 3B. Calculated mean absolute error was 1.4124317746614325 and calculated root mean square error was found to be 1.800150000427347. Hence compounds were chosen for docking studies.
Fig. 3.
A (left) Scatter plot before feature based selection 3B (right)Regression line plotted for the predicted and actual values of the phytocompounds based on their ADMET predictions.
3.5. Protein target identification and preparation
The target protein chosen for further studies was the crystal structure of CoViD-19 main protease in complex with an inhibitor N3 (6LU7). The energy of the target was −1415.91 Kcal/mol after hydrogen bond optimization, removal of water molecules, minimised and processed using OPLS 2005 force field [33]. Fig. 4 is a representation of Ramachandran plot constructed after the energy minimization step (Left Fig. 4A) and the site map module predicted six binding pockets of the prepared protein (Right Fig. 4B). Binding site 1 had high score of 0.759 and volume of 247.30 with the residues 3 Val, 4 Leu, 26 Thr, 27 Leu, 41 His, 45 Thr, 46 Ser, 49 Met, 140 Phe, 141 Leu, 142 Asn, 143 Gly, 144 Ser, 145 Cys, Asn151, Ser 158, 164 His, 165 Met, 166 Glu, 192 Glu containing the co-crystallized inhibitor hence receptor grid was generated for the site 1 and was set for molecular docking. The other sitemaps had comparatively less volume and scores and were eliminated from docking
Fig. 4.
A Ramachandran plot of energy minimised M Pro target and 4B the binding site residues predicted from sitemap.
3.6. Molecular docking analysis of the original ligand N3
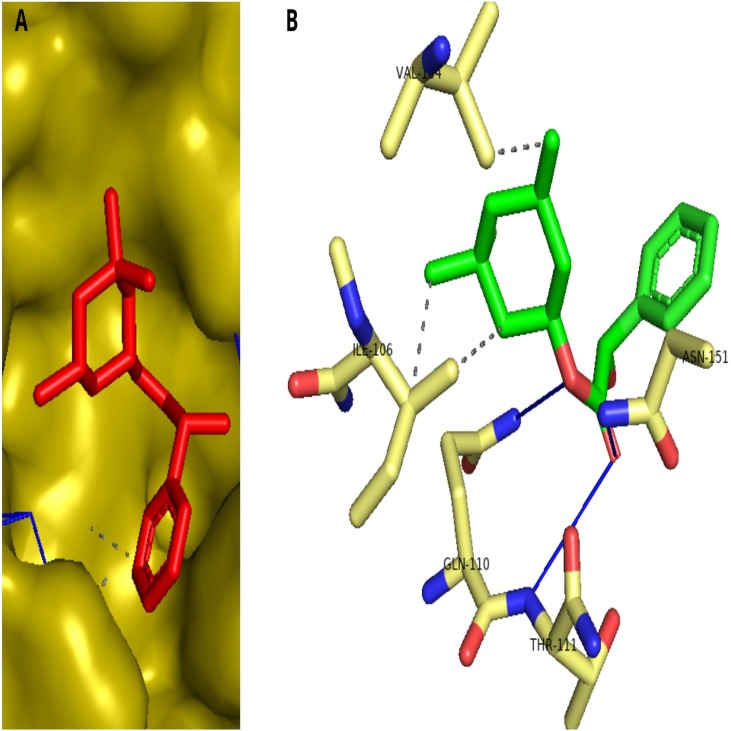
Molecular docking was performed using the Glide module of Schrodinger for the optimized original ligand of the target using XP mode. Table 3 represents the glide energy score and interactions of all the best docked inhibitors of Covid-19 main Protease. The binding energy obtained was −82.29 Kcal/mol and was found to interact with the binding site residues of Gly 143, and His 164, with good hydrogen bond distances of 2.80A0 and 3.07A0, hydrophobic bonds with Thr25 and Thr 26 at a distance of 3.73A0 and 3.81A0, respectively. The docked co-crystallized ligand and the interaction of co-crystallized ligand with binding sites are represented in Fig. 5 A Dotted representation of the ligand with target, 5B- docked ligand complex and 5C interaction of active site residues
Table 3.
Glide XP docking results of drug and phytocompounds.
| S. No | Compound | Nature of drug/Plant source | Glide Energy | Glide Score | H Bond |
Hydrophobic Bond |
||
|---|---|---|---|---|---|---|---|---|
| Residues | Distance A0 | Residues | Distance A0 | |||||
| 1 | Ligand-N3 | Peptide like Inhibitor N3 | −82.229 | −7.784 | (HO…O)GLY143 (HO…O)HIS164 |
2.80 3.07 |
THR 25 THR 26 |
3.73 3.81 |
| Drugs | ||||||||
| 2 | Bitolterol | Bronchodilator | −84.529 | −6.967 | LYS102(NH…O) ARG105(NH…O) (O…OH)ARG105 (NH…O)ASN151 (OH…O)SER 158 |
3.25 3.04 2.74 3.71 2.95 |
PHE 294 PHE 294 |
3.61 3.49 |
| 3 | Cyclandelate | Vasodilator | −83.966 | −6.741 | (NH…O) GLN110 (NH…O) THR 111 (NH…O)ASN151 |
3.02 3.20 3.51 |
VAL 104 ILE 106 ILE 106 |
3.51 3.83 3.81 |
| 4 | Atropine | Nerve agent |
−81.63 | −6.123 | (NH…O)GLN110 (NH…O) THR 111 THR 111 (OH…O) |
2.29 2.14 2.19 |
VAL 104 ILE106 GLN110 |
3.77 3.72 3.72 |
| 5 | Alosetron | Irritable Bowel syndrome | −80.892 | −5.746 | ARG105(NH…O) (NH…N) GLN 107 ASN151 (NH…O) 158 SER (HO…O) |
2.51 3.68 3.35 3.25 |
ILE106 GLN110 |
3.81 3.70 |
| 6 | Cloperastine | Antitussive and antihistamine | −80.294 | −3.673 | Gly143 (HO…O) | 3.37 | – | |
| Phytocompounds | ||||||||
| 7 | Davidigenin | Glycyrrhiza Glabra | −88.785 | −7.57 | (OH…O) LYS102 GLN110 (OH…O) (OH…O)ASP153 (OH…O)SER 158 SER 158 (OH…O) |
3.12 2.88 2.17 2.11 2.19 |
ASN 151 ASP 153 |
3.77 3.89 |
| 8 | Agarol | Aquilaria agallocha | −86.34 | −6.78 | (OH…0)SER 158 | 2.28 | ILE 106 ASN 151 PHE 294 |
3.79 3.92 3.45 |
| 9 | Eugenin | Pisonia aculeata | −86.22 | −7.32 | GLN110 (NH…O) THR 111 (NH…O) (OH…O)THR 111 (OH…O)ASN151 ASN151 (NH…O) |
2.94 3.21 2.80 3.42 3.93 |
– | |
| 10 | 7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one | Glycyrrhiza Glabra | −80.86 | −6.31 | (HO…O) Glu166 Leu 24 (HO…O) |
2.43 3.12 |
– | |
Fig. 5.
(A) Ligand N3 in complex with target in dotted representation; 5(B) Ligand N3 in the active site of Covid-19 protease; 5(C) Interactions of Ligand N3 with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.7. Drug repurposing through molecular docking of FDA approved generic drugs
A total of 18,000 drug compounds were optimized with the OPLS2005 force field and were screened using high throughput virtual screening (HTVS) against the target. There were 2000 compounds that passed HTVS and were subsequently screened using SP mode, and the resulting compounds of SP were docked in XP mode and the final 50 compounds were filtered. Among them, the top five drugs were filtered based on energy less than 80Kcal/mol to aid comparison with the co crystallized ligand. Bitolterol, cyclandelate, atropine, alosetron, and cloperastine exhibited lesser energy and good interaction with the binding sites of the target. The top lead bitolterol docked into the protein active site, and residues interacting are represented in Fig. 6 A(left) and 6B(right). Bitolterol showed an energy of −84.529KCal/mol with H bonds at a distance of 3.25A0, 3.04A0, 2.74A0, 3.71A0 and 2.95A0 with the residues Lys102, Arg 105, Arg105, Asn151 and Ser158 respectively. The drug also exhibited hydrophobic interactions of 3.61A0 and 3.49 A0 with Phe 294. Cyclandelate was the next drug that showed an energy of −83.96 KCal/mol with H Bond interactions with the residues of Gln110, Thr111 and Asn151 at a distance of 3.02A0, 3.20A0 and 3.51A0 respectively shown in Fig. 7 A and B. Hydrophobic interactions of Val 104 at 3.51A0, Ile106 at 3.83A0 and 3.81A0 were also found. From Fig. 8 A,B Atropine which is a nerve agent exhibited an energy of −81.63KCal/mol with Hbonds at the residues Gln110, Thr111, with a distance of 2.29A0, 2.14A0 and 2.19A0 and hydrophobic interactions of 3.74A0 with Val104, 3.72A0 with Ile106 and 3.72A0 with Gln110. Alosetron an irritable bowel syndrome drug showed an energy of −80.892 and four H Bonds with Arg105, Gln107, Asn151 and Ser158 at distances of 2.51A0, 3.68A0, 3.35A0 and 3.25A0 respectively.Hydrophobic interactions of 3.81 with Ile106 and 3.70 with Gln110 was also observed and is represented in the Fig. 9 A,B. Finally the antitussive drug cloperastine showed an energy of −80.294 with only one hydrogen bond with Gly143 at 3.37A0.
Fig. 6.
(A) Bitolterol in the active site of Covid-19 protease; 6(B) Interactions of Bitolterol with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 7.
(A) Cyclandelate in the active site of Covid-19 protease; 7(B) Interactions of cyclandelate with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 8.
.(A) Atropine in the active site of Covid-19 protease; 8(B) Interactions of atropine with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 9.
(A) Alosetron in the active site of Covid-19 protease; 9(B) Interactions of alosetron with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.8. Highthroughput virtual screening of natural phytocompounds
Molecular docking in the XP mode of the phytocompounds [46] revealed 4 prominent compounds Davidigenin, Agarol, Eugenin and 7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one, with energy values of −88.78 Kcal/mol, −86.34Kcal/mol, −86.22Kcal/mol, and −80.86Kcal/mol respectively. Fig. 10 A,B show Davidigenin(C15H14O4), a hydrogenated metabolite of liquiritigenin (C15H12O4) found in licorice roots [43] showed good interactions with Lys102, Gln110, Asp153, Ser158 showing a distance of 3.12A0, 2.88A0, 2.17A0, 2.11A0 and 2.19A0 respectively along with the hydrophobic bonds at Asn151 and Asp153 at 3.77 and 3.89 respectively. The phytocompound agarol from Aquilaria agallocha represented in Fig. 11 A, B showed a hydrogen bond at Ser158 with 2.28A0 and hydrophobic interactions at Ile106, Asn151 and Phe294 at 3.79A0, 3.92A0 and 3.45A0 respectively. From Fig. 12 A,B, a phytocompound Eugenin of Pisonia aculeata exhibited hydrogen bonds with residues Gln110, Thr111, and Asn151 at distances of 2.94A0, 3.21A0, 2.80A0, 3.42A0 and 3.93A0. One more compound from licorice roots, 7-hydroxy-2-(4-hydroxyphenyl)chroman-4-one with Cys145 and Gly143 showed up in the top list. Other compounds though passed SD screening, did not yield satisfactory energy and interactions.
Fig. 10.
(A) Davidigenin in the active site of Covid-19 protease; 10(B) Interactions of Davidigenin with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 11.
(A) Agarol in the active site of Covid-19 protease; 11(B) Interactions of agarol with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines and hydrophobic interactions in dashed grey lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 12.
(A) Eugenin in the active site of Covid-19 protease; 12(B) Interactions of eugenin with active site residues of covid 19 main protease. Hydrogen bonds are represented in solid blue lines. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Davidigenin also followed Lipinski’s rule of 5. The molecular weight and its lipophilicity of davidigenin were 258.27 g/mol and 1.97, the Hydrogen bond acceptors were 4, and donors were 3 and had a very high absorption rate of 80%. ADMET studies of davidigenin proved that it is an efficient lead compound and is exhibiting drug likeness.
3.9. Molecular dynamics simulations
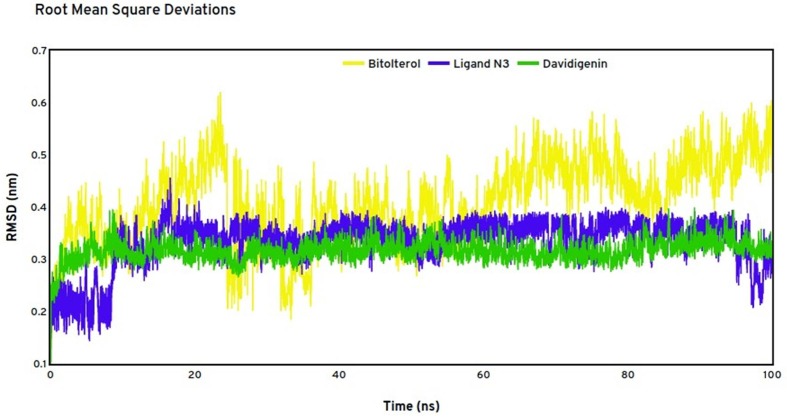
The dynamics and stability of the docked complexes are evident only from the molecular dynamics simulations. The conformational changes in the biological environment of the protein upon ligand binding is identified by simulating the top drug and the natural compound with the target using the Charmm 36 force fields. Structural perturbations and Root mean square deviations of the backbone atom of target protein with ligand N3, drug bitolterol and the natural compound davidigenin when bound with the target MPro is represented in the Fig. 13 . The average deviations of the target protein with the ligand N3 was identified as 0.35 nm. The ligand was very stable throughout the simulation time of 100 ns. The average deviation of drug bitolterol was found to be 0.5 nm. With exception to the maximum deviations of 0.6 nm around 20, 70 and 90 ns, the drug was stable. The natural phytocompound davidigenin was found to be the most stable ligand with the protein and shows an average deviation of 0.3 nm. There were no higher deviations found in the trajectory and hence the compound davidigenin is found to have strong binding affinity to the target M protease. The negligible deviations of the bitolterol-protease complex is significant and states that the drug is more stable with the target and is close to be compared with the experimental results.
Fig. 13.
RMSD calculations of protease 6LU7 in complex Bitolterol in yellow, ligand N3 in blue and davidigenin in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.10. Hydrogen bond analysis
Hydrogen Bonds are the predominant elements responsible for the stability and interactions of the ligands and target. We estimated the number of hydrogen bond interactions that were formed during the simulation of 100 ns and are represented in the Fig. 14 . The original ligand N3, exhibited an average of 3 hydrogen bonds with a minimum of 0 and maximum of 8 bonds. The ligand formed a maximum of 8 hydrogen bonds around 93 ns. Bitolterol was found to be stable with an average of 2 hydrogen bonds with a minimum of 1 and a maximum of 5. At 5 ns and 97 ns, there were 5 hydrogen bond interactions observed between bitolterol and protease of SARS-CoV2. The average of H bonds was identified to be 5 in case of davidigenin binding with protease. The minimum was 1 and the maximum was 9. The higher numbers of H bonds observed upon davidigenin binding is an indication of the significant stability [4], [66]. On comparing with the original ligand N3, davidigenin and bitolterol formed better hydrogen bonds with the target and are significant. The results signify the prominent interactions of the drug bitolterol and davidigenin throughout the simulation period of 100 ns.
Fig. 14.
Hydrogen Bond analysis of Bitolterol in yellow, ligand N3 in blue and davidigenin in green. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.11. Root mean square fluctuations
Root mean square fluctuations of the individual target and in complex with the drug bitolterol and natural compound davidigenin revealed their flexibility. From the Fig. 15 , it is observed that both bitolterol and davidigenin complex are stable compared to the receptor. The overall RMSF of both is less than 1A0 which represents the stability of the receptor upon ligands binding. The fluctuations were very low and are less than the standard cut off of 3A0 and proves their stability throughout the run.
Fig. 15.
Root mean square fluctuations of Protease receptor (green), Bitolterol (red) and Davidigenin (blue). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.12. Binding free energy calculations
The average free binding energy calculations were performed for the two systems bitolterol and davidigenin to estimate their decomposition energies with standard deviation and errors. A total of five trajectories at every 20 ns were extracted to perform MMPBSA with GROMACS [60]. The binding free energy (ΔG) is composed of the binding energy of the receptor in its bound state and unbound state. Table 4 represents the calculated average free binding energy of the bitolterol and davidigenin complexes. Total binding free energy for bitolterol complexed with protease is observed to be −108.904 ± 22.011 and that of davidigenin is −134.584 ± 31.916. Per energy contribution plot is represented in the Fig. 16 which depicts the decomposition energies of the protease residues upon the ligand binding. The binding site residues like Arg105, Gly145, Ser158, His164 and Gln192 showed least energy contributions of −9.4KJ/mol, −8.9KJ/mol, −10.1KJ/mol, −4.1KJ/mol and −5.6KJ/mol respectively upon binding of bitolterol with the protein. The original ligand inhibitor N3 showed least energies at the residues Leu4 and THR25 with energies −4.3KJ/mol and −14.9KJ/mol respectively. The complex davidigenin significantly had least binding energies of −13.2KJ/mol , −13.9KJ/mol , −11.2KJ/mol, −15.1KJ/mol and −6.3KJ/mol at the residues Lys 102, Gln110, Asp 153, Ser 158 and His 164 respectively. From the binding free energy calculations it is inferred that the drug bitolterol and the compound daviigenin show promising results and stable binding with the active site residues of the target.
Table 4.
Binding free energy calculations of top lead compounds.
| Complex | ΔGbinding (kJ/mol) | SASA (kJ/mol) | ΔEpolar solvation (kJ/mol) | ΔEElectrostatic (kJ/mol) | ΔEVan der Waal (kJ/mol) |
|---|---|---|---|---|---|
| Ligand | −75.895 ± 56.436 | −15.343 ± 5.760 | −23.652 ± 20.12 | 17.553 ± 0.675 | −54.453 ± 31.231 |
| Bitolterol | −108.904 ± 22.011 | −17.231 ± 4.208 | −38.248 ± 8.01 | 13.441 ± 0.837 | −66.866 ± 17.553 |
| Davidigenin | −134.584 ± 31.916 | −10.791 ± 3.787 | −62.696 ± 43.497 | 16.408 ± 2.084 | −77.505 ± 30.658 |
Fig. 16.
Per residue binding free energy contribution plot of the ligand N3 (blue), bitolterol (red) and davidigenin(green). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Viruses integrate into the host, replicate, spread at a faster rate, and build up mutations. With the advent of recent sequencing technologies, it is possible that the nature of viral pathogenesis is estimated. Pangenome analysis is a strong evidence that the core genome of the virus is maintained across strains and is available as perfect targets. The study identified ten core fragments of coronavirus-2 from 942 complete genome sequences of the same. Though the virus sequences acquired mutations and have spread across the globe and caused innumerable deaths, there are few conserved regions called core genes that are mutual to all the different isolates and are identified as Core fragments NSP1, hypothetical protein, NSP3, single stranded poly A binding domain, C3 endo peptidase, NS7 and NS8 replicase, NSP11, RNA polymerase N terminal domain, NSP16, Matrix and S2 glycoprotein and spike receptor binding protein and X4 like protein of SARS. Thirty four Small RNA transcripts with high inhibition efficacy were identified to specifically target the viral genes of core regions such as Nucleocapsid, Memebrane glycoprotein, NSP3, Spike glycoprotein [42], Envelope protein and 3CL Protease with guide strands being predominantly GPC5, PDE4D, ILIRAPL2, NSPAS3, CTNNAI, Zinc finger and FBOX. Covi-19 main protease protein was a chosen target since the sequence of it remained the same all through the isolates. Drug repurposing of FDA approved drugs and antivirals through molecular docking predicted 13 small molecules with good interaction and binding energy to be able to inhibit the target. Bitolterol, a bronchodilator, topped the results of docking and dynamics and turned out to be a good treatment option and needed clinical research. Quantitative structure activity relationships studies enabled identification of potential phytocompounds that are generally less toxic and are most widely formulated drugs [28]. A phytocompound davidigenin metabolite of licorice roots [56], also docked against the target well and resulted in a better lead molecule to target SARS-CoV2. Both the compounds bitolterol and davidigenin upon molecular dynamics simulations exhibited strong stability with the target protease. Bitolterol, with an RMSD 0.4 nm, with hydrogen bonds of 5 and root mean square fluctuations of 0.2 A0, proves its stability with the protease of Covid-19. The phytocompound davidigenin exhibited an RMSD of 0.3, maximum of 9 hydrogen bonds and RMSF of 0.2 A0, signifying its stability with the active site residues of target. MMPBSA results also show the preferential least binding energy values of both the drug and phytocompound to be taken to further research in the design of attractive CoViD-19 suppressors. The results throughout were compared with the co crystallised ligand N3 and the better leads were identified as davidigenin, a natural compound and the existing drug bitolterol can be repurposed to treat covid-19. The current study proves to target the SARS-CoV2 pandemic efficiently with not just a single approach but with multiple efforts simultaneously.
5. Conclusion
Severe Acute Respiratory Syndrome –Coronavirus 2 is creating havoc all over the world, though there are claims with proven drug efficacy and vaccines in trials, no prominent cure has been made so far [3]. The pandemic has not only blown down the health and survival of millions of people but also has stalked the economic situations of nations [39]. Intrusions at multiple levels will be beneficial and the current study has engaged a novel approach to tackle the pandemic and halt down its spread. From the study, we conclude that the core genes identified are significant in understanding the viral pathogenesis [63]. Small interfering RNAs of corona viruses can intrude the viral targets and can be used in therapeutics and diagnosis as well. Through molecular docking, ADMET predictions and dynamics approaches we have enumerated both repurposable drugs and natural phytocompounds that can be used as therapeutics upon clinical research. Thus, bioinformatics research will be more subtle in bringing out an eminent solution. The main Protease enzyme of the SARS-CoV 2 is a strong, conserved and viable target which can be efficiently targeted by atropine, bitolterol, and also by a natural compound davidigenin. The studies show promising features that can be taken further for invitro and clinical evaluations.
6. Declaration
Funding
The project did not receive any funding from government or non governmental organisations.
6.2. Data availability
All the data used in the work are publicly available in public databases like NCBI, PDB, Drug bank, and Pubchem.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
The authors are obligated to the management of Stella Maris College Chennai, Tamil Nadu, India. The facility provided by seed research grant is immensely accredited. The Schrodinger software used in the study was procured from the grant of Department of Science and Technology (DST), New Delhi, India through FIST Programme (Level -0, No: SR/FIST/College-252-C.Dy.No.3555/IFD/2016-2017), to Stella Maris College, Chennai, India. DST-FIST grant is greatly acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comtox.2021.100156.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Aarthy M., Panwar U., Selvaraj C., Singh S.K. Advantages of structure-based drug design approaches in neurological disorders. Curr. Neuropharmacol. 2017;15:1136–1155. doi: 10.2174/1570159x15666170102145257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adhikari S.P., Meng S., Wu Y.-J., Mao Y.-P., Ye R.-X., Wang Q.-Z., Sun C., Sylvia S., Rozelle S., Raat H., Zhou H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect. Dis. Poverty. 2020;9(1) doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aouidate A., Ghaleb A., Chtita S., et al. Identification of a novel dual-target scaffold for 3CLpro and RdRp proteins of SARS-CoV-2 using 3D-similarity search, molecular docking, molecular dynamics and ADMET evaluation. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1779130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano T., Ishihara K., Morota T., Takeda S., Aburada M. Permeability of the flavonoids liquiritigenin and its glycosides in licorice roots and davidigenin, a hydrogenated metabolite of liquiritigenin, using human intestinal cell line Caco-2. J. Ethnopharmacol. 2003;89(2-3):285–289. doi: 10.1016/j.jep.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Assis F., Bajrai L., Abrahao J., Kroon E., Dornas F., Andrade K., Boratto P., Pilotto M., Robert C., Benamar S., Scola B., Colson P. Pan-genome analysis of brazilian lineage A amoebal mimiviruses. Viruses. 2015;7(7):3483–3499. doi: 10.3390/v7072782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balaji Hange V. A Narrative literature review of global pandemic novel coronavirus disease 2019 (COVID-19): epidemiology, virology, potential drug treatments available. Arch. Med. 2020;12(3) doi: 10.36648/1989-521610.36648/1989-5216.12.3.310. [DOI] [Google Scholar]
- 7.Banerjee K., Gupta U., Gupta S., Wadhwa G., Gabrani R., Sharma S.K., Jain C.K. Molecular docking of glucosamine-6-phosphate synthase in Rhizopus oryzae. Bioinformation. 2011;7(6):285–290. doi: 10.6026/Bioinformation10.6026/007-0610.6026/007/97320630007285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford J., Enria D., Giesecke J., Heymann D.L., Ihekweazu C., Kobinger G., Lane H.C., Memish Z., Oh M.-D., Sall A.A., Schuchat A., Ungchusak K., Wieler L.H. COVID-19: towards controlling of a pandemic. Lancet. 2020;395(10229):1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Binnicker M.J. Emergence of a novel coronavirus disease (COVID-19) and the Importance of diagnostic testing: why partnership between clinical laboratories, public health agencies, and industry is essential to control the outbreak. Clin. Chem. 2020;66:664–666. doi: 10.1093/clinchem/hvaa071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bösl K., Ianevski A., Than T.T., Andersen P.I., Kuivanen S., Teppor M., Zusinaite E., Dumpis U., Vitkauskiene A., Cox R.J., Kallio-Kokko H., Bergqvist A., Tenson T., Merits A., Oksenych V., Bjørås M., Anthonsen M.W., Shum D., Kaarbø M., Vapalahti O., Windisch M.P., Superti-Furga G., Snijder B., Kainov D., Kandasamy R.K. Common nodes of virus-host interaction revealed through an integrated network analysis. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.0218610.3389/fimmu.2019.02186.s00110.3389/fimmu.2019.02186.s002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhuri S., Symons J.A., Deval J. Innovation and trends in the development and approval of antiviral medicines: 1987–2017 and beyond. Antiviral Res. 2018;155:76–88. doi: 10.1016/j.antiviral.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinnasamy S., Selvaraj G., Kaushik A.C., Kaliamurthi S., Chandrabose S., Singh S.K., Thirugnanasambandam R., Gu K., Wei D.-Q. Molecular docking and molecular dynamics simulation studies to identify potent AURKA inhibitors: assessing the performance of density functional theory, MM-GBSA and mass action kinetics calculations. J. Biomol. Struct. Dyn. 2020;38(14):4325–4335. doi: 10.1080/07391102.2019.1674695. [DOI] [PubMed] [Google Scholar]
- 13.Comess K.M., McLoughlin S.M., Oyer J.A., Richardson P.L., Stöckmann H., Vasudevan A., Warder S.E. Emerging approaches for the identification of protein targets of small molecules – a practitioners’ perspective. J. Med. Chem. 2018;61(19):8504–8535. doi: 10.1021/acs.jmedchem.7b01921. [DOI] [PubMed] [Google Scholar]
- 14.De Clercq E., Li G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016;29(3):695–747. doi: 10.1128/CMR.00102-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding S.-W., Lu R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr Opin Virol. 2011;1(6):533–544. doi: 10.1016/j.coviro.2011.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Asnaoui K., Chawki Y. Using X-ray images and deep learning for automated detection of coronavirus disease. J. Biomol. Struct. Dyn. 2020;1–12 doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elokely K.M., Doerksen R.J. Docking challenge: protein sampling and molecular docking performance. J. Chem. Inf. Model. 2013;53(8):1934–1945. doi: 10.1021/ci400040d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enayatkhani M., Hasaniazad M., Faezi S., et al. Reverse vaccinology approach to design a novel multi-epitope vaccine candidate against COVID-19: an in silico study. J. Biomol. Struct. Dyn. 2020;1:1–16. doi: 10.1080/07391102.2020.1756411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enmozhi S.K., Raja K., Sebastine I., Joseph J. Andrographolide as a potential inhibitor of SARS-CoV-2 main protease: an in silico approach. J. Biomol. Struct. Dyn. 2020;1–7 doi: 10.1080/07391102.2020.1760136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fakhr E., Zare F., Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 2016;23(4):73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- 21.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., Sonnhammer E.L.L., Tate J., Punta M. Pfam: the protein families database. Nucleic Acids Res. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friesner R.A., Murphy R.B., Repasky M.P., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 23.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Biorxiv. 2020 doi: 10.1101/2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., Tan K.-S., Wang D.-Y., Yan Y. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status. Military Med Res. 2020;7(1) doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gupta M.K., Vemula S., Donde R., et al. In-silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J. Biomol. Struct. Dyn. 2020;Apr:1–11 doi: 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyebi G.A., Ogunro O.B., Adegunloye A.P., et al. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CLpro): an in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020;1–13 doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han Y., Zhang J., Hu C.Q., Zhang X., Ma B., Zhang P. In silico ADME and toxicity prediction of ceftazidime and its impurities. Front. Pharmacol. 2019;10 doi: 10.3389/fphar.2019.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasan A., Paray B.A., Hussain A., et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. J. Biomol. Struct. Dyn. 2020;1–9 doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasin Y., Seldin M., Lusis A. Multi-omics approaches to disease. Genome Biol. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendaus M.A. Remdesivir in the treatment of coronavirus disease 2019 (COVID-19): a simplified summary. J. Biomol. Struct. Dyn. 2020;1–6 doi: 10.1080/07391102.2020.1767691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioakimidis L., Thoukydidis L., Mirza A., Naeem S., Reynisson J. Benchmarking the reliability of QikProp. Correlation between experimental and predicted values. QSAR Comb. Sci. 2008;27(4):445–456. doi: 10.1002/(ISSN)1611-021810.1002/qsar.v27:410.1002/qsar.200730051. [DOI] [Google Scholar]
- 32.Jin Z., Du X., Xu Y., et al. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Biorxiv. 2020 doi: 10.1101/2020.02.26.964882. [DOI] [Google Scholar]
- 33.Kausar S., Falcao A.O. An automated framework for QSAR model building. J. Cheminformatics. 2018;10 doi: 10.1186/s13321-017-0256-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kemp C., Imler J.-L. Antiviral immunity in drosophila. Curr. Opin. Immunol. 2009;21(1):3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keretsu S., Bhujbal S.P., Cho S.J. Rational approach toward COVID-19 main protease inhibitors via molecular docking, molecular dynamics simulation and free energy calculation. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-74468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S., Cho K. PyQSAR: A Fast QSAR modeling platform using machine learning and jupyter notebook. Bull. Korean Chem. Soc. 2018;40:39–44. doi: 10.1002/bkcs.11638. [DOI] [Google Scholar]
- 37.Kumar D., Kumari K., Jayaraj A., et al. Understanding the binding affinity of noscapines with protease of SARS-CoV-2 for COVID-19 using MD simulations at different temperatures. J. Biomol. Struct. Dyn. 2020;1–14 doi: 10.1080/07391102.2020.1752310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurdekar V., Jadhav H.R. A new open source data analysis python script for QSAR study and its validation. Med. Chem. Res. 2015;24(4):1617–1625. doi: 10.1007/s00044-014-1240-5. [DOI] [Google Scholar]
- 39.Lata S., Akif M. Structure-based identification of natural compound inhibitor against M. tuberculosis thioredoxin reductase: insight from molecular docking and dynamics simulation. J. Biomol. Struct. Dyn. 2020;1–10 doi: 10.1080/07391102.2020.1778530. [DOI] [PubMed] [Google Scholar]
- 40.Levanova A., Poranen M.M. RNA Interference as a Prospective Tool for the Control of Human Viral Infections. Front. Microbiol. 2018;9:2151. doi: 10.3389/fmicb.2018.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lionta E., Spyrou G., Vassilatis D., Cournia Z. Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr. Top. Med. Chem. 2014;14:1923–1938. doi: 10.2174/1568026614666140929124445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu R., Zhao X., Li J., Niu P., Yang B.o., Wu H., Wang W., Song H., Huang B., Zhu N.a., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L.i., Chen J., Meng Y., Wang J.i., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahanta S., Chowdhury P., Gogoi N., et al. Potential anti-viral activity of approved repurposed drug against main protease of SARS-CoV-2: an in silico based approach. J. Biomol. Struct. Dyn. 2020;1–15 doi: 10.1080/07391102.2020.1768902. [DOI] [PubMed] [Google Scholar]
- 44.Miller B.T., Singh R.P., Klauda J.B., Hodošček M., Brooks B.R., Woodcock H.L. CHARMMing: A new, flexible web portal for CHARMM. J. Chem. Inf. Model. 2008;48(9):1920–1929. doi: 10.1021/ci800133b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra P., Günther S. New insights into the structural dynamics of the kinase JNK3. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-27867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mittal L., Kumari A., Srivastava M., et al. Identification of potential molecules against COVID-19 main protease through structure-guided virtual screening approach. J. Biomol. Struct. Dyn. 2020;1–19 doi: 10.1080/07391102.2020.1768151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ozer E.A. ClustAGE: a tool for clustering and distribution analysis of bacterial accessory genomic elements. BMC Bioinform. 2018;19:150. doi: 10.1186/s12859-018-2154-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan W.-J., Chen C.-W., Chu Y.-W., Jeang K.T. siPRED: predicting siRNA efficacy using various characteristic methods. PLoS One. 2011;6(11):e27602. doi: 10.1371/journal.pone.0027602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ponzoni I., Sebastián-Pérez V., Martínez M.J., Roca C., De la Cruz Pérez C., Cravero F., Vazquez G.E., Páez J.A., Díaz M.F., Campillo N.E. QSAR classification models for predicting the activity of inhibitors of beta-secretase (BACE1) associated with Alzheimer’s disease. Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-45522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramachandran B., Kesavan S., Rajkumar T. Molecular modeling and docking of small molecule inhibitors against NEK2. Bioinformation. 2016;12(2):62–68. doi: 10.6026/bioinformation10.6026/012.0210.6026/97320630012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Salamov A.A. Ab initio gene finding in drosophila genomic DNA. Genome Res. 2000;10:516–522. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salmaso V., Moro S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: an overview. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lakshmi Selvaraj Alagu, Shafreen Raja Mohamed Beema, Priya Arumugam, Shunmugiah Karutha Pandian. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J. Struct. Dyn. Biomol. 2020 doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Selvaraj C. Molecular modeling and drug design techniques in microbial drug discovery. Essent. Bioinform. 2019;2:185–231. doi: 10.1007/978-3-030-18375-2_11. [DOI] [Google Scholar]
- 55.Selvaraj C., Sakkiah S., Tong W., Hong H. Molecular dynamics simulations and applications in computational toxicology and nanotoxicology. Food Chem. Toxicol. 2018;112:495–506. doi: 10.1016/j.fct.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 56.Singh Jatav V., Singh S., Khatri P., Sharma A. Recent pharmacological trends of Glycyrrhiza glabra Linn. Unani Res. 2011;1(2):1–11. doi: 10.5530/ur10.5530/ur.2.201110.5530/ur.2.2011.3. [DOI] [Google Scholar]
- 57.Singh R.K., Lee J.-K., Selvaraj C., Singh R., Li J., Kim S.-Y., Kalia V.C. Protein engineering approaches in the post-genomic era. Curr. Protein Pept. Sci. 2017;19(1) doi: 10.2174/1389203718666161117114243. [DOI] [PubMed] [Google Scholar]
- 58.Singh S.P., Selvaraj C., Knowar B.K., Singh S.K., Singh C.B., Sahoo D. Competitive inhibition of quercetin and apigenin at the ATP binding site of D-alanine-D-alanine ligase of Helicobacter pylori – A molecular modeling approach. Curr Biotechnol. 2019;7(5):340–348. doi: 10.2174/2211550107666180612100441. [DOI] [Google Scholar]
- 59.Söderholm S., Fu Y.u., Gaelings L., Belanov S., Yetukuri L., Berlinkov M., Cheltsov A., Anders S., Aittokallio T., Nyman T., Matikainen S., Kainov D. Multi-omics studies towards novel modulators of influenza A virus-host interaction. Viruses. 2016;8(10):269. doi: 10.3390/v8100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun H., Duan L., Chen F., Liu H., Wang Z., Pan P., Zhu F., Zhang J.Z.H., Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 7. Entropy effects on the performance of end-point binding free energy calculation approaches. PCCP. 2018;20(21):14450–14460. doi: 10.1039/c7cp07623a. [DOI] [PubMed] [Google Scholar]
- 61.Tai K., Shen T., Börjesson U., Philippopoulos M., McCammon J.A. Analysis of a 10-ns molecular dynamics simulation of mouse Acetylcholinesterase. Biophys. J. 2001;81(2):715–724. doi: 10.1016/S0006-3495(01)75736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thakur N., Qureshi A., Kumar M. VIRsiRNAdb: a curated database of experimentally validated viral siRNA/shRNA. Nucleic Acids Res. 2011;40:D230–D236. doi: 10.1093/nar/gkr1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(5):305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 64.Titov D.V., Liu J.O. Identification and validation of protein targets of bioactive small molecules. Bioorg. Med. Chem. 2012;20(6):1902–1909. doi: 10.1016/j.bmc.2011.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vijayakumar S., Manogar P., Prabhu S., Sanjeevkumar Singh R.A. Novel ligand-based docking; molecular dynamic simulations; and absorption, distribution, metabolism, and excretion approach to analyzing potential acetylcholinesterase inhibitors for Alzheimer’s disease. J. Pharm. Anal. 2018;8(6):413–420. doi: 10.1016/j.jpha.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang B.o., Li L., Hurley T.D., Meroueh S.O. Molecular recognition in a diverse set of protein-ligand interactions studied with molecular dynamics simulations and end-point free energy calculations. J. Chem. Inf. Model. 2013;53(10):2659–2670. doi: 10.1021/ci400312v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang C., Greene D., Xiao L.i., Qi R., Luo R. Recent Developments and Applications of the MMPBSA Method. Front. Mol. Biosci. 2018;4 doi: 10.3389/fmolb.2017.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu Y., Zaiden N., Cao B. The core- and pan-genomic analyses of the genus comamonas: from environmental adaptation to potential virulence. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xiao J., Zhang Z., Wu J., Yu J. A brief review of software tools for pangenomics. Genom. Proteom. Bioinf. 2015;13(1):73–76. doi: 10.1016/j.gpb.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yadav D.K., Khan F. QSAR, docking and ADMET studies of camptothecin derivatives as inhibitors of DNA topoisomerase-I. J. Chemom. 2013;27(1-2):21–33. doi: 10.1002/cem.v27.1-210.1002/cem.2488. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data used in the work are publicly available in public databases like NCBI, PDB, Drug bank, and Pubchem.