Autophagy is a eukaryotic intracellular degradation pathway which can act as an innate immune response to eliminate pathogens. Conversely, pathogens can evolve proteins which modulate the autophagy pathway to subvert degradation and establish an infection. Wolbachia, a vertically transmitted obligate endosymbiont which infects up to 40% of insect species, is negatively regulated by autophagy in whole animals, but the specific molecular mechanism and tissue which govern this interaction remain unknown.
KEYWORDS: Wolbachia, autophagy, effector functions, host-pathogen interactions, innate immunity, symbiosis
ABSTRACT
Autophagy is an intracellular degradation pathway involved in innate immunity. Pathogenic bacteria have evolved several mechanisms to escape degradation or exploit autophagy to acquire host nutrients. In the case of endosymbionts, which often have commensal or mutualistic interactions with the host, autophagy is not well characterized. We utilized tissue-specific autophagy mutants to determine if Wolbachia, a vertically transmitted obligate endosymbiont of Drosophila melanogaster, is regulated by autophagy in somatic and germ line cell types. Our analysis revealed core autophagy proteins Atg1 and Atg8 and a selective autophagy-specific protein Ref(2)p negatively regulate Wolbachia in the hub, a male gonad somatic cell type. Furthermore, we determined that the Wolbachia effector protein, CifB, modulates autophagy-Wolbachia interactions, identifying a new host-related pathway which these bacterial proteins interact with. In the female germ line, the cell type necessary for inheritance of Wolbachia through vertical transmission, we discovered that bulk autophagy mediated by Atg1 and Atg8 positively regulates Wolbachia density, whereas Ref(2)p had no effect. Global metabolomics of fly ovaries deficient in germ line autophagy revealed reduced lipid and carbon metabolism, implicating metabolites from these pathways as positive regulators of Wolbachia. Our work provides further understanding of how autophagy affects bacteria in a cell type-dependent manner.
INTRODUCTION
Wolbachia is a widespread obligate endosymbiont, estimated to infect up to 40% of terrestrial arthropods as well as filarial nematodes (1, 2). Recently, Wolbachia has been employed as a biocontrol agent to reduce the spread of several human diseases, including Zika virus infection, dengue virus infection, and malaria (3–5). This is accomplished through artificial transfection of Wolbachia-uninfected mosquitoes with a Wolbachia strain, wMel, originating from the fruit fly Drosophila melanogaster (6, 7). While the exact mechanisms eliciting pathogen blocking by Wolbachia remain unknown, Wolbachia density has been correlated with the effectiveness of this process (8–10). Several studies have begun to elucidate host-related pathways which modulate intracellular Wolbachia density, postulating an indirect means to boost pathogen blocking (11–15). Utilizing genetic analysis in Brugia malayi and the autophagy-inducing drug rapamycin in Drosophila melanogaster larvae, autophagy was identified as a negative regulator of Wolbachia (16). Interestingly, when Wolbachia was investigated in the female germ line after adult flies were fed rapamycin, Wolbachia levels were increased (17). This indicates a possible cell type-dependent effect that autophagy may have in regulating Wolbachia density. It should be noted that rapamycin inhibits target of rapamycin (TOR) signaling, which regulates several downstream signaling pathways, including those that mediate cell growth, protein translation, and autophagy (18). Genetic manipulations to confirm that autophagy is responsible for modulating Wolbachia densities in this cell type-dependent manner remain incomplete.
Macroautophagy, here referred to as autophagy, is a well-conserved eukaryotic degradation and recycling pathway used to protect the cell from different stresses, including nutritional starvation, mitochondrial damage, and intracellular pathogens (19, 20). Autophagy is characterized by the formation of an autophagosome, a double-membrane vesicle, which sequesters and shuttles cargo to the lysosome for degradation (21). There are two main forms of autophagy, selective and bulk, where the former is active under physiological normal conditions and maintains cellular homeostasis and the latter responds to nutrient starvation to recycle cytoplasmic constituents for the cell to use (21–23). Selective and bulk autophagy utilize a similar set of core autophagy genes to form an autophagosome (24). Atg1, a serine/threonine kinase, is the most upstream protein and is known to induce the formation of an autophagosome (25). Atg8 is a ubiquitin-like protein found conjugated to phosphatidylethanolamine (PE) and incorporated into autophagosome membranes and is essential for autophagosome formation (26). Drosophila have two Atg8 genes: the well-characterized and ubiquitously expressed Atg8a and its paralog Atg8b, which is restricted in expression to larval developmental stages and adult male testis (27). During selective autophagy, Atg8 is capable of binding to the adaptor protein Ref(2)p (p62 in mammals), which can target specific cargo to autophagosomes for degradation (28, 29). Selective autophagy has been well characterized as an innate immune response to pathogenic bacteria and viruses, but its role in regulating endosymbionts is less understood.
Bacteria have evolved several ways to evade host autophagy and ensure survival. One method, deubiquitination, allows for bacteria to escape recognition by host selective autophagy adapter proteins and thus survive in the cell. Salmonella and Chlamydia secrete the deubiquitinating (DUB) enzymes Ssel and ChlaOTU and ChlaDUB, respectively, which target and remove ubiquitin (30–32). Interestingly, Wolbachia has been shown to harbor a deubiquitinase named cytoplasmic incompatibility factor B (CifB), which is sufficient to drive the reproductive phenotype of cytoplasmic incompatibility (33–35).
Cytoplasmic incompatibility (CI) is a reproductive phenomenon observed when a Wolbachia-infected male mates with an uninfected female, rendering the eggs sterile. It has been discovered that when Drosophila uninfected with Wolbachia express two Wolbachia proteins, CifA and CifB, in the male germ line, CI can be recapitulated (33, 36). Biochemical studies utilizing the CifB homologue, CidB, from Wolbachia infecting the mosquito Culex pipiens (wPip) showed that a functional DUB domain was necessary to drive CI when expressed in Drosophila melanogaster (34). Interestingly, CifA can act as a rescue factor, whereby it inhibits CI when expressed in the female germ line. This reveals a complex and incomplete mechanism, where CifA aids CifB in driving CI when expressed in the male germ line but CifA rescues this effect when expressed in the female germ line.
Here, we utilized genetic manipulation of core autophagy genes (atg1 and atg8a) and the selective autophagy gene ref(2)p to determine if Wolbachia density is affected by autophagy in an opposite cell type-dependent manner as previously suggested (16, 17). We first analyzed autophagy’s effect on Wolbachia density in the hub, a nondividing somatic cell type in which Wolbachia density has been extensively characterized (37). The hub is established during embryonic development, anchors both the somatic and germ line stem cells, and regulates stem cell division and differentiation (38, 39). We determined that Wolbachia is negatively regulated in the hub by Ref(2)p-dependent selective autophagy in a strain-dependent manner. Additionally, overexpression of the two Wolbachia effector proteins, CifA and CifB, implicate CifB, a bacterial DUB, in positively regulating bacterial density in the hub. Epistasis analysis between CifB and autophagy protein Atg1 provides evidence they are acting in the same pathway.
Conversely, in the female germ line, Wolbachia density is positively affected by autophagy in a strain-independent manner, and Ref(2)p-mediated selective autophagy has no effect in regulating Wolbachia density. This suggests that Wolbachia utilizes a bulk autophagy program to increase its bacterial load. To begin to identify what metabolites autophagy modulates to aid in Wolbachia growth, we utilized a global metabolomics analysis. This analysis allows us to identify changes in host metabolic pathways at the level of metabolites, whereas RNA sequencing or proteomics may not directly reflect metabolic shifts caused by a knockdown of autophagy. Global metabolomics of autophagy mutants in the female germ line reveal a downregulation of glycolysis and glycerolipid metabolism, implicating metabolites from this pathway as positive regulators of Wolbachia. Together, our findings demonstrate a mechanism by which a Wolbachia effector protein and host autophagy proteins act in regulating bacterial density in a cell type- and strain-dependent manner.
RESULTS
Autophagy negatively regulates Wolbachia wMel but not wMelCS density in the hub.
To determine the effect autophagy has in regulating Wolbachia density in somatic tissues, we knocked down core autophagy proteins Atg1 and Atg8a in the Drosophila hub, a cell type previously shown to have high levels of Wolbachia tropism (37). Knockdown of Atg1 resulted in a marked increase in Wolbachia wMel accumulation in the hub (Fig. 1A and B). Quantification of relative Wolbachia density showed a 2.87-fold increase in average Wolbachia density upon Atg1 knockdown (Fig. 1G). To confirm the autophagy pathway was involved in regulating hub cell Wolbachia density, we knocked down Atg8a. We specifically targeted atg8a because it is much more extensively characterized and ubiquitously expressed in the male and female tissues we were investigating than atg8b. Upon knockdown of Atg8a, we saw an increase in the density of wMel (Fig. 1C and D). Quantification of relative Wolbachia density showed a 1.77-fold increase upon Atg8a knockdown (Fig. 1G).
FIG 1.
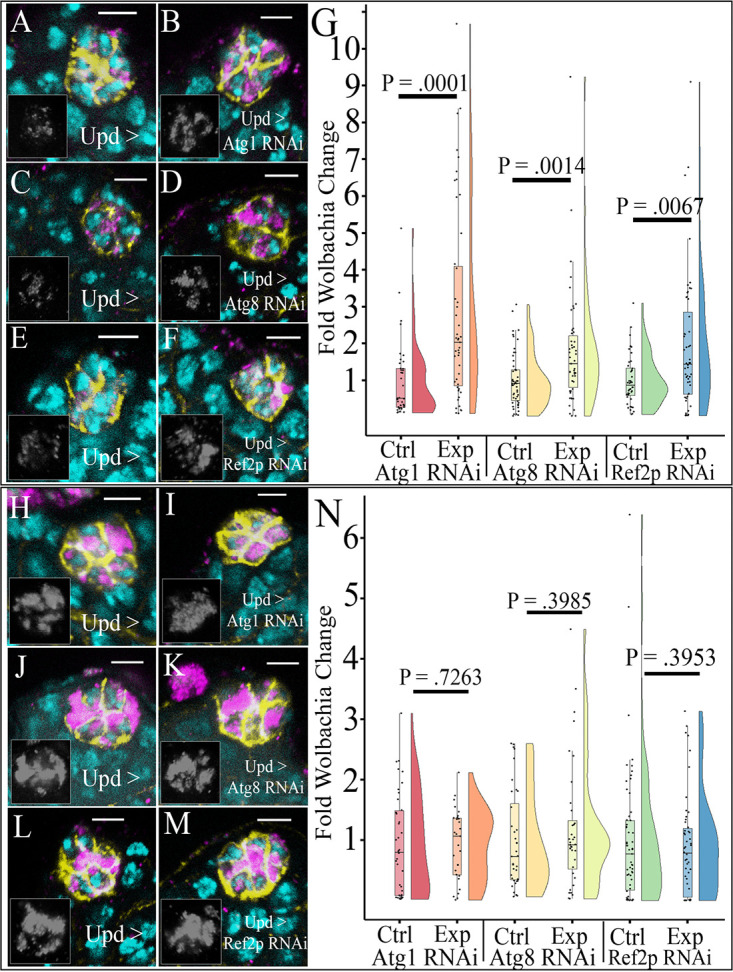
Knockdown of selective autophagy increased density of Wolbachia strain wMel but not wMelCS in the hub. Representative confocal z-stacks of hubs expressing RNAi against autophagy genes. Unpaired (Upd) was used to express small interfering RNAs (siRNAs) specifically in the hub and not male germ line and soma in the testis. DNA is colored cyan, D cadherin (labeling the hub) is yellow, and the HSP60 antibody detecting Wolbachia is in magenta. The insets display grayscale images of only the Wolbachia channel from the respective image. (A) Sibling control hub of wMel-infected male testis displaying Wolbachia at a low density. (B) Knockdown of Atg1 increased wMel density in the hub. (C) Control Atg8 RNAi wMel-infected male hub displaying low Wolbachia density. (D) Knockdown of Atg8 in the hub increased wMel density. (E) Control Ref(2)p RNAi hub of wMel-infected male testis where Wolbachia is at a low density. (F) Knockdown of Ref(2)p with RNAi increases Wolbachia wMel density in the hub. (G) Vertical raincloud plots display each quantified value overlaid on a box and whisker plot showing the median value, upper and lower quartiles (box), and upper and lower extremes (whiskers, 1.5× interquartile range). A split violin plot accompanies each box and whisker plot which displays the probability density function of the data set. Quantification of relative Wolbachia density reveals a significant increase in Wolbachia density upon knockdown of either Atg1 (Ncont = 37, Nexp = 47), Atg8 (Ncont = 54, Nexp = 50), or Ref(2)p (Ncont = 44, Nexp = 52) in the hub. (H) Control Atg1 RNAi hub of wMelCS (CS)-infected male testis where Wolbachia is at a moderate/high density. (I) Knockdown of Atg1 has no significant effect on moderating the density of CS. (J) Control Atg8a RNAi hubs of CS-infected male testis display a moderate/high density. (K) Atg8a knockdown displays a moderate/high density of CS similar to that for the control. (L) Control Ref(2)p hub of wMelCS-infected hubs displays moderate density. (M) Knockdown of Ref(2)p does not result in any change in wMelCS density, displaying moderately infected hub densities. (N) Quantification of relative CS density in the hub shows no difference in density upon knockdown of Atg1 (Ncont = 39, Nexp = 31), Atg8 (Ncont = 36, Nexp = 36), or Ref(2)p (Ncont = 59, Nexp = 50). Scale bars, 5 µM. Mann-Whitney U tests were performed for statistical analysis.
wMelCS, a closely related strain of Wolbachia which resides at higher density than wMel, has been shown to decrease Drosophila susceptibility to death compared to that with wMel from the Drosophila C virus, while both strains induce similar levels of cytoplasmic incompatibility (40, 41). These observations highlight strong phenotypic variability even in closely related bacterial strains. We hypothesized that wMelCS would also be affected by autophagy due to the minimal genomic differences from wMel (41). Upon knockdown of Atg1, we saw no noticeable difference in wMelCS density in the hub (Fig. 1H and I). Quantification showed no discernible difference in densities infecting the hubs, where there was a near even 1.02-fold decrease in average wMelCS density (Fig. 1N). Atg8a knockdown also resulted in no change in wMelCS density within the hub (Fig. 1J and K). Quantification of hubs expressing Atg8a RNA interference (RNAi) showed a nonsignificant 1.18-fold increase in average density of wMelCS (Fig. 1N). It should be noted that there was no difference in Wolbachia tropism (the preferential accumulation of Wolbachia in a specific cell type or tissue) upon modulation of either Atg1 or Atg8a (see Fig. S2A and B in the supplemental material). Lastly, knockdown of Atg1 and Atg8 resulted in increased levels of Ref(2)p protein in the hub compared to that for control staining, indicating an efficient knockdown of autophagy (Fig. S1A to D).
Ref(2)P staining confirms RNAi knockdown of autophagy genes. Ref(2)p is increased in tissues with autophagy knocked down. Image of hubs labeled with genotypes (control on the left, experimental on right) antibody stained for D-cadherin (yellow) and Ref(2)p (magenta). Prime-labeled panels are greyscale of Ref(2)p staining. (A) Control hub displays few small Ref(2)p puncta within the hub. (B) Atg1 RNAi expressed in the hub causes several smaller Ref(2)p puncta. (C) Control flies display several small Ref(2)p puncta within the hub. (D) Expression of Atg8a RNAi in the hub causes an increase in Ref(2)p puncta and size, indicating a block in autophagy. (E, E′) Control flies show a few small Ref(2)p puncta in the hub. (E′) Grayscale of the Ref(2)p channel is shown. (F) Ref(2)p RNAi knocks down expression of Ref(2)p in the hub. (F′) Grayscale image of Ref(2)p channel shows little to no expression of Ref(2)p in the hub. Scale bars, 5 µM. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Autophagy does not affect wMel and CS hub tropism. (A) Knockdown of either Atg1 or Atg8a does not affect the proportions of hubs displaying tropism for strain wMel. (B) Knockdown of Atg1 or Atg8a does not affect the proportions of hubs displaying tropism for the strain CS. (C) Ref(2)p knockdown does not affect hub tropism proportions in either wMel or CS. P values represent proportions test between control and knockdown. Error bars represent 95% confidence intervals. Download FIG S2, PDF file, 2.0 MB (2.1MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The selective autophagy adapter protein Ref(2)p negatively regulates Wolbachia wMel but not wMelCS in the hub.
Selective autophagy has been implicated in regulating several mammalian intracellular bacterial infections (42, 43). We wanted to test if Ref(2)p-mediated selective autophagy was involved in regulating Wolbachia densities in the hub. Upon knockdown of Ref(2)p in the hub, we saw an increase in wMel density (Fig. 1E and F). Quantification of relative Wolbachia density in the hub showed an average 1.94-fold increase in wMel density upon knockdown of Ref(2)p (Fig. 1G).
While our previous data revealed no significant effect of autophagy in regulating wMelCS density, we wanted to confirm that selective autophagy does not affect wMelCS as well. Indeed, Ref(2)p knockdown did not affect wMelCS density in the hub. Representative images displayed no change in hub density in the control or Ref(2)p knockdown (Fig. 1J and K). Quantification of hubs showed an average 1.13-fold decrease in wMelCS density (Fig. 1N). To verify the knockdown of Ref(2)p in the hub, we performed antibody staining and saw reduced Ref(2)p puncta formation compared to that in control hubs (Fig. S1E and F). It should be noted that there was no difference in Wolbachia tropism upon modulation of Ref(2)p (Fig. S2C).
Wolbachia effector protein CifB modulates bacterial density of wMel in the hub.
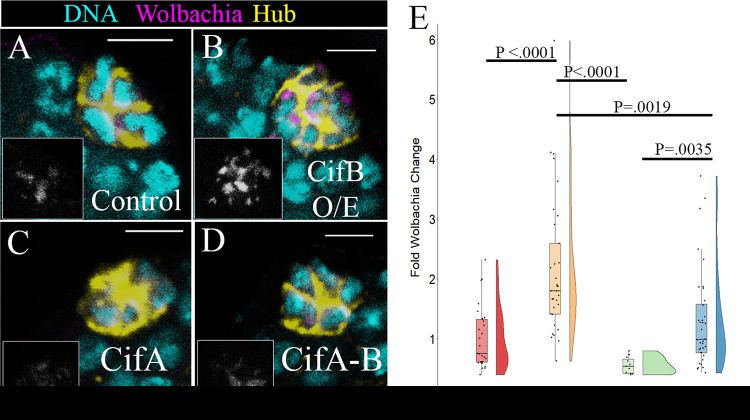
To determine if wMel harbored factors could play a role in regulating wMel density, and because Wolbachia is currently not able to be transformed, we used the Gal4-upstream activation sequence (UAS) system to overexpress (OE) either CifA, CifB, or CifA and CifB together in hub cells. We hypothesized that the similar deubiquitinating activities of CifB and CidB would aid Wolbachia in escape from ubiquitination, detection, and thus destruction by the autophagy system (34, 35, 44).
Representative images show CifB overexpression significantly increased wMel density compared to that in the control (Fig. 2A and B). Quantification of wMel density revealed a 2.39-fold increase in average hub Wolbachia density compared to that in the control (Fig. 2E). Overexpression of CifA resulted in a trend toward reduced Wolbachia density compared to that in control flies but did not reach significance (Fig. 2A and C). Quantification showed a decrease of approximately 1.41-fold in Wolbachia density in the hub (Fig. 2E). Lastly, coexpression of CifA and CifB resulted in no difference in Wolbachia density compared to that in the control (Fig. 2A and D). Quantification showed a 1.31-fold increase in density, but this was not significantly different from the control (Fig. 2E). Interestingly, CifB was significantly different compared to coexpression of CifB and CifA in the hub (Fig. 2B, D, and E). Overall, these results show that CifB acts to positively regulate Wolbachia density in the hub and suggests CifA may partially negatively regulate Wolbachia density.
FIG 2.
Expression of Wolbachia cytoplasmic incompatibility genes CifA and CifB modulate wMel density in the hub. Representative confocal z-stacks of hubs with overexpressed Wolbachia Cif genes. Unpaired (Upd) was used to overexpress Cif constructs specifically in the hub and not male germ line and soma in the testis. DNA is cyan, D cadherin (labeling the hub) is yellow, and a fluorescently labeled DNA probe to detect Wolbachia is in magenta. Grayscale insets display the Wolbachia-only channel. (A) Sibling control hubs displayed low relative wMel density. (B) Overexpression of CifB resulted in higher relative wMel density. (C) Overexpression of CifA in the hub resulted in a trend for lower relative wMel density. (D) Overexpression of both CifA and CifB resulted in wMel hub density similar to that of the control. (E) A Kruskal-Wallis test of significance revealed a significant difference in the data set (P < 0.0001). Quantification of relative wMel hub density showed overexpression of CifA results in a nonsignificant trend for lower wMel densities (Ncont = 29, NCifA = 11, P < 0.0905). Overexpression of CifB results in a significant increase in wMel density compared to that in the control (Ncont = 29, NCifB = 33, P < 0.00001) and to those with both CifA and CifB overexpression (NCifB = 33, NcifA-B = 36, P < 0.0019). Overexpression of both CifA and CifB resulted in no difference from the control (Ncont = 29, NCifA-B = 36, P = 1.0). P values reported for individual comparisons were from a Kruskal-Wallis post hoc Dunn’s test with Bonferroni correction. Scale bars, 5 µM.
Autophagy-CifB epistasis reveals they function in the same pathway.
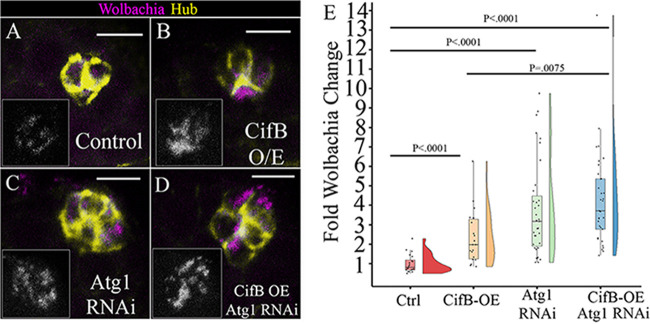
CifB contains a deubiquitinating domain, suggesting it may aid in removal of ubiquitin and help subvert autophagy. To characterize how Wolbachia cifB expression regulates intracellular density, we used epistasis to determine if CifB modulates density through autophagy or an independent host pathway. An additive epistasis model was tested between the Wolbachia gene CifB and Atg1. As previously observed, we expected an approximately 2-fold increase when only CifB was expressed and a 3-fold increase when only Atg1 RNAi was expressed. If both are expressed in the same hub and we see an approximately 5-fold increase, then both constructs are working in independent pathways to modulate Wolbachia density. If the hub Wolbachia density phenotype is similar to that of one of the constructs, then that would reveal that these genes work in the same pathway.
When we overexpressed CifB, there was a significant 2.33-fold increase in average wMel hub density (Fig. 3A, B, and E). When Atg1 RNAi was expressed in hubs, there was a significant 3.89-fold increase in Wolbachia density (Fig. 3A, C, and E). When both constructs were expressed in the hub, we again saw an increase in wMel density (Fig. 3A and D), and quantification revealed a 4.3-fold increase in wMel density, which was significantly different from that in the control (Fig. 3E). Compared to that with CifB alone, coexpression of Atg1 RNAi-CifB OE was also significantly different (Fig. 3E). Compared to that when Atg1 RNAi was expressed alone, there was no statistical difference between the groups (Fig. 3E). This genetic analysis supports that CifB acts in the autophagy pathway. If they acted in different pathways, we would expect to see an additive effect of an approximately 6.22-fold increase from the control, as that would be the sum of CifB (2.33-fold) and Atg1 RNAi expression (3.89-fold) together, and this would lead to CifB-Atg1 RNAi coexpression being significantly different than both CifB and Atg1 RNAi alone.
FIG 3.
Epistasis analysis of Wolbachia CifB and Atg1 genes reveal Wolbachia effector CifB acts in the autophagy pathway. Representative confocal z-stacks of hubs. Unpaired (Upd) was used to express siRNAs or Cif constructs specifically in the hub and not male germ line and soma in the testis. DNA was not acquired for this experiment; D cadherin labeling the hub is yellow, and a fluorescently conjugated DNA probe to detect Wolbachia is in magenta. Grayscale insets display the Wolbachia-only channel. (A) Control hubs displayed low relative wMel density. (B) Overexpression of CifB resulted in higher relative wMel density. (C) Expression of Atg1 RNAi in the hub resulted in higher relative wMel density. (D) Overexpression of both CifB and Atg1 RNAi resulted in high wMel hub density, similar to that with Atg1 RNAi. (E) A Kruskal-Wallis test of significance revealed a significant difference in the data set (P < 0.0001). Quantification of relative wMel hub density showed overexpression of CifB results in a statistically significant increase in wMel density (Ncont = 20, NCifB = 22, P < 0.0001). Expression of Atg1 RNAi result in a significant increase in wMel density (Ncont = 29, NAtg1RNAi = 35, P < 0.0001). Coexpression of CifB and Atg1 RNAi results in a significant increase in wMel density compared to that in the control (NCont = 29, NCifB-Atg1RNAi = 33, P < 0.0001) and with CifB alone (NCifB-Atg1RNAi = 33, NCifB = 22, P < 0.0075) and a density similar to that with Atg1 RNAi expression alone (NAtg1RNAi = 35, NCifB-Atg1RNAi = 33, P = 1.0). P values reported for individual comparisons were from a Kruskal-Wallis post hoc Dunn’s test with Bonferroni correction. Scale bars, 5 µM.
Autophagy positively regulates Wolbachia density in the female germ line.
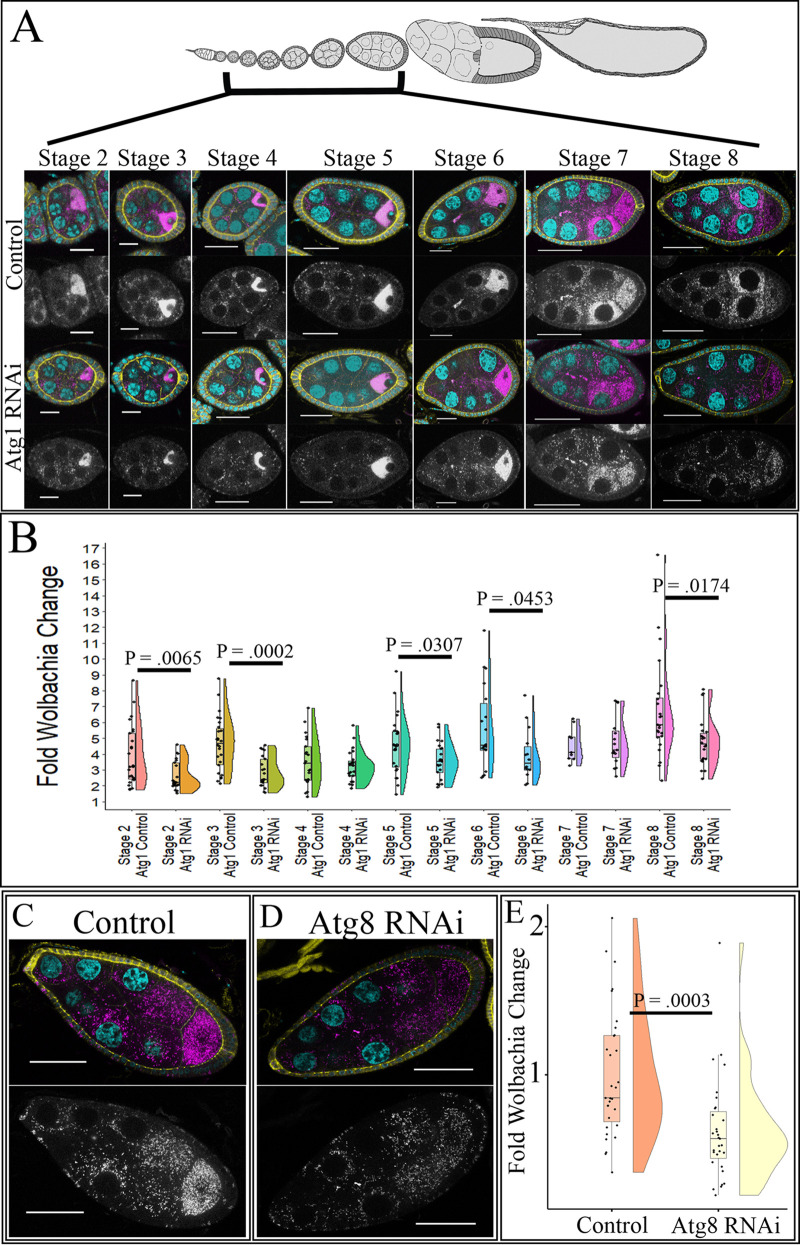
To investigate the effects of autophagy on Wolbachia density in the female germ line, we knocked down Atg1 and Atg8a in only the germ line and quantified relative Wolbachia density in the germ line and in the surrounding follicle cells. Quantitative reverse transcriptase PCR (RT-qPCR) of whole ovaries determined that Atg1 was knocked down 77% when Atg1 RNAi was expressed under the NGT;nos-Gal4 driver (see Fig. S3A). Using confocal microscopy, the Wolbachia density in egg chambers between stages 2 and 8 was quantified. In wMel-infected ovaries, stages 2, 3, 5, and 8 displayed 1.47-, 1.60-, 1.28-, and 1.4-fold decreases in average Wolbachia density, respectively (Fig. 4A and B). To confirm the role of autophagy in regulating wMel density in the female germ line, we knocked down Atg8a. In this experiment, we used a stronger germ line driver, the maternal triple driver (MTD), because the knockdown efficiency utilizing NGT;nos was not >50% (Fig. S3B). Upon knockdown of Atg8a in the female germ line, we saw a 1.60-fold decrease in average relative Wolbachia density in stage-8 egg chambers (Fig. 4C to E). Lastly, as starvation has been shown to increase autophagy, we investigated if starvation could drive a larger difference in wMel density in Atg1 RNAi ovaries. Whole-ovary qPCR analysis revealed that there was a significant decrease in the density of wMel compared to that in the control, but the difference was similar to what was observed for well-fed flies (see Fig. S4).
FIG 4.
Knockdown of autophagy in the female germ line decreases wMel density. Stage-specific confocal analysis reveals a decrease in relative Wolbachia density upon knockdown of autophagy genes during multiple stages of development. NGT;nos was used to knockdown Atg1, and MTD was used to knockdown Atg8a. DNA is colored cyan, D cadherin (labeling the follicle cells) is yellow, and a fluorescently conjugated DNA probe detecting Wolbachia is in magenta. Grayscale images display the Wolbachia-only channel. (A) Representative confocal z-stack images of control and Atg1 knockdown in the germ line of wMel-infected flies. (B) Quantification of wMel-infected stage-specific egg chambers upon Atg1 knockdown. Statistically significant P values are reported only. (C) Representative confocal z-stack images of wMel-infected stage-8 control egg chambers show high levels of germ line Wolbachia. (D) Representative confocal z-stack image of stage-8 egg chambers with Atg8 RNAi expression displays reduced wMel density in the germ line. (E) Quantification of wMel-infected stage-8 egg chambers for the control and Atg8 knockdown. Scale bars, 10 µM for stages 2 and 3, 20 µM for stages 4 to 6, and 40 µM for stages 7 and 8. Student’s t tests were conducted to determine significance.
Knockdown efficiency of autophagy-related mRNA in the ovaries upon RNAi expression. RT-qPCR used to determine knockdown efficiency of different RNAi constructs in whole female ovaries. Genes were normalized to RPL32 expression. (A) Two replicates of 20 ovary pools show a knockdown efficiency of 64%. (B) Three replicates of 20 ovary pools shows an Atg8a knockdown efficiency of 47% when the NGT;nos driver was used. (C) Three replicates of 20 ovary pools show a Ref(2)p knockdown efficiency of 87%. Download FIG S3, PDF file, 0.8 MB (782.4KB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Starvation does not modify the effect of autophagy knockdown on Wolbachia density in the germline. Quantitative PCR of WSP normalized to 14-3-3 of 20 whole ovaries from 10 females. (A) Whole-ovary quantitative PCR of Wolbachia density revealed significant decreases in Wolbachia density upon knockdown of Atg1 under well fed and 2-day starvation conditions. Four-day starvation resulted in a similar trend in reduced Wolbachia density but was not significant. P values represent paired Student’s t test results. Download FIG S4, PDF file, 1.9 MB (2.3MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
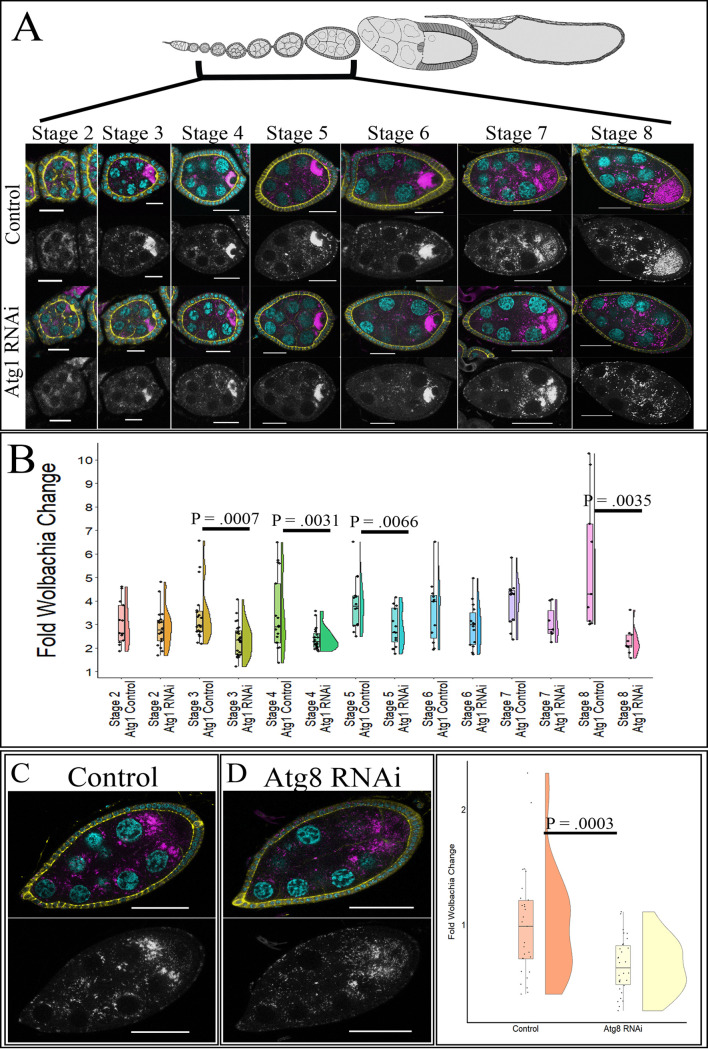
Unlike the hub, autophagy knockdown in the female germ line affected wMelCS density as well. Upon expression of Atg1 RNAi in the germ line by the NGT;nos driver, we saw a stage-specific reduction in wMelCS density. Stages 3, 4, 5, and 8 displayed 1.44-, 1.44-, 1.37-, and 2.47-fold decreases in average Wolbachia density, respectively (Fig. 5A and B). To confirm that autophagy positively regulated wMelCS density in the female germ line, we used the MTD driver to express Atg8a RNAi in the germ line and quantified relative wMelCS levels in stage-8 egg chambers. There was a 1.55-fold reduction in average Wolbachia density upon Atg8a knockdown in the germ line (Fig. 5C to E).
FIG 5.
Knockdown of autophagy in the female germ line decreases wMelCS density. Stage-specific confocal analysis reveals a decrease in relative Wolbachia density upon knockdown of autophagy genes during multiple stages of development. NGT;nos was used to knockdown Atg1, and MTD was used to knockdown Atg8a. DNA is colored cyan, D cadherin (labeling the follicle cells) is yellow, and a fluorescently labeled DNA probe detecting Wolbachia is in magenta. Grayscale images display the Wolbachia-only channel. (A) Representative confocal z-stack images of control and Atg1 knockdown in the germ line of wMelCS-infected flies. (B) Quantification of wMelCS-infected stage-specific egg chambers upon Atg1 knockdown. Statistically significant P values are reported only. (C) Representative confocal z-stack images of wMelCS-infected stage-8 egg chambers for control flies display a high Wolbachia density. (D) Representative confocal z-stack images of wMelCS-infected stage-8 egg chambers with Atg8 RNAi expressed display reduced germ line Wolbachia density. (E) Quantification of wMelCS-infected stage-8 egg chambers for the control and Atg8 knockdown reveal decreased density upon germ line expression of Atg8 RNAi. Scale bars, 10 µM for stages 2 and 3, 20 µM for stages 4 to 6, and 40 µM for stages 7 and 8. Student’s t tests were conducted to determine significance.
Ref(2)p-dependent selective autophagy does not affect Wolbachia density in the female germ line.
To determine if Ref(2)p-dependent selective autophagy regulates Wolbachia density in the female germ line, similar to that in the hub, we knocked down Ref(2)p in the germ line and quantified relative Wolbachia density for both wMel and wMelCS. Utilizing the NGT;nos driver, we were able to obtain an 86% knockdown efficiency, indicating a robust knockdown of Ref(2)p (Fig. S3C). Upon knockdown of Ref(2)p in the germ line, we saw no change in wMel density in stage-8 egg chambers (Fig. 6A and B). Quantification showed a nonsignificant 1.11-fold increase in Wolbachia density (Fig. 6C). Whole-ovary qPCR showed a nonsignificant 1.05-fold decrease in wMel density, confirming our image analysis (see Fig. S5A).
FIG 6.
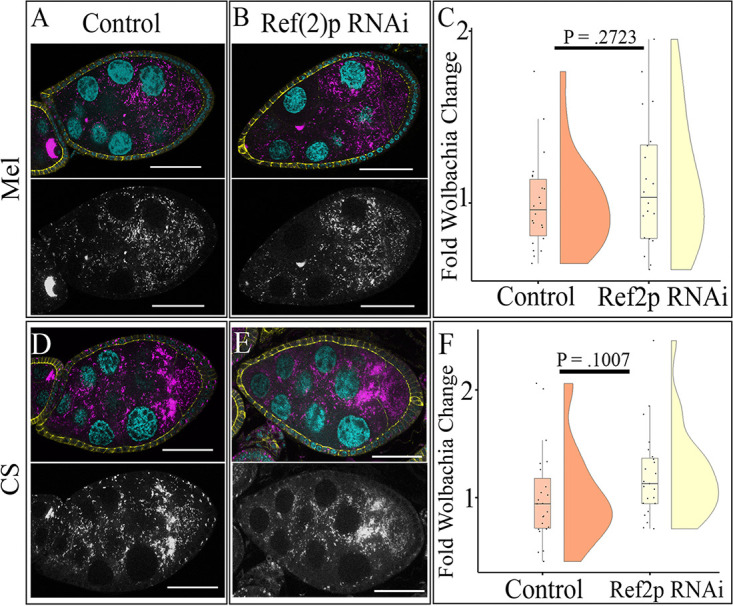
Ref(2)p does not regulate Wolbachia density in stage-8 egg chambers. (A) Representative confocal z-stack image of wMel-infected control stage-8 egg chamber shows moderate density of Wolbachia. NGT;nos was used to drive knockdown of Ref(2)p. DNA is colored cyan, D cadherin (labeling the follicle cells) is yellow, and a fluorescently labeled DNA probe to detect Wolbachia is in magenta. Grayscale images display the Wolbachia-only channel. (B) Representative confocal z-stack image of wMel-infected stage-8 egg chamber with Ref(2)p knocked down shows similar moderate density to that of the control. (C) Quantification of relative germ line Wolbachia density reveals no difference in density upon the expression of Ref(2)p RNAi. (D) Representative confocal z-stack image of wMelCS-infected control stage-8 egg chamber shows high germ line density. (E) Representative confocal z-stack image of wMelCS-infected stage-8 egg chamber with Ref(2)p knocked down displays similar germ line density to that of the control. (F) Scale bars, 40 µM. P values determined by Student’s t test.
Quantitative PCR of Ref(2)p RNAi mutants reveals no change in Wolbachia density in the female germline. (A) Quantitative PCR of wMel-infected ovaries reveals no change in density upon the knockdown of Ref(2)p. Five replicates of 20 ovary pools were used to determine average Wolbachia density. (B) Quantitative PCR of wMelCS-infected ovaries reveals no change in density upon the knockdown of Ref(2)p. Two replicates of 20 ovary pools were used to determine average Wolbachia density. Paired Student’s t tests were used for statistical analysis. Download FIG S5, PDF file, 0.6 MB (600KB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We also tested the effect of a knockdown of Ref(2)p on wMelCS. Image analysis revealed a nonsignificant 1.21-fold increase in wMelCS density in the female germ line (Fig. 6D to F). Furthermore, qPCR of whole ovaries with Ref(2)p knockdown showed a 1.07-fold decrease in wMelCS density (Fig. S5B). These data support a mechanism by which Ref(2)p does not influence either wMel or wMelCS density in the germ line cell autonomously.
Wolbachia cif genes do not impact Wolbachia density in the female germ line.
We explored whether CifA and CifB regulate wMel density in the female germ line, a tissue functionally relevant for cytoplasmic incompatibility. Previous results from the hub indicated that CifB modulates wMel density through autophagy. Since Ref(2)p has no effect in the female germ line, we expected no changes in density upon overexpression of either cifA or cifB. We overexpressed either cifA or cifB independently or together to determine their role in modulating wMel density in stage-8 egg chambers. Overexpression of cifB resulted in a nonsignificant reduction of wMel density of approximately 1.29-fold. Of note, this was similar to the level seen in autophagy knockdowns (see Fig. S6A, B, and E). Overexpression of either cifA alone or cifA and cifB together resulted in no change in Wolbachia density compared to that in the control. cifA alone displayed a fold change of 1, indicating no change in density (Fig. S6A, C, and E). cifA and cifB coexpression resulted in no significant increase in density (Fig. S6A, D, and E).
CifA and CifB overexpression do not affect wMel density in the female germline. NGT;nos was used to overexpress Cif constructs. DNA is colored cyan, D cadherin (labeling the follicle cells) is yellow, and a fluorescently conjugated DNA probe detecting Wolbachia is in magenta. (A) Control stage-8 egg chambers show high germline Wolbachia densities. (B) Stage-8 egg chambers overexpressing CifB result in a nonsignificant trend for lower relative wMel density. (C) Stage-8 egg chambers with overexpressed CifA result in a Wolbachia density similar to that in control egg chambers. (D) Stage-8 egg chambers overexpressing both CifA and CifB result in germline Wolbachia densities similar to that of the control. (E) A one-way ANOVA revealed a significant difference in the data set (P = 0.0171). A post hoc Tukey’s analysis revealed a significant difference between CifB overexpression and CifA-CifB overexpression groups (NCifB = 30, NCifA-CifB = 26, P = 0.024). Of note, CifB showed a trend for reduced wMel density in the germline compared to that in the control (NCont = 27, NCifB = 30, P = 0.0704). Grayscale panels display the Wolbachia-only channel. Scale bars, 40 µM. Download FIG S6, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Global metabolomics identifies dysregulation of carbohydrate and glycerolipid metabolism in Wolbachia-infected autophagy mutant ovaries.
To attempt to address how autophagy may regulate Wolbachia density in the female germ line, we performed global metabolic profiling of autophagy knockdown and control ovaries in wMel-infected and uninfected flies. Three replicates of four different samples, including uninfected wild type (w_Ctrl), uninfected Atg1 RNAi (w_mut), wMel-infected wild type (mel_ctrl), and wMel-infected Atg1 RNAi (mel_mut), were subject to liquid-liquid extraction tandem solid-phase microextraction (LLE-SPME) and run on Boston University’s Center for Network Systems Biology nanoscale liquid chromatography-mass spectrometry (nanoLC/MS) platform. Positive ion mode was used for sample comparison, and 10,251 features were detected in total displaying a metabolome drift (see Fig. S7A; Table S1). Principal-component analysis (PCA) revealed that PC1 accounts for 40% of the variation and clustered infected and uninfected ovaries well. PC2 accounted for 10% of the variation, showing a weak clustering of wMel-infected ovaries with and without autophagy knocked down (Fig. S7B). The PCA showed that infection contributed mostly to the variance rather than the autophagy knockdown. Features were then implemented into MetaboAnalyst’s fast gene set enrichment analysis (fGSEA) platform to identify dysregulated pathways between samples (Table S1).
Wolbachia infection and autophagy knockdown drive different metabolic profiles. (A) Heat map indicating Z-score values for each detected positive ion mode metabolite. (B) Principal-component analysis of all positive ion mode-detected features. Download FIG S7, PDF file, 2.4 MB (2.4MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Positive ion metabolites and differentially regulated pathways. Download Table S1, XLSX file, 6.9 MB (8.2MB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
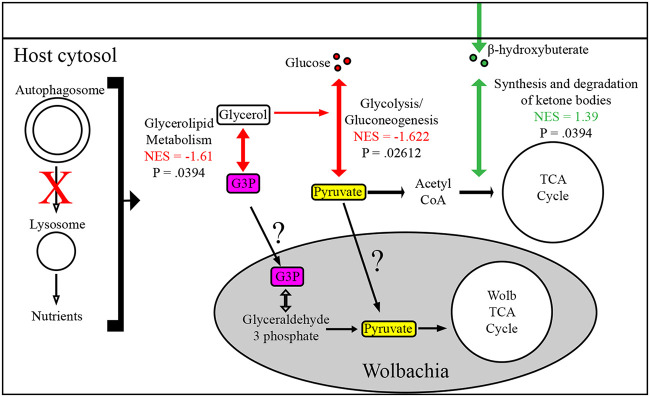
Nine pathways were significantly dysregulated between Wolbachia-infected and uninfected ovaries, with seven pathways positively enriched and two pathways negatively enriched in Wolbachia-infected ovaries (Table S1). Pathways which can support central carbon metabolism were positively enriched and include pyruvate metabolism (normalized enrichment score [NES] = 1.946, P = 0.001), glycine, serine, and threonine metabolism (NES = 1.722, P = 0.005), butanoate metabolism (NES = 1.706, P = 0.006), propanoate metabolism (NES = 1.623, P = 0.017), citrate cycle (NES = 1.644, P = 0.019), d-glutamine and d-glutamate metabolism (NES = 1.562, P = 0.024), and glyoxylate and dicarboxylate metabolism (NES = 1.538, P = 0.026). Sphingolipid metabolism was negatively enriched (NES = −1.958, P = 0.003) and the pentose phosphate pathways was negatively enriched (NES = −1.510, P = 0.035). Interestingly, these data suggest central carbon metabolism is altered in Wolbachia-infected ovaries, with an increase in pyruvate metabolism and pathways which can support both pyruvate metabolism and the citrate cycle while reducing the pentose phosphate pathway, which compete with glycolysis for glucose-6-phosphate to generate nucleotides and NADPH (45).
We investigated how knocking down autophagy in the presence of Wolbachia altered metabolic pathways which could lead to a decrease in Wolbachia density as previously seen. Under these circumstances, there were only three significantly dysregulated pathways, with one carbohydrate and two lipid metabolism pathways dysregulated (Table S1). Glycolysis (Fig. 7) (NES= −1.622, P = 0.026) was negatively enriched in autophagy mutant ovaries. For lipid metabolism, the synthesis and degradation of ketone bodies was positively enriched (Fig. 7) (NES = 1.39, P = 0.039) and glycerolipid metabolism was negatively enriched (Fig. 7) (NES = 1.61, P = 0.039). Interestingly, data suggest Wolbachia may compete for glycerol-3-phosphate and/or pyruvate from the host for energy, which are metabolites prevalent in our observed downregulated pathways (46, 47). Overall, these data reveal that autophagy mutants, when in the presence of Wolbachia, reduce glycolysis and glycerolipid metabolism, which could restrict Wolbachia density through limited accumulation of essential metabolites.
FIG 7.
Differentially regulated metabolic pathways in Atg1 RNAi mutant ovaries of wMel-infected flies in the context of a metabolic model. All significantly differentiated pathways are reported between wMel-infected ovaries with autophagy knocked down (Atg1 RNAi) and the wild type. Downregulated metabolic pathways are highlighted with red, while upregulated pathways are green. Results are reported in the context of a proposed model of how they interact and could affect Wolbachia. NES, normalized enrichment score from fGSEA MetaboAnalyst results.
DISCUSSION
Autophagy can act as an innate immune response aiding in the removal of pathogenic bacteria and viruses, but its role in host-endosymbiont interactions remains less understood. Previous results indicated that autophagy negatively regulated Wolbachia in systems predominantly composed of somatic cells, while known autophagy-inducing drugs increased Wolbachia density in the female germ line, leading to confusion in understanding the role of autophagy in modulating Wolbachia levels (16, 17). Here, we used systematic and comparative genetic approaches to address this discrepancy and determined that autophagy does modulate Wolbachia levels differentially in male somatic cells and female germ line cells. Moreover, we have identified bacterial proteins which modulate the interaction between Wolbachia and autophagy.
Our results show that Ref(2)p-mediated selective autophagy is responsible for negatively regulating wMel but not wMelCS in the hub. Since wMelCS resides at naturally higher densities than wMel, this result suggests that wMelCS evolved a mechanism to subvert host autophagy to aid in survival (discussed further below). These results do not completely agree with previous data that described wMelPop, a pathogenic strain more closely related to wMelCS than wMel, being negatively related by autophagy. In that study, rapamycin treatment reduced Wolbachia density in whole larva, and Atg1 RNAi reduced density in Drosophila PC15 cells (16). For whole larval in vivo studies performed by Voronin et al. (16), the use of rapamycin, a TORC1 inhibitor, affects several different host-related pathways, including ribosome biogenesis, translation, and nutrient import, and has also been shown to stimulate ubiquitin-proteasome degradation (48). Proteasome degradation has been shown to support increased Wolbachia density, and this mechanism could possibly explain additional mechanisms by which rapamycin is capable of modulating several aspects of host biology which regulate Wolbachia accumulation (13). The close ties of autophagy and the ubiquitin proteasome should be teased apart in future studies to determine how each pathway exerts changes in Wolbachia density.
Beyond host biological mechanisms which could cause Wolbachia strain-specific density changes, bacterial derived differences could explain our observed strain-dependent subversion of host autophagy, including higher expression levels of bacterial effector proteins, which have been shown to aid in bacterial escape from autophagy (43). We tested a known functional effector of wMel, CifB, which contains a ubiquitin-like protease domain (Ulp1) and has a moderate preference for lysine 63 ubiquitin chains (K63) over K48 (34, 49). Of note, K63 has been associated with P62-mediated selective autophagy in mammalian systems compared to K48, which is well characterized to be involved in proteolysis (50). Overexpression of CifA and CifB proteins individually showed CifB expression positively regulated wMel density, while CifA expression showed a trend toward reducing wMel density. When coexpressed, CifA and CifB showed no difference from the control. These results suggest that the deubiquitinating function of CifB could be protective for Wolbachia in cells in which autophagy negatively restricts higher densities, while CifA is antagonistic to CifB function, since CifA on its own showed a modest reduction in density and eliminated the benefit CifB expression had on wMel density. These results partially agree with what has been reported previously (33). In the study by LePage et al. (33), qPCR of male testis overexpressing CifA or CifB in the germ line independently increased Wolbachia density, while overexpression of both constructs recapitulated CifA density increases. This observation disagrees with our CifA observation but agrees with our CifB observation and could be driven by a cell type-dependent interaction of host-bacterial proteins which can modify bacterial density that is still currently unknown.
Our follow-up CifB-autophagy epistasis analysis revealed that CifB functions in the autophagy pathway. When we expressed CifB or Atg1 RNAi individually, wMel density increased. When coexpressed, density recapitulated Atg1 RNAi levels and not a summation of CifB and Atg1 RNAi levels. It should be noted that the hub can sustain extremely high densities of Wolbachia when infected with wMelPop (∼50× more than in surrounding tissue), even to the point of rupturing the hub cell plasma membrane, and so we believe modulating these pathways does not create a scenario where the hub cannot support an additive model density (37).
Biochemically, our data partially agree with what has been shown with the mosquito CifB homologue, CidB, for which extensive biochemical characterization has been done (34). Beckmann et al. (34) showed that when CidB is expressed in Drosophila males, it drives the cytoplasmic incompatibility (CI) phenotype, and this is abolished if CidB has a single amino acid change creating a catalytically dead DUB mutation. To biochemically characterize this protein’s function, in vitro assays and yeast studies were performed. CidB was shown to drive toxicity in yeast, and this was DUB domain dependent, as a catalytically dead mutant did not drive toxicity. CidA, the CifA homologue, was shown to bind CidB biochemically, and when expressed in yeast, it rescued CidB toxicity. This agrees with our model that CifB may rescue wMel through its deubiquitinase activity and that CifA can block this. Interestingly, when in vitro ubiquitin cleavage assays were performed, CidB either alone or when coexpressed with CidA was capable of cleaving ubiquitin 48 (K48) or 63 chains, providing evidence that CidA does not directly block CidB DUB activity. Conversely, in yeast, the ubiquitin profile was not changed drastically when CidB was overexpressed, showing that further analysis must be completed to understand the exact function and that the result may be host or experiment specific (34, 35, 44, 49).
Autophagy has been implicated in male-derived sterility, with Ref(2)p homozygous mutants in Drosophila being described as male sterile (51). Our data implicating CifB in modulating Ref(2)p-mediated selective autophagy highlight a possible host-derived mechanism by which CI proteins drive sterility (33, 34, 36). From our studies, we are not proposing that CifA and CifB expression in the male hub directly affects male germ line Ref(2)p-mediated selective autophagy, but rather, our hub studies highlight CifB interacting with selective autophagy and that this should be explored in the male germ line. It should be recognized that the PD-(D/E)xK nuclease domains found in CifB, CidB, and other gene paralogs found in several Wolbachia strains are thought to be sufficient to drive CI in flies, indicating ubiquitinase activity may not be the only mechanism Wolbachia utilizes to drive CI (52).
Previous literature showed that wMelCS and wMelPop reside at higher densities than wMel. wMelPop’s pathogenic overreplication has been elegantly shown to have a strong correlation with the octomom region of its genome, but it may still possess the capabilities to subvert autophagy in vivo similar to wMelCS (53). Since wMelCS is not regulated by Ref(2)p-dependent selective autophagy, this suggests that these closely related strains have evolved additional mechanisms to subvert the autophagy pathway. NCBI’s conserved domain database and previous genomic annotations of Wolbachia highlight wMel as having several effector proteins which contain operational taxonomic unit (OTU) and Ulp1 domains, which have been shown to modulate host ubiquitination (31, 32, 41). WD0443, a protein which harbors an OTU domain, has a nonsynonymous amino acid change (R119C) between wMel and wMelCS which could influence protein function (41). Additionally, an interesting candidate to study is the hypothetical Wolbachia protein WD0026, which has been predicted to be a secreted effector of Wolbachia and contains a Ulp1 domain (54). Expression level and/or coding differences in Wolbachia effector proteins may drive changes in host-Wolbachia interactions and should further be validated to begin to elucidate Wolbachia-host interactions and may be involved in host ubiquitin modulation and possibly autophagy subversion.
In the female germ line, a nutrient-sensitive tissue, autophagy positively regulated both wMel and wMelCS. This was independent of Ref(2)p, implicating an association between bulk autophagy and Wolbachia. It should be noted that Ref(2)p RNAi achieved an 86% knockdown efficiency, and the remaining 14% expression could rescue an observed phenotype or indicate only modest reduction at the protein level; thus, this negative result should be interpreted with extra caution. Overall though, these results support previous data where flies fed rapamycin displayed higher density in the female germ line (17). Genetic analysis in that paper implicated a mechanism by which TOR signaling and possibly autophagy in surrounding follicle cells may modulate germ line Wolbachia density. Direct genetic modulation of autophagy was never performed in the female germ line or soma to determine if follicle cell autophagy modulated germ line Wolbachia density. Our results show direct evidence that cell-autonomous female germ line autophagy is capable of supporting strain-independent Wolbachia growth. Even though evidence pointed to bulk autophagy regulating Wolbachia in the germ line, we tested if CifA and CifB effectors could modulate density. Expression of CifA and CifB alone had no effect on Wolbachia density, but CifB expression did result in a trend for reduced Wolbachia density similar to autophagy knockdown. Coexpression of CifA and CifB also resulted in no difference in Wolbachia density. Since selective autophagy does not affect Wolbachia in the female germ line and CifB overexpression trends toward recapitulating the effect of autophagy knockdown, this may suggest CifB disrupts ubiquitin signaling involved in bulk autophagy (or a non-Ref(2)p-dependent form of selective autophagy) to target substrates for degradation and thus disrupts the beneficial effects autophagy provides for Wolbachia. Extensive biochemical analysis needs to be conducted to confirm which host ubiquitin substrate may be targeted by CifB.
Lastly, we performed global metabolomics to begin to identify what metabolic pathways may be dysregulated upon autophagy knockdown that are responsible for a reduction in Wolbachia density. It should be noted in the Drosophila female germ line autophagy is dispensable for proper egg formation, does not change fecundity, and does not influence egg hatching but remains active in the germ line, postulating that it may contribute to optimal metabolism in a metabolically demanding tissue (55). Previous metabolomics analysis of Drosophila ovaries revealed high levels of phosphoarginine, an energy reserve metabolite used to regulate ATP levels as well as increases in various lipid metabolites compared to that in other organs, highlighting increased lipid metabolism (56). In Drosophila, autophagy has been associated with glycogen breakdown in the fat body and amino acid metabolism through epigenetic modulation by a histone methyltransferase under starvation conditions in whole flies, but direct measurement of the metabolome in autophagy mutants has yet to be reported (57, 58). In RAS-driven cancer cell lines, glycolysis has been positively linked to autophagy, while in liver cancer cell lines, selective autophagy negatively regulates glycolysis through selective degradation of hexokinase 2 (57, 59, 60). These results highlight a complex relationship between these pathways which may be system and cell type dependent.
In Wolbachia-infected ovaries with autophagy knocked down, metabolomics analysis revealed a reduction in glycolysis/gluconeogenesis and glycerolipid metabolism. Interestingly, the density of Wolbachia infecting Brugia malayi has been coupled to host glycolysis and pyruvate levels, indicating that autophagy-induced reduction in glycolysis could lead to an unfavorable growth environment for Wolbachia (16, 47). It should also be noted that glycerol-3-phosphate is a member of the glycerolipid metabolism pathway, and Wolbachia has a predicted transporter and ability to convert glycerol-3-phosphate into a functional metabolite in Wolbachia glycolysis (46). Remarkably, there were no differentially regulated amino acid metabolism pathways, and Wolbachia has been shown to have extensive metabolism related to host amino acid sequestration. This does not rule out Wolbachia utilizing host amino acids as an energy source but, rather, supports additional host metabolites which Wolbachia may utilize.
The data described here support a working model for fundamental understandings that cell type can define the role autophagy has in interacting with intracellular microbes (Fig. 8). In static cell types where selective autophagy is the predominant autophagy pathway, bacteria may be negatively impacted if they are recognized by the host and subsequently targeted for degradation. In dynamically growing cell types which are highly nutrient sensitive, such as the developing eggs of Drosophila females, the bulk autophagy pathway may be predominant. These underlying biological characteristics of cell types have rarely been addressed both in the host-pathogen and host-endosymbiont fields. We provide additional evidence for the role of deubiquitinating enzymes in bacterial survival within a host cell. Specific to the Wolbachia field, CifA and CifB are essential proteins to study because of their ability to drive CI, a highly parasitic reproductive phenotype which is imperative for establishing Wolbachia-infected mosquitoes in the wild. CI provides a selective advantage for the establishment of Wolbachia-infected mosquitoes which are then capable of reducing human pathogens such as Zika and dengue viruses. Our characterization of CifA and CifB and identification of CifB functions in the autophagy pathway are important to further understand how these proteins could interact with Ref(2)p in driving male sterility and to modulate intracellular bacterial titers, which may indirectly play a role in pathogen blocking through boosting Wolbachia density rather than interacting directly with the pathogens (3–5, 51).
FIG 8.
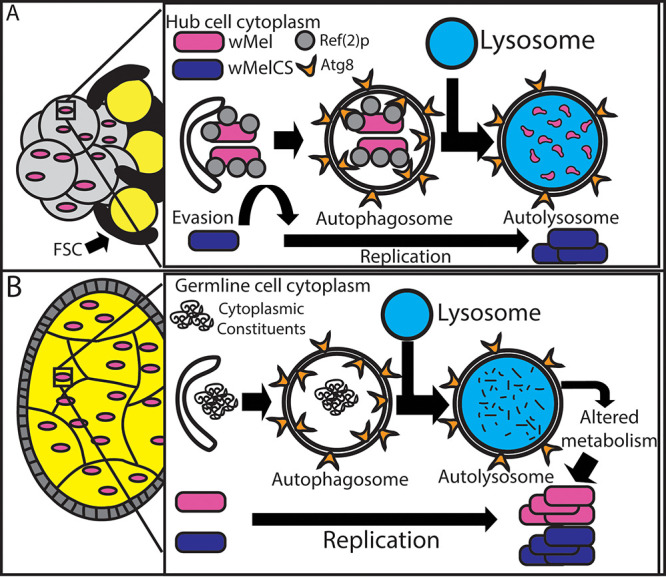
Model of how autophagy regulates Wolbachia density differently in the germ line and somatic cell types. (A) Depiction of how autophagy regulates Wolbachia density in the hub in a strain-dependent manner. wMel is shown to be negatively regulated by selective autophagy. (B) In the germ line, Wolbachia is able to subvert Ref(2)p-mediated selective autophagy. Knockdown of autophagy proteins involved in bulk autophagy results in a decrease in Wolbachia density. This indicates a possible mechanism by which Wolbachia utilizes autophagy-derived nutrients for energy.
MATERIALS AND METHODS
Fly maintenance and stocks.
For information on specific fly strains, including their full genotype and source reference, see Table S2 in the supplemental material. All fly crosses were maintained at 25°C and reared on a mixture of molasses, cornmeal yeast, and agar supplemented with active dry yeast pellets. For knockdown of autophagy in the hub and polar cells, Wolbachia-infected virgin females with the unpaired (Upd) Gal4 driver (Upd;;) were crossed to males with an autophagy RNAi construct. Autophagy RNAi constructs were balanced with either curly (CyO), tubby humeral (TM6B), or MKRS as indicated in Table S2. In the germ line, Wolbachia-infected females had the nanos Gal4-tubulin and nanos Gal4 drivers on the second and third chromosomes, respectively (;NGT;nos). Virgin females were crossed to males which had UAS-autophagy RNAi constructs balanced as previously described. In both soma and germ line crosses, F1 flies inheriting the balancer were used as a control to which the autophagy knockdown siblings were compared, allowing for flies from the same parental cross to be compared. F1 offspring were allowed to age 7 days before testis or ovary dissection. In germ line analysis, 10 control and 10 experimental female flies were housed in vials with 10 male flies, and flies were flipped to new food on day 4. Subsequently, tissue was fixed, stained, and imaged according to descriptions below.
Drosophila fly stock genotypes used in experiments. Download Table S2, XLSX file, 0.01 MB (10KB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Dissection, fixation, and staining protocols for cell types and detecting Wolbachia.
Tissues were dissected and fixed for 20 min in a 4% formaldehyde solution (EMS) with Grace’s insect medium (Lonza catalog no. 04-457F) and 0.2% Triton X-100 (Sigma-Aldrich). For the autophagy knockdown hub analysis (Fig. 1), antibody staining was performed according to references 37 and 61. Mouse anti-HSP60 antibody (1:100; Sigma-Aldrich) was used to detect Wolbachia, and rat anti-D cadherin antibody (DCAD2, concentrated, 1:200; DSHB) was used to visualize the hub. Alexa Fluor secondary antibodies were used to visualize primary antibodies (Invitrogen). Cif hub experiments and female germ line experiments utilized a modified antibody in situ protocol previously described in reference 62. The rat anti-D cadherin antibody was used to mark the hub and, in the female germ line, the boundary between somatic cells and germ line cells. In germ line staining, control and experimental tissues were dissected and fixed and underwent antibody staining in separate tubes. Different secondary antibody fluorophores were used (goat anti-rat IgG 488 [experimental], goat anti-rat IgG 633 [control]) to later identify experimental and control tissues. After antibody staining, control and experimental tissue were combined to be subject to the same exact in situ protocol for Wolbachia detection. Two Wolbachia probes labeled with Cy3 at the 5′ end were used: Wpan16S887 5′-ATCTTGCGACCGTAGTCC-3′ and Wpan16S450 5′-CTTCTGTGAGTACCGTCATTATC-3′. Hybridization was performed at 37°C in 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 250 mg/liter salmon sperm DNA, 0.5× Denhardt’s solution, 20 mM Tris-HCl, and 0.1% SDS. After a 30-min preincubation period, tissue was incubated in 100 ng of each probe for 3 h. Tissue was then washed twice for 15 min at 37°C in a 1× SSC wash with 0.1% SDS and 20 mM Tris-HCl and then twice for 15 min in a 0.5× SSC wash with 0.1% SDS and 20 mM Tris-HCl. Hoechst stain was added to all the posthybridization washes at a concentration of 10 µg/ml. Tissue was then washed in phosphate-buffered saline (PBS), mounted in Prolong Gold antifade solution, and imaged as described below.
Image acquisition and quantification of autophagy’s effect on Wolbachia.
A FluoView FV1000 confocal microscope system (Olympus) was utilized to acquire images for subsequent analysis. Laser power, sensitivity (HV), gain, offset, and Kalman filter paramters were the same for control and experimental images of the same data set. One-micron z-stack images were acquired of entire hubs or egg chambers. Images were taken at ×600 magnification (60× lens objective) For hubs; a 2.6 digital zoom was implemented for better visualization. Relative Wolbachia density was quantified by imaging the cell type of interest (COI; hub or female germ line) and surrounding cells (SC). The SC in the hub consist of the mitotic region of the germ line. The SC of the germ line were the surrounding follicle cell layer. For hubs, all middle z-stack slices were quantified and normalized to the surrounding cell type density. This was to ensure correct quantification of all Wolbachia organisms within our COI, since Wolbachia infection and density can vary cell to cell and from individual to individual. For the germ line, stages 2 to 8 were identified, and the middle 5 z-stack planes were used for analysis (63). The calculation was as follows:
Nucleic acid purification.
For all nucleic acid purifications, ovaries were dissected and placed in an empty 1.5-ml Eppendorf tube and stored at −80°C until extraction. DNA was purified utilizing the Qiagen blood and tissue kit (catalog number [no.] 69506) per the manufacturer’s protocol. In brief, dissected and homogenized tissues were treated with proteinase K for 3 h, column purified, and eluted in 100 μl of molecular-grade water. RNA was purified by either the Qiagen miRNeasy minikit (catalog no. 217004) to test Atg1 RNAi knockdown or TRIzol for all subsequent experiments. For TRIzol extractions, 150 μl of TRIzol was added to a 1.5-ml Eppendorf tube with samples and homogenized. Pestles were washed with 850 μl of TRIzol to recover any sample tissue attached to the pestle. Samples were spun at 4°C for 15 s at 12,000 × g. Afterwards, 200 μl of 100% chloroform was added and briefly vortexed. Samples were centrifuged at 4°C for 15 min at 20,000 × g. The transparent supernatant was removed and transferred to a new 1.5-ml Eppendorf tube. Isopropanol (100%) was added 1:1 (vol/vol) and mixed by inversion. Subsequently, 1 μl of 20 μg/μl GlycoBlue was added to each sample and incubated for 30 min at −80°C. Samples were immediately centrifuged at 4°C for 15 min at 20,000 × g. The supernatant was removed, and 200 μl of 75% ethanol was used to resuspend the pellet. Samples were centrifuged at 4°C for 5 min at 7,500 × g. The supernatant was removed, and pellets were able to air dry for no longer than 10 min. Samples were resuspended in 50 μl of RNase/DNase-free sterile molecular-grade water and incubated for 10 min at 55°C. RNA was then treated with a Turbo DNA-free kit (AM1907) according to the manufacturer’s protocol. Samples were stored at −80°C.
Quantitative PCR and quantitative reverse transcriptase PCR.
qPCR was used to detect Wolbachia density and RT-qPCR was used to determine knockdown efficiency of RNAi constructs in ovaries. Pools of 5 female ovary pairs were combined per experimental replicate for all quantitative experiments. An EXPRESS one-Step SYBR GreenER kit with premixed ROX from Life Technologies, Inc., was used (catalog no. 1179001k) to detect DNA (reverses transcriptase removed) and mRNA (reverse transcriptase included). Ten nanograms of either DNA or mRNA was added to reaction mixtures. Primers for Wolbachia detection were wsp_F, 5′-TTGGAACCCGCTGTGAATGA-3′, and WSP_R, 5′-CCGAAATAACGAGCTCCAGCA-3′, which were normalized to the host gene 14-3-3 using primers 14-3-3F, 5′-CATGAACGATCTGCCACCAAC-3′, and 14-3-3R, 5′-CTCTTCGCTCAGTGTATCCAAC-3′. For autophagy knockdown confirmation, autophagy-specific primers were picked from reported primers designed by the Drosophila RNAi Screening Center (DRSC). The following primers were used: Atg1_F, 5′-CGTCAGCCTGGTCATGGAGTA-3′; Atg1_R, 5′-TAACGGTATCCTCGCTGAG-3′ (DRSC, PA60369); ATG8a_03F, 5′-GGTCAGTTCTACTTCCTCATTCG-3′; ATG8a_03R, 5′-GATGTTCCTGGTACAGGGAGC-3′; Ref(2)p_F, 5′-AATCGAGCTGTATCTTTTCCAGG-3′; and Ref(2)p_R, 5′-AACGTGCATATTGCTCTCGCA-3′. For internal normalization, primers Rpl32_F, 5′-ATGCTAAGCTGTCGCACAAATG-3′, and Rpl32_R, 5′-GTTCGATCCGTAACCGATGT-3′, were used (64). Technical replicate average threshold cycle (CT) values for all qPCR experiments can be referenced in Table S3.
Average CT values in qPCR experiments. Download Table S3, XLSX file, 0.01 MB (11.1KB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Metabolomic sample preparation, LC-MS analysis, and data processing.
Forty total ovaries (20 female flies under each condition) were extracted by cold methanol-acetonitrile-water (MeOH-ACN-H2O; 40/40/20 [vol/vol]), cleaned up/enriched by SPME, and dried using a vacuum concentrator. Afterwards, metabolites were reconstituted in 2% ACN before injection. A C18 precolumn (3 μm, 100 Å, 75 μm by 2 cm) hyphenated to a rapid separation liquid chromatography (RSLC) C18 analytical column (2 μm, 100 Å, 75 μm by 25 cm) was used to separate metabolites. LC-MS/MS analyses were completed using an EASY nLC 1200 system (Thermo Scientific) coupled to a Q Exactive HF mass spectrometer. Full MS spectra were collected at a unit resolution of 60,000 with an automatic gain control (AGC) of 3 × 106 or maximum injection time of 25 ms and a scan range of 67 to 1,000 m/z. MS2 scans were performed at a resolution of 15,000 and using stepped normalized collision energy of 10, 20, and 40 (%). Source ionization parameters were optimized with the spray voltage at 1.8 kV, dynamic exclusion was set to 10 s. Raw data were converted to mzML files with msConvert, and peak detection, deconvolution, and retention time alignment were performed using OpenMS. Subsequently, the m/z to pathway was enriched by MetaboAnalyst 4.0.
Statistics and graphing.
All statistics were performed in R, and a P value of <0.05 was considered significant. For hub analysis, data were checked for normality by performing the Shapiro-Wilks test and determined to not be normally distributed. For pairwise comparisons of two unrelated samples, the Mann-Whitney U test was performed. For multigroup comparisons, a Kruskal-Willis test was performed, and if found significant, a post hoc Dunn’s test with Bonferroni corrections was performed to determine which groups were significantly different. For the germ lines, all data were checked for normality with the Shapiro-Wilks test and found to be normally distributed. Unpaired Student’s t tests were performed to compare two independent samples. In the case of multigroup comparisons, a one-way analysis of variance (ANOVA) was performed, and when significant, a post hoc Tukey’s test was performed to identify statistically significant groups. All graphs were made in R using the raincloud theme.
ACKNOWLEDGMENTS
We thank the following individuals and labs for their help and expertise: Kim McCall and the McCall lab for input and suggestions throughout several stages of the project, reagents, and fly stocks, Seth Bordenstein and the Bordenstein lab for the wMel CifA and CifB fly stocks and Cif protein expertise, John Beckmann for providing expertise and background related to Cif proteins, Eric Baehrecke for providing autophagy expertise, Ramakrishna Simhadri and Ajit Kamath for methodology and protocol implementation, the Developmental Studies Hybridoma Bank for antibodies, and the Bloomington Drosophila Stock Center for fly stocks.
Footnotes
Citation Deehan M, Lin W, Blum B, Emili A, Frydman H. 2021. Intracellular density of Wolbachia is mediated by host autophagy and the bacterial cytoplasmic incompatibility gene cifB in a cell type-dependent manner in Drosophila melanogaster. mBio 12:e02205-20. https://doi.org/10.1128/mBio.02205-20.
Contributor Information
Luis Teixeira, Instituto Gulbenkian De Ciencia.
Margaret J. McFall-Ngai, University of Hawaii at Manoa.
REFERENCES
- 1.Zug R, Hammerstein P. 2012. Still a host of hosts for Wolbachia: analysis of recent data suggests that 40% of terrestrial arthropod species are infected. PLoS One 7:e38544. doi: 10.1371/journal.pone.0038544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor MJ, Bandi C, Hoerauf A. 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol 60:245–284. doi: 10.1016/S0065-308X(05)60004-8. [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann AA, Montgomery BL, Popovici J, Iturbe-Ormaetxe I, Johnson PH, Muzzi F, Greenfield M, Durkan M, Leong YS, Dong Y, Cook H, Axford J, Callahan AG, Kenny N, Omodei C, McGraw EA, Ryan PA, Ritchie SA, Turelli M, O’Neill SL. 2011. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 476:454–457. doi: 10.1038/nature10356. [DOI] [PubMed] [Google Scholar]
- 4.Dutra HL, Rocha MN, Dias FB, Mansur SB, Caragata EP, Moreira LA. 2016. Wolbachia blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti mosquitoes. Cell Host Microbe 19:771–774. doi: 10.1016/j.chom.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourtzis K, Dobson SL, Xi Z, Rasgon JL, Calvitti M, Moreira LA, Bossin HC, Moretti R, Baton LA, Hughes GL, Mavingui P, Gilles JRL. 2014. Harnessing mosquito-Wolbachia symbiosis for vector and disease control. Acta Trop 132 Suppl:S150–S163. doi: 10.1016/j.actatropica.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Aliota MT, Peinado SA, Velez ID, Osorio JE. 2016. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci Rep 6:28792. doi: 10.1038/srep28792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan PA, Turley AP, Wilson G, Hurst TP, Retzki K, Brown-Kenyon J, Hodgson L, Kenny N, Cook H, Montgomery BL, Paton CJ, Ritchie SA, Hoffmann AA, Jewell NP, Tanamas SK, Anders KL, Simmons CP, O'Neill SL. 2019. Establishment of wMel Wolbachia in Aedes aegypti mosquitoes and reduction of local dengue transmission in Cairns and surrounding locations in northern Queensland. Gates Open Res 3:1547. doi: 10.12688/gatesopenres.13061.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walker T, Johnson PH, Moreira LA, Iturbe-Ormaetxe I, Frentiu FD, McMeniman CJ, Leong YS, Dong Y, Axford J, Kriesner P, Lloyd AL, Ritchie SA, O'Neill SL, Hoffmann AA. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476:450–453. doi: 10.1038/nature10355. [DOI] [PubMed] [Google Scholar]
- 9.Lu P, Bian G, Pan X, Xi Z. 2012. Wolbachia induces density-dependent inhibition to dengue virus in mosquito cells. PLoS Negl Trop Dis 6:e1754. doi: 10.1371/journal.pntd.0001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amuzu HE, McGraw EA. 2016. Wolbachia-based dengue virus inhibition is not tissue-specific in Aedes aegypti. PLoS Negl Trop Dis 10:e0005145. doi: 10.1371/journal.pntd.0005145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan X, Pike A, Joshi D, Bian G, McFadden MJ, Lu P, Liang X, Zhang F, Raikhel AS, Xi Z. 2018. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J 12:277–288. doi: 10.1038/ismej.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rances E, Johnson TK, Popovici J, Iturbe-Ormaetxe I, Zakir T, Warr CG, O'Neill SL. 2013. The Toll and Imd pathways are not required for Wolbachia-mediated dengue virus interference. J Virol 87:11945–11949. doi: 10.1128/JVI.01522-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White PM, Serbus LR, Debec A, Codina A, Bray W, Guichet A, Lokey RS, Sullivan W. 2017. Reliance of Wolbachia on high rates of host proteolysis revealed by a genome-wide RNAi screen of Drosophila cells. Genetics 205:1473–1488. doi: 10.1534/genetics.116.198903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopez-Madrigal S, Duarte EH. 2019. Titer regulation in arthropod-Wolbachia symbioses. FEMS Microbiol Lett 366:fnz232. doi: 10.1093/femsle/fnz232. [DOI] [PubMed] [Google Scholar]
- 15.Yen PS, Failloux AB. 2020. A review: Wolbachia-based population replacement for mosquito control shares common points with genetically modified control approaches. Pathogens 9:404. doi: 10.3390/pathogens9050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voronin D, Cook DA, Steven A, Taylor MJ. 2012. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc Natl Acad Sci U S A 109:E1638–1646. doi: 10.1073/pnas.1203519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serbus LR, White PM, Silva JP, Rabe A, Teixeira L, Albertson R, Sullivan W. 2015. The impact of host diet on Wolbachia titer in Drosophila. PLoS Pathog 11:e1004777. doi: 10.1371/journal.ppat.1004777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loewith R, Hall MN. 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189:1177–1201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ravanan P, Srikumar IF, Talwar P. 2017. Autophagy: the spotlight for cellular stress responses. Life Sci 188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Kimmey JM, Stallings CL. 2016. Bacterial pathogens versus autophagy: implications for therapeutic interventions. Trends Mol Med 22:1060–1076. doi: 10.1016/j.molmed.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen T, Lamark T. 2011. Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. 2005. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell 120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 23.Onodera J, Ohsumi Y. 2005. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J Biol Chem 280:31582–31586. doi: 10.1074/jbc.M506736200. [DOI] [PubMed] [Google Scholar]
- 24.Zirin J, Nieuwenhuis J, Samsonova A, Tao R, Perrimon N. 2015. Regulators of autophagosome formation in Drosophila muscles. PLoS Genet 11:e1005006. doi: 10.1371/journal.pgen.1005006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott RC, Juhasz G, Neufeld TP. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 17:1–11. doi: 10.1016/j.cub.2006.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirisako T, Ichimura Y, Okada H, Kabeya Y, Mizushima N, Yoshimori T, Ohsumi M, Takao T, Noda T, Ohsumi Y. 2000. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, Kaufman TC, Calvi BR, FlyBase Consortium . 2019. FlyBase 2.0: the next generation. Nucleic Acids Res 47:D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 29.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rytkonen A, Poh J, Garmendia J, Boyle C, Thompson A, Liu M, Freemont P, Hinton JC, Holden DW. 2007. SseL, a Salmonella deubiquitinase required for macrophage killing and virulence. Proc Natl Acad Sci U S A 104:3502–3507. doi: 10.1073/pnas.0610095104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misaghi S, Balsara ZR, Catic A, Spooner E, Ploegh HL, Starnbach MN. 2006. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 61:142–150. doi: 10.1111/j.1365-2958.2006.05199.x. [DOI] [PubMed] [Google Scholar]
- 32.Furtado AR, Essid M, Perrinet S, Balana ME, Yoder N, Dehoux P, Subtil A. 2013. The chlamydial OTU domain-containing protein ChlaOTU is an early type III secretion effector targeting ubiquitin and NDP52. Cell Microbiol 15:2064–2079. doi: 10.1111/cmi.12171. [DOI] [PubMed] [Google Scholar]
- 33.LePage DP, Metcalf JA, Bordenstein SR, On J, Perlmutter JI, Shropshire JD, Layton EM, Funkhouser-Jones LJ, Beckmann JF, Bordenstein SR. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543:243–247. doi: 10.1038/nature21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beckmann JF, Ronau JA, Hochstrasser M. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol 2:17007. doi: 10.1038/nmicrobiol.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindsey ARI, Rice DW, Bordenstein SR, Brooks AW, Bordenstein SR, Newton ILG. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol 10:434–451. doi: 10.1093/gbe/evy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shropshire JD, Bordenstein SR. 2019. Two-by-one model of cytoplasmic incompatibility: synthetic recapitulation by transgenic expression of cifA and cifB in Drosophila. PLoS Genet 15:e1008221. doi: 10.1371/journal.pgen.1008221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toomey ME, Frydman HM. 2014. Extreme divergence of Wolbachia tropism for the stem-cell-niche in the Drosophila testis. PLoS Pathog 10:e1004577. doi: 10.1371/journal.ppat.1004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Cuevas M, Matunis EL. 2011. The stem cell niche: lessons from the Drosophila testis. Development 138:2861–2869. doi: 10.1242/dev.056242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spradling AC, Nystul T, Lighthouse D, Morris L, Fox D, Cox R, Tootle T, Frederick R, Skora A. 2008. Stem cells and their niches: integrated units that maintain Drosophila tissues. Cold Spring Harbor Symp Quant Biol 73:49–57. doi: 10.1101/sqb.2008.73.023. [DOI] [PubMed] [Google Scholar]
- 40.Reynolds KT, Hoffmann AA. 2002. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res 80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- 41.Chrostek E, Marialva MS, Esteves SS, Weinert LA, Martinez J, Jiggins FM, Teixeira L. 2013. Wolbachia variants induce differential protection to viruses in Drosophila melanogaster: a phenotypic and phylogenomic analysis. PLoS Genet 9:e1003896. doi: 10.1371/journal.pgen.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bah A, Vergne I. 2017. Macrophage autophagy and bacterial infections. Front Immunol 8:1483. doi: 10.3389/fimmu.2017.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu YW, Li F. 2019. Bacterial interaction with host autophagy. Virulence 10:352–362. doi: 10.1080/21505594.2019.1602020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckmann JF, Markowski TW, Witthuhn BA, Fallon AM. 2013. Detection of the Wolbachia-encoded DNA binding protein, HU beta, in mosquito gonads. Insect Biochem Mol Biol 43:272–279. doi: 10.1016/j.ibmb.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin-Sandoval V, Grüning N-M, Krüger A, Tauqeer Alam M, Keller MA, Breitenbach M, Brindle KM, Rabinowitz JD, Ralser M. 2015. The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc 90:927–963. doi: 10.1111/brv.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu M, Sun LV, Vamathevan J, Riegler M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O'Neill SL, Eisen JA. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol 2:E69. doi: 10.1371/journal.pbio.0020069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voronin D, Schnall E, Grote A, Jawahar S, Ali W, Unnasch TR, Ghedin E, Lustigman S. 2019. Pyruvate produced by Brugia spp. via glycolysis is essential for maintaining the mutualistic association between the parasite and its endosymbiont, Wolbachia. PLoS Pathog 15:e1008085. doi: 10.1371/journal.ppat.1008085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wullschleger S, Loewith R, Hall MN. 2006. TOR signaling in growth and metabolism. Cell 124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 49.Shaid S, Brandts CH, Serve H, Dikic I. 2013. Ubiquitination and selective autophagy. Cell Death Differ 20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan JMM, Wong ESP, Kirkpatrick DS, Pletnikova O, Ko HS, Tay S-P, Ho MWL, Troncoso J, Gygi SP, Lee MK, Dawson VL, Dawson TM, Lim K-L. 2008. Lysine 63-linked ubiquitination promotes the formation and autophagic clearance of protein inclusions associated with neurodegenerative diseases. Hum Mol Genet 17:431–439. doi: 10.1093/hmg/ddm320. [DOI] [PubMed] [Google Scholar]
- 51.Wyers F, Petitjean AM, Dru P, Gay P, Contamine D. 1995. Localization of domains within the Drosophila Ref(2)P protein involved in the intracellular control of sigma rhabdovirus multiplication. J Virol 69:4463–4470. doi: 10.1128/JVI.69.7.4463-4470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen H, Ronau JA, Beckmann JF, Hochstrasser M. 2019. A Wolbachia nuclease and its binding partner provide a distinct mechanism for cytoplasmic incompatibility. Proc Natl Acad Sci U S A 116:22314–22321. doi: 10.1073/pnas.1914571116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chrostek E, Teixeira L. 2015. Mutualism breakdown by amplification of Wolbachia genes. PLoS Biol 13:e1002065. doi: 10.1371/journal.pbio.1002065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice DW, Sheehan KB, Newton ILG. 2017. Large-scale identification of Wolbachia pipientis effectors. Genome Biol Evol 9:1925–1937. doi: 10.1093/gbe/evx139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barth JM, Szabad J, Hafen E, Kohler K. 2011. Autophagy in Drosophila ovaries is induced by starvation and is required for oogenesis. Cell Death Differ 18:915–924. doi: 10.1038/cdd.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chintapalli VR, Al Bratty M, Korzekwa D, Watson DG, Dow JA. 2013. Mapping an atlas of tissue-specific Drosophila melanogaster metabolomes by high resolution mass spectrometry. PLoS One 8:e78066. doi: 10.1371/journal.pone.0078066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.An PNT, Shimaji K, Tanaka R, Yoshida H, Kimura H, Fukusaki E, Yamaguchi M. 2017. Epigenetic regulation of starvation-induced autophagy in Drosophila by histone methyltransferase G9a. Sci Rep 7:7343. doi: 10.1038/s41598-017-07566-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zirin J, Nieuwenhuis J, Perrimon N. 2013. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLoS Biol 11:e1001708. doi: 10.1371/journal.pbio.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiao L, Zhang H-L, Li D-D, Yang K-L, Tang J, Li X, Ji J, Yu Y, Wu R-Y, Ravichandran S, Liu J-J, Feng G-K, Chen M-S, Zeng Y-X, Deng R, Zhu X-F. 2018. Regulation of glycolytic metabolism by autophagy in liver cancer involves selective autophagic degradation of HK2 (hexokinase 2). Autophagy 14:671–684. doi: 10.1080/15548627.2017.1381804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, Debnath J. 2011. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell 22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Frydman HM, Li JM, Robson DN, Wieschaus E. 2006. Somatic stem cell niche tropism in Wolbachia. Nature 441:509–512. doi: 10.1038/nature04756. [DOI] [PubMed] [Google Scholar]
- 62.Kamath AD, Deehan MA, Frydman HM. 2018. Polar cell fate stimulates Wolbachia intracellular growth. Development 145:dev158097. doi: 10.1242/dev.158097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.King RC 1970. The meiotic behavior of the Drosophila oocyte. Int Rev Cytol 28:125–168. doi: 10.1016/S0074-7696(08)62542-5. [DOI] [PubMed] [Google Scholar]
- 64.Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. 2011. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol 57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ref(2)P staining confirms RNAi knockdown of autophagy genes. Ref(2)p is increased in tissues with autophagy knocked down. Image of hubs labeled with genotypes (control on the left, experimental on right) antibody stained for D-cadherin (yellow) and Ref(2)p (magenta). Prime-labeled panels are greyscale of Ref(2)p staining. (A) Control hub displays few small Ref(2)p puncta within the hub. (B) Atg1 RNAi expressed in the hub causes several smaller Ref(2)p puncta. (C) Control flies display several small Ref(2)p puncta within the hub. (D) Expression of Atg8a RNAi in the hub causes an increase in Ref(2)p puncta and size, indicating a block in autophagy. (E, E′) Control flies show a few small Ref(2)p puncta in the hub. (E′) Grayscale of the Ref(2)p channel is shown. (F) Ref(2)p RNAi knocks down expression of Ref(2)p in the hub. (F′) Grayscale image of Ref(2)p channel shows little to no expression of Ref(2)p in the hub. Scale bars, 5 µM. Download FIG S1, PDF file, 1.3 MB (1.3MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Autophagy does not affect wMel and CS hub tropism. (A) Knockdown of either Atg1 or Atg8a does not affect the proportions of hubs displaying tropism for strain wMel. (B) Knockdown of Atg1 or Atg8a does not affect the proportions of hubs displaying tropism for the strain CS. (C) Ref(2)p knockdown does not affect hub tropism proportions in either wMel or CS. P values represent proportions test between control and knockdown. Error bars represent 95% confidence intervals. Download FIG S2, PDF file, 2.0 MB (2.1MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Knockdown efficiency of autophagy-related mRNA in the ovaries upon RNAi expression. RT-qPCR used to determine knockdown efficiency of different RNAi constructs in whole female ovaries. Genes were normalized to RPL32 expression. (A) Two replicates of 20 ovary pools show a knockdown efficiency of 64%. (B) Three replicates of 20 ovary pools shows an Atg8a knockdown efficiency of 47% when the NGT;nos driver was used. (C) Three replicates of 20 ovary pools show a Ref(2)p knockdown efficiency of 87%. Download FIG S3, PDF file, 0.8 MB (782.4KB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Starvation does not modify the effect of autophagy knockdown on Wolbachia density in the germline. Quantitative PCR of WSP normalized to 14-3-3 of 20 whole ovaries from 10 females. (A) Whole-ovary quantitative PCR of Wolbachia density revealed significant decreases in Wolbachia density upon knockdown of Atg1 under well fed and 2-day starvation conditions. Four-day starvation resulted in a similar trend in reduced Wolbachia density but was not significant. P values represent paired Student’s t test results. Download FIG S4, PDF file, 1.9 MB (2.3MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Quantitative PCR of Ref(2)p RNAi mutants reveals no change in Wolbachia density in the female germline. (A) Quantitative PCR of wMel-infected ovaries reveals no change in density upon the knockdown of Ref(2)p. Five replicates of 20 ovary pools were used to determine average Wolbachia density. (B) Quantitative PCR of wMelCS-infected ovaries reveals no change in density upon the knockdown of Ref(2)p. Two replicates of 20 ovary pools were used to determine average Wolbachia density. Paired Student’s t tests were used for statistical analysis. Download FIG S5, PDF file, 0.6 MB (600KB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CifA and CifB overexpression do not affect wMel density in the female germline. NGT;nos was used to overexpress Cif constructs. DNA is colored cyan, D cadherin (labeling the follicle cells) is yellow, and a fluorescently conjugated DNA probe detecting Wolbachia is in magenta. (A) Control stage-8 egg chambers show high germline Wolbachia densities. (B) Stage-8 egg chambers overexpressing CifB result in a nonsignificant trend for lower relative wMel density. (C) Stage-8 egg chambers with overexpressed CifA result in a Wolbachia density similar to that in control egg chambers. (D) Stage-8 egg chambers overexpressing both CifA and CifB result in germline Wolbachia densities similar to that of the control. (E) A one-way ANOVA revealed a significant difference in the data set (P = 0.0171). A post hoc Tukey’s analysis revealed a significant difference between CifB overexpression and CifA-CifB overexpression groups (NCifB = 30, NCifA-CifB = 26, P = 0.024). Of note, CifB showed a trend for reduced wMel density in the germline compared to that in the control (NCont = 27, NCifB = 30, P = 0.0704). Grayscale panels display the Wolbachia-only channel. Scale bars, 40 µM. Download FIG S6, PDF file, 2.5 MB (2.5MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Wolbachia infection and autophagy knockdown drive different metabolic profiles. (A) Heat map indicating Z-score values for each detected positive ion mode metabolite. (B) Principal-component analysis of all positive ion mode-detected features. Download FIG S7, PDF file, 2.4 MB (2.4MB, pdf) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Positive ion metabolites and differentially regulated pathways. Download Table S1, XLSX file, 6.9 MB (8.2MB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Drosophila fly stock genotypes used in experiments. Download Table S2, XLSX file, 0.01 MB (10KB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Average CT values in qPCR experiments. Download Table S3, XLSX file, 0.01 MB (11.1KB, xlsx) .
Copyright © 2021 Deehan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.