Mutations in ANO5 cause a limb-girdle muscular dystrophy characterized by membrane repair defects that make muscles more susceptible to permanent damage from normal use. This work links failed membrane repair in ANO5-deficient muscle to altered trafficking of the annexins, crucial repair proteins.
Abstract
Mutations in ANO5 (TMEM16E) cause limb-girdle muscular dystrophy R12. Defective plasma membrane repair is a likely mechanism. Using myofibers from Ano5 knockout mice, we show that trafficking of several annexin proteins, which together form a cap at the site of injury, is altered upon loss of ANO5. Annexin A2 accumulates at the wound to nearly twice the level observed in WT fibers, while annexin A6 accumulation is substantially inhibited in the absence of ANO5. Appearance of annexins A1 and A5 at the cap is likewise diminished in the Ano5 knockout. These changes are correlated with an alteration in annexin repair cap fine structure and shedding of annexin-positive vesicles. We conclude that loss of annexin coordination during repair is disrupted in Ano5 knockout mice and underlies the defective repair phenotype. Although ANO5 is a phospholipid scramblase, abnormal repair is rescued by overexpression of a scramblase-defective ANO5 mutant, suggesting a novel, scramblase-independent role of ANO5 in repair.
Introduction
Recessive mutations in the ANO5 gene are responsible for a spectrum of myopathies with variable severity. These disorders, characterized by muscle weakness and atrophy, include limb-girdle muscular dystrophy (LGMD) R12 (LGMDR12/LGMD2L), Miyoshi muscular dystrophy type 3, and muscle weakness with myalgia and rhabdomyolysis (Bolduc et al., 2010; Hicks et al., 2011; Liewluck and Milone, 2018; Mahjneh et al., 2010). Since Anoctamin-5 (ANO5)myopathies were discovered (Bolduc et al., 2010), >70 ANO5 variants have been reported to be pathogenic (https://www.ncbi.nlm.nih.gov/clinvar/). In a screen of 35 LGMD genes in 4,656 clinically suspected LGMD patients, ANO5 was the fourth–most likely contributor to LGMD phenotypes in the United States (Nallamilli et al., 2018). ANO5 protein expression is absent in patients with the founder mutation (c.191dupA, p.Asn64Lysfs*15; Hicks et al., 2011; Sarkozy et al., 2013; Vihola et al., 2018), and ANO5 protein is reduced in patients with some point mutations (e.g., c.2101A>G, p.N701D and c.2272C>T, p.R758C; Chandra et al., 2019; Vihola et al., 2018). Although ANO5 myopathies are inherited in a recessive manner, dominant ANO5 mutations are associated with gnathodiaphyseal dysplasia, a skeletal syndrome (Andreeva et al., 2016; Duong et al., 2016; Katoh and Katoh, 2004; Mizuta et al., 2007; Otaify et al., 2018).
ANO5 is a member of the TMEM16/ANO family that includes ion channels and phospholipid scramblases (PLSases; Falzone et al., 2018; Pedemonte and Galietta, 2014; Whitlock and Hartzell, 2017). Within this family, ANO5 is most closely related to the PLSase ANO6 (Gyobu et al., 2016; Mizuta et al., 2007; Suzuki et al., 2013; Suzuki et al., 2010). Phospholipid scrambling (PLS) disrupts the asymmetric distribution of phospholipids that normally exists between the leaflets of the plasma membrane (Bevers and Williamson, 2016). A consequence of PLS is the exposure of anionic phospholipids such as phosphatidylserine (PtdSer), which are normally sequestered on the intracellular face of the plasma membrane, to the extracellular space (Bevers and Williamson, 2016; Whitlock and Hartzell, 2017). Heterologous expression of ANO5 in HEK cells induces PLS (Di Zanni et al., 2020; Di Zanni et al., 2018; Whitlock et al., 2018), and PLS is absent in muscle precursor cells (MPCs) from mice with disruption of Ano5 (ANO5-knockout [KO]). However, the pathological mechanism of ANO5 myopathies is poorly understood, and it remains unclear whether ANO5-linked disorders are a consequence of defective PLS.
ANO5 myopathies are likely caused by defects in the ability of muscle fibers to self-repair (Bolduc et al., 2010; Chandra et al., 2019; Jaiswal et al., 2007; Monjaret et al., 2013). Skeletal muscle endures considerable mechanical stress during normal use, which produces small tears in the plasma membrane (sarcolemma; Proske and Morgan, 2001). Healthy muscle has robust mechanisms to repair torn sarcolemma through the coordinated interplay of various proteins, lipids, organelles, and small molecules (Andrews and Corrotte, 2018; Barthélémy et al., 2018; Blazek et al., 2015; Cooper and McNeil, 2015; Horn and Jaiswal, 2018). Slow or incomplete resealing results in prolonged Ca2+ entry that activates calpain endopeptidases and phospholipases to initiate muscle damage (Gissel and Clausen, 2001). Repair mechanisms are disrupted in several muscular dystrophies, most notably LGMD R2 (LGMDR2/LGMD2B) and Miyoshi muscular dystrophy type 1 (Bansal et al., 2003), caused by mutations in the dysferlin (DYSF) gene (Bashir et al., 1998; Liu et al., 1998). We have shown that ANO5-KO mice display a muscle phenotype that includes defective cell membrane repair (Griffin et al., 2016b; Whitlock et al., 2018). Recently, the pathology of LGMDR12 has been tied to defective membrane repair through a mechanism involving ANO5-regulated Ca2+ handling by the ER (Chandra et al., 2019). While ANO5 myopathies share features with dysferlinopathies, ANO5 and dysferlin probably have nonoverlapping roles because ANO5 overexpression in dysferlin-null mice does not rescue the repair defect (Monjaret et al., 2013).
One of the most rapid responses to membrane injury is the Ca2+-dependent accumulation of annexins ANXA1, ANXA2, ANXA4, ANXA5, ANXA6, and ANXA11 at the wound (Bittel et al., 2020; Bouter et al., 2011; Carmeille et al., 2016; Demonbreun et al., 2019; Demonbreun et al., 2016b; Fuson et al., 2014; Koerdt et al., 2019; Lennon et al., 2003; Middel et al., 2016; Roostalu and Strähle, 2012; Swaggart et al., 2014). Annexins have well-established roles in plasma membrane repair in various cell types (Boye and Nylandsted, 2016; Häger and Nylandsted, 2019; Jaiswal et al., 2014; Lennon et al., 2003; McNeil et al., 2006; Sønder et al., 2019). Membrane repair in vitro and in vivo is enhanced by overexpression of ANXA1, A2, or A6 (Demonbreun et al., 2019) or extracellular application of ANXA5 or A6 protein (Bouter et al., 2011). Loss of ANXA2 is associated with defective membrane repair, and it was recently proposed that ANXA2 plays a role in dysferlin trafficking to the wound (Bittel et al., 2020). Annexins are known to perform a spectrum of diverse membrane-related functions that range from membrane stabilization to membrane deformation and promotion of fusion (Florentsen et al., 2020; Gerke and Moss, 2002; Koerdt et al., 2019; Moreno-Pescador et al., 2019; Simonsen et al., 2020). Various annexins differ in their Ca2+ affinities, binding partners, and quaternary structures, and this is thought to confer each annexin with a unique biological role (Gerke and Moss, 2002; Koerdt et al., 2019). The idea that each annexin may have its own unique function is supported by the observation that the recruitment of annexins to the lesion occurs sequentially and in some cases interdependently (Demonbreun et al., 2016b; Koerdt et al., 2019; Roostalu and Strähle, 2012). Annexins bind anionic phospholipids in a Ca2+-dependent manner, and accumulation of repair proteins at the lesion coincides with local enrichment of PtdSer at the plasma membrane adjacent to the lesion or in membrane “patches” that arise during repair (Demonbreun et al., 2016b; Middel et al., 2016). This PtdSer enrichment is intriguing because both the annexins and dysferlin bind PtdSer; however, the mechanism of PtdSer externalization is unknown but may involve membrane trafficking or phospholipid scrambling. The latter would provide an attractive link to ANO5 in the repair process.
Here, we show that ANO5 rapidly appears at the plasma membrane of muscle fibers damaged with a laser pulse. Furthermore, loss of ANO5 is associated with abnormal trafficking of several annexin proteins to the wound. However, contrary to our initial hypothesis, PtdSer exposure during repair is not ANO5 dependent, and ANO5-mediated PLS is dispensable for repair. Lastly, we demonstrate that a patient-associated point mutation in ANO5 results in defective repair in primary human myocytes, implicating defective membrane repair in the development of LGMDR12.
Results
ANO5-deficient myofibers are defective in membrane repair
A standard method to study membrane repair involves rupturing the cell membrane with an intense laser pulse (Fig. 1 A; Bansal et al., 2003; McNeil et al., 2003). We used two complementary approaches to evaluate sarcolemmal resealing after a laser pulse. First, muscle fibers from WT or ANO5-KO mice were loaded with the Ca2+-sensitive dye Cal-520 (Kd = 320 nM). Upon membrane damage, extracellular Ca2+ entered myofibers, resulting in a transient increase in Cal-520 fluorescence. In WT fibers, maximal fluorescence was observed 24 s after injury and then declined with a t1/2 = 72 s to reach a plateau level that was ∼31.6% greater than baseline (Fig. 1, B and C). In contrast, in ANO5-KO fibers, Ca2+ transients peaked at 28 s after injury and declined with a t1/2 = 56 s to a final value 76.7% greater than baseline (Fig. 1, B and C). To quantitatively compare the response of WT or ANO5-KO muscle to damage, we calculated the area under the curve (AUC) for Ca2+ transients from individual fibers. AUCs were significantly higher in ANO5-KO fibers, indicating increased intracellular Ca2+ levels after injury (Fig. 1 D). It should be noted that a possible role of ANO5 in regulation of Ca2+ homeostasis (Chandra et al., 2019) may also contribute to the changes in the postinjury Ca2+ transients in ANO5-KO myofibers.
Figure 1.
Plasma membrane repair is defective in ANO5-KO mouse muscle fibers. (A) Workflow for laser injury assay. FDB myofibers were isolated and damaged with 405-nm light directed at the plasma membrane on the lateral edge of the fiber. (B) Representative images of cytosolic Ca2+ detected with Cal-520 following membrane damage in WT or ANO5-KO fibers. t = 0 is before injury (WT and ANO5-KO data points superimposed). Scale bars = 10 µm. (C and D) Cal-520 fluorescence time course after injury, normalized to initial fluorescence (C) and AUC values (D) for individual WT or KO fibers. ANO5-KO n = 12 fibers from three mice, WT n = 18 fibers from two mice. (E) Representative images of FM1-43 infiltration following membrane damage in WT or ANO5-KO fibers. Scale bars = 10 µm. (F and G) Time course of FM1-43 fluorescence normalized to average maximal fluorescence in ANO5-KO fibers (FmaxKO; G) and AUC values for individual WT or KO fibers (G). ANO5-KO n = 17 fibers from three mice, WT n = 22 fibers from three mice. (H) Maximum intensity projections of FM1-43 fluorescence generated from z-stacks of WT or ANO5-KO fibers acquired ∼10 min after injury. (I and J) Quantification of FM1-43–labeled blebs in the repair patch following injury (I) and patch volumes (J). Blebbing of the patch in ANO5-KO fibers contributed to a significant increase in overall patch volume. ANO5-KO n = 17 fibers from three mice, WT n = 14 fibers from two mice. Data are mean ± SEM. *, P < 0.05. Scale bar = 10 μm.
Second, we employed a common damage assay based on the cell-impermeant dye FM1-43 (Bansal et al., 2003; Carmeille et al., 2016; Demonbreun et al., 2019; Demonbreun et al., 2016b; Griffin et al., 2016a; Swaggart et al., 2014). FM1-43 is a water-soluble stearyl dye that fluoresces intensely when inserted into lipid-rich membranes. When the sarcolemma is damaged, FM1-43 enters the cell, labels internal membranes, and produces a bright fluorescent spot around the site of injury. If the membrane is quickly repaired, the quantity of dye entering the fiber is attenuated compared with fibers where repair is defective. In our system, FM1-43 fluorescence increased after injury approximately three times more rapidly in ANO5-KO muscle fibers than in WT. Fluorescence intensity doubled at t = 16 s and t = 48 s after injury in ANO5-KO and WT fibers, respectively (Fig. 1, E and F), similar to that previously reported under different experimental conditions (Griffin et al., 2016b). ANO5-KO FM1-43 AUCs were significantly larger than those of WT (Fig. 1 G). These data strongly support the idea that ANO5-deficient myofibers are defective in plasma membrane repair.
The structure of the membrane repair patch is abnormal in the absence of ANO5
Sarcolemmal injuries in the presence of FM1-43 revealed the formation of a large “cap” of fluorescence at the site of injury. This cap, which likely is involved in the repair of the lesion, was compact in WT fibers but was larger and heavily vesiculated in ANO5-KO fibers. While membrane blebs were occasionally observed in repairing WT myofibers as well, these blebs were released as extracellular vesicles (Video 1), consistent with previous reports (Sønder et al., 2019). The volume of FM1-43–positive membrane in the patch was significantly elevated in ANO5-KO fibers, which is plausibly explained by the retention of blebs/vesicles (Fig. 1, H–J and Video 2). This difference in the structure of the repair cap and the difference in extracellular vesicle shedding is also revealed by the structure of the repair cap visualized by annexin accumulation as described in a subsequent section (Fig. 4, Video 4, Video 6, and Video 7).
Video 1.
Patch material is shed in WT myofibers. Plasma membrane of an isolated WT muscle fiber was ablated with 405-nm laser application in the presence of the lipophilic dye FM1-43 (green). Intracellular accumulation of the dye was monitored via time-lapse fluorescence confocal microscopy. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
Video 2.
Prominent blebbing of repair patch in ANO5-KO myofibers. Plasma membrane of an isolated ANO5-KO muscle fiber was ablated with 405-nm laser application in the presence of the lipophilic dye FM1-43 (green). Intracellular accumulation of the dye was monitored via time-lapse confocal fluorescence microscopy. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
Figure 4.
Architecture of the annexin repair cap is lost in ANO5-KO fibers. (A and B) Representative images of coelectroporated ANXA2 and ANXA5 (A) or ANXA2 and ANXA6 (B) in injured muscle fibers. Scale bars = 10 µm. (C and D) Quantification of the spatial organization of annexin proteins derived from line profiles drawn through the repair cap images taken ∼7 min after injury. ANXA2 and ANXA5: ANO5-KO n = 23 fibers from three mice, WT n = 26 fibers from three mice; ANXA2 and ANXA6: ANO5-KO n = 18 fibers from three mice, WT n = 14 fibers from two mice. (E) Orthogonal views of the annexin repair cap generated from z-stacks acquired ∼10 min after injury. Scale bars = 5 µm. (F) Volumes for individual annexin proteins in repair caps following injury. Z-plane images were acquired every 0.5 µm. ANXA2: ANO5-KO n = 35 fibers from four mice, WT n = 36 fibers from four mice; ANXA5: ANO5-KO n = 32 fibers from three mice, WT n = 37 fibers from four mice; ANXA6: ANO5-KO n = 13 fibers from two mice, WT n = 21 fibers from three mice. Data are mean ± SEM. *, P < 0.05.
Video 4.
Repair cap ANXA2 is released in vesicles from WT myofibers. An isolated WT myofiber expressing ANXA2-mEmerald fusion protein was injured and imaged with confocal fluorescence microscopy for several minutes as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A number of ANXA2-positive vesicles are seen to be shed from the repair cap as the fiber repairs itself. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
Video 6.
ANO5 but not ANO5-1-5 overexpression in ANO5-KO fibers reduces ANXA2 accumulation in the repair cap. ANO5-KO fibers left untransfected (left), electroporated with plasmid encoding ANO5 (not shown) and ANXA2 (green; middle), or electroporated with plasmid encoding chimeric ANO5-1-5 (not shown) and ANXA2 (green; right) were laser damaged and imaged with time-lapse confocal microscopy. Substantial ANXA2-positive material is seen being shed from fibers expressing ANO5, but not from untransfected fibers or fibers expressing ANO5-1-5. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
Video 7.
ANO5 or ANO5-1-5 overexpression in ANO5-KO fibers improves ANXA6 trafficking during the wound response. ANO5-KO fibers left untransfected (left), electroporated with plasmid encoding ANO5 (not shown) and ANXA6 (green; middle), or electroporated with plasmid encoding chimeric ANO5-1-5 (not shown) and ANXA6 (green; right) were laser damaged and imaged with time-lapse confocal microscopy. ANXA6 accumulation was improved, both in terms of response time and overall magnitude, in fibers expressing ANO5 or ANO5-1-5. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
ANO5 traffics to the site of injury
Given that muscle fibers lacking ANO5 are repair deficient, we sought to determine if ANO5 localizes to the site of injury using plasmids encoding fluorescently tagged ANO5 electroporated into muscle fibers. We observed fast accumulation of ANO5 at the plasma membrane in WT fibers adjacent to the site of damage (Fig. 2, A and C), a region termed the wound “shoulder” (Demonbreun et al., 2016b). This behavior was recapitulated in ANO5-KO fibers, indicating that the machinery necessary for ANO5 trafficking is intact (Fig. 2, A and C). The time course of accumulation of ANO5 was well fit by a single exponential with a similar τ for WT and KO fibers (WT τ = 19.9 s, ANO5-KO τ = 17.5 s; Fig. S1 A). This trafficking was unlikely to be explained by local, injury-induced muscle contraction because ANO1 did not accumulate like ANO5; furthermore, it cannot be explained by FRAP because we observed little to no recovery of ANO5-tdTomato signal after bleaching with a pulsed 561-nm laser, which does not damage the plasma membrane (Fig. S1, B–F). Because plasma membrane repair is Ca2+ dependent and ANO5 is Ca2+ activated, we measured the kinetics of ANO5 trafficking to the wound shoulder and the injury-induced Ca2+ transient. ANO5-Neon was coelectroporated with the genetically encoded Ca2+ sensor R-GECO1.2 (Kd = 1.2 µM; Wu et al., 2013). As seen with Cal-520 (Fig. 1 B), intracellular Ca2+ increased in a semicircular area immediately after injury (Video 3). The time course of ANO5 accumulation at the wound shoulder paralleled the time course of the Ca2+ transient. As the Ca2+ levels returned to baseline, ANO5 dispersed from the site of injury.
Figure 2.
ANO5 accumulates at the plasma membrane in response to wounding. (A and B) Deconvolved images of ANO5 (A) or dysferlin (B) translocation to wound-adjacent plasma membrane in WT or ANO5-KO fibers after injury. Scale bars = 10 µm. (C) Quantification of ANO5 fluorescence at the site of injury in ANO5-KO or WT fibers, normalized to fluorescence before injury. ANO5-KO n = 16 fibers from four mice, WT n = 15 fibers from four mice. (D) Quantification of dysferlin fluorescence in ANO5-KO (red squares) or WT (black circles) myofibers following injury. The time course of hANO5 accumulation in WT fibers is shown as a blue dotted line. ANO5-KO n = 11 fibers from two mice, WT n = 9 fibers from three mice. Data are mean ± SEM.
Figure S1.
Rapid ANO5 accumulation at injury sites is not explained by FRAP or localized fiber contraction. (A) Time constants and associated errors from single exponential fits of ANO5 or dysferlin accumulation time courses (from Fig. 2). (B) Deconvolved images showing human ANO5-tomato after bleaching with an 8-s 561-nm laser pulse. (C) Quantification of human ANO1 fluorescence at wound-adjacent sarcolemma following 405-nm laser injury or ANO5 fluorescence after 561-nm laser bleaching in WT mouse myofibers. Fluorescence is normalized to prelaser fluorescence values. Time course of human ANO5 accumulation in WT fibers (from Fig. 2) is shown as a blue dotted line for comparison. ANO1 n = 10 fibers pooled from two mice, ANO5 n = 8 fibers from one mouse. (D) Images showing the contribution of localized myofiber contraction after injury to the apparent accumulation of plasma membrane proteins at the wound site. (E and F) Line profile analysis of ANO1 (E) or ANO5 (F) fluorescent intensity before (Initial) or after (Max) laser injury. Fluorescence values are measured from dotted lines as shown in top-row images of D. “Distance” is in units of micrometers along the analysis line, centered on the maximal relative fluorescence. ANO1 n = 8 fibers pooled from two mice, ANO5 n = 9 fibers pooled from four mice. Data are mean ± SEM. Scale bar = 10 μm.
Video 3.
ANO5 accumulates at the wound shoulder throughout the Ca2+ transient. WT myofibers were electroporated with plasmid encoding ANO5-mNeon fusion (green) and the Ca2+-sensitive fluorescent protein RGECO1.2. Relocation of plasma membrane ANO5 to the wound shoulder was observed simultaneously with the injury-induced Ca2+ transient following laser damage via time-lapse confocal fluorescence microscopy. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
ANO5 trafficking after injury resembles that reported for dysferlin, a protein with strong connections to sarcolemmal repair (Bansal et al., 2003; Cárdenas et al., 2016; Demonbreun et al., 2016b; Lennon et al., 2003; Middel et al., 2016). The similarity in clinical presentations of LGMDR2 and LGMDR12 have suggested a link between ANO5 and dysferlin, but a mechanistic relationship has not been established. To test whether dysferlin trafficking is altered upon loss of ANO5, we examined the response of dysferlin-EGFP to laser damage in WT or ANO5-KO myofibers. We conclude that the repair defect in ANO5-deficient muscle is independent of dysferlin because the kinetics of dysferlin accumulation was nearly identical in WT (τ = 29.0 s) and ANO5-KO fibers (τ = 25.8 s; Fig. 2, B and D; and Fig. S1 A).
Annexins require ANO5 for normal trafficking
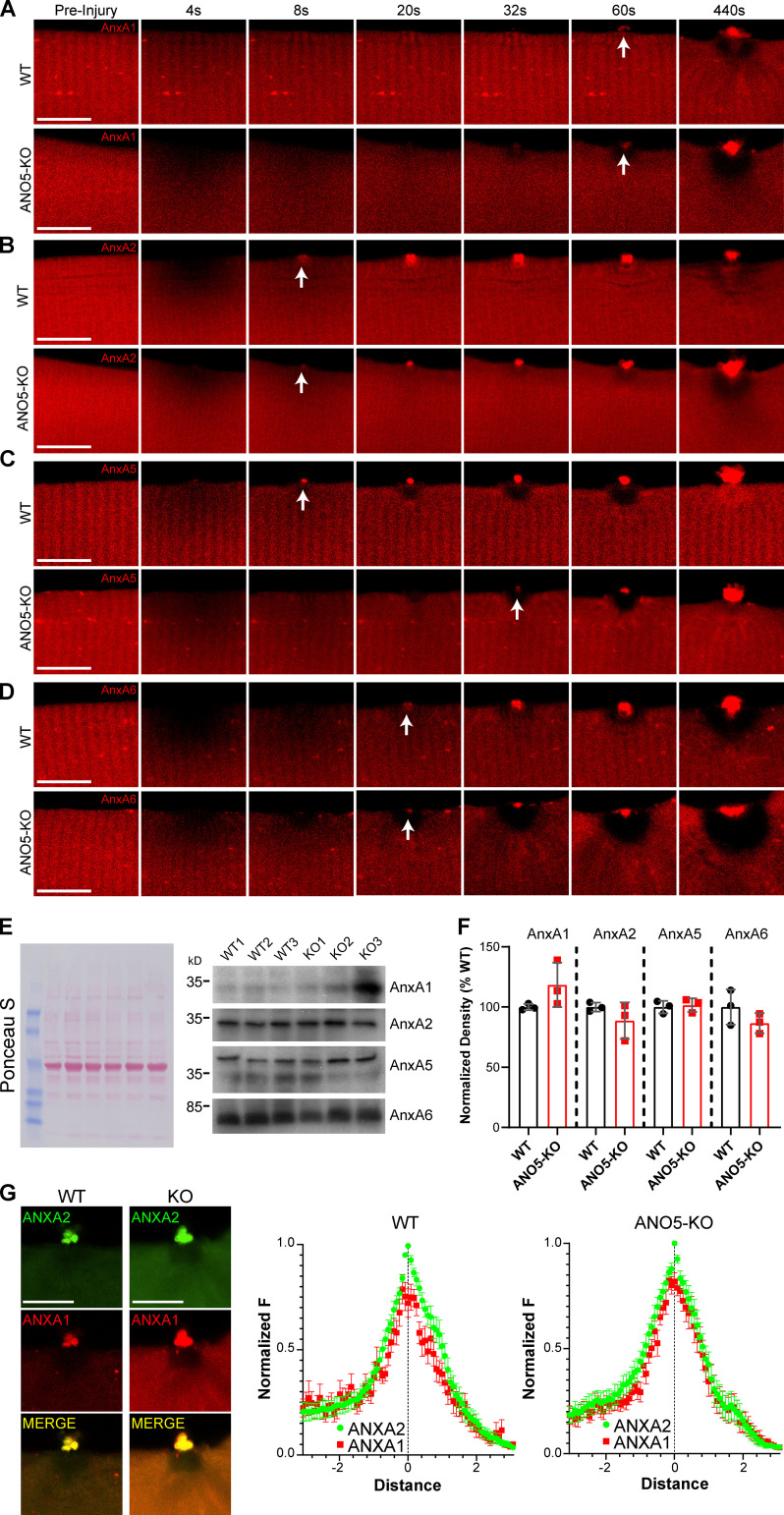
Because annexins play important roles in sarcolemmal repair, we tested whether delayed membrane resealing in ANO5-KO fibers was associated with abnormal trafficking of annexins to the site of injury. We expressed fluorescently tagged ANXA1, ANXA2, ANXA5, and ANXA6 (individually or together) in WT or ANO5-KO fibers and followed cap formation after injury (Fig. S2). All annexins trafficked to the cap with biphasic kinetics (Fig. 3; Table S1). To quantify the kinetics, fluorescence at a given time point (F) divided by fluorescence before injury (F0), F/F0, versus time was fit to the sum of two exponentials (R2 > 0.995). In WT, ANXA5 (τfast = 12.8 s, τslow = 142 s) and ANXA2 (τfast = 24 s, τslow = 118 s) accumulated most rapidly. By comparison, F/F0 for ANXA6 reached a higher plateau but proceeded more slowly (τfast = 32.6 s, τslow = 543 s). ANXA2 and ANXA6 were affected dramatically by loss of ANO5, but in opposite directions. F/F0 amplitude of the slow component of ANXA2 accumulation was increased approximately fourfold, corresponding to significantly greater amounts of ANXA2 in the repair cap at later time points (Fig. 3 B and Fig. S2). In WT fibers, ANXA2 was shed as extracellular vesicles from the cap in a manner as described above for FM1-43 (Video 4). Thus, ANXA2 may reach a steady-state level during normal repair through opposing actions of trafficking and shedding. This balance depends on ANO5. In contrast, loss of ANO5 greatly slows the accumulation of ANXA6: the time constant of the slow component of ANXA6 accumulation was increased approximately threefold in ANO5-KO. AUC values for ANXA2 and ANXA6 time courses also reflected this: ANO5-KO ANXA2 AUCs were significantly larger, and ANXA6 AUCs significantly smaller, than those of WT (Fig. 3, B and D). We performed Western blot analysis on tibialis anterior lysate to determine whether endogenous annexin levels differed in ANO5-KO muscle, which might affect assembly of the repair cap in vivo. However, we found that protein levels of ANXA1, ANXA2, ANXA5, and ANXA6 were nearly equivalent in WT or ANO5-KO muscles (Fig. S2, E and F). We conclude that loss of proper annexin trafficking, but not expression, is a key mechanism underlying deficient repair in ANO5-KO myofibers.
Figure S2.
Abnormal annexin trafficking and accumulation in ANO5-KO mouse muscle fibers. (A–D) Representative images of ANXA1 (A), ANXA2 (B), ANXA5 (C), and ANXA6 (D) in WT or ANO5-KO fibers following laser-induced injury. White arrows mark the first appearance of annexin in the cap. Scale bars = 10 µm. ANXA1: ANO5-KO n = 18 fibers, WT n = 14 fibers; ANXA2: ANO5-KO n = 42 fibers, WT n = 37 fibers; ANXA5: ANO5-KO n = 36 fibers, WT n = 29 fibers; ANXA6: ANO5-KO n = 20 fibers, WT n = 31 fibers. (E) Western blot analysis of annexin expression in WT or ANO5-KO mice. Ponceau S is shown as a loading control. (F) Densitometry of annexin Western blots, normalized to Ponceau S and expressed as %WT expression. WT n = 3 mice, ANO5-KO n = 3 mice. (G) Images and quantification of spatial organization of ANXA1 in the repair cap as in Fig. 4, A–D. WT n = 14 fibers pooled from two mice, KO n = 15 fibers pooled from two mice.
Figure 3.
The kinetics of annexin trafficking are changed in ANO5-KO fibers. (A–D) Time courses of annexin fluorescence as a function of time after injury were fitted to the sum of two exponentials (black lines). Amplitudes, A, and time constants, τ, are shown for the fast and slow components of the curves. AUCs for individual WT or KO fibers are also shown. Data were compared for WT or ANO5-KO fibers expressing ANXA1 (A), ANXA2 (B), ANXA5 (C), and ANXA6 (D). ANXA1: ANO5-KO n = 18 fibers from three mice, WT n = 14 fibers from two mice; ANXA2: ANO5-KO n = 42 fibers from five mice, WT n = 37 fibers from five mice; ANXA5: ANO5-KO n = 36 fibers from four mice, WT n = 29 fibers from three mice; ANXA6: ANO5-KO n = 20 fibers from three mice, WT n = 31 fibers from four mice. Data are mean ± SEM. *, P < 0.05.
Loss of ANO5 disrupts normal annexin repair cap architecture
To quantify the organization of different annexins in the cap, ANXA1, ANXA5, and ANXA6 were each coelectroporated into fibers with ANXA2. After injury, the intensity of each annexin was measured relative to ANXA2, normalized to maximal ANXA2 intensity, and plotted versus distance from this maximal ANXA2 intensity. In WT fibers, ANXA5 was concentrated toward the extracellular space relative to ANXA2, while ANXA6 was concentrated toward the intracellular space, suggesting an in-to-out hierarchy of ANXA6-ANXA2-ANXA5. No such organization was observed in ANO5-KO fibers, where ANXA5/ANXA2 and ANXA6/ANXA2 pairs essentially colocalized (Fig. 4, A–D). ANXA1 colocalized with ANXA2 in both WT and ANO5-KO fibers (Fig. S2 G). Orthogonal views, generated from z-stack images taken ∼10 min after injuring fibers, revealed that ANXA2 extended a greater distance in all three dimensions. Furthermore, calculated ANXA2 repair cap volumes were significantly larger in ANO5-KO than in WT (Fig. 4, E and F). In contrast, ANXA5 and ANXA6 caps were significantly smaller in ANO5-KO muscle (Fig. 4, E and F). This agrees with our findings that in ANO5-KO fibers, ANXA2 accumulates beyond normal physiological levels and ANXA5 and ANXA6 show depressed trafficking.
PtdSer and phosphatidylethanolamine (PtdEtn) are exposed after injury
The inner leaflet of the plasma membrane is normally enriched in PtdSer and PtdEtn. Accumulation of PtdSer at the site of plasma membrane injury has been documented, but the mechanism by which PtdSer is exposed extracellularly during repair is unresolved (Demonbreun et al., 2016b; Middel et al., 2016). We previously showed that ANO5 supports Ca2+-dependent PLS (Whitlock et al., 2018), so we hypothesized that PtdSer exposure after injury is ANO5 dependent. To test this, we laser-injured WT or ANO5-KO fibers in the presence of the PtdSer-binding C2 domain of Lactadherin (LactC2) fused to Clover or mCherry fluorescent proteins (LactC2-FPs). LactC2-FP was applied extracellularly rather than expressed intracellularly as described by others, ensuring that accumulation of the probe is attributable to extracellular exposure of PtdSer. Contrary to our expectations, LactC2-FP accumulated at the site of injury rapidly and robustly in both WT and ANO5-KO fibers, indicating that ANO5 is not required for accumulation of PtdSer (Fig. 5, A–C). Although ANO PLSases are generally considered nonspecific with regard to their phospholipid substrates (Accardi, 2016; Brunner et al., 2016), we also asked whether loss of ANO5 reduces exposure of PtdEtn. We conducted injury experiments in the presence of Cy3-conjugated duramycin, a PtdEtn probe. Duramycin-Cy3 (Dur-Cy3) accumulated rapidly at the patch site, similar to LactC2 (Fig. 5 D). Although PtdEtn accumulation was similar at the end of the 440-s recording period in WT and ANO5-KO fibers, initial PtdEtn accumulation was significantly less in ANO5-KO fibers (Fig. 5, E and F). This difference is not explained by changes in ANO6 expression (Fig. S3; Whitlock et al., 2018). Interestingly, PtdSer and PtdEtn exposure occurred in fibers injured in Ca2+-free solution, suggesting that PtdSer and PtdEtn exposure is not strictly dependent on Ca2+ influx (Video 5). Lipid probe accumulation in our system may thus reflect membrane trafficking or access of the probes to intracellular membranes rather than ANO5-mediated scrambling at the damage site.
Figure 5.
ANO5 is not required for exposure of PtdSer or PtdEtn following injury. (A) Representative images showing accumulation of the PtdSer sensor LactC2-Clover after laser-mediated injury. The cell boundary is indicated by a dotted white line. Scale bars = 10 µm. (B) Time course of LactC2-FP accumulation after damage, normalized to initial fluorescence at the injury site. (C) LactC2 AUC for initial (0–100 s) and late (100–437 s) stages of the repair time course. ANO5-KO n = 23 fibers from six mice, WT n = 20 fibers from two mice. (D) PtdEtn, detected by Cy3-conjugated duramycin, appears rapidly at the repair patch following wounding. The cell boundary is marked by a dotted white line. Scale bar = 10 µm. (E) Quantification of PtdEtn kinetics, indicating rapid, ANO5-independent recruitment of PtdEtn to the extracellular surface of muscle fibers after damage. Inset in top left of plot highlights the difference in initial PtdEtn accumulation. (F) Dur-Cy3 AUC for initial (0–100 s) and late (100–437 s) stages of the repair time course. ANO5-KO n = 9 fibers from two mice, WT n = 21 fibers from two mice. Data are mean ± SEM. *, P < 0.05.
Figure S3.
Ano5 but not Ano6 mRNA is increased during myogenesis, and Ano6 expression is not changed in myocytes or myotubes from ANO5-KO mice. (A) mRNA expression of Ano5 in WT or ANO5-KO mouse MPCs differentiated for 18 h (myocytes, left) or 72 h (myotubes, right). *, P < 0.05; **, P < 0.01. (B) mRNA expression of Ano6 in WT or ANO5-KO mouse MPCs differentiated for 18 h (myocytes, left) or 72 h (myotubes, right). WT n = 3, ANO5-KO n = 2. Data are mean ± SEM.
Video 5.
Rapid accumulation of PtdSer sensor LactC2-Clover and PtdEtn sensor Dur-Cy3 at a wound site in Ca2+-free media (composition described in the Ca2+-free lipid imaging subsection of Materials and methods). WT fibers were isolated and maintained in Ca2+-free buffer for between 10 and 60 min. LactC2-Clover (green) was used at a concentration of 2 µg/ml, and Dur-Cy3 (red) was used at a concentration of 250 nM. Following laser-induced injury, fibers were imaged with time-lapse confocal fluorescence microscopy. Both probes were seen to accumulate rapidly, indicating that lipid accumulation following fiber damage is at least partially Ca2+ independent. Images were collected as described in the Laser injury and imaging of membrane damage and repair subsection of Materials and methods. A timestamp is provided in the top left corner. The playback rate is 6 frames per second.
ANO5 “scrambling domain” is not required for repair
A conserved 34–amino acid region of ANO5 is necessary for ANO5-mediated PLS and ionic currents (Gyobu et al., 2016; Whitlock et al., 2018). Replacement of this scrambling domain with the corresponding sequence from the Cl− channel ANO1 to form chimeric “ANO5-1-5” destroys ANO5-dependent PLS (Whitlock et al., 2018). Given that PtdSer and PtdEtn are exposed during membrane repair in the absence of ANO5, we tested whether muscle membrane repair proceeds independently of ANO5-dependent PLS in ANO5-KO fibers expressing fluorescently tagged ANO5 or ANO5-1-5. Generally, the expression of ANO5-1-5 was lower than WT ANO5 as judged by weak and diffuse fluorescence intensity. Nevertheless, both ANO5 and ANO5-1-5 significantly reduced FM1-43 infiltration after injury (Fig. 6, A–C). Furthermore, using the next-generation Ca2+ indicator Calbryte 520 and a smaller laser injury (see Materials and methods), we confirmed the ability of ANO5 and ANO5-1-5 to reduce postinjury Ca2+ transients. In these experiments, repair occurred in WT but not in ANO5-KO fibers. Both ANO5 and ANO5-1-5 expression in ANO5-KO myofibers restored normal repair (Fig. 6, D–F).
Figure 6.
ANO5 does not require “scrambling domain” to rescue defective repair. (A) Deconvolved images of FM1-43 infiltration in ANO5-KO, ANO5-KO + hANO5, or ANO5-KO + ANO5-1-5 fibers before and following injury. Scale bars = 10 µm. (B and C) FM1-43 accumulation time course (B) and AUC values (C) for individual ANO5-KO, KO + hANO5, KO + ANO5-1-5, or WT fibers. ANO5-KO n = 11 fibers from three mice, ANO5-KO + hANO5 n = 16 fibers from three mice, ANO5-KO + ANO5-1-5 n = 10 fibers from three mice. Blue dotted line represents the average FM1-43 F/FmaxKO value from 11 injured WT fibers. (D) Representative images of Ca2+ transients visualized in damaged fibers with Calbryte 520. (E and F) Calbryte 520 time course (C) and AUC values (F) for individual ANO5-KO, KO + hANO5, KO + ANO5-1-5, or WT fibers. ANO5-KO n = 15 fibers from three mice, KO + ANO5 n = 16 fibers from two mice, KO + ANO5-1-5 n = 13 fibers from two mice, WT n = 9 fibers from three mice. (G and H) ANXA2 F/F0 time course (G) and AUC values (H) for individual ANO5-KO fibers + ANO5 or ANO5-1-5. KO + ANO5 n = 16 fibers from two mice, KO + ANO5-1-5 n = 16 fibers from two mice. Blue and red dotted lines represent ANXA2 accumulation in WT and ANO5-KO fibers, respectively. (I and J) ANXA6 F/F0 time course (I) and AUC values (J) for individual ANO5-KO fibers + ANO5 or ANO5-1-5. KO + ANO5 n = 24 fibers from five mice, KO + ANO5-1-5 n = 18 fibers from three mice. Blue and red dotted lines represent ANXA6 accumulation in WT and ANO5-KO fibers, respectively. Data are mean ± SEM. *, P < 0.05. Scale bar = 10 μm.
To test whether these ANO5 constructs restore normal annexin trafficking, we coexpressed ANO5 or ANO5-1-5 with ANXA2 or ANXA6 in ANO5-KO fibers. ANO5 overexpression reduced ANXA2 accumulation close to WT levels; however, ANO5-1-5 did not have a significant effect (Fig. 6, G and H; Table S2; and Video 6). In contrast, both ANO5 and ANO5-1-5 increased ANXA6 trafficking to the lesion above the level observed in ANO5-KO fibers alone (Fig. 6, I and J; Table S2; and Video 7). Because ANO5-1-5 expression reverses ANXA6 but not ANXA2 phenotypes while also substantially reducing FM1-43 accumulation in ANO5-KO fibers, it appears likely that ANXA6 translocation to the repair cap is crucial for efficient wound resealing. However, our results suggest that the ANO5 scrambling domain enables release of ANXA2-positive extracellular vesicles.
An R58W mutation found in human patients produces defective repair in human myocytes
To determine whether membrane repair is also defective in human patients with ANO5 mutations, we isolated MPCs from a human patient carrying a homozygous c.172C>T (p.R58W) point mutation. This variant is classified as likely pathogenic (https://www.ncbi.nlm.nih.gov/clinvar/) because it is consistent with an autosomal recessive inheritance pattern of LGMDR12 and is predicted to disrupt ANO5 secondary structure (Bohlega et al., 2015; Punetha et al., 2016; Savarese et al., 2015; Schessl et al., 2012; Witting et al., 2013). Differentiated patient myocytes laser damaged in the presence of FM1-43 showed a markedly elevated uptake of FM1-43 relative to healthy control myocytes. Furthermore, FM1-43 accumulation was accompanied by significant plasma membrane blebbing in patient cells (Fig. 7, A–C). After 2 min, FM1-43 fluorescence was 2.5 times greater in the R58W cells than in control cells (Fig. 7 B). We reasoned that if deficient repair in patient cells was caused by ANO5 loss of function associated with the R58W mutation, this variant ought to be incapable of rescuing repair in ANO5-KO mouse myofibers. We electroporated ANO5-R58W-mCherry into ANO5-KO fibers and found that it was well expressed (Fig. S4). Expression of ANO5-R58W in ANO5-KO fibers had no effect on FM1-43 accumulation or patch volumes (Fig. 7, D–G). We therefore conclude that pathogenicity associated with ANO5-R58W is related to plasma membrane repair incompetence. These results differ from those reported for a different mutation (R758C) from a human LGMDR12 patient, which leads to ANO5 degradation and has been suggested to alter Ca2+ signaling (Chandra et al., 2019).
Figure 7.
An R58W mutation in human patients causes defective repair in cultured muscle cells. (A) Representative images of FM1-43 infiltration in control or R58W human patient myocytes following injury. (B and C) FM1-43 accumulation time course following injury (B) and AUC values (C) for individual R58W or control myocytes. Control 1 n = 9 cells, Control 2 n = 8 cells, Patient n = 11 cells. (D) Images showing FM1-43 uptake in injured mouse ANO5-KO (KO), ANO5-KO + R58W ANO5 (KO+R58W), and WT fibers. (E and F) Kinetics of FM1-43 accumulation (E) and FM1-43 time course AUCs (F) from mouse myofibers with or without expression of R58W ANO5. ANO5-KO n = 22 fibers from four mice, ANO5-KO + R58W n = 31 fibers from four mice, WT n = 11 fibers from one mouse. (G) FM1-43 patch volumes obtained by integrating patch areas from individual slices (separated by 0.5 µm) of z-stacks acquired ∼10 min after injury. ANO5-KO n = 12 fibers from three mice, ANO5-KO + R58W n = 16 fibers from two mice. Data are mean ± SEM.
Figure S4.
Expression of ANO5-R58W mutant in human or mouse muscle. (A) Quantitative RT-PCR analysis from biopsied deltoid muscle of a human LGMD2L patient (n = 1) and tibialis anterior muscles of healthy control patients (Con, n = 2) with primers directed at three regions. ANO5-R58W mutant does not produce degraded ANO5 transcript. However, because exon 4 expression is reduced, there is the possibility that the transcript is alternatively spliced or uses an alternative start codon. ANO6 mRNA is shown as a comparison. Data are mean ± SEM. (B) ANO5-R58W-mCherry is detectable when expressed in mouse muscle fibers. Left panel is differential interference contrast (DIC). Middle panel is ANO5-R58W-Cherry fluorescence of the same fibers. White boxes indicate areas enlarged in the right panels. Scale bars = 10 µm.
Discussion
A role for ANO5 in membrane repair
Our results provide additional support for the growing literature implicating ANO5 in muscle membrane repair (Chandra et al., 2019; Griffin et al., 2016a). We show that ANO5-KO myofibers are more permeant to FM1-43 dye and exhibit larger Ca2+ signals after injury. However, these assays do not distinguish between defective repair and increased membrane fragility. The interpretation is further complicated by the fact that ANO5 may regulate Ca2+ homeostasis (Chandra et al., 2019; Phuong et al., 2019). Chandra et al. (2019) demonstrated that injury-induced Ca2+ transients were prolonged in human myoblasts with the R758C mutation. The prolonged transients were explained by defective uptake of Ca2+ into the ER. However, it is not known whether this feature is specific to the R758C mutation. In our experiments with ANO5-KO fibers, we observed an increase in the amplitude of the Ca2+ transient and a very pronounced slowing of its return to baseline, which we believe is consistent with delayed repair and/or increased damage.
We found that ANO5 rapidly condensed along the plasma membrane immediately adjacent to laser-induced lesions (Fig. 2, A and C). We suggest that this is an active process because previous studies using plasma membrane–targeted fluorescent proteins in zebrafish muscle did not find evidence of nonspecific enrichment of plasma membrane proteins at wounds (Middel et al., 2016; Roostalu and Strähle, 2012). The accumulation of ANO5 at the wound requires membrane injury because simple photobleaching of the protein without membrane injury does not stimulate ANO5 accumulation at the shoulder of the wound. Furthermore, we show that sarcolemma-localized ANO1 does not accumulate at wound sites to the same extent that ANO5 does (Fig. S1). However, because we cannot observe what happens to the bleached protein, we cannot say whether the increased fluorescence of ANO5 (and dysferlin) at the site of injury represents an increased concentration of these proteins at the wound compared with the concentration in uninjured fibers. This is an important caveat that applies not only to our experiments reported here but also to other published studies using the laser-induced injury approach. It was previously shown that contraction of cortical actin is involved in trafficking of dysferlin and annexins to wounds (Demonbreun et al., 2016a; Demonbreun and McNally, 2016; Demonbreun et al., 2016b; McDade et al., 2014), but we do not have data addressing this issue for ANO5. Nevertheless, the appearance of ANO5 at wound shoulders, regardless of its mechanism, places it at a favorable location to facilitate repair cap formation. The recent finding that the ANO5 paralog ANO6 is involved in nonconventional annexin secretion (Stewart et al., 2018) favors a model where ANO5 may play an active role in organization of the annexin cap.
ANO5-dependent lipid sorting is not required for muscle membrane repair
Phospholipids are emerging as central players in membrane repair processes. Previous studies have demonstrated rapid accumulation of PtdSer and phosphatidylinositol (4,5)-bisphosphate at or adjacent to damage sites in muscle. However, it is unclear precisely which lipids are required to facilitate repair and where they are needed. For example, Demonbreun et al. (2016b) showed that PtdSer accumulated at the shoulder of the wound in mouse myofibers. These studies employed genetically encoded, intracellularly expressed LactC2-GFP as a PtdSer probe. This approach has disadvantages because binding of the probe to PtdSer before injury could interfere with downstream events. Furthermore, the assay cannot distinguish between PtdSer located in the inner and outer membrane leaflets. Middel et al. (2016) attempted to mitigate this problem by expressing a secreted form of Annexin-V-YFP and found that PtdSer localized to the repair patch itself, rather than the wound shoulder. This result agrees with our finding using a different approach: we bathed fibers in purified LactC2-FP protein before and during injury (Zaitseva et al., 2017). Because LactC2 protein is impermeant, it labels only PtdSer in the external leaflet of the membrane, although it is uncertain whether PtdSer exposure occurs as a result of PLS or some other mechanism, such as vesicular fusion.
We were surprised to find that LactC2-FP and Dur-Cy3 accumulation in both WT and ANO5-KO fibers was robust after injury. Accumulation of these probes was restricted to the repair cap itself rather than the shoulder of the wound. Also, we observed that the accumulation of these probes at damage sites occurred in the absence of extracellular Ca2+ (Video 5), suggesting that PtdSer exposure may be mediated by intracellular vesicle aggregation (Cai et al., 2009) and/or the Ca2+-independent dysferlin-PtdSer trafficking (Middel et al., 2016). Our results exclude ANO5-PLS as a repair mechanism in isolated myofibers It is paradoxical that PtdSer exposure does not occur in patch-clamped cultured MPCs from ANO5-KO mice (Whitlock et al., 2018) while PtdSer exposure triggered by membrane damage is apparently intact in adult ANO5-KO muscle fibers. This may be explained by (i) differences between MPCs and adult muscle fibers or (ii) the spatio-temporal features or magnitude of the Ca2+ signal. Regardless, PtdSer exposure in damaged adult fibers appears to be ANO5 independent.
ANO5 and dysferlin function separately
Several annexin proteins have been clearly linked to myofiber repair (Bouter et al., 2011; Carmeille et al., 2016; Demonbreun et al., 2019; Demonbreun et al., 2016b; Swaggart et al., 2014). ANXA1 and ANXA2 interact with dysferlin to mediate repair in cultured myoblasts/myotubes, and ANXA2 is required for trafficking of dysferlin to the wound (Bittel et al., 2020; Lennon et al., 2003). Furthermore, a trafficking-deficient ANXA6 mutant inhibits dysferlin accumulation in wounded myofibers (Demonbreun et al., 2016b). However, in ANO5-KO mouse myofibers, where both ANXA2 and ANXA6 trafficking are altered (Fig. 3 D and Fig. S2), dysferlin behavior is apparently unchanged (Fig. 2, B and D). Similarly, in zebrafish lacking ANXA6, dysferlin behavior was unaffected (Roostalu and Strähle, 2012). Taken together, these data suggest that dysferlin trafficking may be dependent on ANXA2 but not directly on ANXA6. ANXA6 and dysferlin might share a common subcellular compartment at some step during repair so that certain ANXA6 mutants are capable of preventing progression of dysferlin to the lesion. Additionally, these data likely indicate that repair in muscle may not be a single process so much as several parallel and overlapping processes working in concert to effectively reseal the sarcolemma. We also noted less dysferlin trafficking in WT fibers than we expected from the literature (Bansal et al., 2003; McDade et al., 2014). Since the dysferlin C-terminus (Lek et al., 2013; Middel et al., 2016; Roostalu and Strähle, 2012), rather than full-length dysferlin, seems to accumulate during repair, one possibility is that only some dysferlin expressed in our system is accessible to proteolytic cleavage. It seems likely that ANO5 exerts minimal influence on dysferlin during the repair; whether the converse is true is an interesting point for future study.
A coordinated annexin response reseals damaged myofibers
Annexins possess several different membrane-shaping properties that facilitate membrane bending, constriction, and fusion (Bendix et al., 2020; Boye et al., 2018; Boye et al., 2017; Moreno-Pescador et al., 2019). Recruitment of annexins to the membrane would be expected to produce a bending effect on the membrane edges, as shown recently by Florentsen et al. (2020) for annexin A4 trimers, or a constriction, which has been demonstrated for several annexins (e.g., Boye et al., 2017). Therefore, understanding the fine structure of the annexin cap is important for clarifying how annexins participate in wound healing.
We observed that the trafficking and final localization of ANXA2, ANXA5, and ANXA6 was abnormal in ANO5-KO fibers. First, ANXA5 accumulation was delayed in the absence of ANO5 (Fig. 3 C and Fig. S2). ANXA5 is capable of homo-oligomerizing to form two-dimensional arrays, which are associated with membrane resealing (Bouter et al., 2011; Carmeille et al., 2016). Also, the relative positioning of ANXA5 and ANXA2 was different in WT and ANO5-KO fibers. One possibility is that ANXA5 forms an initial proteolipid “scab” while additional membrane/protein is trafficked to the wound. ANXA2 has been shown to foster membrane blebbing, presumably through binding to the outer surface of membrane lesions, while both ANXA2 and ANXA6 are capable of inducing “membrane folding,” which involves binding and pulling together membranes. ANXA6 in particular has been suggested as a key factor in collapsing membrane lesions because it has two annexin cores and can consequently bind two membranes (Boye et al., 2017; Demonbreun et al., 2019). We identified depressed trafficking of ANXA6 overall in ANO5-KO myofibers, which is consistent with delayed plasma membrane resealing. In addition, whereas ANXA2 translocation to the repair cap plateaued within a few minutes after wounding in WT fibers, it continued to accumulate at the lesion in ANO5-KO fibers. ANXA2 is a driver of membrane bleb formation and marks blebs in the repair cap of ANXA6-deficient muscle (Roostalu and Strähle, 2012). Thus, one intriguing possibility is that prolonged accumulation of ANXA2 in ANO5-KO fibers may be a factor in increased blebbing of the repair patch during FM1-43 experiments (Fig. 8). While altered annexin trafficking might be explained as an effect, rather than a cause, of defective repair in ANO5-KO fibers, our finding that ANO5-1-5 was capable of rescuing repair (according to FM1-43 infiltration) without restoring normal ANXA2 trafficking stands as a counterpoint to this idea.
Figure 8.
ANO5 coordinates annexin-mediated plasma membrane resealing. Top: damage to the plasma membrane leads to Ca2+ influx and initiation of repair processes. Bottom left: in normal muscle, ANO5 and dysferlin are enriched around plasma membrane wounds. Annexins are recruited to the lesion, where they assemble into an ordered cap along with PtdSer and PtdEtn. ANXA2 vesicles are shed from the cap. Bottom right: in the absence of ANO5, ANXA2 overaccumulates while ANXA5 and ANXA6 underaccumulate. Normal hierarchy of annexin species is lost within the repair cap, which features increased blebbing. Created with BioRender.com.
The ANO5 paralog ANO6 was recently proposed to play a role in repair of membranes damaged by bacterial toxins (Wu et al., 2020). Furthermore, ANO6-dependent scrambling of membrane lipids has been reported to be involved in unconventional secretion of ANXA2 and ANXA5 (Stewart et al., 2018). This strengthens the link between ANOs, annexins, and membrane repair, but more importantly it raises the possibility that ANO5 and ANO6 could perform similar (though separate) functions in muscle. For example, ANO5 and ANO6 may have distinct annexin specificities and coordinate the annexin response during repair cooperatively by facilitating transport of particular annexin proteins to the wound. Loss of either ANO could result in a misbalance of annexin species within the repair cap, thus leading to a reduction in sarcolemmal resealing similar to what we have described here.
What is the cell biology of ANO5?
ANO5 is most closely related to ANO6, which is a well-characterized PLSase (Suzuki et al., 2010; Suzuki et al., 2014; Yu et al., 2015). While ANO6-mediated scrambling has an important role in blood clotting (e.g., Suzuki et al., 2010) and the scramblase domain of ANO6 is highly conserved in ANO5 (Whitlock et al., 2018; Yu et al., 2015), a physiological role for ANO5-mediated scrambling has not been unambiguously demonstrated. ANO5-KO MPCs are PLS deficient and are defective in cell fusion to form myotubes (Griffin et al., 2016b; Whitlock et al., 2018). Although a link between PLS and scrambling remains to be established, pharmacological inhibition of ANO-mediated PtdSer exposure reduces fusion of osteoclast precursors (Verma et al., 2018). Here, we propose that while ANO5 plays a role in membrane repair in a process involving annexins, PLS is not required. 72 ANO5 mutations in ClinVar have been described as likely pathogenic or pathogenic, yet only four of these reside specifically in the coding region of the scrambling domain. Here, we show that myocytes from an LGMDR12 patient carrying a homozygous R58W mutation are repair deficient as are myoblasts previously reported from a patient with the C758C mutation (Chandra et al., 2019), but it is unknown whether defective membrane repair is common to all ANO5 mutations. Neither of these mutations tested is located in the scrambling domain. Overall, this suggests that the cell biology of ANO5 extends beyond its putative function as a PLSase. Our data suggest that one such role for ANO5 is as a facilitator of the translocation of key repair proteins following muscle wounding, though additional work is required to clarify whether this is through direct protein–protein interaction or some other mechanism.
Materials and methods
Reagents
FM1-43 (T35356) was purchased from ThermoFisher. Cal-520 AM (Cat #21130) and Calbryte 520 a.m. (Cat #20650) were purchased from AAT Bioquest. 1,000X stocks of the dyes were prepared in distilled deionized water (FM1-43) or DMSO (Cal-520, Calbryte 520) and used within 7 d. Dur-Cy3 (Molecular Targeting Technologies; D-1006) was prepared as a 500-µM (2,000X) stock in 1% DMSO in deionized water.
cDNAs
pEGFP-N1 (Clontech) was the backbone for many of the plasmids used in this study. This plasmid was modified by replacing EGFP with either NeonGreen (pmNeon), mEmerald (pmEmerald), tdTomato (pTomato), or mCherry (pmCherry). In some cases, proteins were tagged with a mutant GFP (A206K), which does not spontaneously homo-dimerize like WT GFP (Zacharias et al., 2002). GFP-A206K was generated through direct mutagenesis of the GFP sequence in the pEGFPN1 vector. A list of gene/tag combinations is provided in Table S3. We note no changes in the behaviors of the proteins studied when using different fluorescent protein tags. In addition to WT human ANO5, two mutants were used: an R58W mutant produced by point mutagenesis of WT ANO5 and chimeric “ANO5-1-5,” in which ANO5 amino acids 530–564 are replaced by amino acids 554–588 from ANO1. Production of ANO5-1-5 by replacing the scrambling domain of hANO5 with the homologous sequence from mouse ANO1 has been described previously (Whitlock et al., 2018).
The cDNA construct for expression of Clover fluorescent protein conjugated to lactadherin C2 domain (LactC2-Clover) in the pET-28 bacterial vector was a gift from Dr. Leonid Chernomordik (National Institutes of Health/National Institute of Child Health and Human Development, Bethesda, MD). Purification of this probe has been described previously (Zaitseva et al., 2017). The plasmid encoding LactC2-mCherry was made by excising Clover with XbaI and EcoRI and ligating in mCherry. This was purified identically to LactC2-Clover. Stock solutions (1–3 mg/ml) in 150 mM imidazole, pH 7.0, plus 0.02% sodium azide were diluted to 2 µg/ml for PtdSer imaging.
Mice
All procedures involving animals were approved by the Emory Institutional Care and Use Committee under protocols 201800130 (Hartzell) and 201700233 (Choo). Ano5 KO mice have been described previously and were bred and maintained as homozygous KOs. Age-matched control C57BL/6 mice were purchased from The Jackson Laboratory. Male mice between the ages of 3 and 6 mo were used for all experiments. Female mice were excluded because ANO5 myopathies appear to disproportionately affect males (e.g., Magri et al., 2012; Penttilä et al., 2012; van der Kooi et al., 2013).
Electroporation of flexor digitorum brevis (FDB) muscle fibers
FDB fibers were transfected by in vivo electroporation (Demonbreun and McNally, 2015; DiFranco et al., 2009). Mice were maintained under isoflurane anesthesia throughout the procedure. Footpads were first injected with 20 µl 0.5 mg/ml hyaluronidase (Sigma-Aldrich; H4272). After 2 h, <20 µl of plasmid was injected into the footpad (total plasmid injected was typically 20 µg, but ranged from 10 to 50 µg). Electroporation was performed with a two-needle array electrode connected to an ECM830 in vivo electro-square porator (BTX Harvard Apparatus). 20 pulses (300 V/cm) lasting 20 ms were delivered at 1-s intervals. FDB muscles were isolated 7 d after electroporation to allow for muscle healing and plasmid expression. Expression of a given plasmid differed among fibers, but we noted no difference in the ability of myofibers of different genotypes to express any specific plasmid, and we did not observe an effect of the level of plasmid expression on the downstream measurements taken.
Isolation of muscle fibers
Mice were euthanized by isoflurane overdose followed by cervical dislocation. FDB muscles were removed, rinsed briefly in Hanks’ balanced salt solution, and transferred to digestion buffer consisting of 0.2% collagenase A (Roche; 10103586001) in Hepes-buffered DMEM. Muscles were digested for 3 h at 37°C, then transferred to DMEM containing 10% BSA. Individual fibers were dissociated via pipetting through a series of pipette tips with decreasing bore size. Following dissociation, fibers were allowed to settle for 15 min and were then resuspended in a vitamin-free modification of DMEM consisting of 1.8 mM CaCl2, 0.8 mM MgSO4, 5.3 mM KCl, 44 mM NaHCO3, 110 mM NaCl, 0.9 mM NaH2PO4, 1 mM sodium pyruvate, 5.6 mM D-Glucose, and MEM amino acids solution (ThermoFisher; 11130051, used at 2X final concentration) supplemented with 0.4 mM L-serine, 0.4 mM glycine, 4 mM L-glutamine, and 100 U/L penicillin/streptomycin (ThermoFisher; 15140122). Fibers were seeded onto Matrigel (Corning; 356234)-coated glass-bottom dishes (MatTek; P35G-0-14-C) and allowed to adhere for at least 30 min before imaging. The skeletal muscle myosin II inhibitor N-benzyl-p-toluene sulphonamide (Tocris; 1870) was added to fiber dishes at a concentration of 15 µM to reduce Ca2+-induced contraction after plasma membrane injury. All experiments were conducted within 24 h of fiber isolation.
Isolation and culture of human MPCs
All studies with human cells were reviewed and approved by the Emory Institutional Review Board (IRB00105208 and IRB00084168) and comply with all recommendations and requirements of the National Institutes of Health. Written informed consent was obtained from all subjects (except anonymous autopsy material). Human muscles were isolated by biopsy or autopsy. Isolated muscle chunks were minced by blades and incubated with 0.25% Trypsin for 20 min and filtered to isolate mononucleated cells including satellite cells. Cells were cultured on 1% porcine gelatin (Sigma-Aldrich; G1890)–coated plates to expand and then sorted by surface markers (CD31−/CD45−/CD56+) using flow cytometry. Sorted human MPCs were grown on gelatin-coated dishes in Ham’s F10 medium supplemented with 10% fetal bovine serum (Sigma-Aldrich; 12303C), 5% heat-inactivated calf-serum (Hyclone; SH3008703), 0.5% chick embryo extract (US Biological; C3999), 100 U/ml penicillin/streptomycin (Gibco; 15140122), and 5 ng/ml basic fibroblast growth factor (Peprotech; 100-18b). Cells were differentiated in low-glucose DMEM with 2% horse serum (Gibco; 16050130) on gelatin-coated plastic imaging dishes (ibidi). For injury experiments, subconfluent cells were differentiated for 14 d.
Laser injury and imaging of membrane damage and repair
Culture dishes containing isolated muscle fibers were placed in a stage-top incubator (Tokai-Hit) heated to 37°C and gassed with 95% air/5% CO2. All images were taken on a Nikon A1R HD25 scanning confocal microscope using a Nikon 60× oil Apo 1.4 NA objective or a Nikon 60× oil CFI60 Apo total internal reflection fluorescence 1.49 NA objective (Calbryte 520 experiments only). Digital zoom was set to 2.5, and a scanning resolution of 1,024 × 1,024 was used for all experiments, resulting in a pixel size of 0.08 µm. Pinhole size was set to 1 (myofiber injury) or 2 (human myocyte injury) Airy units. A 2 µm × 2 µm (24.1 pixel × 24.1 pixel) region of interest (ROI) was specified at the lateral edge of the fiber for laser ablation, except in the case of Calbryte 520 experiments, where a 1 µm × 1 µm ROI was used. Fibers were irradiated with a 405-nm laser set to 100% power (corresponding to ∼0.9–1.1 mW) for 8 s (myofibers) or 2 s (human myocytes) using the ND Stimulation toolbox in Nikon Elements software. Images were taken immediately before and after injury, then for every 4 s for a total of 2 min (31 frames), and finally every 15 s for a total of 5 min (21 frames). Times are reported as time from the start of scanning image #1 (preinjury) t = 0. Green channel images were acquired with a 488-nm laser and 525 ± 50–nm filter cube, while red channel images were acquired with a 561-nm laser and 595 ± 50–nm filter cube.
Z imaging
z-stacks were acquired in Nikon Elements software using the ND Acquisition menu. Highest and lowest focal planes of interest were identified by manually focusing through the depth of the injured region of the fiber. Images were taken every 0.5 µm with microscope and camera settings identical to those used to acquire time lapses after laser-induced injury.
Deconvolution
Images were deconvolved using the deconvolution module in Nikon Elements AR Analysis software. 2D deconvolution without subtraction was used for XY images, and the point spread function was determined automatically by the software. All quantification was performed on original images. Deconvolution is only for the purpose of presentation and clarity and is noted in figure legends where applicable.
Ca2+ imaging
Cal-520 or Calbryte 520 was mixed 1:1 with Pluronic F-127 and diluted to a final concentration of 10 µM or 1 µM, respectively, in imaging medium with 10 mM Hepes. Fibers were loaded with dye in this solution for 30 min at RT, at which point the loading solution was replaced with fresh imaging medium. Fibers were then seeded onto MatTek dishes and allowed to adhere for 30 min before imaging.
FM1-43 repair assay
Imaging medium was replaced with imaging medium containing 2.5 µM FM1-43 at least 10 min before initiation of wounding experiments. For ANO5 rescue experiments, fibers expressing the desired fluorescent constructs were identified in the absence of FM1-43. This was done because FM1-43 has broad excitation/emission peaks in green and red wavelengths that would mask ANO5-expressing fibers if maintained in the bath at a working concentration. When ANO5-expressing fibers were identified, the X and Y positions of the microscope stage were marked using the ND Acquisition panel in Nikon Elements software. 100 µl of a 10X stock solution of FM1-43 in imaging medium was added dropwise to the dish (final buffer volume = 1 ml) with additional mixing by gentle pipetting. Fibers were left in the presence of FM1-43 at least 10 min before imaging.
Quantification
All image analysis was performed on unaltered images using Fiji (ImageJ) software. Analysis was not blinded with respect to experimental group.
Time courses
Elliptical ROIs were placed within the fiber boundaries and aligned to the center of the wound. Mean fluorescent intensity was measured within the ROI for each acquisition time point. Measurements were normalized to F0 and plotted as a function of time in seconds. ROIs of the following sizes were used: 1,000 µm2, mouse myofibers in the presence of FM1-43 or loaded with Cal-520; 500 µm2, fibers loaded with Calbryte 520; 50 µm2, human myocytes in the presence of FM1-43; 17 µm2, annexin electroporated mouse myofibers; and 100 µm2, lipid probes (Fig. S5).
Figure S5.
ROIs used for quantification. ROIs described in Materials and methods are shown (white ovals) on representative images from denoted figures. ROIs were moved if fibers contracted. If necessary, ROIs were reshaped to fit within fiber boundaries, but total ROI size was maintained (see Fig. 1, FM1-43). Dotted line in Fig. 5 image is the cell boundary. Scale bars = 10 µm.
Volumes
Reported volumes were calculated from z-stack images according to the formula
where Ai is the area of an object of interest, determined by manually encircling the object in ImageJ, from slice i of a stack of n images taken at 0.5-µm intervals.
Membrane blebs
Blebs were manually annotated from z-stack images in ImageJ using the multipoint tool. After the entire stack was scanned, images were checked to ensure that blebs spanning more than one slice were not counted more than once.
Spatial localization of annexins in repair cap
ANXA1, ANXA5, and ANXA6 were coelectroporated into fibers with ANXA2. The final imaging frames from time-lapse experiments were used for analysis (∼7.5 min after injury as described above). In ImageJ, the line tool was used to draw a line through the center of the ANXA2 cap. This line was set as an ROI so that its X and Y coordinates within the viewing frame in ImageJ were stored. The intensity profile in the ANXA2 channel was plotted for this line, and then the intensity profile in the channel for the coelectroporated annexin along the same line was plotted. The absolute intensity data were transformed according to the formula
so that all values for each line intensity profile ranged between 0 and 1. Fnorm values for both annexin proteins were plotted as a function of a variable termed here as “distance.” Units of distance are in micrometers and reflect points along the length of the line used to generate intensity profiles. For each replicate, distance = 0 was defined as the point where Fnorm for ANXA2 [Fnorm(A2)] = 1. Fnorm values for the coelectroporated annexin are plotted relative to the distance scale defined by ANXA2. Fnorm values are shown for distances of −3 µm (toward myofiber center relative to Fnorm(A2) =1) to 3 µm (away from the myofiber center relative to Fnorm(A2) = 1). As an example, if the maximal Fnorm value for the coelectroporated annexin protein was closer to the center of the fiber than the maximal Fnorm(A2) value, you would expect to see the curve of the coelectroporated annexin shifted to the left in a plot of Fnorm versus distance. The point (0,1) appears in every plot for ANXA2 because distance = 0 is defined by Fmax(A2), but because of variability from fiber to fiber, Fnorm(A[X]) may not equal 1.
Curve fitting
Mean ± SEM F/F0 values were fit either to a single exponential equation or the sum of two exponentials using the fit functions in Origin 2019.
ANO5 and dysferlin were fit to an equation of the form
where A represents the amplitude (Yplateau − Y0) and τ is the time constant.
Annexins were fit to an equation of the form
where Afast and Aslow represent amplitude (Yplateau − Y0) contributed by the fast and slow components, respectively; τfast and τslow are the time constants of the fast and slow components, and d is a time offset for the second exponential. R2 was >0.995 in all cases.
Protein isolation and Western blotting
Protein was isolated from whole tibialis anterior. For individual fibers, FDB muscles were digested in collagenase and triturated to release fibers from the tendon. Fibers were examined under a dissecting scope, and 25–50 healthy fibers were transferred to 30 µl lysis buffer containing 50 mM Tris, 150 mM NaCl, 1% NP-40 alternative (Calbiochem), and 0.5% sodium deoxycholate supplemented with protease inhibitors (Millipore inhibitor cocktail set III). Tibialis anterior and gastrocnemius muscles were homogenized via Teflon/glass Dounce homogenizer in 1 ml lysis buffer. Homogenates were rotated at 4°C for 1 h, then spun at 21,000g for 15 min. Supernates were stored at −20°C before use.
15 µg protein was denatured in 1X Laemmli sample buffer with 2.5% β-mercaptoethanol and run through precast 4–15% SDS-PAGE gels (Bio-Rad). Protein was transferred to a polyvinylidene difluoride membrane. Membranes were blocked in 5% BSA in Tris-buffered saline plus 0.1% Triton X-100 (TBS-T) for 1 h at RT, then placed in primary antibody at 4°C overnight. Antibodies were diluted as follows in 5% BSA in TBS-T: rabbit anti-ANXA1 (Abcam; AB214486) 1:2,000; rabbit anti-ANXA2 (Abcam; AB178677) 1:2,000; rabbit anti-ANXA5 (Abcam; AB108194) 1:2,500; and mouse anti-ANXA6 (BD Biosciences; 610300) 1:10,000. Membranes were washed three times for 5 min in TBS-T, then placed in goat anti-mouse or goat anti-rabbit secondary antibody (Amersham) diluted 1:25,000 or 1:10,000, respectively, in 5% BSA in TBS-T for 1 h at RT. Membranes were washed three more times for 5 min in TBS-T, developed in ECL substrate (Pierce) according to the manufacturer’s instructions, and imaged with film.
Blot quantification was performed in ImageJ. A box was drawn around each band of interest, and mean intensity was measured. This value was divided by mean Ponceau S signal from the same lane as the band (loading control). Values were quantified from three WT and three ANO5-KO mice from two technical replicates. For each protein of interest, the average of the WT replicates was set at 100%, and ANO5-KO values were adjusted to this scale.
Statistics
Statistical tests were performed in Prism 8 software (GraphPad). Data were analyzed via Student’s t test or one-way analysis of variance with Sidak’s multiple comparison test, as appropriate. Data distribution was assumed to be normal, but this was not formally tested. Spatial localizations of repair cap annexins were analyzed through repeated-measures two-way analysis of variance with Geisser-Greenhouse correction and Sidak’s posttest correction for multiple comparisons. AUCs were calculated according to the formula
P values <0.05 were considered statistically significant. All data are presented as mean ± SEM.
Isolation and differentiation of mouse primary MPCs
Mononucleated cells were isolated from the hindlimb muscles as described previously (Jansen and Pavlath, 2006). Isolated MPCs were cultured in Ham’s F-10, 20% fetal bovine serum, 5 ng/ml basic fibroblast growth factor, 100 U/ml penicillin, and 100 µg/ml streptomycin on collagen-coated plates for 3 or 5 d. Medium was changed every 2 d. For differentiation, MPCs were seeded at ECL-coated tissue culture plates and cultured until 90% confluence. Then, culture medium was changed to differentiation medium (2% horse serum in low glucose DMEM, 100 U/ml penicillin, and 100 µg/ml streptomycin) and further cultured for 18 h (myocytes) or 72 h (myotubes).
Real-time PCR analyses
Total RNA was isolated from primary cultured myocytes or myotubes, and deltoid muscle tissues were biopsied from an ANO5 R58W patient using Trizol according to the manufacturer’s protocol. The reverse transcription reaction was performed using 250 ng of total RNA/sample using random hexamers and M-MLV reverse transcription (Invitrogen). cDNA was amplified using the SYBR select master mix (Applied Biosystems) and 2.5 µM of each primer. All RNA samples were tested for DNA contamination by PCR. Amplified cDNA signals were detected and analyzed by StepOne software v2.2.2 (Applied Biosystems) using Hprt as internal control for mouse samples and GAPDH as an internal control for human samples. Fold change of gene expression was determined using the ΔΔCt method (Schmittgen and Livak, 2008). Three to four independent experiments were performed. Human primer sequences were ANO5_Ex4-5F: 5′-GCGGCGGCTTATGTTTCAAAA-3′; ANO5_Ex4-5R: 5′-CGCCTTTAACTCTGCGTCTTTC-3′; ANO5_Ex7F: 5′-TATTCCCCGCCCTAAGCACA-3′; ANO5_Ex7R: 5′-AGAAGGTTGCCTGATCTTCGAT-3′; ANO5_Ex13F: 5′-TTTTGGAAACAACGACAAGCCA-3′; ANO5_Ex13R: 5′-ACCATACTGGTGACGACAAGAG-3′; ANO6F: 5′-AGCAAAGAAGTTTGTCATCC-3′; ANO6R: 5′-GAATGGACAAAGCCTATCAC-3′; GAPDHF: 5′-CTTTTGCGTCGCCAG-3′; and GAPDHR: 5′-TTGATGGCAACAATATCCAC-3′. Mouse primer sequences were Ano5F: 5′-TCTTCCCACTGAGCACTTTC-3′; Ano5R: 5′-TGAGCATTCCTACACCAACC-3′; Ano6F: 5′-CTTATCAGGAAGTATTACGGC-3′; Ano6R: 5′-AGATATCCATAGAGGAAGCAG-3′; HprtF: 5′-TCAGTCAACGGGGGACATAAA-3′; and HprtR: 5′-GGGGCTGTACTGCTTAACCAG-3′.
Ca2+-free lipid imaging
Fiber isolation, injury, and imaging were similar to methods described elsewhere in the manuscript except for the imaging buffer used. Specifically, FDB muscle fibers were isolated from WT and seeded on uncoated glass-bottom dishes in extracellular solution containing (in millimolars): 140 NaCl, 5 KCl, 1 MgCl2, 1 pyruvate, 25 glucose, 25 Hepes, and 10 EGTA. Fibers were maintained in this Ca2+-free solution for at least 10 min before imaging, but for no more than 60 min total.
Data availability
All data referenced in the manuscript are included within the presented figures. Unique reagents, including cDNAs, and laboratory protocols will be promptly provided upon request.
Online supplemental material
Fig. S1 shows the specificity of ANO5 accumulation at the wound shoulder using the plasma membrane–localized ANO/TMEM16 family member ANO1 as a comparison. ANO5 translocates to the wound more quickly and to a greater extent than ANO1, and this is not explained by FRAP. Fig. S2 is a companion to Fig. 3 and Fig. 4 in the manuscript. Fig. S2, A–D provides representative images of annexin accumulation in wounded myofibers. These data are described quantitatively in manuscript Fig. 3. Fig. S2, E and F shows protein expression analysis of the annexin species evaluated in this study in WT or ANO5-KO mice. Fig. S2 G shows that ANO5 does not influence the relative positioning of ANXA1 within the repair cap (unlike ANXA5 and ANXA6, shown in Fig. 4). Fig. S3 shows induction of ANO5 during MPC differentiation and demonstrates that ANO5 message is completely lost in MPCs isolated from ANO5-KO mice. It also provides an indication that ANO6, a homologue of ANO5, is not up-regulated as a compensatory mechanism for loss of ANO5. Fig. S4 provides evidence that transcript for mutant ANO5-R58W is expressed. ANO5 message is detectable from human R58W patient muscle, while ANO5-R58W is visibly detectable when expressed in mouse myofibers and fused to mCherry fluorescent protein. Fig. S5 shows examples of how parameters in this study were quantified. Video 1 shows shedding of patch material during repair in WT myofibers, while Video 2 shows patch blebbing and retention of patch material in ANO5-KO myofibers. Video 3 shows that ANO5 accumulation at the wound shoulder coincides with an injury-induced Ca2+ transient. Video 4 shows release of ANXA2-positive vesicles from a repair cap in WT muscle. Video 5 shows accumulation of PtdSer and PtdEtn in injured WT muscle in the absence of Ca2+. Video 6 shows that expression of ANO5, but not ANO5-1-5, reduces levels of ANXA2 in the repair cap. Video 7 shows that expression of ANO5 or ANO5-1-5 increases ANXA6 accumulation in the repair cap. Table S1 provides numerical values for fitted parameters presented in Fig. 3. Table S2 provides numerical annexin trafficking values in ANO5-KO myofibers expressing ANO5 or ANO5-1-5, expressed as a percentage of WT or ANO5-KO values from Fig. 3. Table S3 lists the genes expressed in mature mouse myofibers and the fluorescent tags attached to them.
Supplementary Material
presents the fitted parameters of annexin time courses.
compares F/F0 or AUC values for ANXA2 or ANXA6 in ANO5-KO fibers rescued with ANO5 or ANO5-1-5.
lists the genes expressed in mature mouse myofibers and the fluorescent tags attached to them.
Acknowledgments
We thank Drs. Matthew Wicklund (University of Colorado), Samya Chakravorty, and Steven Goudy (Emory University) for assistance in procuring muscle biopsies, Dr. Eunhye Kim for assistance with human MPCs isolation, and Dr. Fang Wu for technical assistance. The LactC2-Clover cDNA was a gift from Dr. Leonid Chernomordik (National Institute of Child Health and Human Development).
This research was supported in part by the Emory University Integrated Cellular Imaging Microscopy Core. This research was supported by grants from the National Institutes of Health: the National Institute of Arthritis and Musculoskeletal and Skin Diseases awards R01AR067786 (to H. Criss Hartzell), R01AR071397 (to H.J. Choo), and F32AR074249 (to S.J. Foltz); National Eye Institute award R01EY114852 (to H. Criss Hartzell); and National Institute of General Medical Sciences grant R01GM132598 (to H. Criss Hartzell). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The authors declare no competing financial interests.
Author contributions: S.J. Foltz conceptualized and performed the research, acquired and analyzed data, and wrote the manuscript. YY. Cui provided essential resources for the project. H.J. Choo performed the research and wrote the manuscript. H. Criss Hartzell conceptualized the research, analyzed data, secured funding, and wrote the manuscript.
References
- Accardi, A. 2016. Mechanisms of Ion and Lipid Transport by TMEM16 Scramblases. Biophys. J. 110:174a. 10.1016/j.bpj.2015.11.968 [DOI] [Google Scholar]
- Andreeva, T.V., Tyazhelova T.V., Rykalina V.N., Gusev F.E., Goltsov A.Y., Zolotareva O.I., Aliseichik M.P., Borodina T.A., Grigorenko A.P., Reshetov D.A., et al. 2016. Whole exome sequencing links dental tumor to an autosomal-dominant mutation in ANO5 gene associated with gnathodiaphyseal dysplasia and muscle dystrophies. Sci. Rep. 6:26440. 10.1038/srep26440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, N.W., and Corrotte M.. 2018. Plasma membrane repair. Curr. Biol. 28:R392–R397. 10.1016/j.cub.2017.12.034 [DOI] [PubMed] [Google Scholar]
- Bansal, D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., and Campbell K.P.. 2003. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 423:168–172. 10.1038/nature01573 [DOI] [PubMed] [Google Scholar]
- Barthélémy, F., Defour A., Lévy N., Krahn M., and Bartoli M.. 2018. Muscle Cells Fix Breaches by Orchestrating a Membrane Repair Ballet. J. Neuromuscul. Dis. 5:21–28. 10.3233/JND-170251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir, R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., et al. 1998. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 20:37–42. 10.1038/1689 [DOI] [PubMed] [Google Scholar]
- Bendix, P.M., Simonsen A.C., Florentsen C.D., Häger S.C., Mularski A., Zanjani A.A.H., Moreno-Pescador G., Klenow M.B., Sønder S.L., Danielsen H.M., et al. 2020. Interdisciplinary Synergy to Reveal Mechanisms of Annexin-Mediated Plasma Membrane Shaping and Repair. Cells. 9:1029. 10.3390/cells9041029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevers, E.M., and Williamson P.L.. 2016. Getting to the Outer Leaflet: Physiology of Phosphatidylserine Exposure at the Plasma Membrane. Physiol. Rev. 96:605–645. 10.1152/physrev.00020.2015 [DOI] [PubMed] [Google Scholar]
- Bittel, D.C., Chandra G., Tirunagri L.M.S., Deora A.B., Medikayala S., Scheffer L., Defour A., and Jaiswal J.K.. 2020. Annexin A2 Mediates Dysferlin Accumulation and Muscle Cell Membrane Repair. Cells. 9:1919. 10.3390/cells9091919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazek, A.D., Paleo B.J., and Weisleder N.. 2015. Plasma Membrane Repair: A Central Process for Maintaining Cellular Homeostasis. Physiology (Bethesda). 30:438–448. 10.1152/physiol.00019.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlega, S., Monies D.M., Abulaban A.A., Murad H.N., Alhindi H.N., and Meyer B.F.. 2015. Clinical and genetic features of anoctaminopathy in Saudi Arabia. Neurosciences (Riyadh). 20:173–177. 10.17712/nsj.2015.2.20140547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolduc, V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., et al. 2010. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86:213–221. 10.1016/j.ajhg.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouter, A., Gounou C., Bérat R., Tan S., Gallois B., Granier T., d’Estaintot B.L., Pöschl E., Brachvogel B., and Brisson A.R.. 2011. Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat. Commun. 2:270. 10.1038/ncomms1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye, T.L., and Nylandsted J.. 2016. Annexins in plasma membrane repair. Biol. Chem. 397:961–969. 10.1515/hsz-2016-0171 [DOI] [PubMed] [Google Scholar]
- Boye, T.L., Maeda K., Pezeshkian W., Sønder S.L., Haeger S.C., Gerke V., Simonsen A.C., and Nylandsted J.. 2017. Annexin A4 and A6 induce membrane curvature and constriction during cell membrane repair. Nat. Commun. 8:1623. 10.1038/s41467-017-01743-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boye, T.L., Jeppesen J.C., Maeda K., Pezeshkian W., Solovyeva V., Nylandsted J., and Simonsen A.C.. 2018. Annexins induce curvature on free-edge membranes displaying distinct morphologies. Sci. Rep. 8:10309. 10.1038/s41598-018-28481-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, J.D., Schenck S., and Dutzler R.. 2016. Structural basis for phospholipid scrambling in the TMEM16 family. Curr. Opin. Struct. Biol. 39:61–70. 10.1016/j.sbi.2016.05.020 [DOI] [PubMed] [Google Scholar]
- Cai, C., Weisleder N., Ko J.K., Komazaki S., Sunada Y., Nishi M., Takeshima H., and Ma J.. 2009. Membrane repair defects in muscular dystrophy are linked to altered interaction between MG53, caveolin-3, and dysferlin. J. Biol. Chem. 284:15894–15902. 10.1074/jbc.M109.009589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cárdenas, A.M., González-Jamett A.M., Cea L.A., Bevilacqua J.A., and Caviedes P.. 2016. Dysferlin function in skeletal muscle: Possible pathological mechanisms and therapeutical targets in dysferlinopathies. Exp. Neurol. 283(Pt A):246–254. 10.1016/j.expneurol.2016.06.026 [DOI] [PubMed] [Google Scholar]
- Carmeille, R., Bouvet F., Tan S., Croissant C., Gounou C., Mamchaoui K., Mouly V., Brisson A.R., and Bouter A.. 2016. Membrane repair of human skeletal muscle cells requires Annexin-A5. Biochim. Biophys. Acta. 1863:2267–2279. 10.1016/j.bbamcr.2016.06.003 [DOI] [PubMed] [Google Scholar]
- Chandra, G., Defour A., Mamchoui K., Pandey K., Mishra S., Mouly V., Sreetama S., Mahad Ahmad M., Mahjneh I., Morizono H., et al. 2019. Dysregulated calcium homeostasis prevents plasma membrane repair in Anoctamin 5/TMEM16E-deficient patient muscle cells. Cell Death Discov. 5:118. 10.1038/s41420-019-0197-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, S.T., and McNeil P.L.. 2015. Membrane Repair: Mechanisms and Pathophysiology. Physiol. Rev. 95:1205–1240. 10.1152/physrev.00037.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A.R., and McNally E.M.. 2015. DNA Electroporation, Isolation and Imaging of Myofibers. J. Vis. Exp. 10.3791/53551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A.R., and McNally E.M.. 2016. Plasma Membrane Repair in Health and Disease. Curr. Top. Membr. 77:67–96. 10.1016/bs.ctm.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A.R., Allen M.V., Warner J.L., Barefield D.Y., Krishnan S., Swanson K.E., Earley J.U., and McNally E.M.. 2016a. Enhanced Muscular Dystrophy from Loss of Dysferlin Is Accompanied by Impaired Annexin A6 Translocation after Sarcolemmal Disruption. Am. J. Pathol. 186:1610–1622. 10.1016/j.ajpath.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A.R., Quattrocelli M., Barefield D.Y., Allen M.V., Swanson K.E., and McNally E.M.. 2016b. An actin-dependent annexin complex mediates plasma membrane repair in muscle. J. Cell Biol. 213:705–718. 10.1083/jcb.201512022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun, A.R., Fallon K.S., Oosterbaan C.C., Bogdanovic E., Warner J.L., Sell J.J., Page P.G., Quattrocelli M., Barefield D.Y., and McNally E.M.. 2019. Recombinant annexin A6 promotes membrane repair and protects against muscle injury. J. Clin. Invest. 129:4657–4670. 10.1172/JCI128840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Zanni, E., Gradogna A., Scholz-Starke J., and Boccaccio A.. 2018. Gain of function of TMEM16E/ANO5 scrambling activity caused by a mutation associated with gnathodiaphyseal dysplasia. Cell. Mol. Life Sci. 75:1657–1670. 10.1007/s00018-017-2704-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Zanni, E., Gradogna A., Picco C., Scholz-Starke J., and Boccaccio A.. 2020. TMEM16E/ANO5 mutations related to bone dysplasia or muscular dystrophy cause opposite effects on lipid scrambling. Hum. Mutat. 41:1157–1170. 10.1002/humu.24006 [DOI] [PubMed] [Google Scholar]
- DiFranco, M., Quinonez M., Capote J., and Vergara J.. 2009. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J. Vis. Exp. (32):1520. 10.3791/1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong, H.A., Le K.T., Soulema A.L., Yueh R.H., Scheuner M.T., Holick M.F., Christensen R., Tajima T.L., Leung A.M., and Mallya S.M.. 2016. Gnathodiaphyseal dysplasia: report of a family with a novel mutation of the ANO5 gene. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 121:e123–e128. 10.1016/j.oooo.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzone, M.E., Malvezzi M., Lee B.C., and Accardi A.. 2018. Known structures and unknown mechanisms of TMEM16 scramblases and channels. J. Gen. Physiol. 150:933–947. 10.1085/jgp.201711957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentsen, C.D., Kamp-Sonne A., Moreno-Pescador G., Pezeshkian W., Hakami Zanjani A.A., Khandelia H., Nylandsted J., and Bendix P.M.. 2020. Annexin A4 trimers are recruited by high membrane curvatures in giant plasma membrane vesicles. Soft Matter. 10.1039/D0SM00241K [DOI] [PubMed] [Google Scholar]
- Fuson, K., Rice A., Mahling R., Snow A., Nayak K., Shanbhogue P., Meyer A.G., Redpath G.M., Hinderliter A., Cooper S.T., and Sutton R.B.. 2014. Alternate splicing of dysferlin C2A confers Ca2+-dependent and Ca2+-independent binding for membrane repair. Structure. 22:104–115. 10.1016/j.str.2013.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke, V., and Moss S.E.. 2002. Annexins: from structure to function. Physiol. Rev. 82:331–371. 10.1152/physrev.00030.2001 [DOI] [PubMed] [Google Scholar]
- Gissel, H., and Clausen T.. 2001. Excitation-induced Ca2+ influx and skeletal muscle cell damage. Acta Physiol. Scand. 171:327–334. 10.1046/j.1365-201x.2001.00835.x [DOI] [PubMed] [Google Scholar]
- Griffin, D.A., Johnson R.W., Whitlock J.M., Pozsgai E.R., Heller K.N., Grose W.E., Arnold W.D., Sahenk Z., Hartzell H.C., and Rodino-Klapac L.R.. 2016a. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum. Mol. Genet. 25:1900–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin, D.A., Johnson R.W., Whitlock J.M., Pozsgai E.R., Heller K.N., Grose W.E., Arnold W.D., Sahenk Z., Hartzell H.C., and Rodino-Klapac L.R.. 2016b. Defective membrane fusion and repair in Anoctamin5-deficient muscular dystrophy. Hum. Mol. Genet. 25:1900–1911. 10.1093/hmg/ddw063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyobu, S., Miyata H., Ikawa M., Yamazaki D., Takeshima H., Suzuki J., and Nagata S.. 2016. A role of TMEM16E carrying a scrambling domain in sperm motility. Mol. Cell. Biol. 36:645–659. 10.1128/MCB.00919-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häger, S.C., and Nylandsted J.. 2019. Annexins: players of single cell wound healing and regeneration. Commun. Integr. Biol. 12:162–165. 10.1080/19420889.2019.1676139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks, D., Sarkozy A., Muelas N., Köehler K., Huebner A., Hudson G., Chinnery P.F., Barresi R., Eagle M., Polvikoski T., et al. 2011. A founder mutation in Anoctamin 5 is a major cause of limb-girdle muscular dystrophy. Brain. 134:171–182. 10.1093/brain/awq294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn, A., and Jaiswal J.K.. 2018. Cellular mechanisms and signals that coordinate plasma membrane repair. Cell. Mol. Life Sci. 75:3751–3770. 10.1007/s00018-018-2888-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal, J.K., Marlow G., Summerill G., Mahjneh I., Mueller S., Hill M., Miyake K., Haase H., Anderson L.V., Richard I., et al. 2007. Patients with a non-dysferlin Miyoshi myopathy have a novel membrane repair defect. Traffic. 8:77–88. 10.1111/j.1600-0854.2006.00505.x [DOI] [PubMed] [Google Scholar]
- Jaiswal, J.K., Lauritzen S.P., Scheffer L., Sakaguchi M., Bunkenborg J., Simon S.M., Kallunki T., Jäättelä M., and Nylandsted J.. 2014. S100A11 is required for efficient plasma membrane repair and survival of invasive cancer cells. Nat. Commun. 5:3795. 10.1038/ncomms4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen, K.M., and Pavlath G.K.. 2006. Mannose receptor regulates myoblast motility and muscle growth. J. Cell Biol. 174:403–413. 10.1083/jcb.200601102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, M., and Katoh M.. 2004. GDD1 is identical to TMEM16E, a member of the TMEM16 family. Am. J. Hum. Genet. 75:927–928, author reply:928–929. 10.1086/425341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerdt, S.N., Ashraf A.P.K., and Gerke V.. 2019. Annexins and plasma membrane repair. Curr. Top. Membr. 84:43–65. 10.1016/bs.ctm.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Lek, A., Evesson F.J., Lemckert F.A., Redpath G.M., Lueders A.K., Turnbull L., Whitchurch C.B., North K.N., and Cooper S.T.. 2013. Calpains, cleaved mini-dysferlinC72, and L-type channels underpin calcium-dependent muscle membrane repair. J. Neurosci. 33:5085–5094. 10.1523/JNEUROSCI.3560-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, N.J., Kho A., Bacskai B.J., Perlmutter S.L., Hyman B.T., and Brown R.H. Jr. 2003. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278:50466–50473. 10.1074/jbc.M307247200 [DOI] [PubMed] [Google Scholar]
- Liewluck, T., and Milone M.. 2018. Untangling the complexity of limb-girdle muscular dystrophies. Muscle Nerve. 58:167–177. 10.1002/mus.26077 [DOI] [PubMed] [Google Scholar]
- Liu, J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B., et al. 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20:31–36. 10.1038/1682 [DOI] [PubMed] [Google Scholar]
- Magri, F., Del Bo R., D’Angelo M.G., Sciacco M., Gandossini S., Govoni A., Napoli L., Ciscato P., Fortunato F., Brighina E., et al. 2012. Frequency and characterisation of anoctamin 5 mutations in a cohort of Italian limb-girdle muscular dystrophy patients. Neuromuscul. Disord. 22:934–943. 10.1016/j.nmd.2012.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjneh, I., Jaiswal J., Lamminen A., Somer M., Marlow G., Kiuru-Enari S., and Bashir R.. 2010. A new distal myopathy with mutation in anoctamin 5. Neuromuscul. Disord. 20:791–795. 10.1016/j.nmd.2010.07.270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade, J.R., Archambeau A., and Michele D.E.. 2014. Rapid actin-cytoskeleton-dependent recruitment of plasma membrane-derived dysferlin at wounds is critical for muscle membrane repair. FASEB J. 28:3660–3670. 10.1096/fj.14-250191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, P.L., Miyake K., and Vogel S.S.. 2003. The endomembrane requirement for cell surface repair. Proc. Natl. Acad. Sci. USA. 100:4592–4597. 10.1073/pnas.0736739100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil, A.K., Rescher U., Gerke V., and McNeil P.L.. 2006. Requirement for annexin A1 in plasma membrane repair. J. Biol. Chem. 281:35202–35207. 10.1074/jbc.M606406200 [DOI] [PubMed] [Google Scholar]
- Middel, V., Zhou L., Takamiya M., Beil T., Shahid M., Roostalu U., Grabher C., Rastegar S., Reischl M., Nienhaus G.U., and Strähle U.. 2016. Dysferlin-mediated phosphatidylserine sorting engages macrophages in sarcolemma repair. Nat. Commun. 7:12875. 10.1038/ncomms12875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta, K., Tsutsumi S., Inoue H., Sakamoto Y., Miyatake K., Miyawaki K., Noji S., Kamata N., and Itakura M.. 2007. Molecular characterization of GDD1/TMEM16E, the gene product responsible for autosomal dominant gnathodiaphyseal dysplasia. Biochem. Biophys. Res. Commun. 357:126–132. 10.1016/j.bbrc.2007.03.108 [DOI] [PubMed] [Google Scholar]
- Monjaret, F., Suel-Petat L., Bourg-Alibert N., Vihola A., Marchand S., Roudaut C., Gicquel E., Udd B., Richard I., and Charton K.. 2013. The phenotype of dysferlin-deficient mice is not rescued by adeno-associated virus-mediated transfer of anoctamin 5. Hum. Gene Ther. Clin. Dev. 24:65–76. 10.1089/humc.2012.217 [DOI] [PubMed] [Google Scholar]
- Moreno-Pescador, G., Florentsen C.D., Østbye H., Sønder S.L., Boye T.L., Veje E.L., Sonne A.K., Semsey S., Nylandsted J., Daniels R., and Bendix P.M.. 2019. Curvature- and Phase-Induced Protein Sorting Quantified in Transfected Cell-Derived Giant Vesicles. ACS Nano. 13:6689–6701. 10.1021/acsnano.9b01052 [DOI] [PubMed] [Google Scholar]
- Nallamilli, B.R.R., Chakravorty S., Kesari A., Tanner A., Ankala A., Schneider T., da Silva C., Beadling R., Alexander J.J., Askree S.H., et al. 2018. Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann. Clin. Transl. Neurol. 5:1574–1587. 10.1002/acn3.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaify, G.A., Whyte M.P., Gottesman G.S., McAlister W.H., Eric Gordon J., Hollander A., Andrews M.V., El-Mofty S.K., Chen W.S., Veis D.V., et al. 2018. Gnathodiaphyseal dysplasia: Severe atypical presentation with novel heterozygous mutation of the anoctamin gene (ANO5). Bone. 107:161–171. 10.1016/j.bone.2017.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedemonte, N., and Galietta L.J.. 2014. Structure and function of TMEM16 proteins (anoctamins). Physiol. Rev. 94:419–459. 10.1152/physrev.00039.2011 [DOI] [PubMed] [Google Scholar]
- Penttilä, S., Palmio J., Suominen T., Raheem O., Evilä A., Muelas Gomez N., Tasca G., Waddell L.B., Clarke N.F., Barboi A., et al. 2012. Eight new mutations and the expanding phenotype variability in muscular dystrophy caused by ANO5. Neurology. 78:897–903. 10.1212/WNL.0b013e31824c4682 [DOI] [PubMed] [Google Scholar]
- Phuong, T.T.T., An J., Park S.H., Kim A., Choi H.B., and Kang T.M.. 2019. Deficiency of Anoctamin 5/TMEM16E causes nuclear positioning defect and impairs Ca2+ signaling of differentiated C2C12 myotubes. Korean J. Physiol. Pharmacol. 23:539–547. 10.4196/kjpp.2019.23.6.539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proske, U., and Morgan D.L.. 2001. Muscle damage from eccentric exercise: mechanism, mechanical signs, adaptation and clinical applications. J. Physiol. 537:333–345. 10.1111/j.1469-7793.2001.00333.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punetha, J., Kesari A., Uapinyoying P., Giri M., Clarke N.F., Waddell L.B., North K.N., Ghaoui R., O’Grady G.L., Oates E.C., et al. 2016. Targeted Re-Sequencing Emulsion PCR Panel for Myopathies: Results in 94 Cases. J. Neuromuscul. Dis. 3:209–225. 10.3233/JND-160151 [DOI] [PubMed] [Google Scholar]
- Roostalu, U., and Strähle U.. 2012. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell. 22:515–529. 10.1016/j.devcel.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Sarkozy, A., Hicks D., Hudson J., Laval S.H., Barresi R., Hilton-Jones D., Deschauer M., Harris E., Rufibach L., Hwang E., et al. 2013. ANO5 gene analysis in a large cohort of patients with anoctaminopathy: confirmation of male prevalence and high occurrence of the common exon 5 gene mutation. Hum. Mutat. 34:1111–1118. 10.1002/humu.22342 [DOI] [PubMed] [Google Scholar]
- Savarese, M., Di Fruscio G., Tasca G., Ruggiero L., Janssens S., De Bleecker J., Delpech M., Musumeci O., Toscano A., Angelini C., et al. 2015. Next generation sequencing on patients with LGMD and nonspecific myopathies: Findings associated with ANO5 mutations. Neuromuscul. Disord. 25:533–541. 10.1016/j.nmd.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schessl, J., Kress W., and Schoser B.. 2012. Novel ANO5 mutations causing hyper-CK-emia, limb girdle muscular weakness and Miyoshi type of muscular dystrophy. Muscle Nerve. 45:740–742. 10.1002/mus.23281 [DOI] [PubMed] [Google Scholar]
- Schmittgen, T.D., and Livak K.J.. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Simonsen, A.C., Boye T.L., and Nylandsted J.. 2020. Annexins Bend Wound Edges during Plasma Membrane Repair. Curr. Med. Chem. 27:3600–3610. 10.2174/0929867326666190121121143 [DOI] [PubMed] [Google Scholar]
- Sønder, S.L., Boye T.L., Tölle R., Dengjel J., Maeda K., Jäättelä M., Simonsen A.C., Jaiswal J.K., and Nylandsted J.. 2019. Annexin A7 is required for ESCRT III-mediated plasma membrane repair. Sci. Rep. 9:6726. 10.1038/s41598-019-43143-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, S.E., Ashkenazi A., Williamson A., Rubinsztein D.C., and Moreau K.. 2018. Transbilayer phospholipid movement facilitates the translocation of annexin across membranes. J. Cell Sci. 131:jcs217034. 10.1242/jcs.217034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, J., Umeda M., Sims P.J., and Nagata S.. 2010. Calcium-dependent phospholipid scrambling by TMEM16F. Nature. 468:834–838. 10.1038/nature09583 [DOI] [PubMed] [Google Scholar]
- Suzuki, J., Fujii T., Imao T., Ishihara K., Kuba H., and Nagata S.. 2013. Calcium-dependent phospholipid scramblase activity of TMEM16 protein family members. J. Biol. Chem. 288:13305–13316. 10.1074/jbc.M113.457937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., Suzuki J., and Nagata S.. 2014. Functional swapping between transmembrane proteins TMEM16A and TMEM16F. J. Biol. Chem. 289:7438–7447. 10.1074/jbc.M113.542324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaggart, K.A., Demonbreun A.R., Vo A.H., Swanson K.E., Kim E.Y., Fahrenbach J.P., Holley-Cuthrell J., Eskin A., Chen Z., Squire K., et al. 2014. Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair. Proc. Natl. Acad. Sci. USA. 111:6004–6009. 10.1073/pnas.1324242111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Kooi, A.J., Ten Dam L., Frankhuizen W.S., Straathof C.S., van Doorn P.A., de Visser M., and Ginjaar I.B.. 2013. ANO5 mutations in the Dutch limb girdle muscular dystrophy population. Neuromuscul. Disord. 23:456–460. 10.1016/j.nmd.2013.03.012 [DOI] [PubMed] [Google Scholar]
- Verma, S.K., Leikina E., Melikov K., Gebert C., Kram V., Young M.F., Uygur B., and Chernomordik L.V.. 2018. Cell-surface phosphatidylserine regulates osteoclast precursor fusion. J. Biol. Chem. 293:254–270. 10.1074/jbc.M117.809681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihola, A., Luque H., Savarese M., Penttilä S., Lindfors M., Leturcq F., Eymard B., Tasca G., Brais B., Conte T., et al. 2018. Diagnostic anoctamin-5 protein defect in patients with ANO5-mutated muscular dystrophy. Neuropathol. Appl. Neurobiol. 44:441–448. 10.1111/nan.12410 [DOI] [PubMed] [Google Scholar]
- Whitlock, J.M., and Hartzell H.C.. 2017. Anoctamins/TMEM16 Proteins: Chloride Channels Flirting with Lipids and Extracellular Vesicles. Annu. Rev. Physiol. 79:119–143. 10.1146/annurev-physiol-022516-034031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock, J.M., Yu K., Cui Y.Y., and Hartzell H.C.. 2018. Anoctamin 5/TMEM16E facilitates muscle precursor cell fusion. J. Gen. Physiol. 150:1498–1509. 10.1085/jgp.201812097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witting, N., Duno M., Petri H., Krag T., Bundgaard H., Kober L., and Vissing J.. 2013. Anoctamin 5 muscular dystrophy in Denmark: prevalence, genotypes, phenotypes, cardiac findings, and muscle protein expression. J. Neurol. 260:2084–2093. 10.1007/s00415-013-6934-y [DOI] [PubMed] [Google Scholar]
- Wu, J., Liu L., Matsuda T., Zhao Y., Rebane A., Drobizhev M., Chang Y.F., Araki S., Arai Y., March K., et al. 2013. Improved orange and red Ca2± indicators and photophysical considerations for optogenetic applications. ACS Chem. Neurosci. 4:963–972. 10.1021/cn400012b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, N., Cernysiov V., Davidson D., Song H., Tang J., Luo S., Lu Y., Qian J., Gyurova I.E., Waggoner S.N., et al. 2020. Critical Role of Lipid Scramblase TMEM16F in Phosphatidylserine Exposure and Repair of Plasma Membrane after Pore Formation. Cell Rep. 30:1129–1140.e5. 10.1016/j.celrep.2019.12.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, K., Whitlock J.M., Lee K., Ortlund E.A., Cui Y.Y., and Hartzell H.C.. 2015. Identification of a lipid scrambling domain in ANO6/TMEM16F. eLife. 4:e06901. 10.7554/eLife.06901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias, D.A., Violin J.D., Newton A.C., and Tsien R.Y.. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916. 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zaitseva, E., Zaitsev E., Melikov K., Arakelyan A., Marin M., Villasmil R., Margolis L.B., Melikyan G.B., and Chernomordik L.V.. 2017. Fusion Stage of HIV-1 Entry Depends on Virus-Induced Cell Surface Exposure of Phosphatidylserine. Cell Host Microbe. 22:99–110.e7. 10.1016/j.chom.2017.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
presents the fitted parameters of annexin time courses.
compares F/F0 or AUC values for ANXA2 or ANXA6 in ANO5-KO fibers rescued with ANO5 or ANO5-1-5.
lists the genes expressed in mature mouse myofibers and the fluorescent tags attached to them.
Data Availability Statement
All data referenced in the manuscript are included within the presented figures. Unique reagents, including cDNAs, and laboratory protocols will be promptly provided upon request.