Abstract
Background
The current COVID-19 pandemic is caused by SARS-CoV-2 which belongs to coronaviridae family. Despite the global prevalence, there are currently no vaccines or drugs. Dietary plant derived exosome-like vesicles are known as edible nanoparticles (ENPs). ENPs are filled with microRNAs (miRNAs), in bioavailable form. Recently, cross-kingdom regulation of human transcripts by plant miRNAs have been demonstrated. However, ENP derived miRNAs targeting SARS-CoV-2 has not been described.
Study design
Mature ENP-derived miRNA sequences were retrieved from small RNA sequencing datasets available in the literature. In silico target prediction was performed to identify miRNAs that could target SARS-CoV-2. ENPs were isolated from ginger and grapefruit plants and the expression of SARS-CoV-2 targeting miRNAs were confirmed by qRT-PCR.
Results
From a total of 260 ENP-derived miRNAs, we identified 22 miRNAs that could potentially target SARS-CoV-2 genome. 11 miRNAs showed absolute target specificity towards SARS-CoV-2 but not SARS-CoV. ENPs from soybean, ginger, hamimelon, grapefruit, tomato and pear possess multiple miRNAs targeting different regions within SARS-CoV-2. Interestingly, osa/cme miR-530b-5p specifically targeted the ribosomal slippage site between ORF1a and ORF1b. We validated the relative expression of six miRNAs (miR-5077, miR-6300, miR-156a, miR-169, miR-5059 and miR-166 m) in ginger and grapefruit ENPs by RT-PCR which showed differential enrichment of specific miRNAs in ginger and grapefruit ENPs.
Conclusion
Since administration of ENPs leads to their accumulation into lung tissues in vivo, ENP derived miRNAs targeting SARS-CoV-2 genome has the potential to be developed as an alternative therapy.
Keywords: Edible nanoparticles, microRNAs, SARS-CoV-2, COVID-19, Extracellular vesicles
Abbreviations: Angiotensin-converting enzyme II, ACEII; Coronavirus disease – 2019, COVID-19; Edible nanoparticles, ENP; Gastrointestinal, GI; microRNAs, miRNAs; Minimum free energy, MFE; polyethylene glycol, PEG; Quantitative real-time polymerase chain reaction, qRT-PCR; Severe acute respiratory syndrome coronavirus, SARS-CoV
Graphical abstract
1. Introduction
Coronaviridae is a family of enveloped viruses with a positive-sense RNA as the viral genome. Coronavirus disease −19 (COVID-19), is caused by one of the subspecies of this family, namely severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The SARS-CoV-2 genome is non-segmented positive-sense RNA genome with 29,903 bases in length, very similar to eukaryotic mRNA. The genome encodes four structural proteins (spike protein S, envelope protein E, membrane protein M and nucleocapsid protein, N) along with the non-structural protein complex of viral replicase (RNA dependent RNA polymerase, ORF1ab) which is located in the 5′ end of the genome (Masters et al., 2006). Besides, several accessory open reading frames (ORFs) are also encoded, which has both structural and non-structural role during viral life cycle. Infection is initiated by binding of the virion to the cognate cellular receptor/s. For SARS-CoV-2, Angiotensin converting enzyme II (ACEII) has been reported as the cognate receptor, which is expressed in type II alveolar cells of the lungs, epithelial cells of the intestine, kidney, oral cavity and mucosa (Xu Zhong et al., 2020). Once the virion enters the cytoplasm, the genome is translated by host cell ribosomes to produce replicase. The genome acts as a template for subsequent rounds of replication/transcription via negative sense RNA intermediates to produce both genomic as well as sub-genomic RNAs coding for other viral proteins.
As of 12th January 2021, there are more than 89 million confirmed COVID-19 cases and ~ 1.9 million COVID-19 related deaths have been reported worldwide (source: https://www.who.int/emergencies/diseases/novel-coronavirus-2019). Currently, there are no vaccines or drugs are available to combat this disease. Even though antiviral drugs such as chloroquine, remdesivir, favipiravir and arbidol are currently under clinical trials against COVID-19, the results are not encouraging and the cost associated with the treatment is high (Babadaei et al., 2020; Dong et al., 2020). Considering the high infectivity rates of COVID-19, fast-paced development of targeted therapeutics against COVID-19 at an affordable cost and large scale feasibility is the need of the hour (To et al., 2020).
Non-coding RNAs are endogenously made RNA transcripts, which do not have protein-coding potential. MicroRNAs (miRNAs), the classical example of non-coding RNAs, are highly conserved, ~18-23 nt long single-stranded RNA molecules. MiRNAs are known to regulate a wide variety of cellular processes during development and diseases (O'Brien et al., 2018). miRNAs bind to target mRNAs based on sequence complementarity which leads either to transcript degradation or translational repression (O'Brien et al., 2018). In plants, target messenger RNAs (mRNAs) possess near-perfect complementary binding sites to miRNAs while animal miRNAs share partial sequence complementarity with target mRNAs. Also, the 2’-O- methylation of the last nucleotide at the 3′ end of mature miRNA is a unique feature in plant miRNAs, which is not present in animal miRNAs (O'Brien et al., 2018). An earlier in silico study predicted hsa-miR-27b as a potential human miRNA that may specifically target Indian isolate of SARS-CoV-2, though this needs to be experimentally validated (Sardar et al., 2020). However, it remains to be studied if dietary plant-derived miRNAs can execute suppressive functions against viral infections. The physiological relevance of such a cross-kingdom regulation has been debated due to the decreased stability of plant-derived miRNAs during the cooking process as well as during their passage through gastrointestinal (GI) digestion.
Nevertheless, an alternative route by which such cross-kingdom regulation can be achieved is via miRNAs present within exosome like-vesicles isolated from dietary plants. These vesicles are known as edible nanoparticles (ENPs). ENPs from several edible plants have been isolated earlier, ranging in size between 100 and 900 nm (Di Gioia et al., 2020). ENPs share both structural and functional similarities with mammalian exosomes, though their exact intra/extracellular origin is currently unclear (Zhang et al., 2016b). The biological cargo within ENPs, including nucleic acids, exhibit excellent bioavailability and resistance to GI digestion process (Yang et al., 2018). ENPs are non-toxic and lack immunogenicity which has led to their exploitation as a nanocarrier for in vivo delivery of biological macromolecules such as plasmid DNA, antibodies, phytochemicals, siRNAs and chemotherapeutic drugs (Rome, 2019; Wang et al., 2015; Wang et al., 2013).
Interestingly, ENPs also contain numerous plant-derived miRNAs which has been predicted to target human transcripts. Xiao et al. (2018) isolated ENPs from 11 edible plants and profiled miRNAs in these ENPs using small RNA sequencing . The authors identified between 32 and 127 miRNAs per plant species. Further, the human target mRNAs of these ENP-derived miRNAs were predicted and validated through chimeric luciferase reporter assay (Xiao et al., 2018). Moreover, ginger ENPs have been extensively studied and have been proven to have versatile therapeutic benefits against inflammatory diseases. Ginger ENPs have also been used as a nanocarrier for delivery of biological cargo. Ginger ENP contains 125 different miRNAs within which 124 miRNAs have been predicted to have putative human targets (Zhang et al., 2016a).
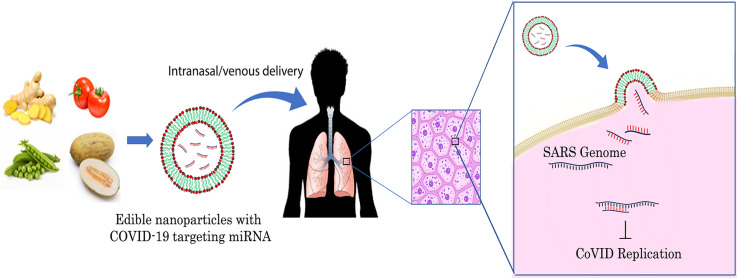
It has earlier been demonstrated that intranasal as well as intravenous administration of ginger ENPs led to their homing in mice lung tissues and was detectable up to 72 h post-administration. Since lung tissue is one of the major replication sites for SARS-CoV-2, antiviral bioactives in ginger ENPs can be used as an effective therapeutic modality against COVID-19 (Wang et al., 2013). In coronaviruses, even though the viral genome is encapsidated by the N protein, the sub-genomic RNAs produced during active viral infection provides an excellent target for plant miRNAs to act upon. Hence, it is also likely that ENP with specific miRNAs against SARS-CoV-2 genes, will decrease disease burden by its spontaneous intracellular uptake and delivery of COVID-19 targeting miRNAs in alveolar epithelial cells.
However, it remains to be determined if edible plant ENP-derived miRNAs can target SARS-CoV-2 genes. Due to the relative abundance and diversity of miRNAs present within the various ENPs isolated so far, we hypothesize that ENP/s may also contain SARS-CoV-2 targeting miRNAs which can be used as a cost-effective and target-specific therapeutic modality against COVID-19 infection.
To this end, we first extracted the mature miRNA sequences that have been identified in ENPs from prior literature. Using in silico analysis with stringent selection criteria, we predicted SARS-CoV-2 targeting miRNAs that are present in high abundance in ENPs. Target specificity of SARS-CoV-2 targeting miRNAs was compared by sequence alignment of miRNA binding site with different isolates of SARS-CoV-2 and SARS-CoV. As a proof of concept, ENPs were isolated from grapefruit and ginger rhizomes and SARS-CoV-2 targeting miRNAs were validated in these ENPs by quantitative real-time PCR (qRT-PCR).
2. Materials and methods
2.1. Sequences of viral genomes
The genome sequences of SARS-CoV-2 isolated from different geological locations across the world were downloaded from NCBI as described earlier (Sardar et al., 2020). The accession numbers of different SARS-CoV-2 isolates used in this study were: SARS-CoV-2 Wuhan isolate; NC_045512.2, India; MT050493.1, Italy; MT066156.1, USA; MN985325.2, Nepal; MT072688.1. We also used the nucleotide sequence of SARS-CoV isolate from the 2003 SARS outbreak (NC_004718.3) for comparative analysis.
2.2. Source files of miRNAs and processing
The studies that were considered for shortlisting ENP-associated miRNAs are listed in Supplementary file 1. The data from Xiao et al. (2018) contains a comprehensive miRNA expression profile across 11 edible plant ENPs (Grapefruit, Hami melon, Pea, Coconut, Blueberry, Tomato, Pear, Ginger, Kiwifruit, Orange, Soybean). Since the small RNA sequencing analysis was performed simultaneously for all 11 plant ENPs, the actual read counts can be compared to calculate the relative abundance of miRNAs within different ENP sources. It is worth noting that in Xiao et al. (2018), the top 20 miRNAs constitute more than 90% of total ENP-associated miRNAs and hence, we chose the top 20 miRNAs, based on their read count from each ENP for our study. We also included the top 20 miRNAs for ginger and grapes from two other independent studies (Ju et al., 2013; Teng et al., 2018).The ENP associated miRNAs from grapes were identified by microarray analysis and hence the actual read counts could not be established. In addition, the read counts of ginger ENP miRNAs cannot be compared between Teng et al. (2018) and Xiao et al. (2018) due to the experimental variability, library construction and the exact sequencing pipeline used. The final list of 260 ENP-miRNAs selected from all the three studies along with their relative abundances is presented in Supplementary file 1. Since some of the miRNA sequences represented in Xiao et al. (2018) were partial, we also downloaded the actual miRNA sequence for each accession no/miRNA name from miRbase version 22 and used for target prediction.
2.3. Determination and alignment of ENP-derived miRNA binding sites in SARS-CoV-2 with global COVID-19 isolates and SARS-CoV
Target prediction was carried out using the RNA hybrid software with stringent prediction criteria using MT050493.1 as a template sequence (Rehmsmeier et al., 2004). The following critical parameters were set during target prediction to identify physiologically relevant SARS-CoV-2 targeting miRNAs. The minimum free energy (MFE) was fixed at -25 kcal/mol to ensure that the binding of miRNA to target mRNA can supersede the local secondary structures (Kertesz et al., 2007; Mückstein et al., 2006). In animal miRNAs, seed sequence complementarity is a major determinant for efficient miRNA binding to target mRNA. Hence, helix constraint was placed in seed sequences between 2 and 8 bases from the 5′ end of the miRNA. Since non-canonical G-U base pairing is well tolerated in miRNA mediated target mRNA suppression in animal miRNAs, this was allowed (Saxena et al., 2003). The maximum internal bulge and bulge loop length were restricted between 0 and 2 bases to ensure specific binding and to avoid false positives. Conservation of miRNA binding site across homologous species is a parameter used to assess the specificity of miRNA target interaction. However, RNA hybrid does not allow the evaluation of binding site conservation within different viral genomes. Hence, to ensure the specificity of miRNA binding to SARS-CoV-2 genome, we aligned the miRNA binding site sequence within the SARS-CoV-2 genome (MT050493.1) with global isolates of SARS-CoV-2 as well as with SARS-CoV sequence using MEGA software, version 10.1 (Stecher et al., 2020). Local RNA secondary structures that may be present in miRNA target sites were predicted using the MFOLD web server (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Zuker, 2003).
2.4. Ginger and grapefruit ENP purification and total RNA extraction
ENPs were isolated from fresh ginger rhizomes and edible portion of grapefruits using a polyethylene glycol (PEG)-based precipitation method developed in the laboratory (Kalarikkal et al., 2020). We have earlier demonstrated that this method yields ENPs of similar biophysical and biochemical characteristics in comparison to the gold standard differential ultracentrifugation protocol. Total RNA was isolated from 100 mg equivalent of ENPs using TRI reagent (Sigma) as per manufacturer's instructions. Total RNA was quantified by Nanodrop spectrophotometer and RNA quality was assessed by resolving it through 1.5% Agarose gel electrophoresis as per Kalarikkal et al. (2020). To ascertain the RNA nature of the total RNA isolated, 1 μg of total RNA was incubated with 0.5 μg of RNAse A for 10 min at 37 °C before resolving them through 1.5% Agarose gel electrophoresis.
2.5. MicroRNA quantification by qRT-PCR
Quantification of mature miRNAs using oligonucleotide primers was carried out as described earlier (Balcells et al., 2011). Briefly, mature miRNAs present in total RNA (100 ng) were first polyadenylated in a total volume of 10 μl containing 1 μl of 10× poly (A) polymerase buffer, 1 μl of 10 mM ATP, 2.5 units of poly (A) polymerase (New England Biolabs) for 30 min at 37 °C. The polyadenylated miRNAs were reverse transcribed using a universal reverse (Rev) primer which contains an anchored oligo (dT) sequence and an adapter sequence at the 5′ end. To the reaction mix (10 μl), 4 μl of 5× RT buffer, 1 μl of 25 mM dNTP, 1 μl of 10 μM universal Rev. primer and 200 units of M-MuLV reverse transcriptase were added and the volume was made up to 20 μl with nuclease-free water. Reverse transcription was performed by incubation at 42 °C for 45 min followed by incubation at 85 °C for 5 min to inactivate the reverse transcriptase. The complementary DNA (cDNA) thus obtained was diluted 10 fold with RNAse free water and 5 μl of the diluted cDNA was used in qRT-PCR reactions containing 10 μl of 2× iTaq Universal SYBR green master mix (BioRad) and 250 nM each of miRNA specific forward and universal real-time PCR reverse (qRT) primer in a total volume of 20 μl. QRT-PCR was carried out in BioRad CFX96 PCR system under the following conditions; initial denaturation at 95 °C for 3 min followed by 40 cycles of denaturation (95 °C for 10 s), annealing (55 °C for 30 s) and extension (72 °C for 30 s). Automatic threshold option was used for each miRNA primer sets for the acquisition of Ct values. No template controls (NTC) lacking the cDNA were amplified to verify the specificity of the primers. The sequences of miRNA specific forward primers, universal Rev. primer and universal qRT primer are provided in Table 1 .
Table 1.
Sequences of primers used in this study (anchored oligodT sequence shown in bold).
| miRNA name | Sequence (5′-3′) |
|---|---|
| 3′ universal Rev. primer | 5′GATTACGCCAAGCTTGTGTGGCCCTGGTGTTGTGAAGTAGCCGTTTTTTTTTTTTTTTTTTTTTTVN3′ |
| 3′ universal qRT primer | 5′ GAT TAC GCC AAG CTT GTG TGG 3′ |
| gma-miR-166 m For | 5′ CGG ACC AGG CTT CAT TCC CC 3′ |
| gma-miR-6300 For | 5′ GTC GTT GTA GTA TAG TGG 3′ |
| aqc-miR-159 For | 5′ TTT GGA CTG AAG GGA GCT CT 3′ |
| osa-miR-5077 For | 5′ GTT CGC GTC GGG TTC ACC A 3′ |
| bdi-miR-5059 For | 5′ GCC TGG GCA GCA CCA CCA 3′ |
| mtr-miR-156a For | 5′ TGA CAG AAG AGA GAG AGC AC 3′ |
2.6. Statistical analysis
qRT-PCR data are presented as mean ± S.D. Statistical significance was determined by unpaired Student's two-tailed t-test analysis, with p < 0.05 considered to be statistically significant. The experiments were repeated at least three times with two independent batches of ENPs..
3. Results
3.1. Edible plant-derived ENPs contain SARS-CoV-2 targeting miRNAs
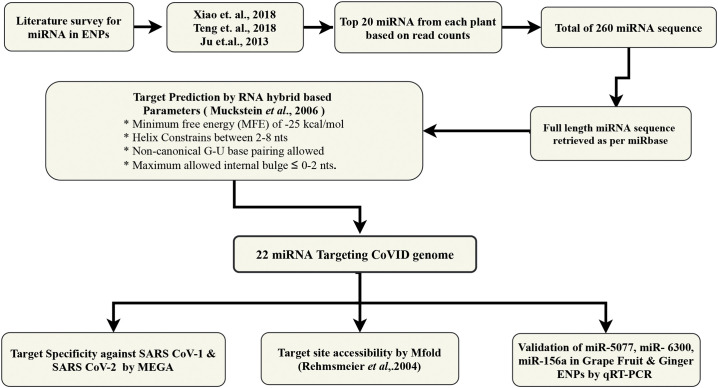
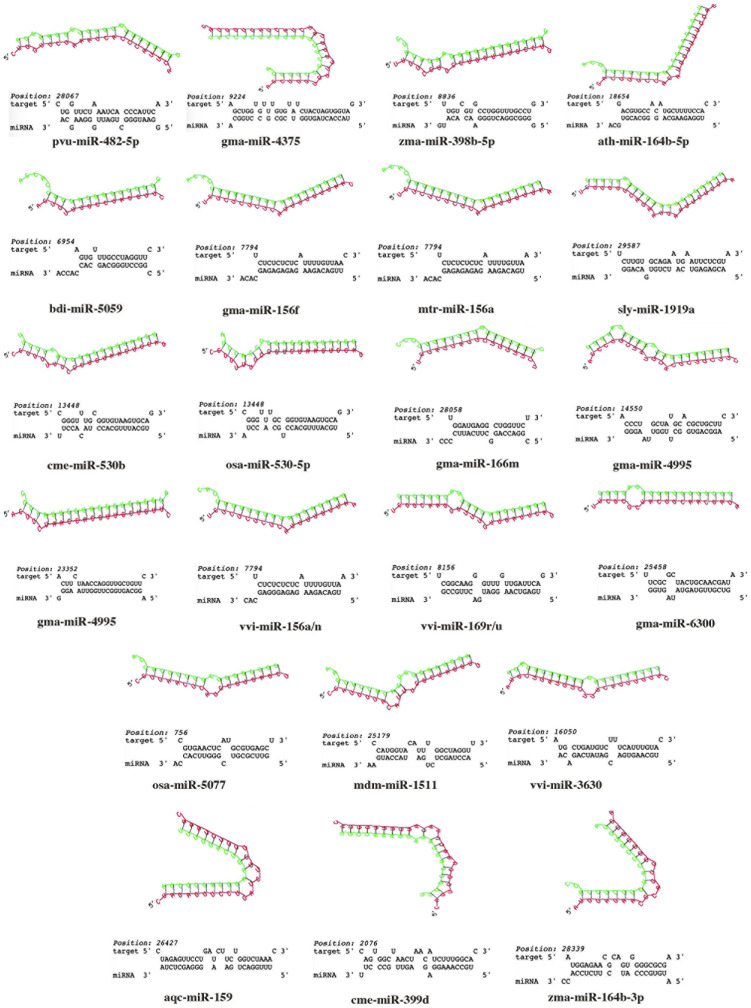
The entire process of miRNA selection from different ENPs, mature miRNA sequence retrieval, target prediction, the specificity of miRNA-target interaction and secondary structure analysis is depicted as a flow chart in Fig. 1. First, we shortlisted top 20 miRNAs from 13 edible plant ENPs based on their relative read counts obtained from small RNA sequencing data sets for each plant species (Supplementary file 1). A total of 260 ENP-associated miRNAs were taken as input for this study and subjected to target site prediction against SARS-CoV-2 genome sequence with RNAhybrid software. With the stringent selection criteria (described in methods), we obtained a sum of 22 ENP-derived miRNAs that could target SARS-CoV-2 genome (Table 2). The secondary structures of miRNA-target mRNA duplex for all 22 miRNAs along with target site sequences are shown in Fig. 2. All the structures obtained passed the selection criteria (≥ −25 kcal/mol MFE). Notably, the vast majority of ENP-derived miRNAs (14 out of 22 miRNAs) had target sites within ORF1ab. We identified both spike and ORF8 genes were targeted by at least two different miRNAs (gma-miR-4995, mdm-miR_1511 targeting spike transcript and pvu-miR_482-5p, gma-miR-166 m targeting ORF8 transcript). Membrane, Nucleocapsid and ORF10 mRNAs were targeted by aqc-miR-159, zma-miR-164-3p and sly-miR-1919a, respectively (Table 2).
Fig. 1.
Flow chart depicting the outline of miRNA target prediction and analysis using in silico approach.
Table 2.
List of miRNAs targeting SARS-CoV-2 genome.
| miRNA name | Sequence (5′-3′) | Target sequence (5′-3′) | Target Position COVID-19 | Target gene in COVID-19 | MFE (kcal/mol) |
|---|---|---|---|---|---|
| gma-miR-4995 | AGGCAGUGGCUUGGUUAAGGG | ACUUCUAACCAGGUUGCUGUUC | 23,352 | Spike | −26.9 |
| mdm-miR-1511 | ACCUAGCUCUGAUACCAUGAA | CCAUGGUACAUUUGGCUAGGUU | 25,179 | Spike | −25.4 |
| pvu-miR-482-5p | GGAAUGGGCUGAUUGGGAAGCA | CUGGUUCUAAAUCACCCAUUCA | 28,067 | ORF8 | −26.6 |
| gma-miR-166 m | CGGACCAGGCUUCAUUCCCC | UGGAUGAGGCUGGUUCU | 28,058 | ORF8 | −25.5 |
| gma-miR-6300 | GUCGUUGUAGUAUAGUGG | UUCGCGCUACUGCAACGAUA | 25,458 | ORF3a | −26 |
| ath-miR-164b-5p | UGGAGAAGCAGGGCACGUGCA | GACGUGCCACAUGCUUUUCCAC | 18,654 | ORF1ab | −33.2 |
| gma-miR-4375 | UACCACUAGUGGUCGCGCCUGGCA | AGCUGGUGUUUGUGUAUCUACUAGUGGUA | 9224 | ORF1ab | −28.6 |
| zma-miR-398b-5p | GGGGCGGACUGGGAACACAUG | UUGUCGUGCCUGGUUUGCCUG | 8836 | ORF1ab | −27 |
| bdi-miR-5059 | CGGCCUGGGCAGCACCACCA | AGUGUUUGCCUAGGUUC | 6954 | ORF1ab | −25.3 |
| gma-miR-156f | UUGACAGAAGAGAGAGAGCACA | UCUCUCUCUCAUUUUGUUAA | 7794 | ORF1ab | −27.6 |
| mtr-miR-156a | UGACAGAAGAGAGAGAGCACA | UCUCUCUCUCAUUUUGUUAA | 7794 | ORF1ab | −27 |
| cme-miR-530b | UGCAUUUGCACCUACACCUU | CGGGUUUGCGGUGUAAGUGCAG | 13,448 | ORF1ab | −28 |
| osa-miR-530-5p | UGCAUUUGCACCUGCACCUA | CGGGUUUGCGGUGUAAGUGCAG | 13,448 | ORF1ab | −27.5 |
| vvi-miR-3630 | UGCAAGUGACGAUAUCAGACA | AUGCUGAUGUCUUUCAUUUGUAC | 16,050 | ORF1ab | −25 |
| gma-miR-4995 | AGGCAGUGGCUUGGUUAAGGG | ACCCUGCUAUGCACGCUGCUUC | 14,550 | ORF1ab | −28.2 |
| vvi-miR-156a/n | UGACAGAAGAGAGGGAGCAC | UCUCUCUCUCAUUUUGUUAA | 7794 | ORF1ab | −26.1 |
| cme-miR-399d | UGCCAAAGGAGAGUUGCCCUU | CAGUGGCUAACUAACAUCUUUGGCAC | 2076 | ORF1ab | −25.1 |
| osa-miR-5077 | GUUCGCGUCGGGUUCACCA | CGUGAACUCAUGCGUGAGCU | 756 | ORF1ab | −26.7 |
| vvi-miR-169r/u | UGAGUCAAGGAUGACUUGCCG | UCGGCAAGGGUUUGUUGAUUCAG | 8156 | ORF1ab | −25.4 |
| sly-miR-1919a | ACGAGAGUCAUCUGUGACAGG | UCUUGUGCAGAAUGAAUUCGCGUA | 29,587 | ORF10 | −25.6 |
| zma-miR-164b-3p | AUGUGCCCAUCUUCUCCACC | AUGGAGAACGCAGUGGGGCGCGA | 28,339 | N | −25.6 |
| aqc-miR-159 | UUUGGACUGAAGGGAGCUCUA | CUAGAGUUCCUGAUCUUCUGGUCUAAAC | 26,427 | M | −25.4 |
Fig. 2.
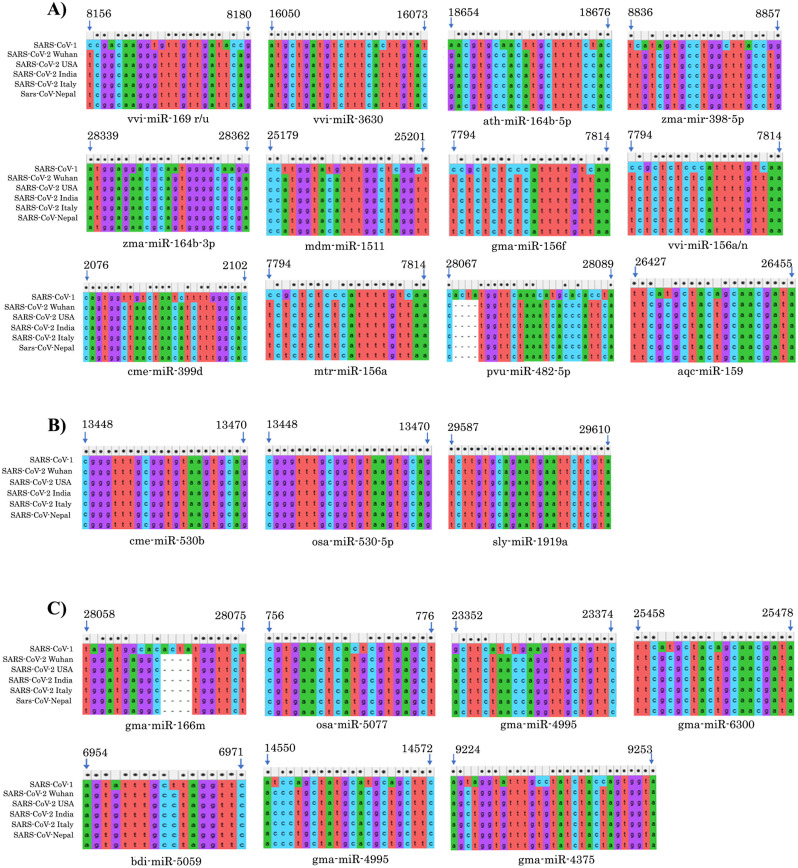
Schematic depiction of stable RNA hybrid structures formed between 22 ENP derived miRNAs (in green) with target sites in SARS-CoV-2 (in red). The relative nucleotide position of target sites for each miRNA in SARS-CoV-2 genome is shown above the alignments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Cme/osa-miR-530b targets ribosomal slippage site in ORF1ab gene
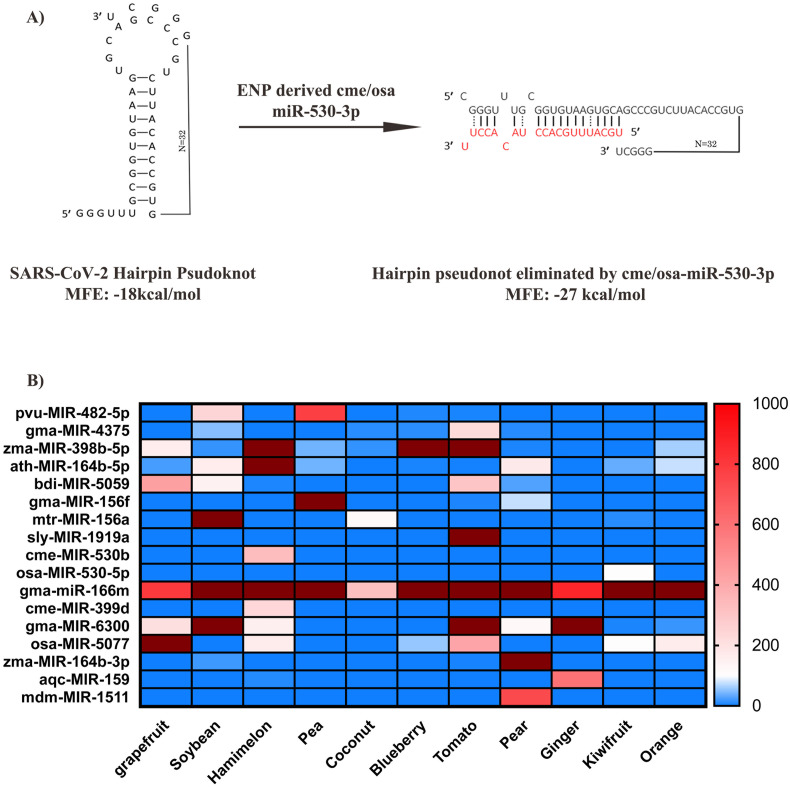
In coronaviruses, the replicase complex is encoded by the ORF1ab gene which occupies the first two thirds of the viral genome from the 5′ end. ORF1ab codes for two different ORFs, namely ORF1a and ORF1b, arranged in tandem manner. Intact synthesis of both ORF1a and ORF1b requires a ribosomal frameshift at the junction between ORF1a and ORF1b. This is achieved by a sequence and structure dependent ribosomal slippage event which occurs during the translation of ORF1ab mRNA. This phenomenon in coronaviruses requires two cis RNA elements, namely, a slippery sequence (UUUAAAC, 13442 to 13,448 nt in SARS-CoV-2) and a hairpin type pseudoknot structure located at the downstream sequence (Baranov et al., 2005). It has been proposed, that pausing of ribosomes in this hairpin pseudoknot allows ribosomal slippage by one base at the −1 direction leading to the in-frame translation of ORF1b. Disruption of ribosomal slippage event at this site will lead to the premature termination of translation at the stop codon located within the hairpin pseudoknot, thereby inhibiting the synthesis of ORF1b. Interestingly, cme-miR-530b and osa-miR-530-5p both of which share similar mature miRNA sequence was found to target ribosomal slippage region between two overlapping ORFs, ORF1a and b (Fig. 3A). The osa/cme-miR-530-5p binding region overlaps with the pseudoknot structure encompassing the stop codon (13,448 to 13,469). The MFE of miR-530-5p binding far exceeds the MFE of the pseudoknot structure (−27 vs −18 kcal/mol). Hence, we posit that even in the absence of a canonical miRNA mediated suppression, this unwinding of pseudoknot by miR-530-5p may indirectly inhibit the synthesis of ORF1b by not allowing ribosomal slippage and thus preventing the replication of SARS-CoV-2 (Fig. 3A).
Fig. 3.
A) The binding site for cme-miR-530b and osa-miR-530-5p overlaps with the ribosomal slippage site in ORF1ab gene. Binding of cme/osa-miR530-5p is expected to unwind the hairpin pseudoknot present in ribosomal slippage site. B) Relative levels of SARS-CoV-2 targeting miRNAs in edible plants displayed as a heatmap. Absolute read counts of the 22 miRNAs in all the 11 plants were extracted from Xiao et al., (2018) and plotted as a heat map using Graphpad Prism software. Expression of miRNAs less than 100 reads are shown in blue and miRNAs from 100 to 1000 reads are shown in shades of white to orange. Dark red shade indicates values beyond the range specified in the graph suggesting very high expression. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Plant ENPs contain multiple miRNAs targeting SARS-CoV-2
To extrapolate our results for COVID-19 treatment, we plotted the relative abundance of the SARS-CoV-2 targeting miRNAs as a heatmap in 11 edible plant ENPs, using the raw read counts obtained from Xiao et al. (2018). Hami melon, Soybean, tomato and pear ENPs has 6–7 miRNAs while grapefruit, pea and blueberry ENPs have 3–4 miRNAs that could target SARS-CoV-2 (Fig. 3B). Gma-miR-166 m which targets ORF8 in SARS-CoV-2 belongs to a highly conserved miR-166 family of miRNAs with 21 sub-members encoded within various genomic locations in soybean (Li et al., 2017). Although, the relative expression of gma-miR-166 m was <1000 reads in ginger ENPs as per the dataset from Xiao et al. (2018), its expression exceeds 100,000 reads in ginger ENPs as per Teng et al. (2018) (Fig. 3B & Supplementary file 1). This could be due to the technical variation during library preparation, sequencing platform or the variety of ginger used for ENP isolation. It is also worth noting that, aly-miR-166a-3p, which share identical sequence with gma-miR-166 m with an additional U at the 5′ end was also found to be highly enriched in ginger ENPs (Supplementary file 1).
3.4. Target specificity of ENP-derived miRNAs to SARS-CoV-2 and SARS-CoV
To investigate the specificity of the selected miRNAs against SARS-CoV-2, we aligned the miRNA target sequences identified using the template strain (MT050493.1) with sequences of SARS-CoV-2 isolated from other countries such as USA, Nepal, France, Italy and China (Wuhan isolate) and SARS-CoV. It has been observed that global strains of SARS-CoV-2 share 99% sequence similarity with the original Wuhan isolate (Sardar et al., 2020). Due to this, all the 22 miRNA target sites were highly conserved across all isolates of SARS-CoV-2 (Fig. 4 ). Even though SARS-CoV and SARS-CoV-2 are highly homologous, six regions of differences (RD) has been identified earlier which are located within ORF1ab, S, ORF7b and ORF8 genes (Xu et al., 2020). Based on this, we segregated the miRNA target sites into 3 groups. Group I consisted of 12 miRNA target sites in which nucleotide mismatch/s were found between SARS-CoV-2 isolates and SARS-CoV in seed sequences (Fig. 4A). Given that complete complementarity at the seed sequences is essential for miRNA mediated target suppression, miRNAs targeting these regions are expected to be specific against SARS-CoV-2 but may not bind to SARS-CoV (Witkos et al., 2011). Group II consisted of 3 miRNA target sites which are conserved between SARS-CoV-2 and SARS-CoV suggesting that these miRNAs can equally suppress both species of viruses (Fig. 4B). On the other hand, 7 miRNA target sites had full conservation at the seed sequence with partial mismatches at the 5′ end and these are classified as Group III (Fig. 4C). However, successful binding of miRNAs from Group III to target sites in SARS-CoV-2 may depend on the miRNA-target mRNA duplex strength and the context of the intracellular environment in virus infected cells.
Fig. 4.
Segregation of ENP miRNAs based on specificity against SARS-CoV-2 and SARS-CoV. A) MicroRNA binding sites that are specific to SARS-CoV-2 but not SARS-CoV based on seed sequence (2–7/8 nt) mismatch. Note that the seed sequences are located at the 3′ end of the mRNA sequence where 5′ end of miRNA binds. B) MicroRNAs binding site conserved in both SARS-CoV and SARS-CoV-2. C) MicroRNA binding sites with intact conserved seed sequence with partial mismatch across the 3′ end.
3.5. Target site accessibility of miRNA binding sites
The accessibility of the target site to miRNA binding is one of the critical factors determining successful miRNA mediated target mRNA suppression (Didiano and Hobert, 2006; Long et al., 2007; Witkos et al., 2011). It has been experimentally validated that the local secondary structure between 17 nt upstream and 13 nt downstream of the miRNA binding site can influence target site accessibility to miRNAs (Kertesz et al., 2007). Hence, we assessed the local secondary structure formation at the miRNA target site along with 17 nt upstream and 13 nt downstream sequences from SARS-CoV-2 using MFOLD webserver. Out of the 22 ENP-miRNA binding sites in SARS-CoV-2, we found 4 miRNA target sequences had at least one predicted secondary structure with MFE ≥ 15 kcal/mol (Supplementary file 2). The target sites of pvu-miR-482-5p and zma-miR-398b-5p overlapped with local secondary structures with MFE of −15.4 kcal/mol and 20.8 kcal/mol, respectively. As discussed earlier, the target sites of cme-miR-530b and osa-miR-530-3p displayed secondary structures with an MFE of 17.5 kcal/mol due to hairpin pseudoknot involved in ribosomal slippage. Since all the 22 miRNA binding sites within SARS-CoV-2 genome formed secondary structures with MFE < −25 kcal/mol, we anticipate that the ENP-derived miRNAs could supersede the local secondary structures to allow its own binding.
3.6. Validation of SARS-CoV-2 targeting miRNAs in ginger and grapefruit ENPs
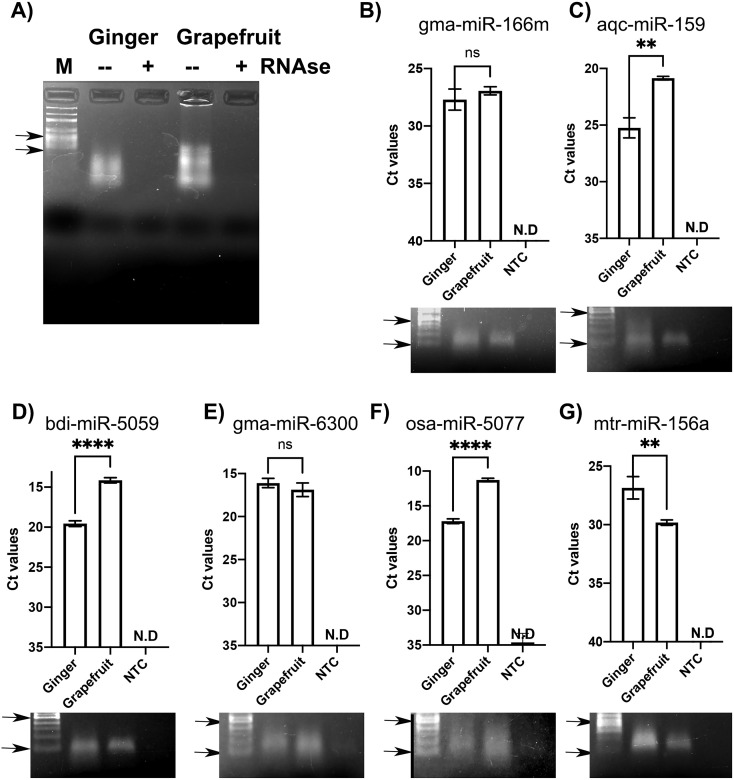
To further confirm that the SARS-CoV-2 targeting miRNAs are present in high abundance within ENPs, we first purified ENPs from ginger rhizomes and grapefruit using a cost-effective PEG method we have developed earlier (Kalarikkal et al., 2020). Total RNA was isolated and resolved through agarose gel electrophoresis. As shown in Fig. 5A, both ginger and grapefruit ENPs showed the presence of small RNA population that are less than 100 bp in size, visualized by ethidium bromide staining. Treatment with RNAse A led to the disappearance of these bands confirming that these are indeed small RNAs likely representing miRNAs extracted from ENPs. Further, relative expression of mature miRNAs were confirmed by qRT-PCR in ginger and grapefruit ENPs. To confirm the differential enrichment of miRNAs in ginger and grapefruit ENPs, we selected six miRNAs based on the heat map in Fig. 3B. These include, miRNA that are expressed equally in both ginger and grapefruit ENP (gma-miR-166 m & mtr-miR-156a), miRNAs that show higher enrichment in grapefruit ENPs (bdi-miR-5059 & osa-miR-5077) and miRNAs that show higher enrichment in ginger ENPs (aqc-miR-159 & gma-miR-6300). In agreement with the heatmap, we observed similar Ct values (between 26 and 28) for gma-miR-166 m in both ginger and grapefruit ENPs suggesting equal expression levels (Fig. 5B). Both bdi-miR-5059 and osa-miR-5077 showed significantly lower Ct values in grapefruit compared to ginger ENPs suggesting their higher expression in grapefruit ENPs, corroborating with their relative expression in the heatmap (Fig. 5D & F). However, very high levels of gma-miR-6300 was detected in both ginger and grapefruit ENPs (Ct values <20) without a significant difference in enrichment (Fig. 5E). Aqc-miR-159 showed higher expression in grapefruit ENPs compared to ginger ENPs, evidenced by significantly lower Ct values (Fig. 5C). On the other hand, mtr-miR-156a showed higher expression in ginger ENPs compared to grapefruit ENPs, evidenced by significantly lower Ct values in ginger ENPs (Fig. 5G). The discrepancy observed between our qRT-PCR results and heatmap data generated from Xiao et al. (2018) in Fig. 3B, for aqc-miR-159, gma-miR-6300 and mtr-miR-156a, could be due to intrinsic differences in miRNA expression levels caused by variation in demographic and climate conditions in which ginger and grapefruit are grown. Reaction mix lacking template cDNA did not show detectable qRT-PCR amplification in all the cases suggesting that the products formed are the result of specific amplification of mature miRNAs. This is also confirmed by resolving the PCR products through Agarose gel electrophoresis which showed single amplicon (size >100 bp) in the gel images shown below the bar graphs in Fig. 5B to G.
Fig. 5.
Validation of relative miRNA expression levels in ginger and grapefruit ENPs validated by qRTPCR. A) Total RNA (~1 μg) isolated from ginger and grapefruit ENPs were resolved through 1.5% Agarose gel electrophoresis and detected by Ethidium bromide staining. To confirm the RNA nature of the samples, it was pre-treated with or without RNAse A for 10 min before loading on the gel. M- 100 bp DNA ladder. B) to F). Bar graph shows the cycle threshold values (Ct) obtained for each miRNA in ginger and grapefruit ENPs. RT PCR reactions without template cDNA served as a negative control (NTC). Results shown are representative of three independent experiments. Error bars represent standard deviation. t-test was used for pairwise comparison between the Ct values for each miRNA in ginger and grapefruit ENPs. **** P < 0.0001, ** P < 0.01, n.s- non significant. N·D: Ct values not determined due to lack of any amplification. The qRTPCR reaction amplicons were resolved through 1.6% Agarose gel and detected by Ethidium Bromide staining. 100 bp ladder is run along with the samples to accurately detect the product size. In all gel images top and bottom arrowheads point towards 500 bp and 100 bp band sizes, respectively. In all cases, a single band of ~150 bp were obtained with no signal detected in NTC samples suggesting specific amplification.
4. Discussion
The cross-kingdom regulation of human mRNAs by dietary plant-derived miRNAs is an interesting hypothesis with far-reaching implications. One such example is the treatment of devastating diseases with a diet enriched with candidate miRNA/s that can suppress disease-causing mRNAs. Although many recent studies have provided a hint at this cross-kingdom regulation of human transcriptome via diet/plant-derived miRNAs, this has also been a subject of intense controversy (Sundaram, 2019). Cross-kingdom regulation between plant miRNAs and human transcriptome was first reported for rice derived miR-156a/miR-168 and soybean miR-159 with human targets (Chin et al., 2016; Zhang et al., 2012). These reports were later disputed due to the lack of data reproducibility, the sequence similarity between some of the plant miRNAs with human counterpart and the stability of plant miRNAs during GI digestion process (Sundaram, 2019). However, this is possible when these plant miRNAs are protected within ENPs during GI digestion, for which several recent reports provide substantial evidence. In a study by Teng et al. (2018), miR-7267-3p derived from ginger ENP was stable during GI digestion process and was shown to suppress Lactobacillus rhamnosus monooxygenase ycnE mRNA, in the gut microbiome, thereby modulating the gut immunity (Teng et al., 2018). Recently, the presence of miR-156c and miR-159a have been demonstrated in ENPs derived from walnuts and these miRNAs can regulate mammalian TNF-α signalling pathway in adipocytes and regulate inflammation (Aquilano et al., 2019). MiRNAs have also been found in grape-derived ENPs in high abundance and whether these miRNAs target human genes, is yet to be validated (Ju et al., 2013).
In this study, we further extend the possibility of exploiting the cross-kingdom regulation by miRNAs to treat the current COVID-19 pandemic. In this line, we identified ENP-derived miRNAs that could potentially target SARS-CoV-2 genes. So far, ENPs have been isolated and characterized from several edible plants and fruits (Ju et al., 2013; Xiao et al., 2018; Yang et al., 2018; Zhang et al., 2016b). We first mined these datasets to identify miRNAs that are present in high abundance in ENPs. Next, using a target prediction pipeline with stringent selection criteria, we identified 22 miRNAs that bind to SARS-CoV-2 and 12 of these miRNA candidates do not bind to SARS-CoV, a close relative of SARS-CoV-2 which caused the SARS outbreak in 2003. Further, we have also independently verified the presence of six miRNAs in ginger and grapefruit ENPs.
To our knowledge, this the first study which has systematically evaluated the possibility of using plant ENP-derived miRNAs to target viral genome. Plant miRNAs targeting influenza genome has been reported earlier for miR-2911, a miRNA highly expressed in honeysuckle (HS) plant. When mice were administered orally with HS decoction, high levels of miR-2911 was detected in serum and this could suppress influenza virus (H5N1) infection in vitro and in vivo (Zhou et al., 2015). The same authors recently showed SARS-CoV-2 targeting potential of HS derived miR-2911. SARS-CoV-2 possessed twenty eight miR-2911 binding sites within its genome and the specific binding of miR-2911 mimics with SARS-CoV-2 target sites were validated in vitro with chimeric luciferase assays. Exosome encapsulated with miR-2911 mimics, was able to suppress SARS-CoV-2 viral replication in vitro. Further, administration of HS decoction to human volunteers led to a favourable reduction in time taken to become SARS-CoV-2 negative (Zhou et al., 2020). However, in both influenza and SARS-CoV-2 studies, encapsulation of miR-2911 in vesicles, is an essential process for systemic delivery of miR-2911. Interestingly, miR-2911 appears to be intrinsically resistant to nucleases and also have a natural tendency to associate itself within exosomes after absorption in GI tract. However, it is not clear if all plant-derived miRNAs from the diet exhibit nuclease resistant and are encapsulated in serum exosomes. Hence, plant miRNAs targeting SARS-CoV-2 may require additional encapsulation in liposomal or nano formulations for intracellular delivery(Lee et al., 2019). The innate ability of ENPs to fuse with any type of mammalian cells obviates the need for separate miRNA delivery methods to target COVID-19 (Wang et al., 2013). Further, ENPs derived from different host plants appears to have preferential target tissue which is likely attributed to their lipid composition (Yang et al., 2018). It has been shown earlier that intranasal/venous delivery of grapefruit derived ENPs, was detected in murine lung tissue up to 72 h. post-delivery, suggesting that SARS-CoV-2 targeting ENP miRNAs could be delivered safely via the intranasal/venous route for tissues affected in COVID-19. However, further experimental research supporting spontaneous uptake of ENPs in lung epithelial cells in vivo, direct binding of ENP derived miRNAs to SARS-CoV-2 transcriptome and suppression of SARS-CoV-2 infection in vitro and in vivo are warranted in future. Alternatively, candidate miRNAs which efficiently suppresses SARS-CoV-2 replication can be overexpressed in edible plants by transgenic approach and ENPs isolated from the transgenic plants can be used for COVID-19 therapy (Yang et al., 2017).
For the successful therapeutic application of ENPs/ENP derived miRNAs, large scale compatible purification methods are essential. We have recently demonstrated a cost-effective and scalable purification method to purify ginger ENPs using PEG-6000 (Kalarikkal et al., 2020). Ginger ENPs purified by this method was structurally and functionally similar to ENPs purified by the gold standard differential ultracentrifugation method which requires expensive instrumental set up and lack scalability. The PEG method developed was advantageous in multiple aspects. Firstly, food-grade PEG is cheaper (<4USD per Kg), it is generally regarded as safe (GRAS) material and it is also employed as a food additive (Panel et al., 2018). Hence, we speculate that exploiting ENP-derived miRNAs as a therapeutic modality against COVID-19 infection offers a safer, target-specific and cheaper solution. Thus, it is evident from our study that ENP derived miRNAs may be developed as a non-toxic therapeutic modality against COVID-19. However, further validation of ENP derived miRNAs binding to target mRNA (SARS-CoV-2 genome/transcriptome) could not be performed due to the requirement of advanced biosafety level − 3 laboratories to handle virus cultures.
5. Conclusion
In this manuscript, we have provided in silico evidence for the ability of ENP derived miRNAs to target SARS-CoV-2 transcriptome. Further, the presence of SARS-CoV-2 target miRNAs was experimentally confirmed in ginger and grapefruit ENPs. The predicted miRNAs have also been verified for target site accessibility, suggesting their higher likelihood to target SARS-CoV-2 genome/transcriptome within the intracellular environment. By employing SARS-CoV-2 targeting miRNAs in the form of ENPs, these miRNAs can selectively be delivered in primary target tissues of SARS-CoV-2. However, further investigations on the intracellular stability of these miRNAs and their purported anti-viral activity under in vitro and in vivo conditions have to be performed. In future, comprehensive experimental validation of these issues will aid in the development of efficient and non-toxic therapy for COVID-19.
List of miRNAs, sequence, read counts of selected miRNAs in ENPs
Target site accessibility of miRNA binding site in SARS-CoV-2 genome. RNA secondary structures were predicted using MFOLD servers in miRNA sequence plus -17 nt upstream and + 13 nt downstream sequence.
Author contribution
SPK performed sequence retrieval, data analysis and experiments. GMS conceived this study and responsible for the overall experimental design, results interpretation, manuscript preparation and communication.
Declaration of Competing Interest
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
Acknowledgements
The authors would like to acknowledge the Director CSIR-CFTRI for facilities and support. Academy of Scientific and Innovative Research (AcSIR) is acknowledged for the fellowship received by SPK. This work was supported by the Start-up Research Grant, Department of Science and Technology, Science and Engineering Research Board, New Delhi, India (SRG/2019/000584). The authors are grateful to Ms. Anaghapriya for assistance in critical evaluation of the manuscript and suggestions.
References
- Aquilano K., Ceci V., Gismondi A., De Stefano S., Iacovelli F., Faraonio R., Di Marco G., Poerio N., Minutolo A., Minopoli G., Marcone A., Fraziano M., Tortolici F., Sennato S., Casciardi S., Potestà M., Bernardini R., Mattei M., Falconi M., Montesano C., Rufini S., Canini A., Lettieri-Barbato D. Adipocyte metabolism is improved by TNF receptor-targeting small RNAs identified from dried nuts. Commun. Biol. 2019;2:317. doi: 10.1038/s42003-019-0563-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babadaei M.M.N., Hasan A., Vahdani Y., Bloukh S.H., Sharifi M., Kachooei E., Haghighat S., Falahati M. Development of remdesivir repositioning as a nucleotide analog against COVID-19 RNA dependent RNA polymerase. J. Biomol. Struct. Dyn. 2020;1–9 doi: 10.1080/07391102.2020.1767210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells I., Cirera S., Busk P.K. Specific and sensitive quantitative RT-PCR of miRNAs with DNA primers. BMC Biotechnol. 2011;11:70. doi: 10.1186/1472-6750-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov P.V., Henderson C.M., Anderson C.B., Gesteland R.F., Atkins J.F., Howard M.T. Programmed ribosomal frameshifting in decoding the SARS-CoV genome. Virology. 2005;332:498–510. doi: 10.1016/j.virol.2004.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin A.R., Fong M.Y., Somlo G., Wu J., Swiderski P., Wu X., Wang S.E. Cross-kingdom inhibition of breast cancer growth by plant miR159. Cell Res. 2016;26:217–228. doi: 10.1038/cr.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gioia S., Hossain M.N., Conese M. Biological properties and therapeutic effects of plant-derived nanovesicles. Open Med. (Warsaw, Poland) 2020;15:1096–1122. doi: 10.1515/med-2020-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didiano D., Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat. Struct. Mol. Biol. 2006;13:849–851. doi: 10.1038/nsmb1138. [DOI] [PubMed] [Google Scholar]
- Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Ther. 2020;14:58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- Ju S., Mu J., Dokland T., Zhuang X., Wang Q., Jiang H., Xiang X., Deng Z.-B., Wang B., Zhang L., Roth M., Welti R., Mobley J., Jun Y., Miller D., Zhang H.-G. Grape exosome-like nanoparticles induce intestinal stem cells and protect mice from DSS-induced colitis. Mol. Ther. 2013;21:1345–1357. doi: 10.1038/mt.2013.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalarikkal S.P., Prasad D., Kasiappan R., Chaudhari S.R., Sundaram G.M. A cost-effective polyethylene glycol-based method for the isolation of functional edible nanoparticles from ginger rhizomes. Sci. Rep. 2020;10:4456. doi: 10.1038/s41598-020-61358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. The role of site accessibility in microRNA target recognition. Nat. Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- Lee S.W.L., Paoletti C., Campisi M., Osaki T., Adriani G., Kamm R.D., Mattu C., Chiono V. MicroRNA delivery through nanoparticles. J. Control. Release. 2019;313:80–95. doi: 10.1016/j.jconrel.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xie X., Li J., Cui Y., Hou Y., Zhai L., Wang X., Fu Y., Liu R., Bian S. Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s. BMC Plant Biol. 2017;17:32. doi: 10.1186/s12870-017-0983-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long D., Lee R., Williams P., Chan C.Y., Ambros V., Ding Y. Potent effect of target structure on microRNA function. Nat. Struct. Mol. Biol. 2007;14:287–294. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- Masters P.S.B., T A., In V.R. Academic Press; 2006. The Molecular Biology of Coronaviruses; pp. 193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mückstein U., Tafer H., Hackermüller J., Bernhart S.H., Stadler P.F., Hofacker I.L. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. (Lausanne). 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panel E.P., On F.A., N.S. added to F, EFSA A.N.S., Younes M., Aggett P., Aguilar F., Crebelli R., Dusemund B., Filipič M., Frutos M.J., Galtier P., Gott D., Gundert-Remy U., Kuhnle G.G., Lambré C., Lillegaard I.T., Moldeus P., Mortensen A., Oskarsson A., Stankovic I., Waalkens-Berendsen I., Woutersen R.A., Wright M., Boon P., Lindtner O., Tlustos C., Tard A., Leblanc J.-C. Refined exposure assessment of polyethylene glycol (E 1521) from its use as a food additive. EFSA J. 2018;16 doi: 10.2903/j.efsa.2018.5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rome S. Biological properties of plant-derived extracellular vesicles. Food Funct. 2019;10:529–538. doi: 10.1039/c8fo02295j. [DOI] [PubMed] [Google Scholar]
- Sardar R., Satish D., Birla S., Gupta D. Comparative analyses of SAR-CoV2 genomes from different geographical locations and other coronavirus family genomes reveals unique features potentially consequential to host-virus interaction and pathogenesis. bioRxiv. 2020 doi: 10.1101/2020.03.21.001586. 2020.03.21.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S., Jónsson Z.O., Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- Stecher G., Tamura K., Kumar S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020;37:1237–1239. doi: 10.1093/molbev/msz312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram G.M. Dietary non-coding RNAs from plants: fairy tale or treasure? Non-coding RNA Res. 2019 doi: 10.1016/J.NCRNA.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., Hutchins E., Mu J., Deng Z., Luo C., Sundaram K., Sriwastva M.K., Zhang L., Hsieh M., Reiman R., Haribabu B., Yan J., Jala V.R., Miller D.M., Van Keuren-Jensen K., Merchant M.L., McClain C.J., Park J.W., Egilmez N.K., Zhang H.-G. Plant-derived Exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe. 2018;24:637–652. doi: 10.1016/j.chom.2018.10.001. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Zhuang X., Mu J., Deng Z. Bin, Jiang H., Xiang X., Wang B., Yan J., Miller D., Zhang H.G. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013;4:11347. doi: 10.1038/ncomms2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Ren Y., Mu J., Egilmez N.K., Zhuang X., Deng Z., Zhang L., Yan J., Miller D., Zhang H.G. Grapefruit-derived nanovectors use an activated leukocyte trafficking pathway to deliver therapeutic agents to inflammatory tumor sites. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkos T.M., Koscianska E., Krzyzosiak W.J. Practical aspects of microRNA target prediction. Curr. Mol. Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Feng S., Wang X., Long K., Luo Y., Wang Y., Ma J., Tang Q., Jin L., Li X., Li M. Identification of exosome-like nanoparticle-derived microRNAs from 11 edible fruits and vegetables. PeerJ. 2018;6 doi: 10.7717/peerj.5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Zhong L., Deng J., Peng J., Dan H., Zeng X., Li T., Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Zhao, Teng T., Abdalla A.E., Zhu W., Xie L., Wang Y., Guo X. Systematic Comparison of Two Animal-to-Human Transmitted Human Coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12 doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Primo C., Elbaz-Younes I., Hirschi K.D. Bioavailability of transgenic microRNAs in genetically modified plants. Genes Nutr. 2017;12:17. doi: 10.1186/s12263-017-0563-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Zhang M., Merlin D. Advances in plant-derived edible nanoparticle-based lipid nano-drug delivery systems as therapeutic nanomedicines. J. Mater. Chem. B. 2018;6:1312–1321. doi: 10.1039/C7TB03207B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., Yin Y., Wang C., Zhang T., Zhu D., Zhang D., Xu J., Chen Q., Ba Y., Liu J., Wang Q., Chen J., Wang J., Wang M., Zhang Q., Zhang J., Zen K., Zhang C.-Y. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 2012;22:107–126. doi: 10.1038/cr.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Viennois E., Prasad M., Zhang Y., Wang L., Zhang Z., Han M.K., Xiao B., Xu C., Srinivasan S., Merlin D. Edible ginger-derived nanoparticles: a novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials. 2016;101:321–340. doi: 10.1016/j.biomaterials.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Viennois E., Xu C., Merlin D. Plant derived edible nanoparticles as a new therapeutic approach against diseases. Tissue Barriers. 2016;4 doi: 10.1080/21688370.2015.1134415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Li X., Liu Jinxiong, Dong L., Chen Q., Liu Jialing, Kong H., Zhang Q., Qi X., Hou D., Zhang L., Zhang G., Liu Y., Zhang Y., Li J., Wang J., Chen X., Wang H., Zhang J., Chen H., Zen K., Zhang C.-Y. Honeysuckle-encoded atypical microRNA2911 directly targets influenza a viruses. Cell Res. 2015;25:39–49. doi: 10.1038/cr.2014.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L.-K., Zhou Z., Jiang X.-M., Zheng Y., Chen X., Fu Z., Xiao G., Zhang C.-Y., Zhang L.-K., Yi Y. Absorbed plant MIR2911 in honeysuckle decoction inhibits SARS-CoV-2 replication and accelerates the negative conversion of infected patients. Cell Discov. 2020;6:54. doi: 10.1038/s41421-020-00197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of miRNAs, sequence, read counts of selected miRNAs in ENPs
Target site accessibility of miRNA binding site in SARS-CoV-2 genome. RNA secondary structures were predicted using MFOLD servers in miRNA sequence plus -17 nt upstream and + 13 nt downstream sequence.