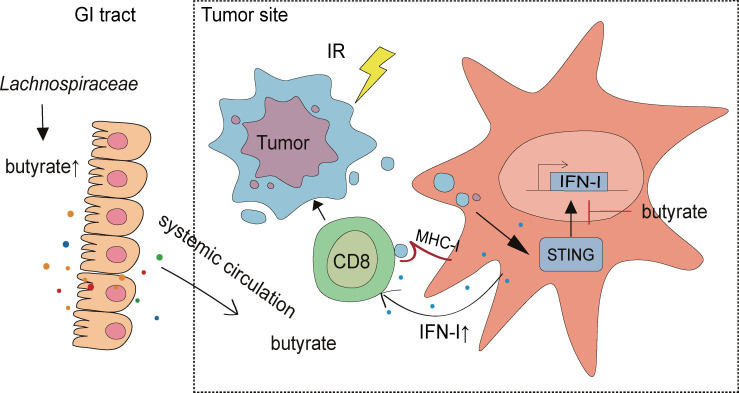
Mechanisms by which specific gut microbiota affect cancer therapies by modulating the antitumor immune response is under intensive investigation. This study reveals selective targeting of butyrate-producing microbiota may provide a therapeutic option to enhance radiotherapy.
Abstract
The antitumor effects of ionizing radiation (IR) are mediated in part through activation of innate and adaptive immunity. Here we report that gut microbiota influences tumor control following IR. Vancomycin decreased the abundance of butyrate-producing gut bacteria and enhanced antitumor responses to IR. Oral administration of Lachnospiraceae, a family of vancomycin-sensitive bacteria, was associated with increased systemic and intratumoral butyric acid levels and impaired the efficacy of IR in germ-free (GF) mice. Local butyrate inhibited STING-activated type I IFN expression in dendritic cells (DCs) through blockade of TBK1 and IRF3 phosphorylation, which abrogated IR-induced tumor-specific cytotoxic T cell immune responses without directly protecting tumor cells from radiation. Our findings demonstrate that the selective targeting of butyrate-producing microbiota may provide a novel therapeutic option to enhance tumor radiation sensitivity.
Graphical Abstract
Introduction
Intestinal microbiota and their metabolites influence the host immune system (Littman and Pamer, 2011; Chung et al., 2012; Nyangale et al., 2012), demonstrated by the fact that germ-free (GF) mice have developmental defects in lymphoid organs and impaired immune responses (Atarashi et al., 2011; Ivanov et al., 2009; Hooper et al., 2012). The microbiome alters antitumor immunity and influences the efficacy of cancer therapies by modulating systemic immunity (Round and Mazmanian, 2009; Zitvogel et al., 2016; Hagan et al., 2019; Scott et al., 2018), bacterial translocation to tumor sites (Yazawa et al., 2000; Yu et al., 2016; Pushalkar et al., 2018; Shi et al., 2020), and the secretion of microbial metabolites (Smith et al., 2013; Donohoe et al., 2014; Wei et al., 2016; Wang et al., 2019; Luu et al., 2018). Microbiome depletion by antibiotics or using GF mice impairs cyclophosphamide efficacy through a reduced Th17 cell inflammatory response (Viaud et al., 2013). Local CpG oligonucleotide and oxaliplatin efficacy is enhanced by the microbiome by modulation of myeloid-derived cell functions in the tumor microenvironment (Iida et al., 2013). In cancer immunotherapy, the microbiota promotes the response to CTLA-4 and PD-1/PDL-1 blockade immunotherapy by enhancing dendritic cell (DC) activity and CD8+ T cell response (Vétizou et al., 2015; Sivan et al., 2015; Routy et al., 2018; Gopalakrishnan et al., 2018; Matson et al., 2018). Conversely, broad-spectrum antibiotics enhance efficacy for anti–PD-1 blockade by upregulating PD-1 expression in pancreatic cancer (Pushalkar et al., 2018). Together, these data suggest that the microbiome can either enhance or dampen antitumor responses in a treatment-specific manner.
Approximately 50% of cancer patients receive radiotherapy (Al-Sarraf et al., 1998; Herman et al., 2015; Kao et al., 2014; Kwon et al., 2014), which stimulates innate and adaptive immunity to augment antitumor response (Qu et al., 2010; Barker et al., 2015; Deng et al., 2016). Radiation alters the antitumor immune response by inducing tumor cell death and the release of danger signals, cytokines, and chemokines (Deng et al., 2016; Laoui et al., 2014; Kachikwu et al., 2011; Qu et al., 2010; Barker et al., 2015). Since the microbiome affects the antitumor immune response to chemotherapies and immunotherapies, we sought to elucidate the role of the commensal microbiome in ionizing radiation (IR)–induced antitumor effects and antitumor immunity.
Here we report gut microbiota–derived butyrate can suppress radiation-induced IFN in the tumor microenvironment. Gram-positive bacteria increased systemic and local butyric acid concentrations and impaired the response to IR. Local sodium butyrate administration is sufficient to impair the antitumor response to IR and reverse the improved IR response following vancomycin-IR combination treatment. Mechanistically, butyrate suppressed type I IFN (IFN-I) production through a decrease in stimulator of IFN genes (STING) downstream activation. Our findings illustrated how specific members of the microbiota can dampen IR-induced antitumor responses and identified potential bacterial and molecular targets for improving the clinical response to radiotherapy.
Results and discussion
Oral vancomycin enhances the antitumor response to IR
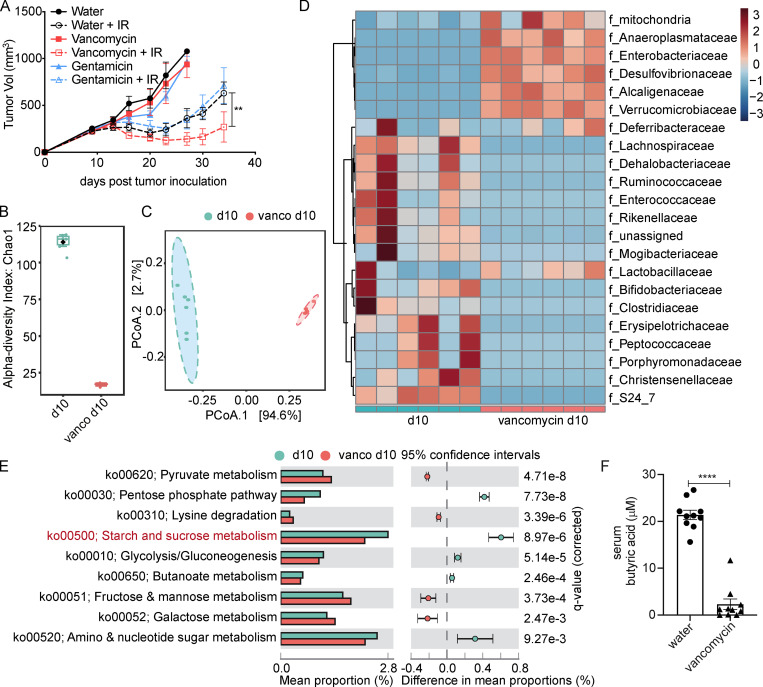
To investigate the potential roles of gram-positive and gram-negative gut bacteria on the response to IR, mice received oral vancomycin or gentamicin 3 wk before tumor inoculation and for the duration of the experiment. As shown in Fig. 1 A, mice receiving vancomycin exhibited increased tumor growth delay following IR as compared with gentamicin-treated or control animals. 10 d after vancomycin administration, fecal samples of vancomycin-treated and control mice were collected for 16S ribosomal RNA (rRNA) sequencing. As identified by α-diversity analysis, vancomycin treatment led to a reduction in the total bacterial diversity (Fig. 1 B). Based on an analysis of group similarities (ANOSIM) test of unweighted UniFrac distances, the control and vancomycin-treated samples showed distinct community structures on feature level (Fig. 1 C). Taxonomic representation was summarized at the family level (Fig. 1 D). Vancomycin treatment did induce dysbiosis of gram-positive and -negative bacteria, among which known butyrate-producing families, Lachnospiraceae (P = 0.0037) and Ruminococcaceae (P = 0.0188), were markedly decreased. Furthermore, 16S rRNA gene-predicted functional profiles, associated with butyrate metabolism, were analyzed using Tax4fun based on the SILVA database (Pruesse et al., 2007; Aßhauer et al., 2015). Comparisons of the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologues for pathway enrichment showed a decrease following vancomycin treatment in the pathway related to starch and sucrose metabolism (ko00500), which associates with butyrate production (Fig. 1 E; Rampelli et al., 2013). Previous studies have demonstrated treating rats with oral vancomycin for 4 wk can alter the composition of the gut microbiome and decrease the concentration of circulating butyrate by 69% (Ho et al., 2015). In addition to the significant decrease of butyrate-producing bacteria, we found vancomycin decreased the concentration of butyric acid in systemic circulation (Fig. 1 F). Together, these results suggest that vancomycin may promote the antitumor effects of IR by decreasing the proportion of butyrate-producing bacteria; i.e., butyrate-producing bacteria may play an opposing role in radiotherapy response.
Figure 1.
Oral administration of vancomycin promotes IR response and eliminates most butyrate-producing bacteria. (A) SPF C57BL/6 mice (n = 5/group) were given oral vancomycin or gentamicin for 1 mo and then injected s.c. with 106 MC38 cells, and established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. The volume of tumors was measured twice a week. Two-way ANOVA tests were used to analyze the tumor growth data. **, P < 0.01. One of three representative experiments is shown. (B) Comparison of within-sample diversity (α-diversity) fecal samples from vancomycin-treated or untreated mice, based on Chao1 index. n = 6/group; P = 0.0042667. (C) Unconstrained principal coordinate analysis (PCoA) of fecal samples from vancomycin-treated or untreated mice, based on unweighted UniFrac dissimilarity matrix between the two groups (n = 6/group). ANOSIM revealed significant differences (P < 0.002). (D) Bacterial taxa observed from vancomycin-treated or untreated mice, shown at the family level. (E) Prediction of changed butyrate production–associated KEGG pathways using Tax4fun analysis based on the SILVA database. Extended error bar plots illustrate differential representation by mean proportion and their differences based on KEGG orthologue groups, ranked by respective effect size of 0.5 thresholds in the Statistical Analysis of Metagenomic Profiles software package (http://kiwi.cs.dal.ca/Software/STAMP). Bar plots on the left side display the mean proportion of each KEGG pathway. Dot plots on the right show the differences in mean proportions between the two indicated groups using q values. (F) Sera from vancomycin-treated and untreated C57BL/6 mice was collected after 1 mo of feeding (control group, n = 10; vancomycin-treated group, n = 10). Unpaired t tests were used to analyze F. ****, P < 0.0001. One representative experiment (out of two experiments in F or three experiments in A) is shown.
Lachnospiraceae species increase systemic butyrate concentration and impair the antitumor response to IR
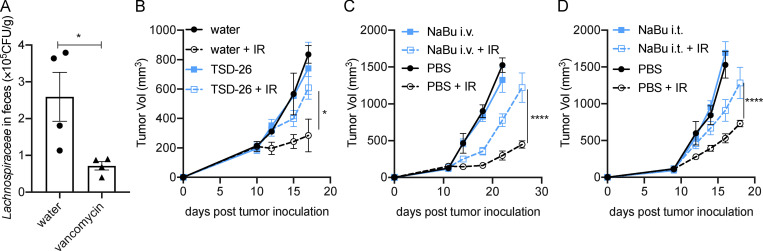
As illustrated in Fig. 1 D, the abundances of operational taxonomic units that belong to the Lachnospiraceae family were decreased after vancomycin treatment. Using Lachnospiraceae genomic DNA as the standard, we verified that the abundance of fecal Lachnospiraceae was decreased following vancomycin treatment by real-time quantitative PCR (qPCR; Fig. S1 A). Lachnospiraceae is a family of anaerobic bacteria that are important butyrate producers residing in the intestinal microbiota (Stackebrandt, 2014). Kineothrix alysoides is reported to be a butyrate-producing species in the Lachnospiraceae family (Haas and Blanchard, 2017). After gavage, K. alysoides (TSD-26; American Type Culture Collection [ATCC])–colonized mice had a decreased antitumor response to IR compared with control mice in both MC38 and B16F1 tumor models (Figs. 2 A and S1 B). To ensure that the observation was due to butyrate production by K. alysoides, we also used heat-inactivated (HI) K. alysoides, which inactivates butyrate production but maintains the pathogen-associated molecular patterns that stimulate the immune response (Zimmermann et al., 2018). HI K. alysoides–administrated mice responded similarly to IR compared with control mice (Fig. 2 B). Importantly, live K. alysoides–inoculated mice had an increase in circulating butyric acid, while mice inoculated with HI K. alysoides had no increase (Fig. 2 C). Following inoculation of live versus HI K. alysoides, our results suggest that the Lachnospiraceae species alone can increase systemic butyrate, which correlated with diminishing IR efficacy. Furthermore, specific pathogen–free (SPF) mice receiving i.v. injections of sodium butyrate (NaBu) on days 1, 4, and 7 after IR had impaired tumor control in MC38 and B16F1 models (Figs. 2 D and S1 C), indicating butyrate may inhibit the antitumor effect of IR.
Figure S1.
Vancomycin decreased Lachnospiraceae in the gastrointestinal tract, while both Lachnospiraceae supplementation and butyrate administration impaired the antitumor effect of IR, in a melanoma model. (A) Verification of the abundance of Lachnospiraceae by qPCR using Lachnospiraceae (TSD-26) genomic DNA as a standard (n = 4/group). Unpaired t tests were used to analyze the data. (B) GF C57BL/6 mice (n = 5/group) were fed Lachnospiraceae and K. alysoides (TSD-26) for 1 mo, 108 CFU/mouse weekly. The control group was fed the same volume of autoclaved water. Then the mice were injected s.c. with 106 B16F1 cells, and established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. (C) C57BL/6 mice (n = 5/group) were injected s.c. with 106 B16F1 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were injected i.v. with 4 μmol of NaBu on days 1, 4, and 7 after IR. (D) C57BL/6 mice (n = 5/group) were injected s.c. with 106 B16F1 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally (i.t.) injected with 2 μmol of NaBu on days 1, 4, and 7 after IR. A tumor growth curve is shown. Two-way ANOVA tests were used to analyze the tumor growth data. *, P < 0.05; ****, P < 0.0001. One representative experiment (out of two experiments) is shown.
Figure 2.
Lachnospiraceae and butyrate restrict the antitumor effect of IR. (A) GF C57BL/6 mice were fed with Lachnospiraceae and K. alysoides (TSD-26) for 1 mo, 108 CFU/mouse weekly. The control group was fed the same volume of autoclaved water. Mice were injected s.c. with 106 MC38 cells, and established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. On day 10 after tumor inoculation, sera were collected for butyrate detection by GC/MS. A tumor growth curve is shown (n = 5/group). (B) GF C57BL/6 mice were fed HI TSD-26 weekly for 1 mo. 108 CFU of TSD-26 were HI at 100°C for 30 min and fed to one mouse. The control group was fed the same volume of autoclaved water. Then the mice were injected s.c. with 106 MC38 cells, and established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. A tumor growth curve is shown (n = 5/group). On day 10 after tumor inoculation, sera were collected for butyrate detection by GC/MS. (C) Butyrate concentration in sera collected from A and B (control group, n = 10; TSD-26 group, n = 10; HI TSD-26 group, n = 12). Data were pooled from two independent experiments. (D) C57BL/6 mice were injected s.c. with 106 MC38 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were injected i.v. with 4 μmol of NaBu on days 1, 4, and 7 after IR. A tumor growth curve is shown (n = 5/group). Two-way ANOVA tests were used to analyze the tumor growth data, and unpaired t tests were used to analyze the other data. n.s., not significant; *, P < 0.05; **, P < 0.01; ****, P < 0.0001. One representative experiment (out of two experiments in A and B or three experiments in D) is shown.
Intratumoral butyrate inhibits the antitumor effect of IR
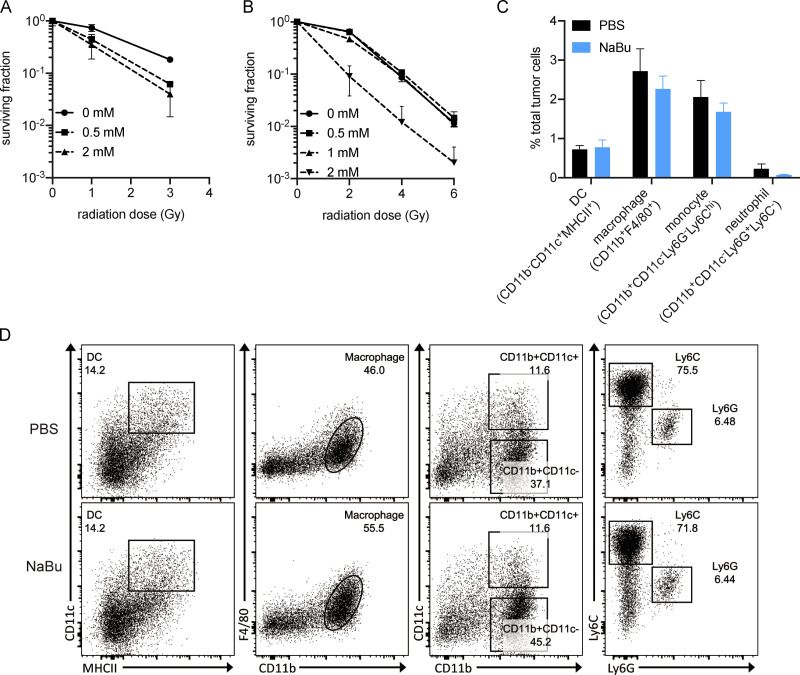
Mounting evidence suggests that the tumor microbiome may influence tumor control (Shi et al., 2020; Nejman et al., 2020). Since live Lachnospiraceae bacteria were not detectable in tumors, we propose that butyric acid may accumulate in tumors and influence the IR-induced antitumor response. Butyric acid was detected not only in serum but also in tumor tissues. Vancomycin decreased, while K. alysoides inoculation increased, the concentration of butyric acid in the tumor microenvironment (Fig. 3, A and B). HI K. alysoides did not affect the intratumoral butyric acid concentration (Fig. S2 A). We do not rule out the contributions from other metabolites or pathogen-associated molecular patterns. Direct injection of butyrate might help to improve the possibility that butyrate participates in this process. Furthermore, tumor control was impaired with intratumoral injection of NaBu on days 1, 4, and 7 after IR in both MC38 and B16F1 models (Figs. 3 C and S1 D). The IR-induced antitumor response was diminished following intratumoral injection of NaBu in vancomycin-treated mice (Fig. 3 D). These results suggest that local butyrate is sufficient to inhibit the antitumor effect after IR. To determine whether intratumoral butyrate directly affects the survival of tumor cells after IR, we performed a clonogenic assay using MC38 and B16F1 cell lines with diluted concentrations of NaBu added to the culture medium. Butyrate did not protect tumor cells from irradiation (Fig. S3, A and B). Since IR stimulates innate and adaptive immunity, we hypothesized that local butyrate would suppress the IR-mediated antitumor immune response.
Figure 3.
Intratumoral (i.t.) butyrate impairs the antitumor response to IR. (A) Established tumors were collected on day 10 after tumor inoculation from vancomycin-treated and untreated C57BL/6 mice after 1 mo of vancomycin feeding. Butyrate concentration in tumor tissues (n = 9/group). (B) Established tumors were collected on day 10 after tumor inoculation from control and TSD-26–fed mice after 1 mo of feeding. Butyrate concentration in tumor tissues (control group, n = 13; TSD-26 group, n = 8). (C) C57BL/6 mice (n = 5/group) were injected s.c. with 106 MC38 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu on days 1, 4, and 7 after IR. (D) C57BL/6 mice (n = 5/group) were fed vancomycin for 1 mo and then injected s.c. with 106 MC38 cells. Established tumors were irradiated (15 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu on days 1, 4, and 7 after IR. Two-way ANOVA tests were used to analyze the tumor growth data. *, P < 0.05; **, P < 0.01; ****, P < 0.0001. One representative experiment (out of two experiments in A, B, and D or three experiments in C) is shown.
Figure S2.
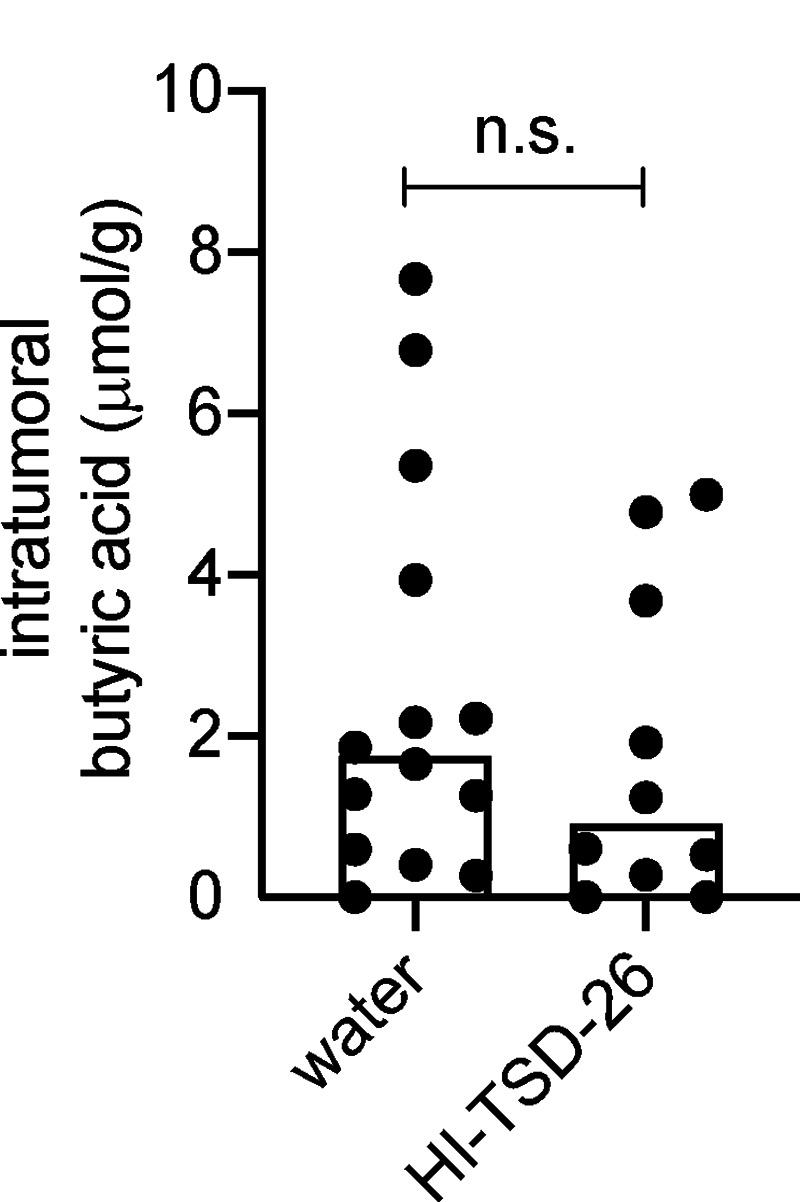
HI Lachnospiraceae species do not affect intratumoral butyrate concentration. GF C57BL/6 mice (n = 5/group) were fed HI Lachnospiraceae and K. alysoides (TSD-26) weekly for 1 mo. 108 CFU of TSD-26 were HI at 100°C for 30 min and fed to one mouse. The control group was fed the same volume of autoclaved water. Then the mice were injected s.c. with 106 MC38 cells. On day 10 after tumor inoculation, tumors (control group, n = 14; HI-TSD-26 group, n = 10) were collected for butyrate detection by GC/MS. Unpaired t tests were used to analyze the data. n.s., not significant. Data were pooled based on two experiments yielding similar results.
Figure S3.
Butyrate does not protect tumor cells from radiation or affect IR-induced intratumoral myeloid cell infiltration. (A) Clonogenic assay of MC38 cells. Radiation doses were 0, 1, and 3 Gy, and NaBu doses were 0, 0.5, and 2 mM. (B) Clonogenic assay of B16F1 cells. Radiation doses were 0, 2, 4, and 6 Gy, and NaBu doses were 0, 0.5, 1, and 2 mM. (C and D) C57BL/6 mice (n = 5/group) were injected s.c. with 106 MC38 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu on day 1 after IR. On day 3 after IR, tumors were harvested and processed for flow cytometry analysis. Percentages of intratumoral myeloid cells from PBS- and NaBu-treated mice. Statistical results are shown in C. Representative dot plots are shown in D. Unpaired t tests were used to analyze the statistical data. One representative experiment (out of two experiments) is shown.
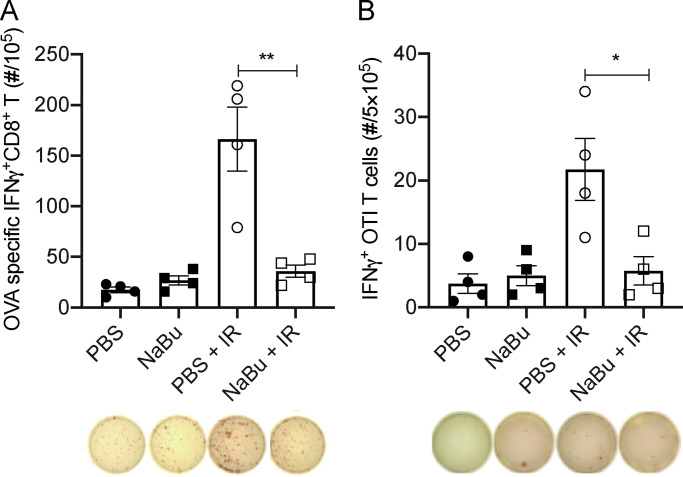
Butyrate inhibits IR-induced tumor-specific T cell response
On day 10 after IR, MC38-OVA tumors were evaluated for intratumoral CD8+ T cell activation using Kb-OVA (SIINFEKL) as the model antigen in an ELISPOT assay. Our data indicated that NaBu prohibited the IR-induced cytotoxic CD8+ T cell response (Fig. 4 A). Based on our previous studies, IR-induced cytotoxic T cell response strongly depends on tumor-associated myeloid cells and STING activation (Mondini et al., 2019; Deng et al., 2016, 2014; Liang et al., 2017). On day 3 after IR, the intratumoral myeloid cells were analyzed by flow cytometry. Unexpectedly, the infiltration of DCs (CD11c−CD11c+MHCII+), macrophages (CD11b+F4/80+), monocytes (CD11b+CD11c−Ly6G−Ly6Chi), and neutrophils (CD11b+CD11c−Ly6G+Ly6C−) were not affected by NaBu treatment (Fig. S3, C and D).
Figure 4.
Butyrate restrains the IR-induced intratumoral innate and adaptive immune response. (A) C57BL/6 mice (n = 4/group) were injected s.c. with 106 MC38-OVA cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu on days 1, 4, and 7 after IR. Tumors were harvested on day 10 after IR and processed for analysis of IFNγ production using Kb-OVA (SIINFEKL) peptide as the model antigen in an ELISPOT assay. (B) C57BL/6 mice (n = 4/group) were injected s.c. with 106 MC38-OVA cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumoral injected with 2 μmol of NaBu on day 1 after IR. Tumors were harvested, and CD11c+ cells were sorted and cocultured with OT-I CD8+ T cells for analysis of their T cell activation ability in an ELISPOT assay. Unpaired t tests were used to analyze the other data. *, P < 0.05; **, P < 0.01. One representative experiment (out of two experiments) is shown.
IR can induce an enhanced tumor-specific cytotoxic T cell response that is DC dependent (Deng et al., 2014). Based on this, since the infiltration of DCs was not affected by local NaBu treatment after IR, we wondered whether butyrate impaired DCs’ function in activating CD8+ T cells. Intratumoral CD11c+ cells were sorted from mice treated with NaBu or without NaBu on day 3 after IR and were incubated with OT-I CD8+ T cells for IFNγ+CD8+ T cell detection in an ELISPOT assay. NaBu prohibited the IR-induced DC function in activating tumor-specific cytotoxic CD8+ T cell response (Fig. 4 B). In summary, intratumoral butyrate inhibited IR-induced tumor-specific T cell response by impairing DCs’ function.
Butyrate inhibits IR-induced IFN-I production
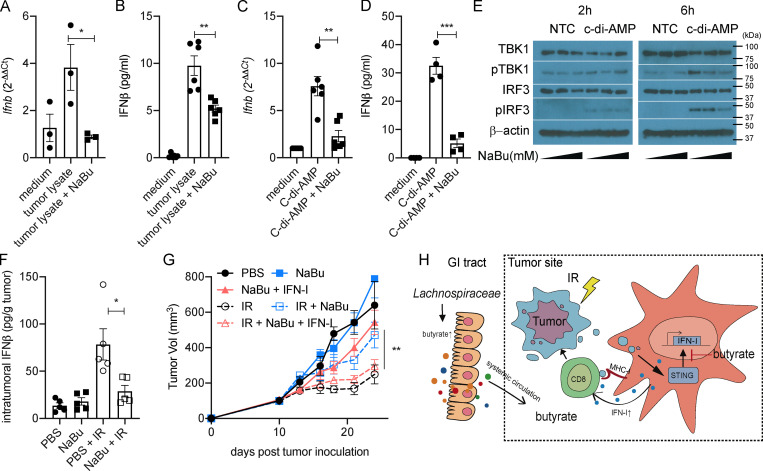
We previously reported that IR induced STING activation in intratumoral DCs and increased IFN-I expression in DCs to enhance the tumor antigen–specific cytotoxic T cell response (Deng et al., 2014). Furthermore, we wondered whether butyrate impaired DCs’ function in producing IFN-I. Upon in vitro stimulation of bone marrow–derived DCs (BMDCs), NaBu inhibited tumor lysate (irradiated tumor cells; Fig. 5, A and B), and a STING agonist (c-di-AMP; Fig. 5, C and D) induced IFNβ expression, on both the mRNA and protein levels. STING activation recruits serine-threonine kinase (TBK1) to phosphorylate the IFN regulatory factor 3 (IRF3), ultimately leading to IFN-I production (Charrel-Dennis et al., 2008; Cai et al., 2014). In BMDCs, upon c-di-AMP stimulation, the phosphorylation of TBK1 and IRF3 were increased, while the increase of pTBK1 and pIRF3 was compromised with NaBu in a dose-dependent manner (Fig. 5 E). These findings demonstrate that NaBu inhibits STING downstream activation by interfering phosphorylation of TBK1 and IRF3.
Figure 5.
Butyrate inhibits the STING activation and restrains the IR-induced intratumoral IFN-I upregulation. (A and B) BMDCs were stimulated with MC38 tumor lysate. MC38 tumor cells were irradiated with 60 Gy. 4 × 104 irradiated cells were cocultured with 106 BMDCs for 24 h. CD11c+ cells were isolated, and mRNA expression of Ifnb was detected by qPCR (A). IFNβ secretion was detected by ELISA (B). (C and D) BMDCs were stimulated with c-di-AMP (1 µg/ml) for 24 h. CD11c+ cells were isolated, and mRNA expression of Ifnb was detected by qPCR (C). IFNβ secretion was detected by ELISA (D). (E) BMDCs were stimulated with or without c-di-AMP (1 µg/ml) and NaBu (0, 0.1, or 1 mM) for 2 or 6 h. Western blot analysis of TBK1, pTBK1, IRF3, pIRF3, and β-actin is shown. (F) C57BL/6 mice (n = 5/group) were injected s.c. with 106 MC38 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu on day 1 after IR. Tumors were collected on day 3 after IR. IFNβ was detected by ELISA. (G) C57BL/6 mice (n = 4/group) were injected s.c. with 106 MC38 cells. Established tumors were irradiated (20 Gy) on day 10 after tumor inoculation. Tumor-bearing mice were intratumorally injected with 2 μmol of NaBu or 10 ng of recombinant IFNβ protein on days 1, 4, and 7 after IR. (H) Gut microbiota–derived butyrate suppresses radiation-induced IFN within the tumor microenvironment. Two-way ANOVA tests were used to analyze the tumor growth data. Unpaired t tests were used to analyze the other data. *, P < 0.05; **, P < 0.01; ***, P < 0.001. One representative experiment (out of two experiments in E and G or three experiments in A–D and F) is shown. GI, gastrointestinal; NTC, no-treatment control.
On day 3 after IR, MC38-OVA tumor-bearing mice from control and NaBu-treated groups were assessed for IFNβ production. NaBu significantly inhibited IR-induced intratumoral upregulation of IFNβ (Fig. 5 F). A low amount of recombinant IFNβ was then complemented intratumorally with simultaneous intratumoral NaBu treatment. IFNβ reversed the effects of NaBu and rescued the efficacy of IR (Fig. 5 G). Taken together, the accumulation of microbiota-derived butyrate can suppress the antitumor activity of IR by abrogating IR-induced IFN-I upregulation and the tumor-specific cytotoxic T cell response.
Upon 16S rRNA amplicon sequencing, we identified that gram-positive butyrate-producing bacteria were proportionally decreased in vancomycin-treated mice with improved tumor control after IR. Vancomycin decreased, while butyrate-producing bacteria Lachnospiraceae increased, the butyrate concentration systemically and locally. Intratumoral butyrate accumulation was sufficient to impair the antitumor radiation response. Local butyrate altered the tumor microenvironment by inhibiting radiation-induced, STING-activated IFN-I upregulation, which is required for tumor-specific cytotoxic T cell response (Fig. 5 H).
We found butyrate inhibited STING-activated TBK1 and IRF3 phosphorylation, which led to decreased IFN-I upregulation and poor antitumor immune responses. How butyrate inhibits the phosphorylation of TBK1 and IRF3 requires further investigation. It has been reported that butyrate attenuates the LPS-induced degradation/phosphorylation of nuclear factor of κ light polypeptide gene enhancer in B cells inhibitor, α (IκBα), DNA binding of NF-κB, and enhanced histone H3 acetylation (Lee et al., 2017). Butyrate affects important cellular processes in part via activation of G protein–coupled receptors (GPCRs) such as GPCR41, GPCR43, and GPCR109A (Ren et al., 2009; Brown et al., 2003; Parada Venegas et al., 2019). The anti-inflammatory effects of butyrate on macrophages were reported independent of GPCRs but were dependent on its inhibition of histone deacetylases (Chang et al., 2014). These mechanisms may act in concert in butyrate suppression of radiation-induced immunity.
The observation that butyrate-producing species K. alysoides can dampen the IR efficacy upon inoculation in mice strongly suggests that butyrate-producing bacteria, such as Lachnospiraceae or Ruminococcaceae, might be a novel therapeutic target after accounting for the complex interplay between the gut microbiota and host immune system. Our data provide a proof of concept and identify potential bacterial targets for subsequent radiotherapy translational clinical trials. One important clinical note is that butyrate supplements have been suggested for patients suffering IR-induced diarrhea, since they can relieve gut-associated inflammation (Ferreira et al., 2014; Mulder et al., 2019; Kelly et al., 2015). However, a reexamination of this clinical strategy is suggested by our findings that butyrate may reduce the efficacy of IR on various cancers.
Recently, the Facciabene group also found that vancomycin administration decreased the systemic concentration of butyrate and modulated the IR-induced antitumor response on DCs in the tumor-draining LN in B16 melanoma and TC-1 lung/cervical tumor models (Uribe-Herranz et al., 2020). Beyond their findings, our results revealed that intratumoral accumulation of butyrate suppressed the activation of STING and production of IFNβ in DCs within the tumor. Our results uncovered and determined the microbiota responsible for butyrate’s effects on antitumor immunity. Taken together, these data suggest butyrate participates in both the systemic and intratumoral IR-induced antitumor immunity.
Our results do not rule out the contribution of other commensal bacterial species in regulating the IR-mediated antitumor response. Our recent published study suggests commensal bacteria migrate to the tumor site and colonize in the tumor microenvironment to participate in an anti-CD47–induced antitumor response (Shi et al., 2020). In this study, we also performed a 16S rRNA amplicon analysis of intratumoral bacteria. Some genera, like Akkermansia and Lactobacillus, had increased proportions following vancomycin treatment both in the gastrointestinal tract and the tumor site. These findings imply that not only antibiotics, but also beneficial bacteria, could be further manipulated for future therapeutic development. The modulation of the gut microbiome should be considered as precision therapy to promote the therapeutic effect of radiotherapy and other cancer therapies. Also, these data suggest that antibiotics administered to patients during radiotherapy may have unexpected effects on clinical outcomes.
In conclusion, a microbiota-derived metabolite, butyrate, appeared to restrain the antitumor effect of radiotherapy. The identification of a specific inhibitor for butyrate-mediated immune-modulatory effects could be considered to enhance radiotherapy responses.
Materials and methods
Mice
All mice used in this work were C57BL/6 background and were purchased from The Jackson Laboratory, Taconic Biosciences, or Envigo. These mice were maintained under SPF conditions. GF C56BL/6 mice were initially obtained from Taconic Biosciences and maintained in flexible-film isolators in the Gnotobiotic Research Animal Facility at the University of Chicago. GF mice were confirmed to be GF via 16S PCR conducted on weekly stool samples. Male and female mice were used at 6–10 wk of age and were sex and age matched in each experiment. Animal care and experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at the University of Chicago.
Cell lines and reagents
MC38 and B16F1 cells were purchased from ATCC and were maintained according to the method of characterization used by ATCC. The cell lines were authenticated by flow cytometry and morphology. In addition, the growth ability of MC38 and B16F1 in immunocompetent syngeneic mice was monitored in each experiment. OVA-expressing tumor cell lines MC38-OVA and B16-OVA were selected as single clones with 5 µg/ml puromycin (InvivoGen) after stable infection with lentivirus-expressing OVA protein. MC38, MC38-OVA, B16F1, and B16-OVA cells were cultured in 5% CO2 and were maintained in vitro in DMEM (Corning) supplemented with 10% HI FBS (Sigma-Aldrich), 2 mM L-glutamine, 0.1 mM MEM nonessential amino acids, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Tumor growth and treatment
Tumor cells (5–10 × 105) were injected s.c. into the backs of 8–10-wk-old mice. The tumor volumes were measured along three orthogonal axes (a, b, and c) and were calculated as follows: tumor volume = (a × b × c)/2. The mice were treated with the standard 15 or 20 Gy of tumor-localized radiation.
Antibiotics and bacteria administration
For antibiotic administration, mice were orally fed with antibiotic suspensions (0.5 mg/ml ampicillin, 0.5 mg/ml gentamicin, 0.5 mg/ml metronidazole, 0.5 mg/ml neomycin, and 0.25 mg/ml vancomycin dissolved in autoclaved water) from 3 wk before tumor inoculation to experimental endpoints. Antibiotics were purchased from Sigma-Aldrich. For bacteria oral administration, GF C57BL/6 mice were gavaged weekly with 108 CFU of Lachnospiraceae family member K. alysoides (TSD-26; purchased from ATCC and cultured with trypticase soya broth + 5% [vol/vol] defibrinated sheep blood in an anaerobic chamber) in 500 µl water per mouse from 3 wk before tumor inoculation to experiment endpoints.
NaBu administration
For systemic administration, mice were intravenously injected with 4 μmol/200 µl of NaBu (Sigma-Aldrich) suspension the day before and every third day after tumor-localized radiation. For intratumoral administration, mice were intratumorally injected with 1–2 μmol/100 µl of NaBu suspension the day before and every third day after tumor-localized radiation.
Quantification of butyric acid
125 µl serum or tissue suspension was added with an equal volume of nuclease-free water and acidified with 50 µl of 50% H2SO4. 2.5 µl of 1 mmol/L isobutyric acid (Sigma-Aldrich) was added as an internal standard. To this solution, 250 µl of diethyl ether was added, vortexed for 30 s, and quickly centrifuged at 5,000 × g for butyric acid extraction. 250 µl of the upper ether layer was transferred to a clean Eppendorf tube, and the extraction was repeated two more times. 500 µl of the extract was transferred to a glass vial and derivatized with 125 µl of N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide (Ho et al., 2015). Samples were stored at −80°C before being analyzed using an Agilent SQ gas chromatograph (GC)/mass spectrometer (MS; 5977A single-quad MS and 7890B GC). Butyric acid (Sigma-Aldrich) was used to generate the standard curve.
Measurement of IFN-γ–secreting T cells by ELISPOT assay
Tumor tissues and LNs from naive mice were harvested and resuspended in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin. For intratumoral T cell detection, CD8+ cells from tumors and CD11c+ cells from naive LNs were purified using EasySep Selection kits (STEMCELL Technologies). Kb-OVA peptides (SIINFEKL; InvivoGen) were used at 10 µg/ml as model antigens. For intratumoral DC function detection, OT-I CD8+ cells from spleens from OT-I–transgenic mice and CD11c+ cells from tumors were purified using EasySep Selection kits (STEMCELL Technologies). A total of 2–4 × 105 CD8+ cells were used for assays. The ratio of CD11c+ cells to CD8+ cells was 1:5 or 1:10. After 48 h of incubation, the IFN-γ production was determined with an IFN-γ ELISPOT assay kit according to the manufacturer’s protocol (BD Biosciences). The visualized cytokine spots were enumerated using an ImmunoSpot Analyzer (Cellular Technology Ltd.).
Measurement of IFN-β secretion by ELISA
Supernatants from BMDC stimulation and tumor tissue lysates were collected and stored at −80°C for ELISA. Protease inhibitor cocktails (Thermo Fisher Scientific) were added according to the manufacturer’s protocol. IFNβ was measured using the VeriKine-HS Mouse Interferon Beta Serum ELISA Kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Quantitative RT-PCR
RNA was extracted using TRIzol or RNeasy Plus Universal Micro kit (Qiagen). cDNA was generated using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Real-time RT-PCR was performed with ABI7500. Primers used were: β-actin: forward, 5′-ACACCCGCCACCAGTTCGC-3′; reverse, 5′-ATGGGGTACTTCAGGGTCAGGGTCAGGATA-3′; Ifnb: forward, 5′-ATGAGTGGTGGTTGCAGGC-3′; reverse, 5′-TGACCTTTCAAATGCAGTAGATTCA-3′; universal bacterial genome: forward, 5′-ACTCCTACGGGAGGCAGC-3′; reverse, 5′-ATTACCGCGGCTGCTGGC-3′; Lachnospiraceae genome: forward, 5′-CCAAGGCGGCCGTACGCTGAAGCAACGCGAAGAACCTTACCA-3′; reverse, 5′-CCGACGTCGACTATCCATCTGAAGGACGACAACCATGCACCACC-3′. Gene expression was normalized to β-actin and calculated according to the 2−ΔΔCt method.
Flow cytometry
Draining LN or tumor cells were digested with 1 mg/ml collagenase IV (Sigma-Aldrich) and 200 µg/ml DNaseI (Sigma-Aldrich) at 37°C for 30 or 60 min. Antibodies used were anti-CD11b (M1/70), anti-CD4 (RM4-5), and anti-CD8α (53-6.7) from eBioscience and anti-CD11c (N418), anti-CD8α (53-6.7), anti-Ly6C (HK1.4), and anti-MHCII (M5/114.15.2) from BioLegend. Dead cells were excluded by LIVE/DEAD Fixable Yellow Dead Cell Stain Kit (Invitrogen). Flow cytometric analysis was acquired using a BD LSR Fortessa and analyzed with FlowJo V10.
BMDC generation and in vitro stimulation
Single-cell suspensions of bone marrow cells were obtained from the indicated mice, plated in 10-cm Petri dishes, and cultured in culture medium (RPMI 1640 + 10% FBS + 1,000 U/ml GM-CSF). Fresh culture medium was added on days 3 and 6. BMDCs were harvested for stimulation assay on day 8. For in vitro stimulation, BMDCs were cocultured with or without 0.1–1 mM NaBu and 1 µg/ml c-di-AMP (InvivoGen) for 2–24 h.
Western blot analysis
Whole-cell protein was extracted with radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific) supplemented with protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific). Protein concentrations were measured by BCA Protein Assay Kit (Pierce). An equal amount of protein (10 µg) was separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane (Invitrogen). Membranes were blocked in 1× TBS, 0.1% Tween 20 Detergent (TBST) (Thermo Fisher Scientific) with 10% nonfat milk for 2 h and incubated with primary antibodies (phospho-IRF3 [Ser396; D6O1M] rabbit mAb, IRF3 [D83B9] rabbit mAb, phospho-TBK1/NAK [Ser172; D52C2] XP Rabbit mAb, TBK1/NAK [D1B4] rabbit mAb; Cell Signaling Technology) at 4°C overnight. After washing with TBST and incubation with donkey antirabbit IgG HRP antibody (Cell Signaling Technology) or anti-β-actin HRP antibody (Cell Signaling Technology) for another 1 h, ECL (enhanced chemiluminescent liquid) and super ECL (Thermo Fisher Scientific) were applied for film development.
Microbiota analyses
Microbial DNA was extracted using a QIAamp Fast Stool Mini Kit (Qiagen) and was submitted to the Environmental Sample Preparation and Sequencing Facility at Argonne National Laboratory for 16S rRNA amplicon sequencing. Amplicon libraries were sequenced on the MiSeq Micro platform (Illumina). The raw sequencing data have been deposited with the National Center for Biotechnology Information under Sequence Read Archive accession no. PRJNA650218. The 16S data were quality filtered and demultiplexed using the same QIIME 1.9.1 scripts (i.e., join_paired_ends.py and split_libraries_fastq.py; Caporaso et al., 2010). The final set of demultiplexed sequences was then selected for exact sequence variant (ESV) picking using the DeBlur pipeline (Amir et al., 2017). ESVs present in <10 samples were removed using the phyloseq package (McMurdie and Holmes, 2013). The final Biological Observation Matrix files (McDonald et al., 2012), comprising 12 murine samples with an average of 30,581 counts per sample, were then used for further analyses. To calculate the relative abundance, we sum up all counts and divide each count with the sum. Unconstrained principal coordinate analysis was employed to reveal β-diversity variations based on unweighted UniFrac (Lozupone et al., 2006) for the 16S rRNA ESV data in the Phyloseq package. The Chao1 index was used to estimate α-diversity, and the variation between groups (β-diversity) was statistically tested using ANOSIM (Anderson, 2017). Analysis of composition of microbiomes was used to identify differentially abundant bacterial ESVs between the groups at a P value cutoff of 0.05 with the Benjamini-Hochberg false discovery rate correction (Mandal et al., 2015). Spearman rank correlations and generalized linear models were used to establish associations between the microbiome and other continuous variables in the metadata by using the microbiomeSeq and glm packages in R (R Foundation for Statistical Computing, 2011; Chong et al., 2020).
Statistical analysis
For tumor growth data, descriptive statistics of tumor size were summarized by treatment group at each time point. Tumor growth curves were plotted over time by treatment. Flow cytometry and T cell function data were summarized, presented using descriptive statistics for each treatment, and compared across treatment groups using two-way ANOVA or two-way Student’s t tests. Statistical figures were prepared using Prism software (version 8.0; GraphPad Software). n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows that vancomycin decreased Lachnospiraceae in the gastrointestinal tract, while both Lachnospiraceae supplementation and butyrate administration impaired the antitumor effect of IR in a melanoma model. Fig. S2 shows that HI Lachnospiraceae species does not affect intratumoral butyrate concentration. Fig. S3 shows that butyrate does not protect tumor cells from radiation or affect IR-induced intratumoral myeloid cell infiltration.
Acknowledgments
We thank Yaoyao Shi, Ainhoa Arina, and Jason Bugno for helpful discussion; Jason Bugno for excellent editing; and Wenbo Han, Xiaona Huang, Melanie Spedale, Candace Cam, and Jason Koval for technical assistance.
This work was supported by a grant from the Ludwig Cancer Research Foundation to R.R. Weichselbaum and by National Institutes of Health/National Cancer Institute Provocative Questions grants R21 CA227528 and R21 CA231273-01 to R.R. Weichselbaum.
Author contributions: K. Yang, Y. Hou, R.R. Weichselbaum, and Y-X. Fu conceived and designed the study. K. Yang, Y. Hou, Y. Zhang, H. Liang, W. Zheng, L. Wang, R. Torres, and K.T. Tatebe performed the experiments. S.J. Chmura and S.P. Pitroda contributed resources. A. Sharma and J.A. Gilbert analyzed the 16S sequencing data. K. Yang, R.R. Weichselbaum, and Y-X. Fu drafted and finalized the manuscript with input from all other authors.
References
- Al-Sarraf, M., LeBlanc M., Giri P.G.S., Fu K.K., Cooper J., Vuong T., Forastiere A.A., Adams G., Sakr W.A., Schuller D.E., and Ensley J.F.. 1998. Chemoradiotherapy versus radiotherapy in patients with advanced nasopharyngeal cancer: phase III randomized Intergroup study 0099. J. Clin. Oncol. 16:1310–1317. 10.1200/JCO.1998.16.4.1310 [DOI] [PubMed] [Google Scholar]
- Amir, A., McDonald D., Navas-Molina J.A., Kopylova E., Morton J.T., Zech Xu Z., Kightley E.P., Thompson L.R., Hyde E.R., Gonzalez A., and Knight R.. 2017. Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems. 2:e00191-16. 10.1128/mSystems.00191-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M.J. 2017. Permutational multivariate analysis of variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online. Balakrishnan N., Colton T., Everitt B., Piegorsch W., Ruggeri F., and Teugels J.L., editors. Wiley Online Library. 10.1002/9781118445112.stat07841 [DOI] [Google Scholar]
- Aßhauer, K.P., Wemheuer B., Daniel R., and Meinicke P.. 2015. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 31:2882–2884. 10.1093/bioinformatics/btv287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi, K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., et al. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 331:337–341. 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker, H.E., Paget J.T.E., Khan A.A., and Harrington K.J.. 2015. The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat. Rev. Cancer. 15:409–425. 10.1038/nrc3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D., Muir A.I., Wigglesworth M.J., Kinghorn I., Fraser N.J., et al. 2003. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 278:11312–11319. 10.1074/jbc.M211609200 [DOI] [PubMed] [Google Scholar]
- Cai, X., Chiu Y.H., and Chen Z.J.. 2014. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 54:289–296. 10.1016/j.molcel.2014.03.040 [DOI] [PubMed] [Google Scholar]
- Caporaso, J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7:335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, P.V., Hao L., Offermanns S., and Medzhitov R.. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 111:2247–2252. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrel-Dennis, M., Latz E., Halmen K.A., Trieu-Cuot P., Fitzgerald K.A., Kasper D.L., and Golenbock D.T.. 2008. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 4:543–554. 10.1016/j.chom.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong, J., Liu P., Zhou G., and Xia J.. 2020. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 15:799–821. 10.1038/s41596-019-0264-1 [DOI] [PubMed] [Google Scholar]
- Chung, H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., et al. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149:1578–1593. 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., Liang H., Xu M., Yang X., Burnette B., Arina A., Li X.-D., Mauceri H., Beckett M., Darga T., et al. 2014. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity. 41:843–852. 10.1016/j.immuni.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, L., Liang H., Fu S., Weichselbaum R.R., and Fu Y.-X.. 2016. From DNA damage to nucleic acid sensing: a strategy to enhance radiation therapy. Clin. Cancer Res. 22:20–25. 10.1158/1078-0432.CCR-14-3110 [DOI] [PubMed] [Google Scholar]
- Donohoe, D.R., Holley D., Collins L.B., Montgomery S.A., Whitmore A.C., Hillhouse A., Curry K.P., Renner S.W., Greenwalt A., Ryan E.P., et al. 2014. A gnotobiotic mouse model demonstrates that dietary fiber protects against colorectal tumorigenesis in a microbiota- and butyrate-dependent manner. Cancer Discov. 4:1387–1397. 10.1158/2159-8290.CD-14-0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, M.R., Muls A., Dearnaley D.P., and Andreyev H.J.N.. 2014. Microbiota and radiation-induced bowel toxicity: lessons from inflammatory bowel disease for the radiation oncologist. Lancet Oncol. 15:e139–e147. 10.1016/S1470-2045(13)70504-7 [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan, V., Spencer C.N., Nezi L., Reuben A., Andrews M.C., Karpinets T.V., Prieto P.A., Vicente D., Hoffman K., Wei S.C., et al. 2018. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 359:97–103. 10.1126/science.aan4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, K.N., and Blanchard J.L.. 2017. Kineothrix alysoides, gen. nov., sp. nov., a saccharolytic butyrate-producer within the family Lachnospiraceae. Int. J. Syst. Evol. Microbiol. 67:402–410. 10.1099/ijsem.0.001643 [DOI] [PubMed] [Google Scholar]
- Hagan, T., Cortese M., Rouphael N., Boudreau C., Linde C., Maddur M.S., Das J., Wang H., Guthmiller J., Zheng N.-Y., et al. 2019. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 178:1313–1328.e13. 10.1016/j.cell.2019.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman, J.M., Chang D.T., Goodman K.A., Dholakia A.S., Raman S.P., Hacker-Prietz A., Iacobuzio-Donahue C.A., Griffith M.E., Pawlik T.M., Pai J.S., et al. 2015. Phase 2 multi-institutional trial evaluating gemcitabine and stereotactic body radiotherapy for patients with locally advanced unresectable pancreatic adenocarcinoma. Cancer. 121:1128–1137. 10.1002/cncr.29161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, K.J., Xiong L., Hubert N.A., Nadimpalli A., Wun K., Chang E.B., and Kibbe M.R.. 2015. Vancomycin treatment and butyrate supplementation modulate gut microbe composition and severity of neointimal hyperplasia after arterial injury. Physiol. Rep. 3:e12627. 10.14814/phy2.12627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper, L.V., Littman D.R., and Macpherson A.J.. 2012. Interactions between the microbiota and the immune system. Science. 336:1268–1273. 10.1126/science.1223490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iida, N., Dzutsev A., Stewart C.A., Smith L., Bouladoux N., Weingarten R.A., Molina D.A., Salcedo R., Back T., Cramer S., et al. 2013. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 342:967–970. 10.1126/science.1240527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov, I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachikwu, E.L., Iwamoto K.S., Liao Y.P., DeMarco J.J., Agazaryan N., Economou J.S., McBride W.H., and Schaue D.. 2011. Radiation enhances regulatory T cell representation. Int. J. Radiat. Oncol. Biol. Phys. 81:1128–1135. 10.1016/j.ijrobp.2010.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao, J., Chen C.-T., Tong C.C.L., Packer S.H., Schwartz M., Chen S.H., and Sung M.W.. 2014. Concurrent sunitinib and stereotactic body radiotherapy for patients with oligometastases: final report of a prospective clinical trial. Target. Oncol. 9:145–153. 10.1007/s11523-013-0280-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., Wilson K.E., Glover L.E., Kominsky D.J., Magnuson A., et al. 2015. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 17:662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, E.D., Drake C.G., Scher H.I., Fizazi K., Bossi A., van den Eertwegh A.J.M., Krainer M., Houede N., Santos R., Mahammedi H., et al. CA184-043 Investigators . 2014. Ipilimumab versus placebo after radiotherapy in patients with metastatic castration-resistant prostate cancer that had progressed after docetaxel chemotherapy (CA184-043): a multicentre, randomised, double-blind, phase 3 trial. Lancet Oncol. 15:700–712. 10.1016/S1470-2045(14)70189-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoui, D., Van Overmeire E., De Baetselier P., Van Ginderachter J.A., and Raes G.. 2014. Functional relationship between tumor-associated macrophages and macrophage colony-stimulating factor as contributors to cancer progression. Front. Immunol. 5:489. 10.3389/fimmu.2014.00489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Kim B.G., Kim J.H.J.S., Chun J., Im J.P., and Kim J.S.. 2017. Sodium butyrate inhibits the NF-κ B signaling pathway and histone deacetylation, and attenuates experimental colitis in an IL-10 independent manner. Int. Immunopharmacol. 51:47–56. 10.1016/j.intimp.2017.07.023 [DOI] [PubMed] [Google Scholar]
- Liang, H., Deng L., Hou Y., Meng X., Huang X., Rao E., Zheng W., Mauceri H., Mack M., Xu M., et al. 2017. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 8:1736. 10.1038/s41467-017-01566-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman, D.R., and Pamer E.G.. 2011. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 10:311–323. 10.1016/j.chom.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone, C., Hamady M., and Knight R.. 2006. UniFrac--an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 7:371. 10.1186/1471-2105-7-371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu, M., Weigand K., Wedi F., Breidenbend C., Leister H., Pautz S., Adhikary T., and Visekruna A.. 2018. Regulation of the effector function of CD8+ T cells by gut microbiota-derived metabolite butyrate. Sci. Rep. 8:14430. 10.1038/s41598-018-32860-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal, S., Van Treuren W., White R.A., Eggesbø M., Knight R., and Peddada S.D.. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. 10.3402/mehd.v26.27663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson, V., Fessler J., Bao R., Chongsuwat T., Zha Y., Alegre M.L., Luke J.J., and Gajewski T.F.. 2018. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 359:104–108. 10.1126/science.aao3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald, D., Clemente J.C., Kuczynski J., Rideout J.R., Stombaugh J., Wendel D., Wilke A., Huse S., Hufnagle J., Meyer F., et al. 2012. The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 1:7. 10.1186/2047-217X-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie, P.J., and Holmes S.. 2013. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 8:e61217. 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondini, M., Loyher P.-L., Hamon P., Gerbé de Thoré M., Laviron M., Berthelot K., Clémenson C., Salomon B.L., Combadière C., Deutsch E., and Boissonnas A.. 2019. CCR2-dependent recruitment of Tregs and monocytes following radiotherapy is associated with TNFα-mediated resistance. Cancer Immunol. Res. 7:376–387. 10.1158/2326-6066.CIR-18-0633 [DOI] [PubMed] [Google Scholar]
- Mulder, W.J.M., Ochando J., Joosten L.A.B., Fayad Z.A., and Netea M.G.. 2019. Therapeutic targeting of trained immunity. Nat. Rev. Drug Discov. 18:553–566. 10.1038/s41573-019-0025-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman, D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E., et al. 2020. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 368:973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyangale, E.P., Mottram D.S., and Gibson G.R.. 2012. Gut microbial activity, implications for health and disease: the potential role of metabolite analysis. J. Proteome Res. 11:5573–5585. 10.1021/pr300637d [DOI] [PubMed] [Google Scholar]
- Parada Venegas, D., De la Fuente M.K., Landskron G., González M.J., Quera R., Dijkstra G., Harmsen H.J.M., Faber K.N., and Hermoso M.A.. 2019. Short chain fatty acids (SCFAs)mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. 10.3389/fimmu.2019.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse, E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., and Glöckner F.O.. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196. 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushalkar, S., Hundeyin M., Daley D., Zambirinis C.P., Kurz E., Mishra A., Mohan N., Aykut B., Usyk M., Torres L.E., et al. 2018. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 8:403–416. 10.1158/2159-8290.CD-17-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, Y., Jin S., Zhang A., Zhang B., Shi X., Wang J., and Zhao Y.. 2010. Gamma-ray resistance of regulatory CD4+CD25+Foxp3+ T cells in mice. Radiat. Res. 173:148–157. 10.1667/RR0978.1 [DOI] [PubMed] [Google Scholar]
- Rampelli, S., Candela M., Turroni S., Biagi E., Collino S., Franceschi C., O’Toole P.W., and Brigidi P.. 2013. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY). 5:902–912. 10.18632/aging.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, N., Kaplan R., Hernandez M., Cheng K., Jin L., Taggart A.K.P., Zhu A.Y., Gan X., Wright S.D., and Cai T.Q.. 2009. Phenolic acids suppress adipocyte lipolysis via activation of the nicotinic acid receptor GPR109A (HM74a/PUMA-G). J. Lipid Res. 50:908–914. 10.1194/jlr.M800625-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Foundation for Statistical Computing . 2011. R: a language and environment for statistical computing. https://www.r-project.org/foundation/ (accessed October 11, 2018).
- Round, J.L., and Mazmanian S.K.. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 9:313–323. 10.1038/nri2515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy, B., Le Chatelier E., Derosa L., Duong C.P.M., Alou M.T., Daillère R., Fluckiger A., Messaoudene M., Rauber C., Roberti M.P., et al. 2018. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 359:91–97. 10.1126/science.aan3706 [DOI] [PubMed] [Google Scholar]
- Scott, N.A., Andrusaite A., Andersen P., Lawson M., Alcon-Giner C., Leclaire C., Caim S., Le Gall G., Shaw T., Connolly J.P.R., et al. 2018. Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci. Transl. Med. 10:eaao4755. 10.1126/scitranslmed.aao4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y., Zheng W., Yang K., Harris K.G., Ni K., Xue L., Lin W., Chang E.B., Weichselbaum R.R., and Fu Y.-X.. 2020. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 217:e20192282. 10.1084/jem.20192282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan, A., Corrales L., Hubert N., Williams J.B., Aquino-Michaels K., Earley Z.M., Benyamin F.W., Lei Y.M., Jabri B., Alegre M.L., et al. 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 350:1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, P.M., Howitt M.R., Panikov N., Michaud M., Gallini C.A., Bohlooly-Y M., Glickman J.N., and Garrett W.S.. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 341:569–573. 10.1126/science.1241165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackebrandt, E. 2014. The family Lachnospiraceae. In The Prokaryotes. Rosenberg E., DeLong E.F., Lory S., Stackebrandt E., Thompson F., editors. Springer, Berlin. 197–201. [Google Scholar]
- Uribe-Herranz, M., Rafail S., Beghi S., Gil-de-Gómez L., Verginadis I., Bittinger K., Pustylnikov S., Pierini S., Perales-Linares R., Blair I.A., et al. 2020. Gut microbiota modulate dendritic cell antigen presentation and radiotherapy-induced antitumor immune response. J. Clin. Invest. 130:466–479. 10.1172/JCI124332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou, M., Pitt J.M., Daillère R., Lepage P., Waldschmitt N., Flament C., Rusakiewicz S., Routy B., Roberti M.P., Duong C.P.M., et al. 2015. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 350:1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud, S., Saccheri F., Mignot G., Yamazaki T., Daillère R., Hannani D., Enot D.P., Pfirschke C., Engblom C., Pittet M.J., et al. 2013. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 342:971–976. 10.1126/science.1240537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Yu Y., Wang Y.Z., Wang J.J., Guan R., Sun Y., Shi F., Gao J., and Fu X.L.. 2019. Role of SCFAs in gut microbiome and glycolysis for colorectal cancer therapy. J. Cell. Physiol. 234:17023–17049. 10.1002/jcp.28436 [DOI] [PubMed] [Google Scholar]
- Wei, W., Sun W., Yu S., Yang Y., and Ai L.. 2016. Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk. Lymphoma. 57:2401–2408. 10.3109/10428194.2016.1144879 [DOI] [PubMed] [Google Scholar]
- Yazawa, K., Fujimori M., Amano J., Kano Y., and Taniguchi S.. 2000. Bifidobacterium longum as a delivery system for cancer gene therapy: selective localization and growth in hypoxic tumors. Cancer Gene Ther. 7:269–274. 10.1038/sj.cgt.7700122 [DOI] [PubMed] [Google Scholar]
- Yu, G., Gail M.H., Consonni D., Carugno M., Humphrys M., Pesatori A.C., Caporaso N.E., Goedert J.J., Ravel J., and Landi M.T.. 2016. Characterizing human lung tissue microbiota and its relationship to epidemiological and clinical features. Genome Biol. 17:163. 10.1186/s13059-016-1021-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, C., Schild M., Kunz C., Zimmermann K., and Kuntz S.. 2018. Effects of live and heat-inactivated E. coli strains and their supernatants on immune regulation in HT-29 cells. Eur. J. Microbiol. Immunol. (Bp.). 8:41–46. 10.1556/1886.2018.00004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitvogel, L., Ayyoub M., Routy B., and Kroemer G.. 2016. Microbiome and anticancer immunosurveillance. Cell. 165:276–287. 10.1016/j.cell.2016.03.001 [DOI] [PubMed] [Google Scholar]