Wieczorek et al. reconstitute the human γ-TuRC using recombinant proteins and examine the contribution of a key structural feature, the lumenal bridge, on the structure and microtubule-nucleating activity of this essential complex.
Abstract
The formation of cellular microtubule networks is regulated by the γ-tubulin ring complex (γ-TuRC). This ∼2.3 MD assembly of >31 proteins includes γ-tubulin and GCP2-6, as well as MZT1 and an actin-like protein in a “lumenal bridge” (LB). The challenge of reconstituting the γ-TuRC has limited dissections of its assembly and function. Here, we report a biochemical reconstitution of the human γ-TuRC (γ-TuRC-GFP) as a ∼35 S complex that nucleates microtubules in vitro. In addition, we generate a subcomplex, γ-TuRCΔLB-GFP, which lacks MZT1 and actin. We show that γ-TuRCΔLB-GFP nucleates microtubules in a guanine nucleotide–dependent manner and with similar efficiency as the holocomplex. Electron microscopy reveals that γ-TuRC-GFP resembles the native γ-TuRC architecture, while γ-TuRCΔLB-GFP adopts a partial cone shape presenting only 8–10 γ-tubulin subunits and lacks a well-ordered lumenal bridge. Our results show that the γ-TuRC can be reconstituted using a limited set of proteins and suggest that the LB facilitates the self-assembly of regulatory interfaces around a microtubule-nucleating “core” in the holocomplex.
Introduction
The γ-tubulin ring complex (γ-TuRC) is an ∼2.3 MD assembly required for proper microtubule network formation in eukaryotes (Knop et al., 1997; Raff et al., 1993; Stearns and Kirschner, 1994; Zheng et al., 1995). In vertebrates, the γ-TuRC contains at least 8 proteins, including γ-tubulin, the γ-tubulin complex proteins (GCPs) 2–6, and the mitotic spindle organizing proteins associated with a ring of γ-tubulin-1 and -2 (MZT1 and MZT2; Murphy et al., 2001; Teixidó-Travesa et al., 2010; Hutchins et al., 2010). The stoichiometry, location, and structures of these proteins in the context of the native human γ-TuRC were recently identified using high-resolution cryo-EM (Wieczorek et al., 2020a, b; Consolati et al., 2020). The γ-TuRC is an asymmetric, cone-shaped structure, in which GCP2-6 orient 14 γ-tubulin molecules in a helical arrangement that is poised to nucleate microtubules. Unexpectedly, the γ-TuRC also contains a prominent feature inside its conical structure, termed the “lumenal bridge” (LB). The LB is composed of two copies of MZT1 that associate with the N-terminal α-helical domains (NHDs) of GCP3 and GCP6 in structurally mimetic “modules,” along with an actin-like protein (Wieczorek et al., 2020a). However, the role of the LB in the assembly and microtubule-nucleating activity of the γ-TuRC is currently unclear.
Much of our biochemical understanding of the γ-TuRC comes from studies of heterotetramers of GCP2, GCP3, and γ-tubulin, also termed the γ-tubulin small complex (γ-TuSC; Oegema et al., 1999). Purified and/or recombinant Saccharomyces cerevisiae and Drosophila melanogaster γ-TuSCs can nucleate microtubules in bulk assays, but with relatively weak efficiencies compared with native γ-TuRCs or multimeric γ-TuSC assemblies (Oegema et al., 1999; Gunawardane et al., 2000; Vinh et al., 2002; Kollman et al., 2010). S. cerevisiae γ-TuSC mutants have been used to demonstrate that GTP binding to γ-tubulin is essential for its microtubule-nucleating activity (Gombos et al., 2013). However, whether the complete γ-TuRC structure is needed for efficient microtubule nucleation, or whether other well-ordered minimal subcomplexes exhibit native γ-TuRC–like activity, are open questions.
Here, we reconstitute the γ-TuRC using a set of 10 recombinant human proteins: γ-tubulin, GCP2, GCP3, GCP4, GCP5, GCP6, NEDD1, MZT1, MZT2, and β-actin. We use EM to show that the reconstituted complex resembles the native γ-TuRC in overall structure. In addition, we generate a subcomplex that lacks the LB components MZT1 and β-actin, and show that it adopts a semi-conical shape partially resembling the native complex. Despite its partial structure, this subcomplex nucleates individual microtubules in a manner analogous to the holocomplex. We further validate our functional assays by introducing a mutation into γ-tubulin that abrogates the subcomplex’s microtubule-nucleating activity. Our results show that the human γ-TuRC can be reconstituted using a limited set of components and suggest that the role of the LB is to build regulatory interfaces around a microtubule-nucleating “core.”
Results and discussion
Reconstitution of a γ-TuRC–sized complex from 10 recombinant human proteins
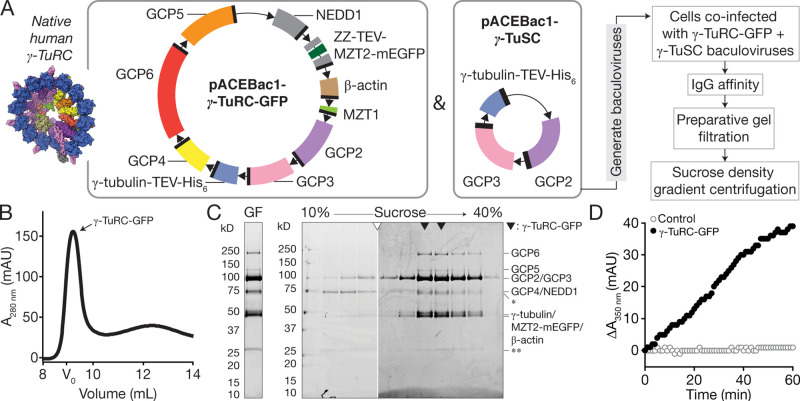
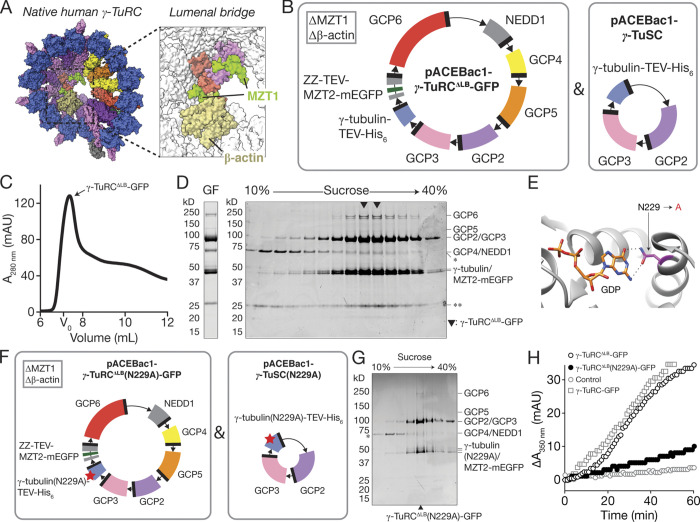
To address the need for a biochemically tractable system with which to study the function and assembly of a vertebrate γ-TuRC, we asked whether the human complex could be reconstituted with recombinant proteins. We tested multiple strategies, including expressing various combinations of γ-TuRC components either in human cells under cytomegalovirus promoters or in insect cells using the baculovirus expression system. Encouraging yields were only obtained when a stoichiometric excess of γ-tubulin, GCP2, and GCP3 were co-overexpressed in insect cells with GCP4, GCP5, GCP6, NEDD1, MZT1, β-actin, and MZT2A (hereafter, MZT2; Wieczorek et al., 2020a;). An IgG binding purification tag (ZZ) was fused to the N-terminus of MZT2 (Fig. 1 A; Huang et al., 2006; see Materials and methods). Based on a previous report that the structured portion of MZT2 (aa 35–80) binds to the N-terminus of GCP2 (GCP2-NHD; Wieczorek et al., 2020a), monomeric enhanced green fluorescent protein (mEGFP) was also fused to MZT2’s unstructured C-terminus (aa 81–158). We collectively refer to this set of 10 recombinant proteins as “γ-TuRC-GFP.” A complex containing all γ-TuRC-GFP proteins could be isolated from infected insect cell lysates using three purification steps (Fig. 1 A): (1) IgG affinity capture and tobacco etch virus (TEV) protease release, (2) gel filtration (Fig. 1, B and C), and (3) sucrose density gradient centrifugation (Fig. 1 C).
Figure 1.
Reconstitution, purification, and characterization of recombinant γ-TuRC-GFP. (A) Strategy for expressing and purifying γ-TuRC-GFP. Left, inset: Surface representation of a model for the native human γ-TuRC (Protein Data Bank accession nos. 6V6S, 6X0U, and 6X0V; Wieczorek et al., 2020a,b). Middle: Schematics of recombinant protein constructs used to express γ-TuRC-GFP in insect cells. Donor plasmids containing 10 γ-TuRC-GFP proteins (pACEBac1-γ-TuRC-GFP) and γ-tubulin-TEV-His6/GCP2/GCP3 (pACEBac1-γ-TuSC) used to generate bacmids are indicated. Coding fragments are colored according to the convention in Wieczorek et al. (2020b) and as in the inset model. Right: γ-TuRC-GFP expression and purification scheme. (B) Superose 6 Increase profile of γ-TuRC-GFP IgG eluate. Fractions containing γ-TuRC-GFP are indicated. The void volume of the column (V0) was estimated by gel filtering Dextran Blue 2000 in gel filtration buffer. mAU, milli-absorbance units. (C) Coomassie-stained SDS-PAGE analysis of γ-TuRC-GFP after gel filtration (GF; left) and sucrose gradient centrifugation (right). Hollow triangle indicates where gels were cropped. Solid triangles indicate peak γ-TuRC-GFP fractions. Single asterisk indicates an ∼70 kD contaminant that does not cosediment with γ-TuRC-GFP components. Double asterisk indicates a contaminant at ∼25 kD likely to be IgG light chain (Murphy et al., 2001). MZT1 (∼8 kD) is not visible on the gel but is detected in liquid chromatography–tandem mass spectrometry (Fig. S1 B). (D) Turbidity-based microtubule nucleation assay of reactions containing tubulin (20 μM) and GTP (1 mM) alone (hollow gray circles) or in the presence of 0.25 nM γ-TuRC-GFP (solid black circles). mAU, milli-absorbance units.
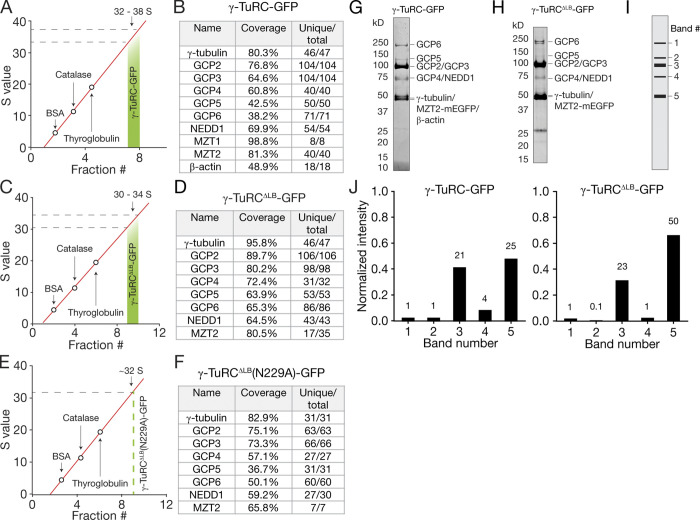
We characterized the biochemical composition of γ-TuRC-GFP in three ways. First, the sucrose density gradient was calibrated with standards of known S values (Fig. 1 C and Fig. S1 A), which indicated that γ-TuRC-GFP sediments to 32–38 S, a range that is larger than individual γ-TuSCs (9.8 S) but comparable to the native D. melanogaster γ-TuRC (35.5 S; Oegema et al., 1999). Notably, the sucrose gradient also removed a contaminant at ∼70 kD, likely corresponding to insect cell Hsp70 (Fig. 1 C). Second, the reconstituted complex was analyzed by mass spectrometry (Fig. S1 B), which confirmed the presence of all 10 overexpressed γ-TuRC-GFP proteins listed above. Third, SDS-PAGE densitometry analysis revealed that reconstituted γ-TuRC-GFP has a higher relative abundance of the γ-TuSC proteins γ-tubulin, GCP2, GCP3, and/or MZT2-mEGFP (Fig. S1, G, I, and J) compared with the native complex (Wieczorek et al., 2020b). We note that γ-TuRC stoichiometry as judged by SDS-PAGE has been reported to vary depending on the affinity handle used (e.g., GCP2 tagged with monomeric blue fluorescent protein [mBFP] and biotin acceptor peptide [BAP] enriches GCP2 relative to GCP3; Choi et al., 2010; Leong et al., 2019; Consolati et al., 2020; Hutchins et al., 2010), yet multiple independent γ-TuRC structures converge on the same core component stoichiometry (Consolati et al., 2020; Liu et al., 2020; Wieczorek et al., 2020a, 2020b). Together, our data suggest that a γ-TuRC–sized complex can be reconstituted with a set of 10 recombinant human proteins expressed in insect cells.
Figure S1.
Biochemical characterization of reconstituted complexes. (A) Standard curve used to estimate an S value range for γ-TuRC-GFP from sucrose gradients. The red line is a linear fit to the peak sedimentation fractions for BSA (4.6 S), catalase (11.3 S), and thyroglobulin (19 S) identified in parallel sucrose gradients. (B) Results from liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis of γ-TuRC-GFP. Coverage represents the percentage of the sequence that was identified by the LC-MS/MS analysis. Unique/total designates the number of unique peptides and total peptides, respectively, identified by the LC-MS/MS analysis. (C) Standard curve used to estimate an S value range for γ-TuRCΔLB-GFP from sucrose gradients. The red line is a linear fit to standard proteins as in A. (D) Results from LC-MS/MS analysis of γ-TuRCΔLB-GFP. (E) Standard curve used to estimate S value of γ-TuRCΔLB(N229A)-GFP from sucrose gradients. The red line is a linear fit to standard proteins as in A. (F) Results from LC-MS/MS analysis of γ-TuRCΔLB(N229A)-GFP. (G) Coomassie-stained SDS-PAGE gel of the peak γ-TuRC-GFP sucrose gradient fraction (cropped from the gel in Fig. 1 C). (H) Coomassie-stained SDS-PAGE gel of the peak γ-TuRCΔLB-GFP sucrose gradient fraction (cropped from the gel in Fig. 3 D). (I) Schematic depicting the five major gel bands (corresponding to recombinant proteins) observed in peak γ-TuRC-GFP (G) and γ-TuRCΔLB-GFP (H) fractions used for densitometry analysis. (J) Plot of the normalized intensity of bands 1–5 as schematized in I for γ-TuRC-GFP (left) and γ-TuRCΔLB-GFP (right). Numbers above bars indicate relative intensity compared with band 1 (see Materials and methods).
To assess the activity of the reconstituted complex, we incubated γ-TuRC-GFP (∼0.25 nM; see Materials and methods) with α/β-tubulin (20 μM) and GTP (1 mM), and measured the change in optical density, or “turbidity” (Gaskin et al., 1974), of the reaction over time. An increase in turbidity (interpreted as the formation of microtubule polymer) was observed only in the presence of both γ-TuRC-GFP and tubulin (Fig. 1 D); no obvious increase in turbidity was detected in tubulin- (Fig. 1 D) or γ-TuRC-GFP–only (data not shown) controls. These results suggest that reconstituted γ-TuRC-GFP is capable of nucleating microtubule assembly.
γ-TuRC-GFP resembles the native γ-TuRC in overall architecture
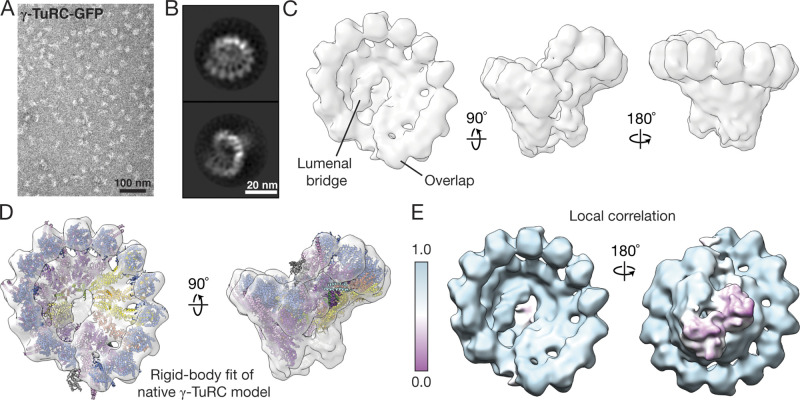
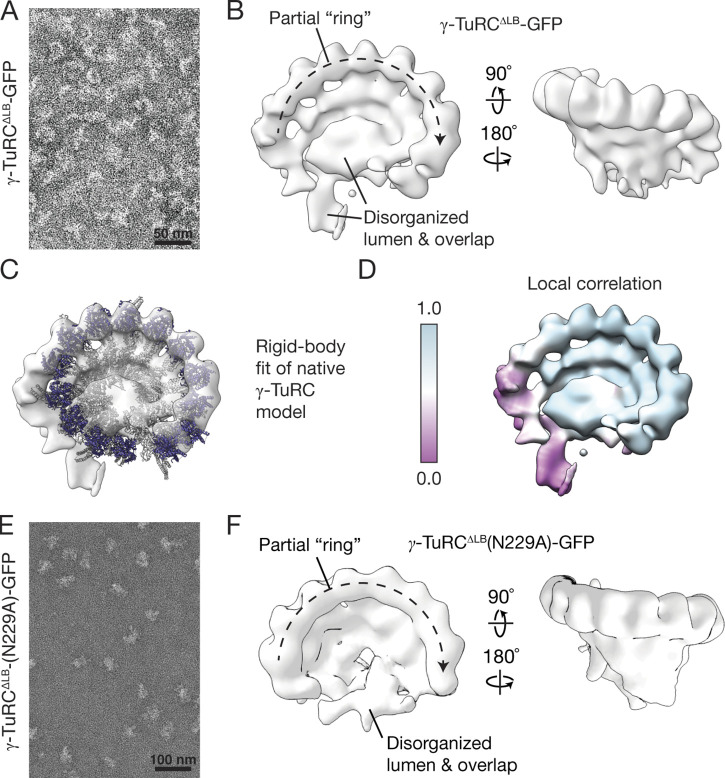
We next examined the structure of γ-TuRC-GFP using negative-stain EM. γ-TuRC-GFP appears as ∼25–30-nm-wide lockwasher-shaped assemblies in EM micrographs (Fig. 2 A). Reference-free 2D classification indicated that γ-TuRC-GFP adopts a ring-shaped assembly, in which multiple ∼5-nm-wide globular densities are laterally associated and supported by stalk-like structures (Fig. 2 B). A 3D reconstruction revealed a cone-shaped structure with ∼14 globular domains (Fourier shell correlation (FSC)0.5 = ∼30 Å; Fig. 2 C and Fig. S2 A), as observed in recent cryo-EM reconstructions of the native human γ-TuRC (Wieczorek et al., 2020b; Consolati et al., 2020; Wieczorek et al., 2020a). Correspondingly, a model of the native complex could be rigid body–fitted into the γ-TuRC-GFP density (Protein Data Bank accession nos. 6V6S, 6X0U, and 6X0V; Wieczorek et al., 2020a,b; Fig. 2 D). A high local correlation score between the fitted model and the γ-TuRC-GFP density (Fig. 2 E) indicates that the asymmetric features (e.g., an LB; Fig. 2 C) in the native complex are also present in γ-TuRC-GFP (Wieczorek et al., 2020b). These data suggest that γ-TuRC-GFP stably assembles into an asymmetric, cone-shaped structure that closely resembles the native complex.
Figure 2.
γ-TuRC-GFP resembles the native human γ-TuRC in overall architecture. (A) Transmission EM micrograph of negatively stained γ-TuRC-GFP. (B) 2D class averages showing two orientations of γ-TuRC-GFP particles. (C) Three views of a 3D reconstruction of γ-TuRC-GFP from negative-stain EM data. The well-ordered LB and overlap regions are labeled. (D) Two views of a rigid body fit of the native human γ-TuRC model from Fig. 1 A into the γ-TuRC-GFP density map. (E) Two views of the γ-TuRC-GFP density map colored according to local correlation of the model fit in D (see Materials and methods).
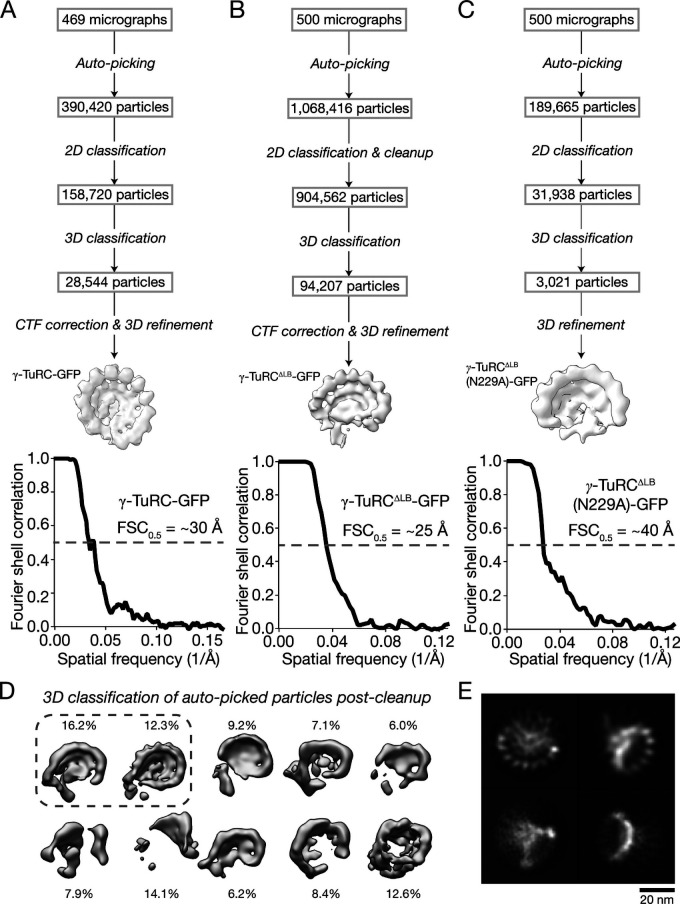
Figure S2.
Supplemental data for negative-stain EM analyses of γ-TuRC-GFP, γ-TuRCΔLB-GFP, and γ-TuRCΔLB(N229A)-GFP. (A) Top: Workflow for generating a negative-stain EM 3D reconstruction of γ-TuRC-GFP. Bottom: Masked FSC curve for the γ-TuRC-GFP reconstruction. FSC = 0.5 is indicated by a dashed gray line, and an estimate of the corresponding resolution is indicated. (B) Top: Workflow for generating a negative-stain EM 3D reconstruction of γ-TuRCΔLB-GFP. Bottom: Masked FSC curve for the γ-TuRCΔLB-GFP reconstruction. FSC = 0.5 is indicated by a dashed gray line, and an estimate of the corresponding resolution is indicated. (C) Top: Workflow for generating a negative-stain EM 3D reconstruction of γ-TuRCΔLB(N229A)-GFP. Bottom: Masked FSC curve for the γ-TuRCΔLB(N229A)-GFP reconstruction. FSC = 0.5 is indicated by a dashed gray line, and an estimate of the corresponding resolution is indicated. (D) 3D classification results of γ-TuRCΔLB-GFP particles after initial cleanup. The percentage of total particles each class comprises is indicated. The dashed line encircles the two 3D classes used for downstream processing, which underwent a second round of 3D classification (classes not shown; this reduced the number of particles from ∼250,000 to 94,207 particles as shown in the workflow) before 3D refinement and final reconstruction of γ-TuRCΔLB-GFP. (E) 2D class averages representing four different views of refined particles used to generate the γ-TuRCΔLB-GFP 3D reconstruction.
Reconstitution of a γ-TuRC subcomplex lacking MZT1 and β-actin
Next, we focused on examining the role of the LB (Fig. 3 A), which spans across the inside of the γ-TuRC cone (Consolati et al., 2020; Liu et al., 2020; Wieczorek et al., 2020a, 2020b). We generated a polycistronic construct containing all γ-TuRC-GFP proteins except for MZT1 and β-actin (“γ-TuRCΔLB-GFP”; Fig. 3 B). γ-TuRCΔLB-GFP proteins were co-overexpressed with a stoichiometric excess of γ-tubulin, GCP2, and GCP3 in insect cells, and the resulting complex was isolated using the same three-step purification procedure as γ-TuRC-GFP (Fig. 1 A). The majority of γ-TuRCΔLB-GFP proteins from peak gel filtration fractions cosedimented together in sucrose density gradients (Fig. 3, C and D), albeit to lower estimated S values when compared with γ-TuRC-GFP (30–34 S, Fig. S1 C; vs. 32–38 S, Fig. S1 A). Mass spectrometry also confirmed the presence of all eight γ-TuRCΔLB-GFP proteins in the peak sucrose density gradient fractions (Fig. S1 D). As observed for γ-TuRC-GFP, SDS-PAGE densitometry analysis also indicated an enrichment of γ-TuSC proteins relative to other γ-TuRCΔLB-GFP subcomponents (Fig. S1, H–J). Additionally, a reduction in the levels of both GCP5 and GCP4/NEDD1 relative to GCP6 was observed (Fig. S1, H–J).
Figure 3.
Reconstitution, purification, and functional characterization of γ-TuRCΔLB-GFP. (A) Surface representation of a model for the native human γ-TuRC, as in Fig. 1 A. Inset shows the LB and highlights β-actin and MZT1. (B) Schematics of recombinant protein constructs used to express γ-TuRCΔLB-GFP in insect cells. Donor plasmids containing eight γ-TuRCΔLB-GFP proteins (pACEBac1-γ-TuRCΔLB-GFP) and γ-tubulin-TEV-His6/GCP2/GCP3 (pACEBac1-γ-TuSC) used to generate bacmids are indicated. (C) Superose 6 elution profile of γ-TuRCΔLB-GFP IgG eluate. Fractions containing intact γ-TuRCΔLB-GFP are indicated. The void volume (V0) of the column was estimated by gel filtering Dextran Blue 2000 in gel filtration buffer. (D) Coomassie-stained SDS-PAGE analysis of γ-TuRCΔLB-GFP after gel filtration (GF; left) and sucrose gradient centrifugation (right). Solid triangles indicate peak γ-TuRCΔLB-GFP fractions. Single asterisk indicates an ∼70 kD contaminant that does not cosediment with γ-TuRC-GFP components. Double asterisk indicates a contaminant at ∼25 kD likely to be IgG light chain (Murphy et al., 2001). (E) View of the nucleotide-binding site in the x-ray crystal structure of GDP-bound human γ-tubulin (Protein Data Bank accession no. 3CB2; gray cartoon representation). Residue N229 (magenta), which was mutated to alanine in this study, and GDP (colored by element; C and P atoms both colored in orange) are indicated. Predicted hydrogen bonds formed between N229 and the guanine moiety of GDP are shown (dotted black lines). (F) Schematics of the recombinant protein constructs used to express γ-TuRCΔLB(N229A)-GFP in insect cells. Donor plasmids containing the eight γ-TuRCΔLB(N229A)-GFP proteins (pACEBac1-γ-TuRCΔLB[N229A]-GFP; left) and γ-tubulin(N229A)-TEV-His6/GCP2/GCP3 (pACEBac1-γ-TuSC[N229A]; right) are indicated. The N229A point mutation is highlighted by a red star in the γ-tubulin(N229A) coding fragments. Coding fragments are colored as in Fig. 1 A. (G) Coomassie-stained SDS-PAGE analysis of γ-TuRCΔLB(N229A)-GFP sucrose gradient centrifugation fractions. Asterisk indicates an ∼70 kD contaminant that does not cosediment with γ-TuRCΔLB(N229A)-GFP components in the sucrose gradient. Peak γ-TuRCΔLB(N229A)-GFP fraction is indicated by a solid triangle at the bottom of the gel. Note: Only odd-numbered gradient fractions were run on this particular gel. (H) Turbidity-based microtubule nucleation assay of reactions containing tubulin (20 μM) and GTP (1 mM) alone (hollow gray circles), in the presence of 0.25 nM γ-TuRCΔLB-GFP (hollow black circles), or in the presence of 0.25 nM γ-TuRCΔLB(N229A)-GFP (solid black circles). Turbidity data for γ-TuRC-GFP from Fig. 1 D (gray squares) are also shown for comparison.
We next asked whether γ-TuRCΔLB-GFP, which assembles in the absence of MTZ1 and β-actin, is capable of nucleating microtubules. As a control, we introduced a point mutation in the nucleotide-binding site of γ-tubulin (N229A; Fig. 3 E). Though the precise role of GTP hydrolysis in γ-TuRC function is not known, this mutation is predicted to significantly reduce human γ-tubulin’s nucleotide affinity and microtubule-nucleating activity based on previous bulk nucleation assays with yeast γ-TuSC assemblies (Gombos et al., 2013). γ-tubulin (N229A) was co-overexpressed along with the other seven γ-TuRCΔLB-GFP components (Fig. 3 F). The resulting subcomplex, γ-TuRCΔLB(N229A)-GFP, was reconstituted using a similar strategy as for γ-TuRC-GFP (Fig. 1 A). γ-TuRCΔLB(N229A)-GFP sedimented to a similar position in sucrose gradients as γ-TuRCΔLB-GFP (∼32 S; Fig. 3 G and Fig. S1 E) and contained all eight overexpressed proteins as judged by mass spectrometry (Fig. S1 F).
Incubation of γ-TuRCΔLB(N229A)-GFP (∼0.25 nM) with tubulin (20 µM) and GTP (1 mM) did not produce a substantial increase in signal in the turbidity assay (Fig. 3 H). Under the same conditions, however, the presence of “wild-type” γ-TuRCΔLB-GFP did result in a turbidity increase (Fig. 3 H). Together, these data indicate that γ-TuRCΔLB-GFP nucleates microtubules despite lacking key components of the LB, and that this activity depends on its ability to bind guanine nucleotide.
γ-TuRCΔLB-GFP adopts a partial ring structure
We next characterized the structure of γ-TuRCΔLB-GFP using negative-stain EM. Individual γ-TuRCΔLB-GFP particles resembled those from grids containing γ-TuRC-GFP (Fig. 4 A vs. Fig. 2 A). However, 3D classification of >900,000 particles revealed that γ-TuRCΔLB-GFP mainly adopts distinct, semi-conical structures displaying only 8–10 γ-tubulin–like globular domains (Fig. S2, B and D). Native γ-TuRC–like structures were not observed in these 3D averages. We generated a 3D reconstruction of the γ-TuRCΔLB-GFP particles that gave the highest resolutions in 3D classifications (FSC0.5 = ∼25 Å; Fig. 4 B and Fig. S2, B, D, and E; see Materials and methods). The resulting density map revealed that γ-TuRCΔLB-GFP contains approximately eight well-defined γ-tubulin–like subunits lining the rim of a semi-conical structure with a helical organization similar to γ-TuRC-GFP (Fig. 4 B). Consistent with this observation, a local correlation analysis of a rigid body–fitted model of the native human γ-TuRC into the γ-TuRCΔLB-GFP density map indicated that γ-TuRCΔLB-GFP closely resembles a portion of the native γ-TuRC corresponding to approximately eight GCP/γ-tubulin subunits (Protein Data Bank accession nos. 6V6S, 6X0U, and 6X0V; Wieczorek et al., 2020a,b; Fig. 4, C and D).
Figure 4.
Reconstituted γ-TuRCΔLB-GFP adopts a partial, cone-shaped structure. (A) Transmission EM micrograph of negatively stained γ-TuRCΔLB-GFP. (B) Two views of a 3D reconstruction of γ-TuRCΔLB-GFP from negative-stain EM data. (C) Rigid body fit of the native human γ-TuRC model from Fig. 1 A into the γ-TuRCΔLB-GFP density map. γ-Tubulin molecules are colored in blue; all other subunits are shown in white. Globular densities in the partial ring are numbered. (D) The γ-TuRCΔLB-GFP density map colored according to local correlation of the model fit in C (see Materials and methods). (E) Transmission EM micrograph of negatively stained γ-TuRCΔLB(N229A)-GFP. (F) Two views of a 3D reconstruction of γ-TuRCΔLB(N229A)-GFP from negative-stain EM data.
We performed a similar negative-stain EM analysis for γ-TuRCΔLB(N229A)-GFP, which revealed that γ-TuRCΔLB(N229A)-GFP also mainly adopts a partial “ring”-like structure exhibiting approximately eight γ-tubulin domains (FSC0.5 = ∼40 Å; Fig. 4, E and F; and Fig. S2 C). Notably, both γ-TuRCΔLB-GFP and γ-TuRCΔLB(N229A)-GFP reconstructions lack well-organized LB and “overlap” regions (positions 1–2 and 13–14 in the native γ-TuRC; Consolati et al., 2020; Liu et al., 2020; Wieczorek et al., 2020a, 2020b; Fig. 4, B, C, and F). These results suggest that γ-TuRCΔLB-GFP takes on a semi-conical structure that organizes approximately eight γ-tubulins into a partial ring that is native γ-TuRC–like. Combined with our reconstitution of γ-TuRC-GFP (Figs. 1 and 2), these data also provide biochemical evidence that β-actin may constitute the “actin-like protein” in the LB (Wieczorek et al., 2020b).
γ-TuRCΔLB-GFP nucleates individual microtubules with kinetics similar to the native γ-TuRC
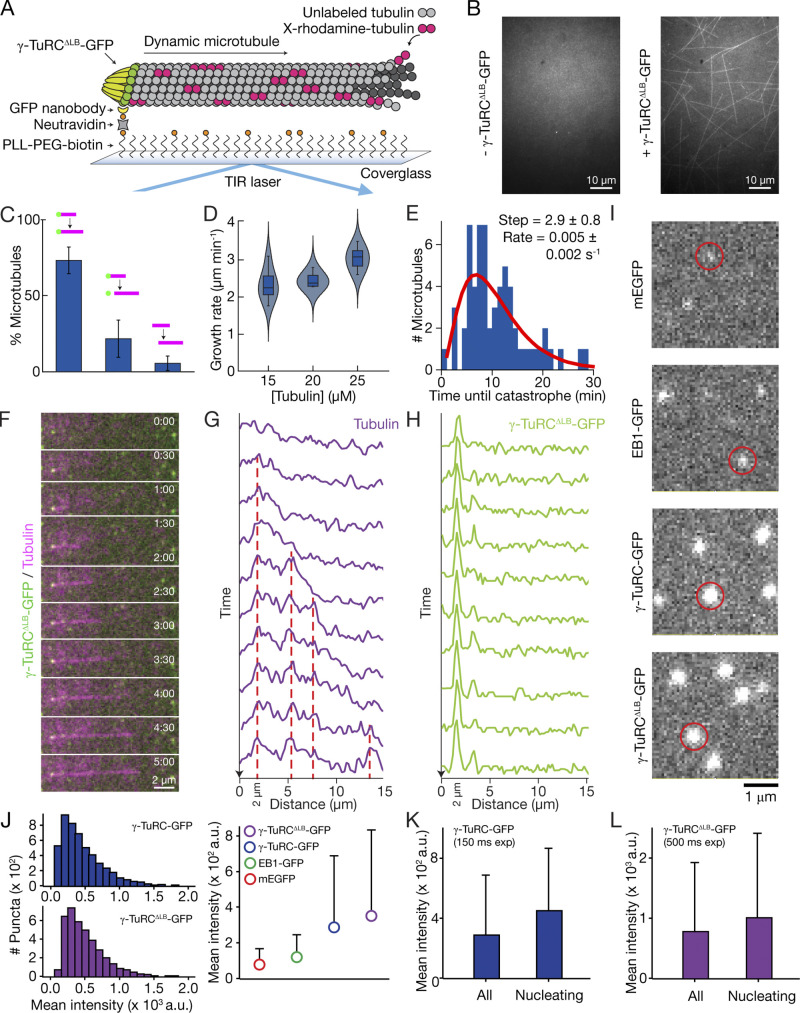
To further examine γ-TuRCΔLB-GFP’s microtubule-nucleating activity, we immobilized the complex to a coverslip surface and imaged individual microtubule nucleation events using total internal reflection fluorescence (TIRF) microscopy (Fig. S3 A; see Materials and methods). Within minutes, the polymerization of individual microtubules on the coverslip surface could be observed in the tubulin channel; in coverslips prepared without γ-TuRCΔLB-GFP, microtubule polymerization was not observed (Fig. S3 B). Approximately 95% of the microtubule nucleation events were observed to initiate from γ-TuRCΔLB-GFP puncta (Fig. 5 A; and Fig. S3, C and F–H). The growth rates and catastrophe frequencies of microtubules nucleated by γ-TuRCΔLB-GFP matched published values for those of plus ends grown off of guanylyl-(α,β)-methylene-diphosphonate (GMPCPP) seeds (Brouhard et al., 2008; Gardner et al., 2011), axonemes (Walker et al., 1988), and native γ-TuRCs (Consolati et al., 2020; Thawani et al., 2020; Fig. S3, D and E). Moreover, the position of fluorescent-tubulin “speckles” along the growing microtubule’s length did not change relative to the position of microtubule end-bound complexes (Fig. S3, F–H; Waterman-Storer et al., 1998), indicating that γ-TuRCΔLB-GFP nucleates microtubules that grow from their distal plus ends.
Figure S3.
Supplementary data for TIRF assays. (A) Schematic of the TIRF-based assay developed to visualize individual microtubule nucleation events from γ-TuRCΔLB-GFP. PLL, poly-L-lysine; PEG, polyethylene glycol; TIR, total internal reflection. (B) Fields of view displaying the tubulin channel after 20 min in the absence (left) or presence (right) of surface-immobilized γ-TuRCΔLB-GFP. (C) The percentage of γ-TuRCΔLB-GFP–nucleated microtubules that remained stably attached in a field of view (left), were temporarily attached (middle), or were not observed to be nucleated from (right) γ-TuRCΔLB-GFP puncta (schematized above each bar). Data are presented as mean ± SEM. n = 733 microtubule nucleation events pooled from five independent experiments. (D) Growth rates of microtubules nucleated by γ-TuRCΔLB-GFP at increasing tubulin concentrations. Solid lines drawn within box plots represent the mean, the boxes represent 25th and 75th percentile data, and the whiskers represent 5th and 95th percentile data. Violin plots show kernel density distributions of the data generated using a bandwidth value of 1 μm/min. n = 19 microtubule plus ends from two independent experiments were measured at each tubulin concentration. (E) Distribution of lifetimes, or the “time until catastrophe,” of microtubules nucleated by γ-TuRCΔLB-GFP. The red line is a fit to the γ distribution (Gardner et al., 2011), which yields a step size of 2.9 ± 0.8 and a rate parameter of 0.005 ± 0.002 s−1 (maximum likelihood estimates ± 95% confidence intervals). n = 61 microtubule catastrophe events pooled from five independent experiments. (F) Two-color overlay time-lapse sequence of a microtubule nucleated by γ-TuRCΔLB-GFP. (G) Line scans of the growing microtubule in F (tubulin channel) over each time frame. Dashed red lines indicate positions of persistent fluorescence intensity fluctuations (“speckles”) that do not appear to change in position over time. (H) Line scans of the growing microtubule in F (γ-TuRCΔLB-GFP channel) over each time frame. The γ-TuRCΔLB-GFP punctum (at ∼2.0 μm) colocalizes with the same position of the minus end of the nucleated microtubule in G. (I) Fluorescence micrographs of, from top to bottom, surface-immobilized mEGFP, EB1-GFP, γ-TuRC-GFP, and γ-TuRCΔLB-GFP. Examples of single molecules are circled in red. (J) Left: Calculated mean intensity distributions of surface-immobilized γ-TuRC-GFP (top) and γ-TuRCΔLB-GFP (bottom) single molecules. Right: Plot of the mean intensities of surface-immobilized mEGFP, EB1-GFP, γ-TuRC-GFP, and γ-TuRCΔLB-GFP fluorescent single molecules imaged using an exposure time of 150 ms. The total numbers of puncta (which includes false positives; see Materials and methods) analyzed from at least three independent experiments are 20,852 (mEGFP), 13,392 (EB1-GFP), 16,639 (γ-TuRC-GFP), and 15,198 (γ-TuRCΔLB-GFP). (K and L) Comparison of the mean intensity of all surface-immobilized γ-TuRC-GFP (I) or v-TuRCΔLB-GFP (J) puncta (labeled as “All”) vs. only ones that are observed to nucleate microtubules (labeled as “Nucleating”) from Fig. 5. The total numbers of microtubule-nucleating puncta measured from at least three independent experiments are 205 (γ-TuRC-GFP) and 112 (γ-TuRCΔLB-GFP). Error bars in J–L represent standard deviations (see Materials and methods).
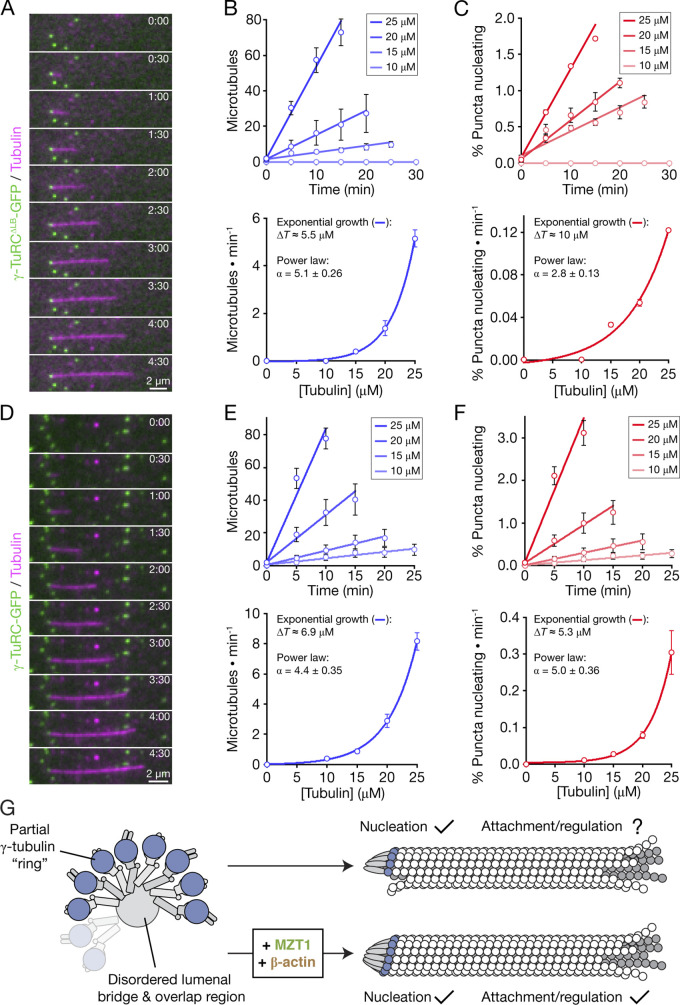
Figure 5.
γ-TuRCΔLB-GFP and γ-TuRC-GFP both nucleate individual microtubules with native γ-TuRC-like efficiency. (A and D) Two-color overlay from a time lapse sequence of a microtubule nucleated by and stably attached to a γ-TuRCΔLB-GFP (A) or γ-TuRC-GFP (D) punctum. Time is reported as minutes:seconds. (B) Top: Plot of the cumulative number of microtubules nucleated by γ-TuRCΔLB-GFP over time at increasing tubulin concentrations. Data were fitted using weighted linear regression (solid lines). Bottom: Plot of the rate of microtubule formation over time as a function of tubulin concentration. Data were fitted to a weighted exponential growth model of the form y = y0 + Aexp(x/t), where x = concentration of tubulin (in micromolar), and ΔT = t/ln(2) is an estimate of the change in tubulin concentration (in micromolar) needed to double the nucleation rate. The nonzero nucleation rates were also fitted to a power law of the form ln(y) = αln(x) + ln(β); the value for the exponent α is indicated. (C) Top: Plot of the cumulative percentage of γ-TuRCΔLB-GFP puncta that nucleated a microtubule over time at increasing tubulin concentrations. Data were fitted using weighted linear regression (solid lines). Bottom: Plot of the rate at which the number of γ-TuRCΔLB-GFP–nucleating puncta increases over time as a function of tubulin concentration. Data were fitted to a weighted exponential growth model of the form y = y0 + Aexp(x/t), where x = concentration of tubulin (in micromolar), and ΔT = t/ln(2) is an estimate of the change in tubulin concentration (in micromolar) needed to double the number of γ-TuRCΔLB-GFP–nucleating puncta. The nonzero nucleation rates were also fitted to a power law of the form ln(y) = αln(x) + ln(β); the value for the exponent α is indicated, and β is a fitting constant. (E) Top: Plot of the cumulative number of microtubules nucleated by γ-TuRC-GFP over time at increasing tubulin concentrations. Data were fitted using weighted linear regression (solid lines). Bottom: Plot of the rate of microtubule formation over time as a function of tubulin concentration. Data were fitted as in B. (F) Top: Plot of the cumulative percentage of γ-TuRC-GFP puncta that nucleated a microtubule over time at increasing tubulin concentrations. Data were fitted using weighted linear regression (solid lines). Bottom: Plot of the rate at which the number of γ-TuRC-GFP–nucleating puncta increases over time as a function of tubulin concentration. Data were fitted as in C. A total of n = 325 microtubule nucleation events and 28,164 γ-TuRCΔLB-GFP puncta from 10 independent experiments was analyzed in B and C. A total of n = 434 microtubule nucleation events and 36,454 γ-TuRC-GFP puncta from 12 independent experiments was analyzed in E and F. Error bars in B (top), C (top), E (top), and F (top) represent SEM from the at least two independent experiments performed per tubulin concentration tested. Error bars in B (bottom), C (bottom), E (bottom), and F (bottom) represent propagated errors generated by the weighted linear regression fits in the top panels. Exponential and power law fit errors were propagated manually (see Materials and methods). (G) Proposed model for the role of the LB in γ-TuRC assembly, microtubule-nucleating function, and downstream regulation. Our reconstitutions suggest that the role of the LB (containing MZT1 and β-actin) is to build regulatory interfaces around a microtubule-nucleating “core” within the γ-TuRC.
The microtubule-nucleating activity of the native human γ-TuRC was previously found to be surprisingly low (<1% nucleation efficiency; Consolati et al., 2020). We also found that many γ-TuRCΔLB-GFPs (>95%) did not nucleate microtubules during the time course of our experiments (Fig. S3 C). To further characterize the microtubule-nucleating efficiency of γ-TuRCΔLB-GFP, we performed the microtubule nucleation assay at increasing tubulin concentrations. The number of γ-TuRCΔLB-GFP-mediated microtubule nucleation events, as well as the percentage of complexes that nucleated microtubules, were both found to increase linearly over time (Fig. 5, B and C, upper panels), suggesting that the majority of surface-immobilized γ-TuRCΔLB-GFP molecules are capable of nucleating microtubules (Consolati et al., 2020). γ-TuRCΔLB-GFP–mediated microtubule nucleation rates were also found to increase exponentially with tubulin concentration (Fig. 5, B and C, lower panels), indicating that microtubule nucleation from γ-TuRCΔLB-GFP is a cooperative process.
Under the same assay conditions, the γ-TuRC-GFP holocomplex also nucleated microtubules that grew from one end (Fig. 5 D). Remarkably, microtubule nucleation from γ-TuRC-GFP followed similar kinetics as observed for γ-TuRCΔLB-GFP (Fig. 5, D–F). Consistent with recent reports using native γ-TuRCs (Consolati et al., 2020), greater than ∼10 µM tubulin was required to observe robust microtubule nucleation from either γ-TuRCΔLB-GFP or γ-TuRC-GFP, a concentration that is greater than that needed to nucleate from preformed microtubule seeds (∼7 µM), but less than that required for spontaneous microtubule assembly (∼20 µM; Wieczorek et al., 2015). Exponential fits to the microtubule nucleation rate data revealed characteristic doubling concentrations of ΔT = ∼5.5 µM tubulin (γ-TuRCΔLB-GFP; Fig. 5 B) and ∼6.9 µM tubulin (γ-TuRC-GFP; Fig. 5 E). Similarly, the percentage of surface-immobilized complexes that nucleate microtubules will double when ΔT = ∼10 µM tubulin (γ-TuRCΔLB-GFP; Fig. 5 C) and ∼5.3 µM tubulin (γ-TuRC-GFP; Fig. 5 F). A power law fit to the microtubule nucleation rate data for both reconstituted complexes generated exponents of α = 5.1 ± 0.26 (γ-TuRCΔLB-GFP; Fig. 5 B) and α = 4.4 ± 0.35 (γ-TuRC-GFP; Fig. 5 E), which we note are similar to the value of 6.7 reported recently in a similar analysis of the native human γ-TuRC (Consolati et al., 2020). Additionally, γ-TuRC-GFP and γ-TuRCΔLB-GFP fluorescent puncta displayed similar intensity distributions and were both brighter than single mEGFP molecules (Fig. S3, I–L), suggesting that both complexes contain a similar number of MZT2-mEGFPs (and hence, γ-TuSCs). Together, these results suggest that γ-TuRCΔLB-GFP nucleates single microtubules in a manner that is analogous to the γ-TuRC holocomplex.
In this study, we have reconstituted the human γ-TuRC (γ-TuRC-GFP) from a set of 10 recombinant proteins co-overexpressed in insect cells. We also reconstitute a subcomplex, γ-TuRCΔLB-GFP, and find that it nucleates single microtubules with a similar efficiency as the reconstituted holocomplex. In contrast to γ-TuRC-GFP, which assembles into a native-like complex, γ-TuRCΔLB-GFP adopts a semi-conical architecture and lacks a well-structured LB.
Previous reconstitution work has focused on the γ-TuSC (Gunawardane et al., 2000; Vinh et al., 2002; Lin et al., 2016; Leong et al., 2019), a stable and evolutionarily conserved heterotetramer containing GCP2, GCP3, and two copies of γ-tubulin (Kollman et al., 2008). In the presence of the accessory factor Spc110, purified S. cerevisiae γ-TuSCs can spontaneously self-assemble into helical assemblies (Kollman et al., 2010). Reconstituted Candida albicans and Schizosaccharomyces pombe γ-TuSCs display analogous properties, though their oligomerization depends on both Spc110 homologues and the microprotein MZT1 (Lin et al., 2016; Leong et al., 2019). In contrast, our work identifies γ-TuRCΔLB-GFP as a partial ring-like subcomplex containing both γ-TuSC proteins and the evolutionarily divergent GCPs. The limited resolution provided by negative-stain EM precludes unambiguous assignment of GCP subunits in the γ-TuRCΔLB-GFP density map (Fig. 4 B). However, our stoichiometry estimates suggest that, in contrast to γ-TuRC-GFP, GCP5 is depleted in γ-TuRCΔLB-GFP (Fig. S1, G–J), but single-molecule analysis of γ-TuRCΔLB-GFP indicates that it still contains a similar number of MZT2-mEGFP (and hence, γ-TuSC) proteins as γ-TuRC-GFP (Fig. S3, I–L). Previously, we identified extensive electrostatic interactions between the N-terminal GCP6 “belt” (aa 130–195) and the lumenal face of at least two γ-TuSCs in the native γ-TuRC (Wieczorek et al., 2020a). Although further work is needed to understand the complete assembly pathway of the human γ-TuRC, our γ-TuRCΔLB-GFP data are consistent with a model in which the GCP6 belt could stabilize the assembly of a finite number of γ-TuSC subunits without necessarily relying on a GCP4/GCP5/GCP4/GCP6 γ-TuSC–like subassembly (Haren et al., 2020). These data also help explain why RNAi silencing of GCP5 in cultured cells results in a milder phenotype than depletion of other GCP components (Cota et al., 2017).
In addition to missing a well-ordered LB, γ-TuRCΔLB-GFP lacks the overlap region, which in the holocomplex is proximal to the binding site for the CM1 motif of CDK5RAP2 (Wieczorek et al., 2020a), a γ-TuRC attachment factor and proposed “activator” of the native complex’s microtubule-nucleating function (Choi et al., 2010). Our data raise the possibility that γ-TuRCΔLB-GFP–like subcomplexes that can retain robust microtubule-nucleating activity may form in cells (Fig. 5 G). Consistent with this idea, depletion of MZT1 in human cells does not eliminate the formation of γ-TuRC–like complexes (Cota et al., 2017; Lin et al., 2016), but does abolish ectopic stimulation of microtubule nucleation by CDK5RAP2 overexpression (Lin et al., 2016). Similarly, most γ-TuRCs in D. melanogaster lack MZT1 (Tovey et al., 2018), yet exhibit robust microtubule-nucleating activity in vitro (Oegema et al., 1999). It is currently unclear whether and how γ-TuRCs might be biochemically activated into efficient microtubule nucleation templates by associated factors or post-translational modifications such as γ-tubulin phosphorylation, as proposed in current models (Kollman et al., 2011). Our reconstitution strategy establishes minimal systems for testing these models using biochemical approaches.
Materials and methods
Expression and purification of γ-TuRC-GFP, γ-TuRCΔLB-GFP, and γ-TuRCΔLB(N229A)-GFP
Plasmids encoding Myc-His6–tagged human γ-tubulin, GCP2, GCP3, GCP4, GCP5, GCP6 (Murphy et al., 2001), GFP-tagged MZT2A (Teixidó-Travesa et al., 2010), and Myc-His6–tagged NEDD1 (Lüders et al., 2006) were kind gifts from Dr. Jens Luders (Institute for Research in Biomedicine, Barcelona, Spain). A plasmid containing the MZT1 coding sequence (pDONR223_C13orf37_WT) was a gift from Jesse Boehm, William Hahn, and David Root (Addgene plasmid 81872; http://n2t.net/addgene:81872; Research Resource Identifier: Addgene_81872; Kim et al., 2016). A plasmid encoding human β-actin was a kind gift from Dr. Gregory Alushin (The Rockefeller University, New York, NY). Coding sequences were individually subcloned into the multiple cloning site of pACEBac1 (Bieniossek et al., 2008) using Gibson assembly (pACEBac1-MZT1 and pACEBac1-β-actin; Gibson et al., 2009) or standard restriction enzyme-based cloning methods (all other coding fragments). BstXI sites within coding sequences were removed by introducing silent mutations using primer-based site-directed mutagenesis. For γ-tubulin, a TEV protease site followed by a His6-tag and a stop codon were introduced at the 3′ end of the coding sequence. For MZT2, a ZZ-tag followed by a TEV cleavage site was PCR-amplified from a dynein expression construct (Steinman et al., 2017) and inserted at the 5′ end of the MZT2 coding sequence. mEGFP with a 5′ PreScission protease site and a 3′ stop codon was PCR-amplified from an in-house mammalian expression vector and inserted at the 3′ end of the MZT2 coding sequence.
Most γ-TuRC-GFP proteins could be individually overexpressed reasonably well, but GCP2 and GCP3 had poor soluble expression levels. Studies of the S. cerevisiae γ-TuSC, comprised of Tub4 (γ-tubulin), Spc97 (GCP2), and Spc98 (GCP3), have shown that their purification yield is enhanced if co-overexpressed (Vinh et al., 2002). It seemed likely to us that a similar strategy could enhance the yield of not just GCP2 and GCP3, but potentially all 10 complex proteins. In initial tests, co-overexpressing all 10 proteins using a single baculovirus did increase soluble yields of GCP2 and GCP3, but only slightly. Further, we noticed that MZT2-mEGFP dominated protein expression levels, especially when codon-optimized for expression in insect cells, but codon optimization of GCP2 and GCP3 had little effect on their expression levels. We therefore chose not to codon-optimize any of the proteins in our final constructs. Finally, to further compensate for the poor expression of GCP2 and GCP3, we also generated a second bacmid containing only the human γ-TuSC proteins (γ-tubulin-TEV-His6, GCP2, and GCP3) and coinfected insect cells with both baculoviruses (Fig. 1 A), which we found significantly enhanced γ-TuRC-GFP yields. pACEBac1 vectors containing all 10 (pACEBac1-γ-TuRC-GFP) or just γ-tubulin-TEV-His6/GCP2/GCP3 (pACEBac1-γ-TuSC) were sequentially constructed using BstXI and I-CeuI restriction digestion and ligation, per the MultiBac system manual (Bieniossek et al., 2008). The final pACEBac1-γ-TuRC-GFP and pACEBac1-γ-TuSC plasmids were analyzed by asymmetric restriction digestion and complete Sanger sequencing of all open reading frames. pACEBac1-γ-TuRCΔLB-GFP was constructed in a similar manner. To construct γ-TuRCΔLB(N229A)-GFP, an N229A point mutation was introduced into the pACEBac1-γ-tubulin-TEV-His6 plasmid using primer-based site-directed mutagenesis. pACEBac1-γ-TuRCΔLB(N229A)-GFP and pACEBac1-γ-TuSC(N229A) donor plasmids were sequentially constructed using BstXI and I-CeuI restriction digestion and ligation, as for pACEBac1-γ-TuRCΔLB-GFP and pACEBac1-γ-TuSC.
Polycistronic donor plasmids were transformed into DH10MultiBacTurbo cells (ATG:biosynthetics GmbH) and transposition-positive colonies were selected and used to generate recombinant bacmids. Bacmids were transfected into Sf9 cells (Novagen) per the Bac-to-Bac manual (Invitrogen), baculoviruses were amplified twice, and fresh P3 virus from γ-TuRC-GFP/γ-TuRCΔLB-GFP/γ-TuRCΔLB(N229A)-GFP and γ-TuSC/γ-TuSC(N229A) bacmids were mixed together at a 1:1 ratio. This virus mixture was used to infect 1.8–2.4 liters of High Five cells (Thermo Fisher Scientific) at a cell density of 3 × 106/ml for 60 h at 27°C. Cells were harvested by centrifugation at 1,000 g, resuspended in 80 ml ice-cold lysis buffer (40 mM Hepes, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol [vol/vol], 0.1% Tween-20, 0.1 mM ATP, 0.1 mM GTP, 1 mM 2-mercaptoethanol, four cOmplete EDTA-free Protease Inhibitor Cocktail tablets (Roche), 500 U benzonase, 2 mM PMSF, and 4 mM benzamidine-HCl), and lysed by dounce homogenization on ice. The lysate was clarified at 322,000 g for 1 h at 4°C, 0.22-µm syringe–filtered, and loaded onto a 1-ml NHStrap column (Cytiva) previously coupled to 10–20 mg rabbit IgG (Innovative Biosciences; IRBIGGAP500MG) following the manufacturer’s instructions. The IgG column was washed with lysis buffer followed by gel filtration buffer (40 mM Hepes, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol [vol/vol], 0.1 mM GTP, and 1 mM 2-mercaptoethanol). An expression vector for TEV protease, pRK793, was a gift from David Waugh (Addgene plasmid 8827; http://n2t.net/addgene:8827; Research Resource Identifier: Addgene_8827; Kapust et al., 2001). TEV was expressed in BL21-CodonPlus (DE3)-RIL and purified using Ni-nitrilotriacetic acid (NTA) and gel filtration following the methods described in Ti et al. (2020). 1 mg of TEV protease (stored in 40 mM Hepes, pH 7.5, 30% [wt/vol] glycerol, 150 mM KCl, 1 mM MgCl2, and 3 mM 2-mercaptoethanol) was diluted into 1 ml of gel filtration buffer and injected onto the NHStrap column, and proteolysis was allowed to proceed for 2 h at 4°C. The digested eluate was pooled, concentrated by dialyzing against dialysis buffer (40 mM Hepes, pH 7.5, 150 mM KCl, 1 mM MgCl2, 60% sucrose [wt/vol], 0.1 mM GTP, and 2 mM 2-mercaptoethanol) for 4 h at 4°C, and gel-filtered over a Superose 6 10/300 GL or a Superose 6 Increase 10/300 GL column (Cytiva) preequilibrated in gel filtration buffer. The peak fractions were identified by SDS-PAGE followed by Coomassie staining and/or negative-stain transmission electron microscopy (TEM), pooled, and loaded onto a 2-ml sucrose gradient composed of 10%, 20%, 30%, and 40% sucrose (wt/vol) in gradient buffer (40 mM Hepes, pH 7.5, 150 mM KCl, 1 mM MgCl2, 0.01% Tween-20 [vol/vol], 0.1 mM GTP, and 1 mM 2-mercaptoethanol). The gradient was centrifuged at 50,000 rpm in a TLS-55 rotor at 4°C for 3 h with minimum acceleration and no break. Fractions were manually collected with a cut-off P1000 pipette tip and analyzed by SDS-PAGE followed by Coomassie staining and/or negative-stain TEM. Peak fractions were aliquoted, snap-frozen, and stored in liquid N2. For estimating sedimentation coefficients, parallel gradients were run with 200 µg each of BSA, catalase, and thyroglobulin (Cytiva). Gradients were fractionated into 250 µl (Fig. S1 A) or 150 µl (Fig. S1, C and E) and analyzed by SDS-PAGE followed by Coomassie staining.
Oligonucleotides
Cloning primers for pACEBac1-γ-tubulin-TEV-His6 (EcoRI/HindIII) were 5′-CGCGGAATTCATGCCGAGGGAAATCATCACCCTACAG-3′, 5′-CGACAAGCTTCTACTGCTCCTGGGTGCCCCAGGAG-3′, and 5′-TCGACAAGCTTCTAGTGGTGATGATGGTGATGGCCCTGAAAATACAGGTTTTCCTGCTCCTGGGTGCCCCAGG-3′; pACEBac1-GCP2 (EcoRI/HindIII), 5′-GCGCGGAATTCATGAGTGAATTTCGGATTCACCATGACG-3′ and 5′-CTCGACAAGCTTTCACTGTGCGGTGACTGCGACCCTGG-3′; pACEBac1-GCP3 (SalI/HindIII), 5′-CCTACGTCGACATGGCGACCCCGGACCAGAAGTCG-3′ and 5′-TCGACAAGCTTTCACGTGTGGGAGCTGCGCCGCC-3′; pACEBac1-GCP4 (SacI/HindIII), 5′-TCGACGAGCTCATGATCCACGAACTGCTCTTGGC-3′ and 5′-TCGACAAGCTTTCACATCCCGAAACTGCCCAGAGTTCC-3′; pACEBac1-GCP5 (SalI/NotI), 5′-CCTACGTCGACATGGCGCGGCACGGGCCACCGTGG-3′ and 5′-CGAAAGCGGCCGCTTAACTTTGTTCCATGCCAGCC-3′; pACEBac1-GCP6 (EcoRI/XbaI), 5′-GCGCGGAATTCATGGCCAGCATCACGCAGCTGTTCG-3′ and 5′-CAGGCTCTAGATCAGGCGTCCTGGTAGTAGTTGTTGAAGTTGATGC-3′; pACEBac1-NEDD1 (SalI/XbaI), 5′-CCTACGTCGACATGCATTTTACAGGCGCAGTCATGC-3′ and 5′-CAGGCTCTAGATCAAAAGTGGGCCCGTAATCTTTTG-3′; pACEBac1-ZZ-TEV-MZT2-mEGFP (EcoRI/HindIII/NotI/XhoI/HindIII), 5′-GCGCGGAATTCATGGCGGCGCAGGGCGTAGGG-3′, 5′-TCGACAAGCTTCTAGGTGCTGCCCTGCGTAGGGC-3′, 5′-GCGCGGAATTCATGGCAGGCCTTGCGCAACACG-3′, 5′-CTCATGAATTCGCCCTGAAAATAAAGATTCTCAGTCG-3′, 5′-CGTAGCGGCCGCATGGCAGGCCTTGCGCAACACGATG-3′, 5′-CGTACTCGAGACCGGTGCTGCCCTGCGTAGGGCTC-3′, 5′-CGTACTCGAGCTGGAGGTGCTGTTCCAGGGACCTATGGTGAGCAAGGGCGAGGAGCTG-3′, and 5′-CGTAAAGCTTTCACTTGTACAGCTCGTCCATGCCGAG-3′; pACEBac1-MZT1 (Gibson assembly), 5′-GGTCCGAAGCGCGCGGAATTCATGGCGAGTAGCAGCGGTGCTG-3′ and 5′-CGCGACAAGTGAGCTCGTCGACCTAGCTTGTCATATTTTCAGCAGCC-3′; and pACEBac1-β-actin (Gibson assembly), 5′-TCCCGGTCCGAAGCGCGCGGAATTCATGGATGATGATATCGCCGCGCT-3′ and 5′-GGCCGCGACAAGTGAGCTCGTCGACCTAGAAGCATTTGCGGTGGACGA-3′.
Primers used for site-directed mutagenesis-mediated removal of BstXI sites in pACEBac1-γ-tubulin-TEV-His6 were 5′-GCCAGCGTGAGGAAGACTACGGTCCTGGATGTCATGAGG-3′ and 5′-CCTCATGACATCCAGGACCGTAGTCTTCCTCACGCTGGC-3′; pACEBac1-GCP2, 5′-GGACATCCTGGCCTCCCTCGCGACCTCGGTGGACAAAGG-3′, 5′-CCTTTGTCCACCGAGGTCGCGAGGGAGGCCAGGATGTCC-3′, 5′-CGATGGCGCACGCCGACCCGACGGAGCTGGCGCTGAGC-3′, and 5′-GCTCAGCGCCAGCTCCGTCGGGTCGGCGTGCGCCATCG-3′; pACEBac1-GCP4, 5′-TCGACGAGCTCATGATCCACGAACTGCTCTTGGC-3′ and 5′-TCGACAAGCTTTCACATCCCGAAACTGCCCAGAGTTCC-3′; pACEBac1-GCP6, 5′-GGCTACGTGCTCATCTCAAAAGAGGTGGAGGACTGTGTTCC-3′, 5′-GGAACACAGTCCTCCACCTCTTTTGAGATGAGCACGTAGCC-3′, 5′-CCTGGAGGTGGCGCTGCAGACAATTAACTTGGACCTGCC-3′, and 5′-GGCAGGTCCAAGTTAATTGTCTGCAGCGCCACCTCCAGG-3′; and pACEBac1-NEDD1, 5′-GAATTGCTCTTTGTAACTATAGGCTTGGATAAAAGAATCATCC-3′ and 5′-GGATGATTCTTTTATCCAAGCCTATAGTTACAAAGAGCAATTC-3′.
Primers used to generate pACEBac1-γ-tubulin(N229A)-TEV-His6 via site-directed mutagenesis were 5′-CCCATCCTTCTCCCAGATCGCCCAGCTGGTGTCTACCATCATGTC-3′ and 5′-GACATGATGGTAGACACCAGCTGGGCGATCTGGGAGAAGGATGGG-3′.
Mass spectrometry
A 40-µl aliquot of reconstituted complex was thawed and mixed with 10 µl of 5× sample buffer (Tris-HCl, pH 6.8, 10% [wt/vol] SDS, 50% [wt/vol] glycerol, 700 mM 2-mercaptoethanol, and 0.25% [wt/vol] bromophenol blue). After boiling, the sample was loaded into a single lane of a 4–20% Tris-glycine precast gel with “wide wells” (Novex) and allowed to migrate ∼1 cm into the stacking gel. A corresponding ∼1 cm × 1 cm gel plug was cut out of the gel and further cut into ~1-mm cubes. Protein gel bands were destained, reduced, alkylated, and digested with endopeptidase Lys-C (Wako) and trypsin (Promega) overnight. Peptides were extracted with 70% acetonitrile in 5% formic acid at 35°C, dried, and redissolved in 10 μl of extraction buffer. 3 μl of the peptide mixture was injected and separated using a gradient increasing from 2% buffer B to 35% buffer B over 70 min (buffer A: 0.1% formic acid; buffer B: 80% acetonitrile in 0.1% formic acid). Peptides were resolved using a 12-cm 75-µm column with 3 µm C18 particles (Nikkyo Technos Co., Ltd.) coupled to a Fusion Lumos mass spectrometer (Thermo Fisher Scientific) operated in high/high mode. Raw data were analyzed with MaxQuant 1.6.2.10.
Quantitative Western blotting
Complex concentrations were estimated using quantitative Western blotting against γ-tubulin standards using anti–γ-tubulin (Millipore Sigma; clone GTU-88), assuming that each complex contains 14 γ-tubulin molecules. γ-Tubulin was expressed in High Five cells using the pACEBac1-γ-tubulin-TEV-His6 plasmid and purified using Ni-NTA affinity followed by gel filtration using the method described in Rice et al. (2008).
Densitometry analysis
Scanned SDS-PAGE gel bands in Fig. S1, G and H, were quantified using Fiji (Schindelin et al., 2012). For each of bands 1–5 (see Fig. S1 I), a region of constant area representing total protein “signal” (including underlying background) was generated, and the integrated intensity was recorded. Assuming the local variation in background signal is negligible, an expression for this intensity was generated as a = x + n, where a = total integrated intensity recorded, x = signal in "a" contributed from protein staining, and n = signal contributed from background. Then, the integrated intensity of a second region exactly three times larger in area than the first (and hence containing ∼3× the amount of “background signal”) was recorded. This intensity can be described by the expression b = x + 3n, where b = total integrated intensity recorded of the larger box, and "x" and "n" are the same as above. By simple algebraic rearrangement, an expression of x = (3a − b)/2 (which is now independent of the background signal "n") could then be derived and used to estimate the density of bands 1–5. To correct for the fact that longer polypeptides collect more Coomassie staining, the resulting band densities were further normalized by the average molecular weight of the protein components expected to comigrate to bands 1–5 on the gels. These intensities are reported relative to the total intensity of bands 1–5 within each gel (Fig. S1 J).
Purification and biotinylation of GFP nanobody
The construct for a recombinant GFP nanobody (clone LaG-16; Fridy et al., 2014) was a kind gift from Dr. Michael Rout. The plasmid was transformed into BL21-CodonPlus (DE3)-RIL (Strategene) cells, and periplasmic overexpression was induced with 0.1 mM IPTG for 16 h at 16°C. Cells from 6 liters of culture were harvested at 5,000 g for 10 min and resuspended in TES buffer (200 mM Tris-HCl, pH 8.0, 0.5 mM EDTA, and 500 mM sucrose). The cells were osmotically shocked on ice for 30 min by diluting them fivefold in 1:4 water/TES buffer. Periplasmic extract was separated from cell debris by centrifugation for 10 min at 6,000 g, and this supernatant was further clarified at 20,000 g for 20 min at 4°C. The final supernatant was supplemented with NaCl to 150 mM and incubated with 3 ml His60 resin (Takara Biosciences) for 30 min. The resin was washed in batch with wash buffer 1 (20 mM Tris-HCl, pH 8.0, and 900 mM NaCl) and wash buffer 2 (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 10 mM imidazole) and loaded onto a disposable column, and the protein was eluted with Ni-NTA elution buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 250 mM imidazole). Peak fractions were identified by Bradford assay, pooled, and concentrated with a 10-kD cut-off spin filter (Millipore). To produce biotinylated GFP nanobody, N-hydroxysuccinimide (NHS) esters of biotin ((NHS)-biotin; Millipore Sigma; H1759-5MG) was dissolved in DMSO and added to the concentrated protein at a molar ratio of 10:1 such that the final DMSO concentration did not exceed 5%. The reaction was incubated at 4°C for 4 h and clarified by centrifugation at 21,000 g for 20 min at 4°C. The nanobody was then gel-filtered over a Superdex 75 10/300 column (Cytiva) equilibrated in coupling buffer (150 mM sodium bicarbonate, pH 8.0, and 150 mM NaCl). Biotinylated antibody was supplemented with glycerol to 10% (vol/vol), flash-frozen in liquid N2, and stored at −80°C.
Purification of mEGFP and EB1-GFP
To obtain mEGFP, 0.5 mg of GFP-γ-TuNA (Wieczorek et al., 2020b) was incubated with 0.5 mg of PreScission protease (Cytiva) on ice for 2 h. The reaction was gel-filtered over a Superdex 75 10/300 column (Cytiva) preequilibrated in 40 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM MgCl2, and 2 mM 2-mercaptoethanol. Purified mEGFP was supplemented with glycerol to 10% (vol/vol), snap-frozen in liquid N2, and stored at −80°C. EB1-GFP in a pET-DUET vector (Novagen) was expressed and purified from BL21(DE3) Rosetta (Novagen) cells using Ni-NTA affinity followed by gel filtration using the methods described in Forth et al. (2014).
Negative-stain EM and data processing
3 µl of sucrose density gradient–purified γ-TuRC-GFP, γ-TuRCΔLB-GFP, or γ-TuRCΔLB(N229A)-GFP was applied to glow-discharged carbon-coated copper grids (EMS; CF-400-Cu) and incubated for 45 s on ice. Protein solution was removed by manual blotting with Whatman No. 1 filter paper. Then 3 µl of protein solution was applied again to improve particle density. This procedure was optionally repeated resulting in a total of two to four applications, depending on the concentration of γ-TuRC complex. After the final application, the protein solution was manually blotted from one side of the grid while freshly filtered 1 or 2% uranyl acetate (wt/vol) was simultaneously pipetted from the opposite side to exchange the solution. Grids were incubated in uranyl acetate for a further 45 s. Stain was removed by manual blotting, and grids were air-dried for >24–48 h in a sealed container layered with desiccant before imaging. TEM micrographs were recorded on an FEI Tecnai G2 microscope operating at 120 kV at a magnification of 23,000× (3.96 Å/pixel) or 30,000× (3.036 Å/pixel) using a BioSprint 29 charge-coupled device camera.
Images were cropped in Fiji to remove micrograph metadata labels (Schindelin et al., 2012), and TIFF formats were converted to MRC using RELION’s “relion_image_handler” function. Contrast transfer function (CTF) parameters were estimated using CTFFIND4 (Rohou and Grigorieff, 2015). All subsequent processing was done in RELION version 3.1 and UCSF Chimera (Zivanov et al., 2018; Pettersen et al., 2004). The main 3D reconstruction workflow steps for γ-TuRC-GFP, γ-TuRCΔLB-GFP, or γ-TuRCΔLB(N229A)-GFP are outlined in Fig. S2, A–C, respectively. Generally, micrographs were imported, and a small set of <1,000 particles was manually picked and subjected to reference-free 2D classification. The resulting set of averages was used as an initial template for RELION’s built-in auto-picking implementation. One or two rounds of auto-picking were performed to yield the best templates for optimal picking. Auto-picked particles were binned by two and subjected to reference-free 2D classification to remove particles likely corresponding to dirt and other contaminants. A random subset of the cleaned, binned particles was used to generate an ab initio model. Then, all cleaned, binned particles were subjected to an intermediate 3D auto-refinement step using the ab initio model as a reference, reextracted using the refined coordinates, and subjected to 3D classification. Particles that generated 3D classes containing the highest level of detail were reextracted from CTF-corrected micrographs at the unbinned pixel size (3.036 Å or 3.96 Å) and subjected to a final round of 3D auto-refinement using one of the classes as a reference model.
A composite model of the native human γ-TuRC was constructed by substituting our recently published models for the LB and MZT2/GCP2-NHD/γ-TuNA (Protein Data Bank accession nos. 6X0U and 6X0V; Wieczorek et al., 2020a) with the corresponding poly-alanine models for the LB, the γ-TuRC "HB," and the γ-TuRC "CC" regions in our previous structure of the native complex (Protein Data Bank accession no. 6V6S; Wieczorek et al., 2020b). A poly-alanine model for the unassigned mitotic spindle organizing proteins associated with a ring of γ-tubulin (MZT) “module” was generated with MZT1/GCP6-NHD and rigid body–fitted into the unassigned MZT module density found at the overlap region, as reported in Wieczorek et al. (2020a). The composite model was then rigid body–fitted into density maps using the “Fit in map” function of Chimera. The local correlation between the model and the density map after fitting was calculated using Chimera’s “vop localCorrelation” command with a window size of 5 voxels. All figures displaying density maps and protein models were generated using UCSF Chimera (Pettersen et al., 2004) or UCSF ChimeraX (Goddard et al., 2018).
Turbidity-based microtubule nucleation assay
For all in vitro assays used in this study, tubulin purified from calf brain (Gell et al., 2011) was quickly thawed at 37°C and centrifuged at 350,000 g for 10 min at 4°C. The concentration of the supernatant was measured spectrophotometrically using an extinction coefficient of 115,000 M−1⋅cm−1 and a molecular weight of 110 kD for the tubulin dimer. Tubulin prepared in this way was kept on ice for no more than 2 h.
Microtubule nucleation was assayed by preparing 75 µl reactions containing 0.25 nM γ-TuRC-GFP, γ-TuRCΔLB-GFP, or γ-TuRCΔLB(N229A)-GFP; 20 µM tubulin; and 1 mM GTP in BRB80 (80 mM K-Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA). An appropriate volume of 30% (wt/vol) sucrose gradient buffer (see Materials and methods section "Expression and purification of γ-TuRC-GFP, γ-TuRCΔLB-GFP, and γ-TuRCΔLB(N229A)-GFP") was included for tubulin-only controls. Components were mixed and incubated on ice for 5 min. Reactions were split and loaded into a prewarmed 384-well, clear-bottomed microplate in duplicate (∼32 µl reaction per well), and 15 µl of fluorescence-free immersion oil (Cargille Laboratories; 16212) was layered over each reaction. This loading time was limited to a total of 5 min. The microplate was immediately placed on a BioTek microplate reader warmed to 37°C, and absorbance measurements at 350 nm were initiated at a frequency of one per minute for a total of 1 h.
Single molecule γ-TuRC-GFP microtubule nucleation assay
The TIRF microscope setup uses a Nikon Eclipse Ti microscope equipped with a 1.49 NA 100× Plan Apo TIRF objective (Nikon) with a 1.5× optovar introduced in the optical path; a three-axis piezo-electric stage (Mad City LabsNano LP-200); a Photometrics Prime 95B sCMOS camera; and two-color TIRF imaging optics (lasers: 488 nm [Spectra-physics] and 561 nm [Cobalt]; filters: Semrock FF01-520/35 and FF01-609/54 [emission] and Semrock Di01-R488/561 [dichroic]). Tubulin purified from calf brains was functionalized with X-rhodamine succinimidyl ester (Thermo Fisher Scientific; C6125) following established protocols (Hyman et al., 1991). Glass coverslips (Gold Seal 18 × 18 mm, No. 1) and glass slides (Buehler 40–80000-01) were rinsed in acetone, sonicated for 20 min in 50% (vol/vol) methanol, sonicated for 20 min in 0.5 M KOH, and rinsed with milliQ water. After drying with nitrogen gas, the coverslips were plasma cleaned for 5 min (Harrick Plasma PDC-32G) and adhered to glass slides via two strips of double-sided tape separated by ∼5 mm. The flow cell was rinsed with BRB80 + 1 mM tris(2-carboxyethyl)phosphine (TCEP), followed by 0.2 mg/ml poly-L-lysine (PLL)-polyethlyne glycol (PEG)-biotin (PLL[20]-g[3.5]-PEG[2]/PEG[3.4]-biotin [20%]; SUSOS AG) prepared in BRB80 + 1 mM TCEP. After 5 min, the flow cell was rinsed with BRB80 + 1 mM TCEP, and a mixture containing 0.5 mg/ml κ-casein (Millipore Sigma) and 0.25 mg/ml neutravidin (Thermo Fisher Scientific) prepared in BRB80 + 1 mM TCEP was flowed in. After 5 min, the flow cell was rinsed with assay buffer (BRB80 + 1 mM TCEP, 50 mM KCl, 0.15% [wt/vol] methylcellulose, 0.2 mg/ml κ-casein, and 1 mM GTP), and 0.02 mg/ml biotinylated GFP nanobody prepared in assay buffer was flowed in. After 5 min, the flow cell was rinsed with assay buffer, and ∼1 pM γ-TuRCΔLB-GFP or γ-TuRC-GFP prepared in assay buffer was flowed in. After 5 min, unbound complexes were rinsed out with assay buffer, and a reaction mixture containing 15 µM tubulin (with ∼5% X-rhodamine–labeled tubulin) and oxygen scavengers (0.035 mg/ml catalase, 0.2 mg/ml glucose oxidase, 2.5 mM glucose, and 10 mM DTT) was prepared in assay buffer and introduced to the flow cell. The flow cell was sealed with VALAP (1:1:1 petroleum jelly/lanolin/paraffin) and placed on the TIRF microscope stage. Two-color images were acquired at 10 s per frame and an exposure time of 500 ms (tubulin and GFP channels; γ-TuRCΔLB-GFP experiments) or 150 ms (GFP channel; γ-TuRC-GFP and intensity distribution experiments; see below). Total imaging times were between 20 and 30 min. The microscope chamber was heated to ∼35°C before image acquisition. Image acquisition was controlled using NIS-Elements AR 4.60.00 (Nikon).
Time-lapse images were drift-corrected using the MultiStackReg plugin in Fiji made by Brad Busse. The number of puncta was measured using the “Spot Intensity Analysis” Fiji plugin built by Nico Stuurman and Ankur Jain. γ-TuRCΔLB-GFP– or γ-TuRC-GFP–mediated microtubule nucleation events were counted and tracked manually in Fiji. Only nucleation events that initiated from green fluorescent puncta were considered in the analysis in Fig. 5, A–F. Microtubule growth rates were calculated by estimating the change in length of microtubules growing from γ-TuRCΔLB-GFP puncta divided by the length of time spent in the growth phase (Fig. S3 D). Growth rates of microtubules that dissociated from GFP puncta were only measured for the time that they were associated with γ-TuRCΔLB-GFP. Fig. S3 C shows the classification of microtubule nucleation events and whether or not they were attached to γ-TuRCΔLB-GFP. The length of time that γ-TuRCΔLB-GFP–associated microtubules spent in the growth phase before undergoing catastrophe was measured and is plotted in Fig. S3 E. A γ distribution was used to fit these data, which produced similar values for the rate (0.005 ± 0.002 s−1) and step (2.9 ± 0.8) parameters as for microtubules nucleated from GMPCPP seeds (Gardner et al., 2011). Fitting was performed in MATLAB (MathWorks) and SciDAVis (http://scidavis.sourceforge.net/).
Collection of single molecule fluorescence intensity distributions shown in Fig. S3, I–L, was performed as follows. Approximate picomolar concentrations of purified mEGFP, EB1-GFP, γ-TuRC-GFP, or γ-TuRCΔLB-GFP prepared in assay buffer were immobilized to coverslip surfaces using the biotinylated GFP nanobody and imaged in assay buffer containing oxygen scavengers, as detailed above. Single-frame images in the GFP channel were collected using 150-ms exposure times. GFP puncta were again identified using the “Spot Intensity Analysis” Fiji plugin. A pixel radius of 4 and median-40-based background subtraction was employed. Due to an inhomogeneous excitation field, and to obtain enough data to generate reliable distributions, the noise tolerance was empirically lowered to values that would “overpick” spots. Resulting intensity values were plotted as a histogram and fitted to a two-peak Gaussian function, which returned mean ± variance estimates for false positives and true GFP-fluorescent puncta (shown in Fig. S3 J, right panel; and Fig. S3, K and L, bars labeled “All”). The distributions for γ-TuRC-GFP and γ-TuRCΔLB-GFP shown in Fig. S3 J (left) were generated by subtracting the estimated false-positive distribution from the total dataset. A distribution of the intensities of microtubule-nucleating γ-TuRC-GFP or γ-TuRCΔLB-GFP puncta (bars labeled “Nucleating” in Fig. S3, K and L) was manually curated by visual inspection of overlaid tubulin/GFP channels from the list of puncta automatically picked by the “Spot Intensity Analysis” Fiji plugin. Note that the higher exposure time for the nucleation experiments using γ-TuRCΔLB-GFP (500 ms vs. 150 ms) is the reason for the higher mean intensity of γ-TuRCΔLB-GFP puncta in Fig. S3 L.
Statistical methods
Data in Fig. 5, B, C, E, and F (top panels), were fitted using weighted linear regression. Data in Fig. 5, B, C, E, and F (bottom panels), were fitted to an exponential growth model of the form y = y0 + Aexp(x/t), where: x = concentration of tubulin (in micromolar); y0 = baseline rate of increase of the number of microtubules (Fig. 5, B and E) or the number of γ-TuRCΔLB-GFP–nucleating puncta (Fig. 5, C and F); A = rate of increase of the number of microtubules (Fig. 5, B and E) or the number of γ-TuRCΔLB-GFP–nucleating puncta (Fig. 5, C and F) at 0 µM tubulin; and ΔT = t/ln(2) is an estimate of the change in tubulin concentration (in micromolar) needed to double the nucleation rate (Fig. 5, B and E) or the number of γ-TuRCΔLB-GFP–nucleating puncta (Fig. 5, C and F). The nonzero nucleation rates were also fitted to a power law of the form ln(y) = αln(x) + ln(β); the values for the exponent α and its uncertainty are indicated. The error in ΔT was propagated manually. Kernel density distributions were generated for data in Fig. S3 D using the violin MATLAB script written by Jasper Fabius (https://www.mathworks.com/matlabcentral/fileexchange/72424-violin) using the normal kernel smoother and a bandwidth value of 1 μm/min.
Data in Fig. S3 E were assumed to follow a Gamma distribution (Gardner et al., 2011). All other distributions were assumed to follow normal distributions, but this was not formally tested.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Online supplemental material
Fig. S1 shows sucrose gradient standard curves, liquid chromatography–tandem mass spectrometry data, and stoichiometry analyses of reconstituted γ-TuRCs. Fig. S2 shows details of the negative-stain analyses used in this study. Fig. S3 shows controls for single-molecule TIRF experiments.
Acknowledgments
The authors acknowledge B. Graczyk for support in generating DNA constructs used in this study, Drs. J. Luders (Institute for Research in Biomedicine, Barcelona, Spain), M. Rout (The Rockefeller University, New York, NY), and G. Alushin (The Rockefeller University, New York, NY) for the generous gift of plasmids, and Dr. S. Liu (The Rockefeller University, New York, NY) for access to a plasma cleaner. The authors also thank Dr. H. A. Pasolli, Dr. N. Soplop, M. Ebrahim, J. Sotiris, and H. Ng for fantastic EM support (The Rockefeller University, New York, NY), and the Rockefeller University Proteomics Resource Center for additional mass spectrometry support.
This work was funded by a National Institutes of Health grant to T.M. Kapoor (R35 GM130234). M. Wieczorek is supported by a Human Frontier Science Program fellowship (LT000025/18-L1). L. Urnavicius is supported by the Rockefeller University’s Pels Center for Biochemistry and Structural Biology.
The authors declare no competing financial interests.
Author contributions: M. Wieczorek, S.-C. Ti, and T.M. Kapoor conceived the experiments. M. Wieczorek, S.-C. Ti, and A. Aher designed constructs. M. Wieczorek and S.-C. Ti purified proteins. K.R. Molloy and B.T. Chait performed and analyzed mass spectrometry experiments. M. Wieczorek and L. Urnavicius performed and analyzed EM experiments. M. Wieczorek performed and analyzed functional assays. M. Wieczorek, S.-C. Ti, L. Urnavicius, A. Aher, and T.M. Kapoor prepared the manuscript.
References
- Bieniossek, C., Richmond T.J., and Berger I.. 2008. MultiBac: multigene baculovirus-based eukaryotic protein complex production. Curr. Protoc. Protein Sci. Chapter 5:Unit 5.20. 10.1002/0471140864.ps0520s51 [DOI] [PubMed] [Google Scholar]
- Brouhard, G.J., Stear J.H., Noetzel T.L., Al-Bassam J., Kinoshita K., Harrison S.C., Howard J., and Hyman A.A.. 2008. XMAP215 is a processive microtubule polymerase. Cell. 132:79–88. 10.1016/j.cell.2007.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.-K., Liu P., Sze S.K., Dai C., and Qi R.Z.. 2010. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191:1089–1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consolati, T., Locke J., Roostalu J., Chen Z.A., Gannon J., Asthana J., Lim W.M., Martino F., Cvetkovic M.A., Rappsilber J., et al. 2020. Microtubule Nucleation Properties of Single Human γTuRCs Explained by Their Cryo-EM Structure. Dev. Cell. 53:603–617.e8. 10.1016/j.devcel.2020.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota, R.R., Teixidó-Travesa N., Ezquerra A., Eibes S., Lacasa C., Roig J., and Lüders J.. 2017. MZT1 regulates microtubule nucleation by linking γTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci. 130:406–419. 10.1242/jcs.195321 [DOI] [PubMed] [Google Scholar]
- Forth, S., Hsia K.-C., Shimamoto Y., and Kapoor T.M.. 2014. Asymmetric friction of nonmotor MAPs can lead to their directional motion in active microtubule networks. Cell. 157:420–432. 10.1016/j.cell.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridy, P.C., Li Y., Keegan S., Thompson M.K., Nudelman I., Scheid J.F., Oeffinger M., Nussenzweig M.C., Fenyö D., Chait B.T., and Rout M.P.. 2014. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods. 11:1253–1260. 10.1038/nmeth.3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, M.K., Zanic M., Gell C., Bormuth V., and Howard J.. 2011. Depolymerizing kinesins Kip3 and MCAK shape cellular microtubule architecture by differential control of catastrophe. Cell. 147:1092–1103. 10.1016/j.cell.2011.10.037 [DOI] [PubMed] [Google Scholar]
- Gaskin, F., Cantor C.R., and Shelanski M.L.. 1974. Turbidimetric studies of the in vitro assembly and disassembly of porcine neurotubules. J. Mol. Biol. 89:737–755. 10.1016/0022-2836(74)90048-5 [DOI] [PubMed] [Google Scholar]
- Gell, C., Friel C.T., Borgonovo B., Drechsel D.N., Hyman A.A., and Howard J.. 2011. Purification of tubulin from porcine brain. Methods Mol. Biol. 777:15–28. 10.1007/978-1-61779-252-6_2 [DOI] [PubMed] [Google Scholar]
- Gibson, D.G., Young L., Chuang R.-Y., Venter J.C., Hutchison C.A. III, and Smith H.O.. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 6:343–345. 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- Goddard, T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., and Ferrin T.E.. 2018. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27:14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gombos, L., Neuner A., Berynskyy M., Fava L.L., Wade R.C., Sachse C., and Schiebel E.. 2013. GTP regulates the microtubule nucleation activity of γ-tubulin. Nat. Cell Biol. 15:1317–1327. 10.1038/ncb2863 [DOI] [PubMed] [Google Scholar]
- Gunawardane, R.N., Martin O.C., Cao K., Zhang L., Dej K., Iwamatsu A., and Zheng Y.. 2000. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol. 151:1513–1524. 10.1083/jcb.151.7.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., Farache D., Emorine L., and Merdes A.. 2020. A stable sub-complex between GCP4, GCP5 and GCP6 promotes the assembly of γ-tubulin ring complexes. J. Cell Sci. 133:jcs244368. 10.1242/jcs.244368 [DOI] [PubMed] [Google Scholar]
- Huang, Qi-Lai, Chen Cheng, Jiang Shu-Han, Pan Xiao, and Hua Zi-Chun. 2006. Immobilized protein ZZ, an affinity tool for immunoglobulin isolation and immunological experimentation. Biotechnol. Appl. Biochem. 45(2):87. [DOI] [PubMed] [Google Scholar]
- Hutchins, J.R.A., Toyoda Y., Hegemann B., Poser I., Hériché J.-K., Sykora M.M., Augsburg M., Hudecz O., Buschhorn B.A., Bulkescher J., et al. 2010. Systematic Localization and Purification of Human Protein Complexes Identifies Chromosome Segregation Proteins. Science. 328:593–599. 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A., Drechsel D., Kellogg D., Salser S., Sawin K., Steffen P., Wordeman L., and Mitchison T.. 1991. Preparation of modified tubulins. Methods Enzymol. 196:478–485. 10.1016/0076-6879(91)96041-O [DOI] [PubMed] [Google Scholar]
- Kapust, R.B., Tözsér J., Fox J.D., Anderson D.E., Cherry S., Copeland T.D., and Waugh D.S.. 2001. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 14:993–1000. 10.1093/protein/14.12.993 [DOI] [PubMed] [Google Scholar]
- Kim, E., Ilic N., Shrestha Y., Zou L., Kamburov A., Zhu C., Yang X., Lubonja R., Tran N., Nguyen C., et al. 2016. Systematic Functional Interrogation of Rare Cancer Variants Identifies Oncogenic Alleles. Cancer Discov. 6:714–726. 10.1158/2159-8290.CD-16-0160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Pereira G., Geissler S., Grein K., and Schiebel E.. 1997. The spindle pole body component Spc97p interacts with the γ-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16:1550–1564. 10.1093/emboj/16.7.1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J.M., Zelter A., Muller E.G.D., Fox B., Rice L.M., Davis T.N., Agard D.A., and Stearns T.. 2008. The structure of the γ-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell. 19:207–215. 10.1091/mbc.e07-09-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J.M., Polka J.K., Zelter A., Davis T.N., and Agard D.A.. 2010. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 466:879–882. 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman, J.M., Merdes A., Mourey L., and Agard D.A.. 2011. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12:709–721. 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, S.L., Lynch E.M., Zou J., Tay Y.D., Borek W.E., Tuijtel M.W., Rappsilber J., and Sawin K.E.. 2019. Reconstitution of Microtubule Nucleation In Vitro Reveals Novel Roles for Mzt1. Curr. Biol. 29:2199–2207.e10. 10.1016/j.cub.2019.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, T.-C., Neuner A., Flemming D., Liu P., Chinen T., Jäkle U., Arkowitz R., and Schiebel E.. 2016. MOZART1 and γ-tubulin complex receptors are both required to turn γ-TuSC into an active microtubule nucleation template. J. Cell Biol. 215:823–840. 10.1083/jcb.201606092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, P., Zupa E., Neuner A., Böhler A., Loerke J., Flemming D., Ruppert T., Rudack T., Peter C., Spahn C., et al. 2020. Insights into the assembly and activation of the microtubule nucleator γ-TuRC. Nature. 10.1038/s41586-019-1896-6 [DOI] [PubMed] [Google Scholar]
- Lüders, J., Patel U.K., and Stearns T.. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147. 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Murphy, S.M., Preble A.M., Patel U.K., O’Connell K.L., Dias D.P., Moritz M., Agard D., Stults J.T., Stearns T., and McIntosh J.R.. 2001. GCP5 and GCP6: two new members of the human γ-tubulin complex. Mol. Biol. Cell. 12:3340–3352. 10.1091/mbc.12.11.3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema, K., Wiese C., Martin O.C., Milligan R.A., Iwamatsu A., Mitchison T.J., and Zheng Y.. 1999. Characterization of two related Drosophila γ-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144:721–733. 10.1083/jcb.144.4.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Raff, J.W., Kellogg D.R., and Alberts B.M.. 1993. Drosophila gamma-tubulin is part of a complex containing two previously identified centrosomal MAPs. J. Cell Biol. 121:823–835. 10.1083/jcb.121.4.823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, L.M., Montabana E.A., and Agard D.A.. 2008. The lattice as allosteric effector: structural studies of alphabeta- and gamma-tubulin clarify the role of GTP in microtubule assembly. Proc. Natl. Acad. Sci. USA. 105:5378–5383. 10.1073/pnas.0801155105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou, A., and Grigorieff N.. 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192:216–221. 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns, T., and Kirschner M.. 1994. In vitro reconstitution of centrosome assembly and function: the central role of gamma-tubulin. Cell. 76:623–637. 10.1016/0092-8674(94)90503-7 [DOI] [PubMed] [Google Scholar]
- Steinman, J.B., Santarossa C.C., Miller R.M., Yu L.S., Serpinskaya A.S., Furukawa H., Morimoto S., Tanaka Y., Nishitani M., Asano M., et al. 2017. Chemical structure-guided design of dynapyrazoles, cell-permeable dynein inhibitors with a unique mode of action. eLife. 6:e25174. 10.7554/eLife.25174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixidó-Travesa, N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P., Caelles C., Roig J., and Lüders J.. 2010. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell. 21:3963–3972. 10.1091/mbc.e10-05-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thawani, A., Rale M.J., Coudray N., Bhabha G., Stone H.A., Shaevitz J.W., and Petry S.. 2020. The transition state and regulation of γ-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. eLife. 9:e54253. 10.7554/eLife.54253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti, S.-C., Wieczorek M., and Kapoor T.M.. 2020. Purification of Affinity Tag-free Recombinant Tubulin from Insect Cells. STAR Protoc. 1:100011. 10.1016/j.xpro.2019.100011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovey, C.A., Tubman C.E., Hamrud E., Zhu Z., Dyas A.E., Butterfield A.N., Fyfe A., Johnson E., and Conduit P.T.. 2018. γ-TuRC Heterogeneity Revealed by Analysis of Mozart1. Curr. Biol. 28:2314–2323.e6. 10.1016/j.cub.2018.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh, D.B.N., Kern J.W., Hancock W.O., Howard J., and Davis T.N.. 2002. Reconstitution and characterization of budding yeast gamma-tubulin complex. Mol. Biol. Cell. 13:1144–1157. 10.1091/mbc.02-01-0607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.A., O’Brien E.T., Pryer N.K., Soboeiro M.F., Voter W.A., Erickson H.P., and Salmon E.D.. 1988. Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol. 107:1437–1448. 10.1083/jcb.107.4.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., Desai A., Bulinski J.C., and Salmon E.D.. 1998. Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol. 8:1227–1230. 10.1016/S0960-9822(07)00515-5 [DOI] [PubMed] [Google Scholar]
- Wieczorek, M., Bechstedt S., Chaaban S., and Brouhard G.J.. 2015. Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 17:907–916. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- Wieczorek, M., Huang T.-L., Urnavicius L., Hsia K.-C., and Kapoor T.M.. 2020a. MZT Proteins Form Multi-Faceted Structural Modules in the γ-Tubulin Ring Complex. Cell Rep. 31:107791. 10.1016/j.celrep.2020.107791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieczorek, M., Urnavicius L., Ti S.-C., Molloy K.R., Chait B.T., and Kapoor T.M.. 2020b. Asymmetric Molecular Architecture of the Human γ-Tubulin Ring Complex. Cell. 180:165–175.e16. 10.1016/j.cell.2019.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., Wong M.L., Alberts B., and Mitchison T.. 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 378:578–583. 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- Zivanov, J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., and Scheres S.H.. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 7:e42166. 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.