Despite being nearly 10 months into the COVID-19 (coronavirus disease 2019) pandemic, the definitive animal host for SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), the causal agent of COVID-19, remains unknown. Unfortunately, similar problems exist for other betacoronaviruses, and no vouchered specimens exist to corroborate host species identification for most of these pathogens.
KEYWORDS: biorepositories, coronaviruses, extended specimen, holistic specimen, museums, zoonoses
ABSTRACT
Despite being nearly 10 months into the COVID-19 (coronavirus disease 2019) pandemic, the definitive animal host for SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2), the causal agent of COVID-19, remains unknown. Unfortunately, similar problems exist for other betacoronaviruses, and no vouchered specimens exist to corroborate host species identification for most of these pathogens. This most basic information is critical to the full understanding and mitigation of emerging zoonotic diseases. To overcome this hurdle, we recommend that host-pathogen researchers adopt vouchering practices and collaborate with natural history collections to permanently archive microbiological samples and host specimens. Vouchered specimens and associated samples provide both repeatability and extension to host-pathogen studies, and using them mobilizes a large workforce (i.e., biodiversity scientists) to assist in pandemic preparedness. We review several well-known examples that successfully integrate host-pathogen research with natural history collections (e.g., yellow fever, hantaviruses, helminths). However, vouchering remains an underutilized practice in such studies. Using an online survey, we assessed vouchering practices used by microbiologists (e.g., bacteriologists, parasitologists, virologists) in host-pathogen research. A much greater number of respondents permanently archive microbiological samples than archive host specimens, and less than half of respondents voucher host specimens from which microbiological samples were lethally collected. To foster collaborations between microbiologists and natural history collections, we provide recommendations for integrating vouchering techniques and archiving of microbiological samples into host-pathogen studies. This integrative approach exemplifies the premise underlying One Health initiatives, providing critical infrastructure for addressing related issues ranging from public health to global climate change and the biodiversity crisis.
INTRODUCTION
The current world focus on COVID-19 (coronavirus disease 2019) has brought considerable interest and urgency to efforts to better understand emerging infectious diseases (EIDs), particularly those that threaten public health. A significant increase in the transmission and emergence of infectious diseases attributed to novel pathogens that spill over from animal populations, including domesticated and wild taxa, has been noted in recent decades (1, 2). SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2) (3), the causative agent of COVID-19, has demonstrated that a previously unknown pathogen can emerge from wildlife species and threaten public health on a global scale within months. SARS-CoV-2 belongs to a clade of betacoronaviruses that have been detected in rhinolophid bats (4–6) and pangolins (4, 7, 8). SARS-CoV-2 was initially recognized by Wu et al. (9) as similar to viruses previously reported from intestinal tissue of Chinese horseshoe bats, Rhinolophus sinicus, despite mitochondrial cytochrome b sequences identifying the bats sampled as least horseshoe bats (Rhinolophus pusillus) elsewhere in the same publication (10). However, the exact animal source of the spillover of SARS-CoV-2 into human populations remains unknown (4, 11).
Despite numerous studies of SARS-CoV-2, the diversity of coronaviruses in bats and other wildlife species remains understudied. Only 19 coronaviruses had been described prior to the SARS epidemic in 2002 (12). Multiple research groups subsequently surveyed wildlife populations for SARS-CoV, the virus that causes the human disease SARS (13–27). Unfortunately, none of these studies included the deposition of vouchered host specimens for SARS-CoV (or any other coronaviruses detected during the surveys) in natural history collections, creating a large gap in our ability to further probe the diversity, distribution, and basic biology responsible for the transmission of SARS and SARS-like viruses in wildlife and human populations and the evolutionary processes by which viruses with pandemic potential evolve in their reservoir hosts. Lack of basic biodiversity infrastructure, in the form of vouchered specimen collections, has limited our ability to respond to the current COVID-19 pandemic yet is critically needed to better understand other zoonotic pathogens with pandemic potential (28).
Host vouchering (i.e., collection and archiving of host species and/or their tissues in a permanent repository) in most recent zoonotic pathogen studies has effectively been nonexistent, including studies relevant to understanding the origins of SARS-CoV-2. For example, Hu et al. (10) euthanized and necropsied over 300 horseshoe bats (genus Rhinolophus) while screening for coronaviruses, yet no host voucher specimen(s) or archived tissue samples were reportedly preserved. None of the cytochrome b sequences from R. pusillus (n = 47) or R. sinicus (n = 206) in GenBank as of June 2020 can be linked to the bats examined by Hu et al. (10). To our knowledge, none of the 30 studies describing sequences of the subgenus Sarbecovirus (genus Betacoronavirus)—which includes SARS-CoV-2—from wild mammalian hosts provided any information concerning permanently vouchered material (i.e., 5, 7, 10, 14–27, 29–41).
The same pattern applies to studies of betacoronaviruses. Few studies describing the recognized viral lineages isolated from wild mammal reservoirs have associated host specimens archived in a natural history collection (but see references 42 and 43). Most published studies of betacoronaviruses used oropharyngeal and rectal swabs collected directly from wild mammals or sampled feces or urine samples from caves or other bat dwellings (34, 44–52). Voucher specimens were either not collected or archived.
We are aware of only a single study that explicitly mentions the use of vouchered host specimens as part of an analysis of coronavirus diversity in bats—a study conducted in the western Indian Ocean (43). The vouchered hosts and corresponding tissue samples were initially deposited in natural history collections as part of previous host-pathogen research (53–56). As shown by Joffrin et al. (43), natural history collections, including associated collections of cryopreserved microbiological samples and other tissues, have the potential to promote powerful interdisciplinary and historical approaches to studying emerging zoonotic pathogens (28). Archived specimens represent a vast, largely untapped, biodiversity infrastructure that can facilitate pathogen identification and insights into host ecology, immunology, and host-pathogen coevolution and dynamics, which may be necessary ingredients for disease prediction (28). In this context, we (i) review the importance of host voucher specimens and microbiological samples in host-pathogen research, (ii) examine vouchering practices in the host-pathogen research community, (iii) provide examples of successful integration of natural history collections in studies of infectious diseases, and (iv) outline recommendations for engaging natural history collections as resources in preventing future pandemics.
IMPORTANCE OF VOUCHERING IN HOST-PATHOGEN STUDIES
Making the most of every sampling event.
Microbiologists and other researchers that conduct pathobiology studies typically collect target samples—bacteria, protozoans, viruses, or fungal pathogens—by capturing and sampling wildlife species. While many microbiological samples do not require sacrificing the host to collect samples (e.g., blood, urine, feces, oral and rectal swabs), others may require lethal sampling (e.g., heart, liver, spleen, brain, and digestive tract) (57). Even when intentional lethal sampling is not a goal of a study, there are frequently accidental deaths of animals during capture and handling (58, 59), and it is poor scientific practice and unethical if resulting carcasses and/or additional microbiological samples are not permanently archived so that additional insights can come from the deceased animal (60). Regardless of the goals, studies involving wildlife species should preserve and archive additional sample types from the host, in addition to the target microbiological sample for pathogen detection, to properly document the symbiotic relationship and identity of the host. This is true even when terminal sampling is not required to achieve the study goals (e.g., when only swabs or blood samples are taken from the host). If only a few voucher specimens or tissue samples (e.g., “representative samples”) are collected and archived during a particular study, these samples provide a means of ensuring correct host identification now and in the future, especially important in species-rich taxa (e.g., bats, rodents), where taxonomy is uncertain and archived materials would provide a resource for future studies beyond the focus of the initial study.
The voucher specimen—a tool for pandemic preparedness.
Voucher specimens have long been the backbone of biodiversity science (61–63). Over the last few decades, natural history collections have embraced the holistic or extended specimen concept (64, 65) whereby associated parasites and diverse cryogenically preserved tissues are associated with each host specimen. The utility of voucher specimens has expanded in novel ways from their traditional use in systematics and taxonomy to studies that now also link together stable isotope analyses, next-generation sequencing, and X-ray computed tomography (CT) scanning (66). Despite these novel uses, the hallmark features of voucher specimens have remained unchanged—to serve as a physical record for biodiversity (67), allowing for verification and documentation of species identifications and distributions, ensuring repeatability in scientific studies, and allowing subsequent researchers to reexamine and reevaluate archived material long after their initial collection (68).
Vouchering in host-pathogen research is necessary for the same reasons that it is important in studies of macroscopic organisms; indeed, it may be more important since hosts effectively represent the habitat of pathogens. Identification of both the pathogen and host is critical to fully understand the biology, ecology, evolution, and potential for transmission of pathogens to other species, including humans and domesticated animals and plants (69). Without depositing host voucher specimens and/or tissue samples in natural history collections or other biorepositories, a host-pathogen study becomes difficult or impossible to repeat and hence is potentially not verifiable by other researchers (68, 70). Furthermore, any possibility of isolating additional coinfecting pathogens is lost when voucher specimens are not collected and properly archived. This problem becomes especially critical in navigating novel viral zoonoses, such as the COVID-19 pandemic, where it is necessary for the scientific community to swiftly and efficiently leverage its collective knowledge and resources to effectively understand and contain the spread of novel pathogens at a time when lockdown restrictions hamper on-going collecting efforts (28).
Despite the clear utility of vouchering in host-pathogen research, especially research focused on EIDs, there are other significant benefits to vouchering that accrue from the process of archiving samples in natural history collections or biorepositories. Vouchered host specimens provide scientific replicability, baselines for temporal and spatial studies, correct taxonomic identification of host species (which may be later revised, e.g., see reference 71), and material for extending research using advanced technologies. Vouchering also integrates an entire community of researchers into the public health sector that otherwise would not typically collaborate (72) and provides critical expertise about the ecology and evolution of host species to support the One Health model for tackling EIDs (73). This proactive approach allows the sharing of data and expertise as well as fostering collaborations to collectively develop novel, creative insights that provide substantial benefits across diverse scientific fields. In essence, vouchering provides both an offensive mechanism to pandemic prevention by expanding the surveillance of wildlife hosts and associated pathogens, in addition to a defensive mechanism by providing a verifiable temporal and spatial archive for baseline comparisons.
INSIGHTS INTO CURRENT VOUCHERING PRACTICES IN HOST-PATHOGEN RESEARCH
We explored current vouchering practices commonly employed by microbiologists (e.g., virologists, bacteriologists, parasitologists) in host-pathogen research and existence of collaborations with natural history collections based on an online survey (see Text S1 in the supplemental material for survey methodology, institutional review board [IRB] approval, and summarized data). Herein we discuss responses from 109 completed surveys (see Data Set S1 in the supplemental material for raw survey results). Distribution of the survey through social media resulted in a broad range of respondents from all continents except Antarctica. Participants were asked to classify their field(s) of research, and 38.5% indicated two or more disciplines, reflecting the multidisciplinary nature of microbiological research today. Not surprising given the diversity of respondents, 39.4% indicated studying more than one taxonomic group. Helminths (e.g., monogeneans, tapeworms, nematodes, leeches) were the most frequently cited focal taxa, followed by bacteria.
Microbiology sampling methods and museums—a human survey. Download Text S1, DOCX file, 0.03 MB (32.4KB, docx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiology sampling methods and museums—a human survey. Download Data Set S1, XLSX file, 0.03 MB (132.8KB, xlsx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Respondents (n = 105) indicated that they collect a number of different microbiological samples using lethal and noninvasive sampling of host species. Of the respondents, 41 (39.0%) indicated that they do not terminally sample hosts as part of any of their research projects. In contrast, 64 (61.0%) respondents indicated that their research does at least sometimes incorporate terminal sampling, including opportunistic sampling (e.g., accidental fatalities during capture) or work piggy-backed on other studies that included lethal sampling (e.g., abattoirs/slaughterhouses, culling of nuisance/pest species). Of the 64 respondents that include terminal sampling in at least some of their research, 36 (56.3%) respondents indicated using only “targeted intentional lethal sampling,” while the remaining respondents use a combination of terminal sampling methods. Importantly, of the 64 respondents who conducted terminal sampling, six did not collect microbiological samples that required terminal sampling of hosts (e.g., collected only nonlethal samples such as ectoparasites, excrement, etc.), and six collected only a single tissue type that required terminal sampling.
When participants were asked about practices related to archiving microbiological samples and host specimens, 75 of 104 respondents (72.1%) permanently archive collected microbiological samples, while only 51 of 107 respondents (47.7%) indicated that they permanently archive host species. Only 41 respondents (39.4%) report vouchering host specimens from which microbiological samples were collected. Of the 75 respondents that archived microbiological samples, 22 (29.3%) reported that they deposited their samples in biobanks/biorepositories and 26 (35.0%) deposited them in natural history collections, both of which typically provide permanent archiving of samples. However, most frequently, samples were stored in institutional laboratory space (60.0%), which is typically not accessible to the broader scientific community. When microbiological samples were reported not to be preserved, the majority of respondents indicated a lack of institutional infrastructure as the reason. These trends were generally reversed for the 51 respondents that reported permanently archiving host specimens in their studies, where 31 of the 51 (61.0%) indicated that their vouchers were deposited at natural history collections. For the 19 (30%) respondents who indicated that they include lethal sampling of hosts in their research but do not preserve voucher specimens, “no tradition of vouchering in my institute” and “no institutional infrastructure” were equally cited as the reason.
When participants were asked if they had ever collaborated with or deposited voucher specimens in a natural history collection, 44 of 105 respondents (41.9%) replied “yes” whereas 50 (47.6%) replied “no.” In addition, 11 (10.5%) indicated that they were unsure how to collaborate with a natural history collection. Of the 48 respondents who addressed the value of collecting host voucher specimens for studies of microorganisms, nearly all (93.8%) spoke to the benefits, often in terms of increased rigor and reproducibility of studies, with one reporting “The host is half of the equation in any host-parasite interaction, why wouldn’t we be vouchering the host?.” Three respondents did not see the value of vouchering, and one respondent expressed concerns about the biosafety of depositing specimens in a collection. Along this line, of the 42 respondents who wrote about their experiences working with natural history collections, 10 (24.4%) discussed problems, including sample types not being accepted, lost samples, and onerous logistical issues.
ROLE OF NATURAL HISTORY COLLECTIONS IN HOST-PATHOGEN RESEARCH
“Extending” the voucher specimen.
Biodiversity data archived in the natural history collections and biorepositories around the world continue to increase as novel questions arise and new research disciplines become aware of their existence (74). The past few decades have seen increasing collaborations between microbiologists and biodiversity scientists as the concept of the “holistic” or “extended” specimen has been adopted by more natural history collections (75). Under this paradigm, host and parasite specimens are collected simultaneously, along with associated cryopreserved tissues and microbiological samples, and all are linked through bioinformatics even if they are stored in different repositories (57, 76, 77). The merits of making these holistic collections are numerous as they naturally promote more integrated science (57, 64, 78). Integration of the “extended specimen” concept into host-pathogen research is now beginning as it is slowly becoming recognized as an essential part of zoonotic disease surveillance (69, 70, 72, 79).
Better integration of natural history collections in infectious disease studies would likely provide more clarity about the various roles of environmental change (e.g., biodiversity loss, climate change, land use change) (80) in the emergence of novel zoonoses (81). Such human-driven disturbances provide increased opportunities for pathogen transmission among host species (i.e., host switching from one wildlife species to others) resulting in heightened risk to public health, but risk assessment is difficult because we lack detailed information regarding naturally occurring host-pathogen relationships (78). Control of potentially zoonotic pathogens through targeted risk management (e.g., reducing contact between humans and reservoir hosts) is likely to be more effective than retroactive efforts aimed at eradicating zoonoses after they have spilled over into human populations (82). However, rigorous and successful prevention strategies require knowledge of historic and contemporary distributions, occurrence, and ecology and evolutionary history of host species in host-pathogen systems. Those critical assessments rely on information that largely resides in natural history collections (78, 83).
Below, we highlight three examples of successful integration of host-pathogen research and natural history collections in collaborative research programs focused on pathogens (i.e., yellow fever, hantaviruses, and helminths), as well as provide an example of a failed integration (i.e., ebolaviruses). Additional examples are provided in Table 1.
TABLE 1.
Selected examples of host-pathogen studies that deposited and/or studied vouchered host specimens and microbiological samples in natural history collections
| Pathogen | Disease | Host(s) | Natural history collection(s) | Reference(s) |
|---|---|---|---|---|
| Viruses | ||||
| Arenaviridae | ||||
| Guanarito virus | Venezuelan hemorrhagic fever | Rodents | American Museum of Natural History | 155 |
| Whitewater Arroyo virus | Hemorrhagic fever syndrome | Rodents | Natural Science Research Laboratory, Texas Tech University | 156, 157 |
| Coronaviridae | ||||
| Alpha- and betacoronaviruses | Pathogenicity unknown | Bats | Field Museum of Natural History | 43 |
| Middle East respiratory syndrome (MERS)-related betacoronavirusa | Pathogenicity unknown | Bats (Neoromicia spp.) | Ditsong National Museum of Natural History | 42 |
| Flaviviridae | ||||
| Flavivirus spp. | Yellow fever | Various | American Museum of Natural History, Smithsonian National Museum of Natural History, and Museu Nacional/Universidade Federal do Rio de Janeiro | 86, 87, 90, 158–163 |
| Filoviridae | ||||
| Ebolavirus | Ebola hemorrhagic fever | Vertebrates | Royal Museum of Central Africa and Museum of Southwestern Biology | 110 |
| Hantaviridae | ||||
| Laguna Negra virus | Hantavirus pulmonary syndrome | Rodents (Calomys laucha) | Field Museum of Natural History | 164 |
| Sin Nombre virus | Hantavirus pulmonary syndrome | Rodents | Museum of Southwestern Biology | 94, 165 |
| Anajatuba, Castelo dos Sonhos viruses | Hantavirus pulmonary syndrome | Rodents (Oligoryzomys spp.) | Museu Nacional/Universidade Federal do Rio de Janeiro | 70, 96 |
| Luteoviridae | ||||
| Barley yellow dwarf virus | Barley yellow dwarf | Grasses | University and Jepson Herbaria, University of California, Berkeley, and Center for Plant Diversity, University of California, Davis | 166 |
| Poxviridae | ||||
| Monkeypox virus | Monkeypox | Squirrels | American Museum of Natural History | 167 |
| Monkeypox virus | Monkeypox | Small mammals | Field Museum of Natural History | 168 |
| Parasites | ||||
| Arthropods | ||||
| Trombiculidae | Scrub typhus (Rickettsia) | Rodents | Smithsonian National Museum of Natural History | 169 |
| Syringophilidae | Ectoparasitic infestation | Birds | Natural History Department of Sarisske Museum | 170 |
| Trichodectidae | Ectoparasitic infestation | Gopher (Cratogeomys spp.) | Museum of Natural Science, Louisiana State University | 171 |
| Ixodoidea | Multiple tick-borne diseases | Mammals | Field Museum of Natural History | 172, 173 |
| Helminths | ||||
| Rictulariidae | ||||
| Paucipectines hymanae | Endoparasitic infection | Shrew opossum | Field Museum of Natural History | 174 |
| Spiruridae | ||||
| Protospirura spp. | Endoparasitic infection | Rodents | Muséum National d'Histoire Naturelle | 175 |
| Philometridae | ||||
| Clavinema marina | Endoparasitic infection | English sole (Parophrys vetulus) | Burke Museum of Natural History & Culture | 108 |
| Metastrongylidae | ||||
| Skrjabingylus spp. | Leptomeningitis | Skunks | Angelo State Natural History Collection | 176 |
| Dactylogyridae | ||||
| Cichlidogyrus spp. | Ectoparasitic infection | Cichlid fish | Royal Museum of Central Africa | 104, 177 |
| Schistosomatidae | ||||
| Schistosoma spp. | Schistosomiasis | Gastropods | Natural History Museum, London | 178 |
| Prokaryotes | ||||
| Trypanosomatidae | ||||
| Trypanosoma lewisi | Chagas disease | Rodents (Rattus spp.) | Natural History Museum, London, Museum of Zoology, University of Cambridge and Oxford University Museum of Natural History | 179 |
| Unicellular eukaryotes | ||||
| Plasmodiidae | ||||
| Plasmodium spp. | Malaria | Bats, primates, rodents | Field Museum of Natural History | 180–183 |
| Hepatocystis sp. | Malaria | Bats | Smithsonian National Museum of Natural History | 184 |
| Nycteria sp. | Malaria | Bats | Smithsonian National Museum of Natural History | 185 |
| Bacteria | ||||
| Leptospiraceae | ||||
| Leptospira spp. | Leptospirosis | Small mammals | Field Museum of Natural History | 186, 187 |
| Mycobacteriaceae | ||||
| Mycobacterium tuberculosis | Tuberculosis | Bison cf. antiquus | Carnegie Museum; University of Kansas Museum of Natural History | 188 |
| Yersiniaceae | ||||
| Yersinia pestis | Plague | Homo sapiens | State Collection for Anthropology and Palaeoanatomy | 189 |
| Yersinia pestis | Plague | Rodents | Museu Nacional/Universidade Federal do Rio de Janeiro | 88 |
| Fungi | ||||
| Peronosporaceae | ||||
| Phytophthora infestans | Potato blight | Potato | Various herbaria | 190–192 |
| Pseudeurotiaceae | ||||
| Pseudogymnoascus destructans | White-nose syndrome | Myotis bechsteinii | National Museum of Natural History | 193 |
| Meripilaceae | ||||
| Rigidoporus microporus | White rot disease | Rubber tree (Hevea brasiliensis) | Botanical Museum, Finnish Museum of Natural History & Botanical Museum, University of Helsinki | 194 |
MERS, Middle Eastern respiratory syndrome.
Yellow fever.
One of the earliest efforts to employ comprehensive host and pathogen collection, vouchering, and data recording was the Yellow Fever Service, an organization maintained in the 1930s by the Brazilian Ministry of Education with the cooperation of the International Health Division of the Rockefeller Foundation (84, 85). The program was truly multidisciplinary, including fieldwork in collaboration with researchers from natural history collections to identify the animal reservoirs of the virus (86–88). As a result of the Yellow Fever Service, tens of thousands of voucher specimens, including amphibians, birds, mammals, and reptiles, were collected and deposited in natural history collections, and these have been employed in subsequent studies on systematics, ecology, evolution, and conservation biology of mammal species. Each sampled specimen was accompanied by extensive and standardized field notes (86), and the samples are accessible to researchers for further study. Among the major outcomes of this collaborative endeavor was the discovery of a sylvatic transmission cycle of the yellow fever virus (88, 89), eradication of Aedes aegypti in Brazil (90, 91) (albeit for just a few decades), and understanding primate population ecology and transmission risk of the yellow fever virus (92, 93).
The idea that voucher specimens provide scientific replicability, baseline data for temporal studies, correct taxonomic identification of host species, and material for future technological advances in host-pathogen research is clear in the seminal publication on results of the Yellow Fever Service by Gilmore (86):
The preservation of specimens for absolute identification in public health investigations is generally an acute problem, because trained field zoologists are often absent, and because the technique is a special one. With this in mind it is suggested that the documentation of every specimen by a protocol, with a few skins temporarily preserved by thorough drying in shape and the saving of even skulls of every different recognizable kind, designated consistently by the same common name, will enable identifications to be made at a later date with a certain amount of surety.
Hantaviruses.
Hantaviruses provide one of the best examples of how to integrate natural history collections with infectious disease research (72, 94). Natural history collections have provided reference libraries for proactive and rapid identification of host species, in addition to temporal and spatial patterns, of new hantavirus strains. For instance, in 1993, pathobiologists were able to quickly identify the reservoir host of a novel hantavirus (Sin Nombre) responsible for a cluster of deaths in the Four Corners region of the southwestern United States by screening archived rodent specimens at the Museum of Southwestern Biology at the University of New Mexico and Natural Sciences Research Laboratory at Texas Tech University and determine that this novel virus had been present in host populations dating back at least as far as samples were available for screening (late 1970s) (94). New analyses or taxonomic revisions have also resulted in a revised understanding of the identities of many pathogen reservoirs subsequent to their original description. For example, recent systematic revisions of pygmy rice rats (genus Oligoryzomys) (i.e., 71, 95–98) have refined our understanding of this taxonomic group and have led to the recognition that many host reservoirs of hantaviruses have restricted geographic distributions. Changes resulting from studies of vouchered host specimens have led to the reassignment of host names for numerous hantaviruses of public health importance (e.g., Anajatuba, Bermejo-Neembucu, Castelo dos Sonhos, Choclo, and Maporal viruses) (99). In addition, the finding that hantaviruses occur in moles, shrews, and bats based on screening archived specimens from natural history collections has falsified the paradigm that this is solely a rodent-borne virus (100–102) and substantially changed our understanding of this family of pathogenic viruses.
Helminths.
Mainly drawing from examples in fish parasitology, an inspection of host specimens originally not intended for parasitological research can serve a range of applications in the study of helminths (parasitic worms). Similar to research on other fish parasites that are unpredictable in prevalence or abundance (e.g., cymothoid isopods) (103), archived host specimens have later been used to describe new helminth species (104). For example, Kmentová et al. (105) characterized gill-infecting monogeneans of freshwater lates perch throughout Africa based on previously collected specimens. However, perhaps the most explicit advantage of voucher specimens to helminthological research is that natural history collections allow the inference of baseline data about parasite communities across various timescales. This potential for tracking disease emergence via shifts in pathogen occurrence and/or abundance and linking with environmental change was, to quote Harmon et al. (106), “hiding in plain sight”—collections of host specimens are indeed heavily underexplored for this purpose. Black (107) confirmed, from archived specimens of the lake trout (Salvelinus namaycush), the presence of the nematode Cystidicola stigmatura which has recently disappeared as a result of dwindling host abundance. Conversely, Howard et al. (108) demonstrated over a 9-decade time span the increased prevalence of the nematode Clavinema mariae infecting Puget Sound English sole (Parophrys vetulus). Invasion biology is another field where natural history collections provide a unique opportunity to establish a historical baseline for often understudied helminth communities. For example, Jorissen et al. (109) compared the monogenean gill parasites of historic and recent tilapia populations in Central Africa. This allowed distinguishing flatworms naturally infecting native fishes from those appearing only after cointroduction with nonnative tilapia species (and subsequent host switching).
Ebolaviruses.
Ebolaviruses represent a group of pathogens where a lack of high-quality vouchered specimens across temporal scales has created a direct impediment to identifying putative reservoir hosts (110). Ranging from insects to antelopes (111), the identification of both reservoir host(s) and susceptible host populations remains largely unknown (112), although bats have been implicated as reservoir hosts for Reston ebolavirus (113), Bombali ebolavirus (114, 115), and Zaire ebolavirus (116, 117) but see Leendertz et al. (118) and Caron et al. (111). The ability to identify wildlife hosts of ebolaviruses is limited by a lack of centralized repositories of historical and modern voucher specimens, especially for mammal species, across the Afrotropics (119, 120).
Impediments in detecting ebolavirus hosts are related to both the distribution of the virus, most notably in the megadiverse and remote regions of the Afrotropics, and the highly unpredictable persistence of the virus in a host (116). Specifically, the prevalence of ebolaviruses circulating in populations of putative hosts, such as bats, can vary greatly across short periods of time (116) and is often lower (118) than prevalences observed in other viruses whose hosts have been identified more rapidly with the aid of voucher specimens (see the “Hantaviruses” section above). Furthermore, for mammals, the most targeted ebolavirus hosts thus far, the Afrotropics represents a hot spot of evolutionary diversity, with one recent study predicting at least 122 undiscovered species (121). Moreover, recent phylogenetic research using voucher specimens from natural history collections has indicated high levels of cryptic diversity in Miniopterus (122), a bat genus speculated to be a reservoir host for Zaire ebolavirus (123). Furthermore, studies on other filoviruses such as Marburg virus have found samples of different organs and body fluids (e.g., blood, colon, and oral-mucosa) collected from bats to be less effective than combined samples of liver and spleen in identifying positively infected bats (i.e., Rousettus aegyptiacus) using quantitative reverse transcription-PCR (Q-RT-PCR) (124, 125). Regardless of sample type collected and low prevalence rates of ebolavirus, limited sample sizes per host species from large and nonrandomized studies offer limited insight into our understanding of ebolaviruses (117).
A lack of historic and recent voucher specimens of mammals from the Afrotropics hinders effective predictions and identification of potential reservoir hosts of ebolaviruses in a number of ways. First, lack of historic material limits the ability to answer historical evolutionary questions regarding virus evolution, such as the consequences of viral population size on lineage extinctions through time (126). Second, a lack of systematically collected and holistic specimens of diverse groups of potential hosts such as bats, combined with myopic sampling of specific taxonomic groups (e.g., fruit bats) has resulted in biased sampling effort that underrepresents the true diversity of hosts (118). Third, a lack of archived samples available for screening pre- and postdisturbance by humans—a process thought to increase ebolavirus spillover between bats and humans (127)—limits our ability to understand the impacts of such processes on viral and host dynamics. Finally, although recent efforts to sample deceased mammals, a potentially important source of ebolavirus infection data (128), across parts of the Congo should be applauded (129), the lack of involvement of natural history collections in disease surveillance efforts is concerning (28).
BEST PRACTICES FOR INTEGRATING NATURAL HISTORY COLLECTIONS IN HOST-PATHOGEN STUDIES
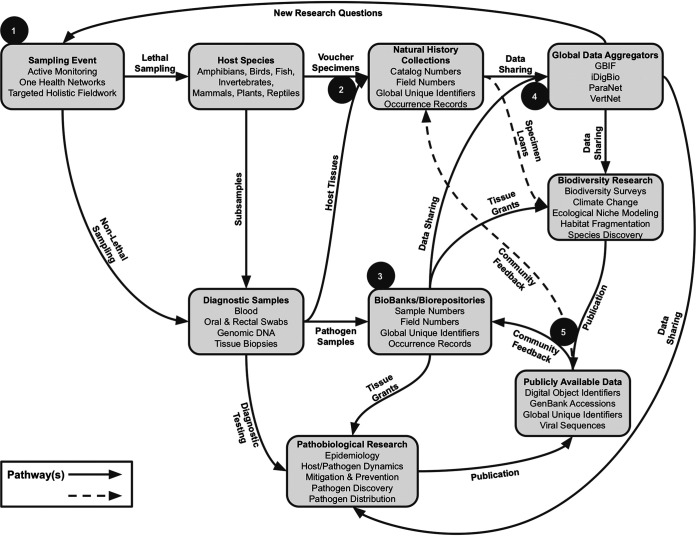
Natural history collections have evolved to incorporate best practices for field collection, vouchering, sample preservation, and data sharing (74), creating a vast resource of globally distributed collections (e.g., 130–132). These best practices revolve around the voucher specimen as the common currency, which has resulted in a predictable workflow (Fig. 1). We highlight five critical steps in this workflow and provide recommendations for integrating vouchering techniques in infectious disease studies and fostering collaboration between natural history collections and the microbiological research community (see also Table S1 in the supplemental material for a nonexhaustive overview of collections with experience in host-pathogen research collaborations).
FIG 1.
Workflow for integrating vouchering practices and natural history collections into host-pathogen research. Light-gray boxes provide descriptions for critical steps of the workflow, and arrows connecting the boxes indicate the pathway for the flow of specimens, samples, data, etc. Gray circles with white numbers represent steps described in the text.
Natural history collections with previous experience and/or interest in voucher-based host-pathogen studies. Abbreviations of natural history collections are as follows: AMNH, American Museum of Natural History; CTUA, Colección Teriológica, Universidad de Antioquia; FMNH, Field Museum of Natural History; GNM, Gothenburg Natural History Museum; LMUSP, Laboratório de Mamíferos da Escola Superior de Agricultura “Luiz de Queiroz,” Universidade de São Paulo; MECN, Museo del Instituto Nacional de Biodiversidad, Ecuador; MN, Museu Nacional, Universidade Federal do Rio de Janeiro; MNHN, Muséum national d’Histoire naturelle; MPEG, Museu Paraense Emilio Goeldi; MSB, Museum of Southwestern Biology, University of New Mexico; NHM, Natural History Museum, London; OMNH, Sam Noble Museum of Natural History, University of Oklahoma; PSUNHM, Princess Maha Chakri Sirindhorn Natural History Museum, Prince of Songkla University; QCAZ, Museo de Zoología de la Pontificia Universidad Católica del Ecuador; RMCA, Royal Museum for Central Africa; TTU, Natural Science Research Laboratory, Texas Tech University; UACH, Universidad Austral de Chile; UMMZ, University of Michigan Museum of Zoology. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Step 1. Sampling event.
Sampling design is one of the most basic components of research. Given the complexity and nuances of wildlife research, early planning is critical, especially when working with pathogens. In addition to standard institutional animal care and biosafety protocols, local, state, and federal permits are often needed to sample and/or collect wildlife species. For international work, import and export permits, material transfer agreements, and compliance with the Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits Arising from their Utilization to the Convention on Biological Diversity may be necessary, as well as Convention on International Trade in Endangered Species (CITES) permits when dealing with threatened and endangered taxa. Natural history collections often have blanket permits and personnel (e.g., collection managers, registrars) that can assist in navigating wildlife research with relative ease. Integrating natural history collections early in sampling design would provide host-pathogen researchers the opportunity to leverage these resources, minimizing problems in downstream applications.
Broad temporal and spatial surveys of host taxa should have sample sizes sufficient to not only detect pathogens (which can occur at widely ranging prevalence rates, e.g., bat-borne viruses can range between 0 and 100% depending on sample type and viral family) (133) but also provide some level of natural history information about the target pathogen (94). For mammals, such as rodents, which tend to have large population sizes and high reproductive rates, targeted lethal sampling has been shown to not have significant adverse effects on population structure or density (134). For taxa that are threatened, long-lived, and/or have low reproductive rates (e.g., many bat and primate species), more limited collection efforts combined with nonlethal sampling methods (e.g., swabs, feces) may be necessary. Animals should be captured, handled, and euthanized following community standards for ethical care and use of vertebrates in research; for mammals, guidelines are provided by Sikes et al. (135).
Capture-sample-release surveys typically do not allow for repeatability or extension of the research (64). However, in some cases, nonlethal sampling may be all that is permitted. In these situations, sampling should still be maximized. Oropharyngeal and rectal swabs, feces, hair samples, ectoparasites, and wing punches or ear clips should be collected. Sample volume should also be maximized to provide sufficient material for multiple tests (i.e., isotopic analyses). Standard measurements and photographs of the host should also be taken to aid in species-level identification. However, measurements and photos are only complementary and alone are often not sufficient to provide unambiguous identification of hosts, particularly for diverse taxonomic groups with numerous cryptic species (e.g., rodents and bats) (120). Voucher specimens are the only reliable option to ensure repeatability and should be considered the gold standard in host-pathogen studies (70).
When hosts are euthanized or incidental deaths occur during nonlethal surveys, it is important to maximize the diversity of tissue types that are collected and preserved. Samples should be collected from the host immediately after death, and ideally, samples should be stored at ultracold temperatures (e.g., liquid nitrogen, dry ice) upon collection. Samples should be archived long-term in an ultracold (–80°C) freezer—but preferably in a vapor-phase nitrogen (–190°C) freezer—at accredited natural history collections and publicly available for use by qualified researchers (136). Alternatively, if tissue collection from hosts is not possible due to limitations in expertise, time, and/or funding, specimens can be preserved in formalin or alcohol (137). These low-cost options allow researchers the ability to obtain a verifiable voucher when euthanizing potential hosts and provide genetic material for species identification (138). An added benefit of formalin fixation of voucher specimens is that it is an approved method for inactivating viruses according to the Centers for Disease Control and Prevention (CDC) (139), thus rendering the deposited specimens noninfectious.
Step 2. Depositing host specimens and tissue samples.
Natural history collections accession voucher specimens and associated tissue samples routinely as part of their mission. Although each institution has its own accession protocols (140), most natural history collections do not charge donors for depositing voucher specimens and tissue samples, but early communication prior to a collection event is recommended in order to address any logistical or financial hurdles. All deposits of voucher specimens and tissues require documentation showing the legal framework under which they were obtained, as well as any data associated with the deposited materials to confirm their legal status.
To facilitate accessibility to the broader scientific community, researchers should shift samples from their personal laboratories to permanent repositories such as natural history collections when samples are no longer needed for the original study. Alternatively, most natural history collections will place temporary embargoes on deposited specimens and samples to allow for the original study to continue while ensuring they are permanently and safely archived. Of course, if a researcher is engaged collaboratively with a curator or collection manager, this process will be part of project development and integrated into the study design.
Full metadata should be included with each specimen and sample. The most basic metadata minimally required by all natural history collections are collection date, geographic location (preferably including both a textual locality and GPS coordinates), collector’s name, and some level of taxonomic identification. In addition, depending on the disposition of the specimens, vouchers often include taxon-specific measurement and life history data. Even when collecting nonlethal samples, metadata should be recorded and shared with the receiving institution when depositing samples.
Last, the receiving institution will formally accession the deposited specimens and/or samples in order to transfer custody to the natural history collection (141). Although custody of the voucher specimens and samples has changed, an embargo can remain in place to allow time for publication and the development of additional projects. This time period also allows collection managers and curators to verify species identifications, curate metadata, and assign formal catalog numbers to the voucher specimens and/or samples (e.g., MSB:Mamm:89863). For researchers engaged in host-pathogen research, this process provides verification of the host vouchers based on current taxonomy, in addition to assignment of a globally unique identifier (GUID) associated with the specimen. GUIDs allow the scientific community the opportunity to digitally curate voucher specimens and link data repositories, literature, and other resources to the specimen (142). Voucher specimens and samples are of no value if they are not given unambiguous unique numbers, thereby preventing data linkage.
Step 3. Archiving microbiological samples.
Archiving microbiological samples from host species that may be infectious requires considering the biosafety capabilities of the receiving collections. Researchers should inform the receiving natural history collection of pathogens detected in deposited samples as well as any treatments to the vouchers and/or tissue samples to deactivate specific pathogens of concern. Concomitantly, the natural history collection should inform the donor of any limitations in their ability to receive potentially pathogenic samples. However, many natural history collections maintain biorepositories to complement their vouchered host collections (143) and utilize best practices for sample curation and personnel safety (136, 144). Specifically, museum personnel should be trained in biosafety protocols for handling pathogenic samples and implementation of increased security measures, such as restricted access to designated areas where pathogenic samples are stored. In addition, if future research is to be conducted using pathogenic samples, many natural history collections are equipped with biosafety level 1 and/or 2 laboratory space. Under certain circumstances, in particular research on designated CDC Select Agents, samples need to be deposited in a biorepository or biobank with higher biosafety level capabilities (145).
Similar to vouchering host specimens and tissues, pathogenic samples should be well-documented with appropriate permits and metadata. Ideally, the metadata should include all data associated with the host species plus pathogen-specific information. When possible, a molecular identification (e.g., nucleic acid sequence) for both the host (symbiotype) and pathogen should be included so that their identities can readily be placed on the Tree of Life and provide a basis for identifying sister species that may serve as potential hosts for related pathogens. GUIDs also should be assigned to microbiological samples and related back to the host specimen record.
Step 4. Specimen access and data sharing.
The archiving institution should maintain a relational collections database (e.g., Arctos, EMu, Specify), which should be linked to biodiversity data aggregators (e.g., GBIF, iDigBio, FishBase, and VertNet) and data repositories (e.g., GenBank, IsoBank, MorphoSource) through reciprocal linkages or via an integrated publishing toolkit (146). By assigning GUIDs during the archiving process, researchers can connect these typically disparate data sets into a single aggregated view of associated vouchered specimens and samples through online searchable databases. This allows all derivative data to be tied back to the original vouchered specimen and/or sample (147), allowing for a more integrated approach to host-pathogen research (72). Any updates made to the vouchered specimen (e.g., species identification, georeferencing, new data sets) would be updated through the network of aggregators and repositories and be made available to the end user. In addition, this makes the vouchers discoverable and available to researchers both remotely online and physically through collection visits and loans.
Step 5. Publication and data repositories.
Citing accession numbers of vouchered specimens and/or samples in publications is essential to link data sets back to natural history collections and biorepositories. A full list of specimens examined and the associated pathogen screening results should be included in publications or minimally available as supplemental material. The individual host specimen from which the novel pathogen is sequenced and/or isolated, and then described, should also be formally designated a symbiotype, along with other specimens from the type series containing the novel pathogen (i.e., symbioparatypes) (148). Symbiotype identity should be confirmed with a DNA sequence (e.g., cytochrome b for mammals) and deposited in National Center for Biotechnology Information (NCBI) BioSample. The biosample attribute “voucher_specimen” is linked to the species name in the biosample. If you are submitting a pathogen sequence (including a full genome), a new attribute, host_specimen_voucher, should be used to store the museum catalog number of host specimen information for SARS-CoV-2 (or any other pathogen). For GenBank submissions, it is recommended to link to a registered BioSample (see reference 149). When naming newly discovered pathogens, the symbiotype catalog number should be included in the name (e.g., Camp Ripley virus, MSB:Mamm:89863) to facilitate linkage between host and pathogen. This same approach should be used when depositing data sets on Dryad and other publicly available data repositories, allowing for linkages back to the physical voucher specimen (147).
CONCLUSIONS
The COVID-19 pandemic has exposed many weaknesses in the current public health approach to zoonotic spillover events and has reinforced the need for shifting to an integrated One Health approach for addressing EIDs (150). In particular, the critical importance of identifying a definitive host of SARS-CoV-2 (4, 11) coupled with the unprecedented potential of current molecular techniques to reinvestigate historic material archived in natural history collections (e.g., human immunodeficiency virus 1 [HIV-1] genome recovered from samples collected in 1966, prior to the discovery of acquired immunodeficiency syndrome [AIDS]) (151) highlights the need for a more integrated approach to surveillance and mitigation of EIDs (70). Vouchering host specimens and associated tissue samples in natural history collections is one of the most effective means of overcoming this hurdle.
To meet their promise of substantially contributing to infectious disease studies, however, natural history collections must grow exponentially in the coming decades (152, 153) and invest in methodology and infrastructure development which meets best practices to maximize the utility of deposited specimens and samples for future research projects (57, 64, 70, 136). When properly cared for (140), natural history collections can last centuries, providing a physical record for current and future pandemics (148). By vouchering and depositing host specimens, associated tissue, and microbiological samples in natural history collections, the host-pathogen research community will contribute to future research endeavors and mobilize a large community of biodiversity scientists into One Health networks (154).
ACKNOWLEDGMENTS
Thanks to W. Addink, A. Casino, and D. Koureas for organizing the Consortium of European Taxonomic Facilities (CETAF) and Distributed System of Scientific Collections (DiSSCo) COVID-19 Task Force (COVID-19 TaF). We also thank the COVID-19 TaF Metadata Practices Working Group for information regarding updates to GenBank deposits. We cordially thank the American Society of Mammalogists (Mammal-L), American Society of Parasitologists (S. Perkins), Belgian One Health Network (P. Huybrechts and H. Keune), Belgian Society of Parasitology and Protistology (G. Caljon), Belgian Wildlife Disease Society (P. Tavernier), British Society for Parasitology (J. Cable and H. Price), Société Française de Parasitologie (F. Grenouillet and J.-L. Justine), World Association for the Advancement of Veterinary Parasitology (R. Wall), and the EvolDir, Taxacom, and WildList mailing lists for distributing the survey. We also extend gratitude to M. De Meyer, E. Gilissen, T. Huyse, and J. Snoeks (Royal Museum for Central Africa, Belgium), B. Coyner (Oklahoma Museum of Natural History, USA), A. Emery (Natural History Museum, UK), A. M. R. Bezerra (Museu Paraense Emílio Goeldi, Brazil), G. D’Elía (Universidad Austral de Chile), J. Brito (Museo del Instituto Nacional de Biodiversidad, Ecuador), S. Solari (Colección Teriológica, Universidad de Antioquia, Colombia), A. R. Percequillo (Laboratório de Mamíferos da Escola Superior de Agricultura “Luiz de Queiroz.” Universidade de São Paulo), and P. Soisook (Princess Maha Chakri Sirindhorn Natural History Museum, Prince of Songkla University, Thailand) for contributing information to Table S1.
K.L.P. was supported by the Defense Threat Reduction Agency (HDTRA11710064), J.A.C. and J.L.D. were supported by the National Science Foundation (NSF2033482), and D.L.P. was supported by the National Science Foundation (DBI-1547229). CETAF and DiSSCo provided funding for this publication.
C.W.T., K.L.P., M.W.A., J.A.C., J.L.D., A.W.F., M.G., F.A.A.K., D.L.P., D.M.R., N.B.S., M.P.M.V., P.W.W., M.W., and C.W.K. contributed to the conceptualization, methodology, and writing of the manuscript. D.M.R. obtained IRB approval for the survey. D.M.R. and K.L.P. curated and analyzed the data from the survey found in Text S1 and Data S1. C.W.T., A.W.F., and M.P.M.V. developed Fig. 1, Table 1, and Table S1, respectively. C.W.T., K.L.P., and C.W.K. provided project administration and supervision.
Footnotes
Citation Thompson CW, Phelps KL, Allard MW, Cook JA, Dunnum JL, Ferguson AW, Gelang M, Khan FAA, Paul DL, Reeder DM, Simmons NB, Vanhove MPM, Webala PW, Weksler M, Kilpatrick CW. 2021. Preserve a voucher specimen! The critical need for integrating natural history collections in infectious disease studies. mBio 12:e02698-20. https://doi.org/10.1128/mBio.02698-20.
REFERENCES
- 1.Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. 2008. Global trends in emerging infectious diseases. Nature 451:990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morens DM, Fauci AS. 2020. Emerging pandemic diseases: how we got to COVID-19. Cell 182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, Rambaut A, Robertson DL. 2020. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nat Microbiol 5:1408–1417. doi: 10.1038/s41564-020-0771-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, Si H-R, Zhu Y, Li B, Huang C-L, Chen H-D, Chen J, Luo Y, Guo H, Jiang R-D, Liu M-Q, Chen Y, Shen X-R, Wang X, Zheng X-S, Zhao K, Chen Q-J, Deng F, Liu L-L, Yan B, Zhan F-X, Wang Y-Y, Xiao G-F, Shi Z-L. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Latinne A, Hu B, Olival KJ, Zhu G, Zhang L, Li H, Chmura AA, Field HE, Zambrana-Torrelio C, Epstein JH, Li B, Zhang W, Wang L-F, Shi Z-L, Daszak P. 2020. Origin and cross-species transmission of bat coronaviruses in China. Nat Commun 11:4235. doi: 10.1038/s41467-020-17687-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Lam TT-Y, Jia N, Zhang Y-W, Shum MH-H, Jiang J-F, Zhu H-C, Tong Y-G, Shi Y-X, Ni X-B, Liao Y-S, Li W-J, Jiang B-G, Wei W, Yuan T-T, Zheng K, Cui X-M, Li J, Pei G-Q, Qiang X, Cheung WY-M, Li L-F, Sun F-F, Qin S, Huang J-C, Leung GM, Holmes EC, Hu Y-L, Guan Y, Cao W-C. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583:282–285. doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- 8.Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou J-J, Li N, Guo Y, Li X, Shen X, Zhang Z, Shu F, Huang W, Li Y, Zhang Z, Chen R-A, Wu Y-J, Peng S-M, Huang M, Xie W-J, Cai Q-H, Hou F-H, Chen W, Xiao L, Shen Y. 2020. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583:286–289. doi: 10.1038/s41586-020-2313-x. [DOI] [PubMed] [Google Scholar]
- 9.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu D, Zhu C, Ai L, He T, Wang Y, Ye F, Yang L, Ding C, Zhu X, Lv R, Zhu J, Hassan B, Feng Y, Tan W, Wang C. 2018. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect 7:154. doi: 10.1038/s41426-018-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau SKP, Woo PCY, Li KSM, Huang Y, Wang M, Lam CSF, Xu H, Guo R, Chan K-H, Zheng B-J, Yuen K-Y. 2007. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology 367:428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, Karesh W, Lipkin WI, Morse SS, PREDICT Consortium, Mazet JAK, Goldstein T. 2017. Global patterns in coronavirus diversity. Virus Evol 3:vex012. doi: 10.1093/ve/vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Müller MA, Deng H, Herrler G, Drosten C. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol 84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, Mazet JK, Hu B, Zhang W, Peng C, Zhang Y-J, Luo C-M, Tan B, Wang N, Zhu Y, Crameri G, Zhang S-Y, Wang L-F, Daszak P, Shi Z-L. 2013. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gouilh MA, Puechmaille SJ, Diancourt L, Vandenbogaert M, Serra-Cobo J, Lopez Roïg M, Brown P, Moutou F, Caro V, Vabret A, Manuguerra J-C, EPICOREM consortium . 2018. SARS-CoV related betacoronavirus and diverse alphacoronavirus members found in western old-world. Virology 517:88–97. doi: 10.1016/j.virol.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y, Xia L, Zhou J, Zhen W, Feng Y, Guo H, Zhang H, Tu C. 2014. Identification of diverse alphacoronaviruses and genomic characterization of a novel SARS-like coronavirus from bats in China. J Virol 88:7070–7082. doi: 10.1128/JVI.00631-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau SKP, Li KSM, Huang Y, Shek C-T, Tse H, Wang M, Choi GKY, Xu H, Lam CSF, Guo R, Chan K-H, Zheng B-J, Woo PCY, Yuen K-Y. 2010. Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events. J Virol 84:2808–2819. doi: 10.1128/JVI.02219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A, Boniotti MB. 2013. Detection of coronaviruses in bats of various species in Italy. Viruses 5:2679–2689. doi: 10.3390/v5112679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X-D, Wang W, Hao Z-Y, Wang Z-X, Guo W-P, Guan X-Q, Wang M-R, Wang H-W, Zhou R-H, Li M-H, Tang G-P, Wu J, Holmes EC, Zhang Y-Z. 2017. Extensive diversity of coronaviruses in bats from China. Virology 507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauly M, Pir JB, Loesch C, Sausy A, Snoeck CJ, Hübschen JM, Muller CP. 2017. Novel alphacoronaviruses and paramyxoviruses cocirculate with type 1 and severe acute respiratory system (SARS)-related betacoronaviruses in synanthropic bats of Luxembourg. Appl Environ Microbiol 83:e01326-17. doi: 10.1128/AEM.01326-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan P-L, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A, Hutchison SK, Egholm M, Osinubi MOV, Niezgoda M, Ogunkoya AB, Briese T, Rupprecht CE, Lipkin WI. 2010. Identification of a severe acute respiratory syndrome coronavirus-like virus in a leaf-nosed bat in Nigeria. mBio 1:e00208-10. doi: 10.1128/mBio.00208-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rihtaric D, Hostnik P, Steyer A, Grom J, Toplak I. 2010. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch Virol 155:507–514. doi: 10.1007/s00705-010-0612-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao Y, Tong S. 2019. Complete genome sequence of a severe acute respiratory syndrome-related coronavirus from Kenyan bats. Microbiol Resour Announc 8:e00548-19. doi: 10.1128/MRA.00548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tong S, Conrardy C, Ruone S, Kuzmin IV, Guo X, Tao Y, Niezgoda M, Haynes L, Agwanda B, Breiman RF, Anderson LJ, Rupprecht CE. 2009. Detection of novel SARS-like and other coronaviruses in bats from Kenya. Emerg Infect Dis 15:482–485. doi: 10.3201/eid1503.081013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Fu S, Cao Y, Zhang H, Feng Y, Yang W, Nie K, Ma X, Liang G. 2017. Discovery and genetic analysis of novel coronaviruses in least horseshoe bats in southwestern China. Emerg Microbes Infect 6:e14. doi: 10.1038/emi.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan J, Hon C-C, Li Y, Wang D, Xu G, Zhang H, Zhou P, Poon LLM, Lam TT-Y, Leung FC-C, Shi Z. 2010. Intraspecies diversity of SARS-like coronaviruses in Rhinolophus sinicus and its implications for the origin of SARS coronaviruses in humans. J Gen Virol 91:1058–1062. doi: 10.1099/vir.0.016378-0. [DOI] [PubMed] [Google Scholar]
- 28.Cook JA, Arai S, Armién B, Bates J, Bonilla CAC, de Souza Cortez MB, Dunnum JL, Ferguson AW, Johnson KM, Khan FAA, Paul DL, Reeder DM, Revelez MA, Simmons NB, Thiers BM, Thompson CW, Upham NS, Vanhove MPM, Webala PW, Weksler M, Yanagihara R, Soltis PS. 2020. Integrating biodiversity infrastructure into pathogen discovery and mitigation of emerging infectious diseases. Bioscience 70:531–534. doi: 10.1093/biosci/biaa064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anthony SJ, Gilardi K, Menachery VD, Goldstein T, Ssebide B, Mbabazi R, Navarrete-Macias I, Liang E, Wells H, Hicks A, Petrosov A, Byarugaba DK, Debbink K, Dinnon KH, Scobey T, Randell SH, Yount BL, Cranfield M, Johnson CK, Baric RS, Lipkin WI, Mazet JAK. 2017. Further evidence for bats as the evolutionary source of Middle East respiratory syndrome coronavirus. mBio 8:e00373-17. doi: 10.1128/mBio.00373-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gouilh MA, Puechmaille SJ, Gonzalez JP, Teeling E, Kittayapong P, Manuguerra J-C. 2011. SARS-coronavirus ancestor’s foot-prints in South-East Asian bat colonies and the refuge theory. Infect Genet Evol 11:1690–1702. doi: 10.1016/j.meegid.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JSM, Poon LLM. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 32.Kan B, Wang M, Jing H, Xu H, Jiang X, Yan M, Liang W, Zheng H, Wan K, Liu Q, Cui B, Xu Y, Zhang E, Wang H, Ye J, Li G, Li M, Cui Z, Qi X, Chen K, Du L, Gao K, Zhao Y-T, Zou X-Z, Feng Y-J, Gao Y-F, Hai R, Yu D, Guan Y, Xu J. 2005. Molecular evolution analysis and geographic investigation of severe acute respiratory syndrome coronavirus-like virus in palm civets at an animal market and on farms. J Virol 79:11892–11900. doi: 10.1128/JVI.79.18.11892-11900.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, Wong SSY, Leung S-Y, Chan K-H, Yuen K-Y. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci U S A 102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang L-F. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 35.Liu P, Chen W, Chen J-P. 2019. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan pangolins (Manis javanica). Viruses 11:979. doi: 10.3390/v11110979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maganga GD, Bourgarel M, Vallo P, Dallo TD, Ngoagouni C, Drexler JF, Drosten C, Nakouné ER, Leroy EM, Morand S. 2014. Bat distribution size or shape as determinant of viral richness in African bats. PLoS One 9:e100172. doi: 10.1371/journal.pone.0100172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, Müller MA, Annan A, Vallo P, Adu-Sarkodie Y, Kruppa TF, Drosten C. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats. Emerg Infect Dis 15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CS, de Jong CE, Meers J, Henning J, Wang L-F, Field HE. 2016. Coronavirus infection and diversity in bats in the Australasian region. Ecohealth 13:72–82. doi: 10.1007/s10393-016-1116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song H-D, Tu C-C, Zhang G-W, Wang S-Y, Zheng K, Lei L-C, Chen Q-X, Gao Y-W, Zhou H-Q, Xiang H, Zheng H-J, Chern S-WW, Cheng F, Pan C-M, Xuan H, Chen S-J, Luo H-M, Zhou D-H, Liu Y-F, He J-F, Qin P-Z, Li L-H, Ren Y-Q, Liang W-J, Yu Y-D, Anderson L, Wang M, Xu R-H, Wu X-W, Zheng H-Y, Chen J-D, Liang G, Gao Y, Liao M, Fang L, Jiang L-Y, Li H, Chen F, Di B, He L-J, Lin J-Y, Tong S, Kong X, Du L, Hao P, Tang H, Bernini A, Yu X-J, Spiga O, Guo Z-M, Pan H-Y, et al. 2005. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc Natl Acad Sci U S A 102:2430–2435. doi: 10.1073/pnas.0409608102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki J, Sato R, Kobayashi T, Aoi T, Harasawa R. 2014. Group B betacoronavirus in rhinolophid bats, Japan. J Vet Med Sci 76:1267–1269. doi: 10.1292/jvms.14-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang XC, Zhang JX, Zhang SY, Wang P, Fan XH, Li LF, Li G, Dong BQ, Liu W, Cheung CL, Xu KM, Song WJ, Vijaykrishna D, Poon LLM, Peiris JSM, Smith GJD, Chen H, Guan Y. 2006. Prevalence and genetic diversity of coronaviruses in bats from China. J Virol 80:7481–7490. doi: 10.1128/JVI.00697-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geldenhuys M, Mortlock M, Weyer J, Bezuidt O, Seamark ECJ, Kearney T, Gleasner C, Erkkila TH, Cui H, Markotter W. 2018. A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa. PLoS One 13:e0194527. doi: 10.1371/journal.pone.0194527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joffrin L, Goodman SM, Wilkinson DA, Ramasindrazana B, Lagadec E, Gomard Y, Le Minter G, Dos Santos A, Schoeman MC, Sookhareea R, Tortosa P, Julienne S, Gudo ES, Mavingui P, Lebarbenchon C. 2020. Bat coronavirus phylogeography in the Western Indian Ocean. Sci Rep 10:6873. doi: 10.1038/s41598-020-63799-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, Madani TA. 2014. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med 370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 45.Corman VM, Kallies R, Philipps H, Göpner G, Müller MA, Eckerle I, Brünink S, Drosten C, Drexler JF. 2014. Characterization of a novel betacoronavirus related to Middle East respiratory syndrome coronavirus in European hedgehogs. J Virol 88:717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang C, Liu WJ, Xu W, Jin T, Zhao Y, Song J, Shi Y, Ji W, Jia H, Zhou Y, Wen H, Zhao H, Liu H, Li H, Wang Q, Wu Y, Wang L, Liu D, Liu G, Yu H, Holmes EC, Lu L, Gao GF. 2016. A bat-derived putative cross-family recombinant coronavirus with a reovirus gene. PLoS Pathog 12:e1005883. doi: 10.1371/journal.ppat.1005883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, AlHakeem R, Al Asmari M, Islam A, Kapoor A, Briese T, Daszak P, Al Rabeeah A, Lipkin WI. 2013. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg Infect Dis 19:1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patrono LV, Samuni L, Corman VM, Nourifar L, Röthemeier C, Wittig RM, Drosten C, Calvignac-Spencer S, Leendertz FH. 2018. Human coronavirus OC43 outbreak in wild chimpanzees, Côte d’Ivoire, 2016. Emerg Microbes Infect 7:118. doi: 10.1038/s41426-018-0121-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woo PCY, Lau SKP, Li KSM, Poon RWS, Wong BHL, Tsoi H-W, Yip BCK, Huang Y, Chan K-H, Yuen K-Y. 2006. Molecular diversity of coronaviruses in bats. Virology 351:180–187. doi: 10.1016/j.virol.2006.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu Z, Yang L, Ren X, Zhang J, Yang F, Zhang S, Jin Q. 2016. ORF8-related genetic evidence for Chinese horseshoe bats as the source of human severe acute respiratory syndrome coronavirus. J Infect Dis 213:579–583. doi: 10.1093/infdis/jiv476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Z, Lu L, Du J, Yang L, Ren X, Liu B, Jiang J, Yang J, Dong J, Sun L, Zhu Y, Li Y, Zheng D, Zhang C, Su H, Zheng Y, Zhou H, Zhu G, Li H, Chmura A, Yang F, Daszak P, Wang J, Liu Q, Jin Q. 2018. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178. doi: 10.1186/s40168-018-0554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yinda CK, Ghogomu SM, Conceição-Neto N, Beller L, Deboutte W, Vanhulle E, Maes P, Van Ranst M, Matthijnssens J. 2018. Cameroonian fruit bats harbor divergent viruses, including rotavirus H, bastroviruses, and picobirnaviruses using an alternative genetic code. Virus Evol 4:vey008. doi: 10.1093/ve/vey008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gomard Y, Dietrich M, Wieseke N, Ramasindrazana B, Lagadec E, Goodman SM, Dellagi K, Tortosa P. 2016. Malagasy bats shelter a considerable genetic diversity of pathogenic Leptospira suggesting notable host-specificity patterns. FEMS Microbiol Ecol 92:fiw037. doi: 10.1093/femsec/fiw037. [DOI] [PubMed] [Google Scholar]
- 54.Wilkinson DA, Mélade J, Dietrich M, Ramasindrazana B, Soarimalala V, Lagadec E, Le Minter G, Tortosa P, Heraud J-M, de Lamballerie X, Goodman SM, Dellagi K, Pascalis H. 2014. Highly diverse morbillivirus-related paramyxoviruses in wild fauna of the southwestern Indian Ocean Islands: evidence of exchange between introduced and endemic small mammals. J Virol 88:8268–8277. doi: 10.1128/JVI.01211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkinson DA, Temmam S, Lebarbenchon C, Lagadec E, Chotte J, Guillebaud J, Ramasindrazana B, Héraud J-M, de Lamballerie X, Goodman SM, Dellagi K, Pascalis H. 2012. Identification of novel paramyxoviruses in insectivorous bats of the Southwest Indian Ocean. Virus Res 170:159–163. doi: 10.1016/j.virusres.2012.08.022. [DOI] [PubMed] [Google Scholar]
- 56.Mélade J, Wieseke N, Ramasindrazana B, Flores O, Lagadec E, Gomard Y, Goodman SM, Dellagi K, Pascalis H. 2016. An eco-epidemiological study of Morbilli-related paramyxovirus infection in Madagascar bats reveals host-switching as the dominant macro-evolutionary mechanism. Sci Rep 6:23752. doi: 10.1038/srep23752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Galbreath KE, Hoberg EP, Cook JA, Armién B, Bell KC, Campbell ML, Dunnum JL, Dursahinhan AT, Eckerlin RP, Gardner SL, Greiman SE, Henttonen H, Jiménez FA, Koehler AVA, Nyamsuren B, Tkach VV, Torres-Pérez F, Tsvetkova A, Hope AG. 2019. Building an integrated infrastructure for exploring biodiversity: field collections and archives of mammals and parasites. J Mammal 100:382–393. doi: 10.1093/jmammal/gyz048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waudby HP, Petit S, Gill MJ. 2019. The scientific, financial and ethical implications of three common wildlife-trapping designs. Wildl Res 46:690–700. doi: 10.1071/WR19084. [DOI] [Google Scholar]
- 59.Lemckert F, Brassil T, Kavanagh R, Law B. 2006. Trapping small mammals for research and management: how many die and why? Aust Mammal 28:201–207. doi: 10.1071/AM06028. [DOI] [Google Scholar]
- 60.Monamy V, Gott M. 2001. Practical and ethical considerations for students conducting ecological research involving wildlife. Austral Ecol 26:293–300. doi: 10.1046/j.1442-9993.2001.01111.x. [DOI] [Google Scholar]
- 61.Culley TM 2013. Why vouchers matter in botanical research. Appl Plant Sci 1:1300076. doi: 10.3732/apps.1300076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huber JT 1998. The importance of voucher specimens, with practical guidelines for preserving specimens of the major invertebrate phyla for identification. J Nat Hist 32:367–385. doi: 10.1080/00222939800770191. [DOI] [Google Scholar]
- 63.Turney S, Cameron ER, Cloutier CA, Buddle CM. 2015. Non-repeatable science: assessing the frequency of voucher specimen deposition reveals that most arthropod research cannot be verified. PeerJ 3:e1168. doi: 10.7717/peerj.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cook JA, Galbreath KE, Bell KC, Campbell ML, Carrière S, Colella JP, Dawson NG, Dunnum JL, Eckerlin RP, Fedorov V, Greiman SE, Haas GMS, Haukisalmi V, Henttonen H, Hope AG, Jackson D, Jung TS, Koehler AV, Kinsella JM, Krejsa D, Kutz SJ, Liphardt S, MacDonald SO, Malaney JL, Makarikov A, Martin J, McLean BS, Mulders R, Nyamsuren B, Talbot SL, Tkach VV, Tsvetkova A, Toman HM, Waltari EC, Whitman JS, Hoberg EP. 2017. The Beringian Coevolution Project: holistic collections of mammals and associated parasites reveal novel perspectives on evolutionary and environmental change in the North. Arctic Sci 3:585–617. doi: 10.1139/as-2016-0042. [DOI] [Google Scholar]
- 65.Lendemer J, Thiers B, Monfils AK, Zaspel J, Ellwood ER, Bentley A, LeVan K, Bates J, Jennings D, Contreras D, Lagomarsino L, Mabee P, Ford LS, Guralnick R, Gropp RE, Revelez M, Cobb N, Seltmann K, Aime MC. 2020. The extended specimen network: a strategy to enhance US biodiversity collections, promote research and education. Bioscience 70:23–30. doi: 10.1093/biosci/biz140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bi K, Linderoth T, Vanderpool D, Good JM, Nielsen R, Moritz C. 2013. Unlocking the vault: next-generation museum population genomics. Mol Ecol 22:6018–6032. doi: 10.1111/mec.12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rocha LA, Aleixo A, Allen G, Almeda F, Baldwin CC, Barclay MVL, Bates JM, Bauer AM, Benzoni F, Berns CM, Berumen ML, Blackburn DC, Blum S, Bolaños F, Bowie RCK, Britz R, Brown RM, Cadena CD, Carpenter K, Ceríaco LM, Chakrabarty P, Chaves G, Choat JH, Clements KD, Collette BB, Collins A, Coyne J, Cracraft J, Daniel T, de Carvalho MR, de Queiroz K, Di Dario F, Drewes R, Dumbacher JP, Engilis A, Erdmann MV, Eschmeyer W, Feldman CR, Fisher BL, Fjeldså J, Fritsch PW, Fuchs J, Getahun A, Gill A, Gomon M, Gosliner T, Graves GR, Griswold CE, Guralnick R, Hartel K, Helgen KM, et al. 2014. Collecting biological specimens is essential in science and conservation. Science 344:814–815. doi: 10.1126/science.344.6186.814. [DOI] [PubMed] [Google Scholar]
- 68.Yates TL 1985. The role of voucher specimens in mammal collections: characterization and funding responsibilities. Acta Zool Fenn 170:81–82. [Google Scholar]
- 69.Mills JN, Childs JE. 1998. Ecologic studies of rodent reservoirs: their relevance for human health. Emerg Infect Dis 4:529–537. doi: 10.3201/eid0404.980403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dunnum JL, Yanagihara R, Johnson KM, Armien B, Batsaikhan N, Morgan L, Cook JA. 2017. Biospecimen repositories and integrated databases as critical infrastructure for pathogen discovery and pathobiology research. PLoS Negl Trop Dis 11:e0005133. doi: 10.1371/journal.pntd.0005133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weksler M, Lemos EMS, D’Andrea PS, Bonvicino CR. 2017. The taxonomic status of Oligoryzomys mattogrossae (Allen 1916) (Rodentia: Cricetidae: Sigmodontinae), reservoir of Anajatuba hantavirus. Am Mus Novit 3880:1–32. doi: 10.1206/3880.1. [DOI] [Google Scholar]
- 72.DiEuliis D, Johnson KR, Morse SS, Schindel DE. 2016. Specimen collections should have a much bigger role in infectious disease research and response. Proc Natl Acad Sci U S A 113:4–7. doi: 10.1073/pnas.1522680112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lajaunie C, Ho CW-L. 2018. Pathogens collections, biobanks and related-data in a One Health legal and ethical perspective. Parasitology 145:688–696. doi: 10.1017/S0031182017001986. [DOI] [PubMed] [Google Scholar]
- 74.McLean BS, Bell KC, Dunnum JL, Abrahamson B, Colella JP, Deardorff ER, Weber JA, Jones AK, Salazar-Miralles F, Cook JA. 2016. Natural history collections-based research: progress, promise, and best practices. J Mammal 97:287–297. doi: 10.1093/jmammal/gyv178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Webster MS 2017. The extended specimen: emerging frontiers in collections-based ornithological research. CRC Press, Boca Rotan, FL. [Google Scholar]
- 76.Brooks DR 1993. Extending the symbiotype concept to host voucher specimens. J Parasitol 79:631–633. doi: 10.2307/3283396. [DOI] [Google Scholar]
- 77.Frey JK, Yates TL, Duszynski DW, Gannon WL, Gardner SL. 1992. Designation and curatorial management of type host specimens (symbiotypes) for new parasite species. J Parasitol 78:930–932. doi: 10.2307/3283335. [DOI] [Google Scholar]
- 78.Brooks DR, Hoberg EP, Boeger WA, Gardner SL, Araujo SBL, Bajer K, Botero-Cañola S, Byrd B, Földvári G, Cook JA, Dunnum JL, Dursahinhan AT, Garamszegi LZ, Herczeg D, Jakab F, Juarrero A, Kemenesi G, Kurucz K, León-Règagnon V, Mejía-Madrid HH, Molnár O, Nisbett RA, Preiser W, Stuart M, Szathmary E, Trivellone V. 2020. Before the pandemic ends: making sure this never happens again. World Complexity Sci Acad J 1:1–10. doi: 10.46473/WCSAJ27240606/15-05-2020-0002//full/html. [DOI] [Google Scholar]
- 79.Peterson AT 2010. The critical role of permanent voucher specimens of hosts and vectors in public health and epidemiology. Emerg Infect Dis 16:341–342. doi: 10.3201/eid1602.091241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ceballos G, Ehrlich PR, Barnosky AD, García A, Pringle RM, Palmer TM. 2015. Accelerated modern human–induced species losses: entering the sixth mass extinction. Sci Adv 1:e1400253. doi: 10.1126/sciadv.1400253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gibb R, Redding DW, Chin KQ, Donnelly CA, Blackburn TM, Newbold T, Jones KE. 2020. Zoonotic host diversity increases in human-dominated ecosystems. Nature 584:398–402. doi: 10.1038/s41586-020-2562-8. [DOI] [PubMed] [Google Scholar]
- 82.Jones SD, Atshabar B, Schmid BV, Zuk M, Amramina A, Stenseth NC. 2019. Living with plague: lessons from the Soviet Union’s antiplague system. Proc Natl Acad Sci U S A 116:9155–9163. doi: 10.1073/pnas.1817339116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McLean BS, Cook JA, Durden LA, Hoberg EP, Guralnick RP. 2019. The next chapter of human-plague science. Proc Natl Acad Sci U S A 116:14411–14412. doi: 10.1073/pnas.1908836116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farley J 2004. To cast out disease: a history of the International Health Division of Rockefeller Foundation (1913–1951). Oxford University Press, New York, NY. [Google Scholar]
- 85.Soper FL 1977. Ventures in world health: the memoirs of Fred Lowe Soper. Pan American Health Organization, Pan American Sanitary Bureau, Regional Office of the World Health Organization, Washington, DC. [Google Scholar]
- 86.Gilmore RM 1943. Mammalogy in an epidemiological study of jungle yellow fever in Brazil. J Mammal 24:144–162. doi: 10.2307/1374796. [DOI] [Google Scholar]
- 87.Kumm HW, Laemmert HW, Jr. 1950. The geographical distribution of immunity to yellow fever among the primates of Brazil. Am J Trop Med Hyg 30:733–748. doi: 10.4269/ajtmh.1950.s1-30.733. [DOI] [PubMed] [Google Scholar]
- 88.de Oliveira JA, Franco SM. 2005. A coleção de mamíferos do serviço nacional de peste no museu nacional, Rio de Janeiro, Brasil. Arq Mus Nac 63:13–20. [Google Scholar]
- 89.Soper FL, Penna H, Cardoso E, Serafim J, Jr, Frobisher M, Jr, Pinheiro J. 1933. Yellow fever without Aedes aegypti: study of a rural epidemic in the Valle do Chanaan, Espirito Santo, Brazil, 1932. Am J Hyg 18:555–587. doi: 10.1093/oxfordjournals.aje.a117967. [DOI] [Google Scholar]
- 90.Löwy I 1997. Epidemiology, immunology, and yellow fever: the Rockefeller Foundation in Brazil, 1923–1939. J Hist Biol 30:397–417. doi: 10.1023/a:1004295806141. [DOI] [PubMed] [Google Scholar]
- 91.Laemmert HW, Jr, De Castro Ferreira L, Taylor RM. 1946. Part II—Investigations of vertebrate hosts and arthropod vectors. Am J Trop Med Hyg S1-26:23–69. doi: 10.4269/ajtmh.1946.s1-26.23. [DOI] [PubMed] [Google Scholar]
- 92.Childs ML, Nova N, Colvin J, Mordecai EA. 2019. Mosquito and primate ecology predict human risk of yellow fever virus spillover in Brazil. Philos Trans R Soc Lond B Biol Sci 374:20180335. doi: 10.1098/rstb.2018.0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laemmert HW, Hughes TP, Causey OR. 1949. The invasion of small forests by yellow fever virus as indicated by immunity in cebus monkeys. Am J Trop Med Hyg 29:555–565. doi: 10.4269/ajtmh.1949.s1-29.555. [DOI] [PubMed] [Google Scholar]
- 94.Yates TL, Mills JN, Parmenter CA, Ksiazek TG, Parmenter RR, Vande Castle JR, Calisher CH, Nichol ST, Abbott KD, Young JC, Morrison ML, Beaty BJ, Dunnum JL, Baker RJ, Salazar-Bravo J, Peters CJ. 2002. The ecology and evolutionary history of an emergent disease: hantavirus pulmonary syndrome: evidence from two El Niño episodes in the American southwest suggests that El Niño–driven precipitation, the initial catalyst of a trophic cascade that results in a delayed density-dependent rodent response, is sufficient to predict heightened risk for human contraction of hantavirus pulmonary syndrome. Bioscience 52:989–998. doi: 10.1641/0006-3568(2002)052[0989:TEAEHO]2.0.CO;2. [DOI] [Google Scholar]
- 95.Agrellos R, Bonvicino CR, Rosa EST, Marques AAR, D’Andrea PS, Weksler M. 2012. The taxonomic status of the Castelo dos Sonhos hantavirus reservoir, Oligoryzomys utiaritensis Allen 1916 (Rodentia: Cricetidae: Sigmodontinae). Zootaxa 3220:1–28. doi: 10.11646/zootaxa.3220.1.1. [DOI] [Google Scholar]
- 96.González-Ittig RE, Rivera PC, Levis SC, Calderón GE, Gardenal CN. 2014. The molecular phylogenetics of the genus Oligoryzomys (Rodentia: Cricetidae) clarifies rodent host–hantavirus associations. Zool J Linn Soc 171:457–474. doi: 10.1111/zoj.12133. [DOI] [Google Scholar]
- 97.Hanson JD, Utrera A, Fulhorst CF. 2011. The delicate pygmy rice rat (Oligoryzomys delicatus) is the principal host of Maporal virus (family Bunyaviridae, genus Hantavirus). Vector Borne Zoonotic Dis 11:691–696. doi: 10.1089/vbz.2010.0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hurtado N, D’Elía G. 2019. Taxonomy of the long-tailed mouse Oligoryzomys destructor (Sigmodontinae: Oryzomyini) with the designation of neotypes for Hesperomys destructor Tschudi, 1844 and Hesperomys melanostoma Tschudi, 1844. J Zool Syst Evol Res 57:127–144. doi: 10.1111/jzs.12232. [DOI] [Google Scholar]
- 99.Firth C, Tokarz R, Simith DB, Nunes MRT, Bhat M, Rosa EST, Medeiros DBA, Palacios G, Vasconcelos PFC, Lipkin WI. 2012. Diversity and distribution of hantaviruses in South America. J Virol 86:13756–13766. doi: 10.1128/JVI.02341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gu SH, Lim BK, Kadjo B, Arai S, Kim J-A, Nicolas V, Lalis A, Denys C, Cook JA, Dominguez SR, Holmes KV, Urushadze L, Sidamonidze K, Putkaradze D, Kuzmin IV, Kosoy MY, Song J-W, Yanagihara R. 2014. Molecular phylogeny of hantaviruses harbored by insectivorous bats in Côte d’Ivoire and Vietnam. Viruses 6:1897–1910. doi: 10.3390/v6051897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kang HJ, Gu SH, Yashina LN, Cook JA, Yanagihara R. 2019. Highly divergent genetic variants of soricid-borne Altai virus (Hantaviridae) in Eurasia suggest ancient host-switching events. Viruses 11:857. doi: 10.3390/v11090857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmitt CJ, Cook JA, Zamudio KR, Edwards SV. 2018. Museum specimens of terrestrial vertebrates are sensitive indicators of environmental change in the Anthropocene. Philos Trans R Soc Lond B Biol Sci 374:20170387. doi: 10.1098/rstb.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Welicky RL, Malherbe W, Hadfield KA, Smit NJ. 2019. Understanding growth relationships of African cymothoid fish parasitic isopods using specimens from museum and field collections. Int J Parasitol Parasites Wildl 8:182–187. doi: 10.1016/j.ijppaw.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vanhove MPM, Volckaert FAM, Pariselle A. 2011. Ancyrocephalidae (Monogenea) of Lake Tanganyika: I: four new species of Cichlidogyrus from Ophthalmotilapia ventralis (Teleostei: Cichlidae), the first record of this parasite family in the basin. Zoologia 28:253–263. doi: 10.1590/S1984-46702011000200016. [DOI] [Google Scholar]
- 105.Kmentová N, Koblmüller S, Van Steenberge M, Artois T, Muterezi Bukinga F, Mulimbwa N’sibula T, Muzumani Risasi D, Masilya Mulungula P, Gelnar M, Vanhove MPM. 2019. Failure to diverge in African Great Lakes: the case of Dolicirroplectanum lacustre gen. nov. comb. nov. (Monogenea, Diplectanidae) infecting latid hosts. J Great Lakes Res 46:1113–1130. [Google Scholar]
- 106.Harmon A, Littlewood DTJ, Wood CL. 2019. Parasites lost: using natural history collections to track disease change across deep time. Front Ecol Environ 17:157–166. doi: 10.1002/fee.2017. [DOI] [Google Scholar]
- 107.Black GA 1983. Taxonomy of a swimbladder nematode, Cystidicola stigmatura (Leidy), and evidence of its decline in the Great Lakes. Can J Fish Aquat Sci 40:643–647. doi: 10.1139/f83-085. [DOI] [Google Scholar]
- 108.Howard I, Davis E, Lippert G, Quinn TP, Wood CL. 2019. Abundance of an economically important nematode parasite increased in Puget Sound between 1930 and 2016: evidence from museum specimens confirms historical data. J Appl Ecol 56:190–200. doi: 10.1111/1365-2664.13264. [DOI] [Google Scholar]
- 109.Jorissen MWP, Huyse T, Pariselle A, Wamuini Lunkayilakio S, Muterezi Bukinga F, Chocha Manda A, Kapepula Kasembele G, Vreven EJ, Snoeks J, Decru E, Artois T, Vanhove MPM. 2020. Historical museum collections help detect parasite species jumps after tilapia introductions in the Congo Basin. Biol Invasions 22:2825–2844. doi: 10.1007/s10530-020-02288-4. [DOI] [Google Scholar]
- 110.Leirs H, Mills JN, Krebs JW, Childs JE, Akaibe D, Woollen N, Ludwig G, Peters CJ, Ksiazek TG. 1999. Search for the Ebola virus reservoir in Kikwit, Democratic Republic of the Congo: reflections on a vertebrate collection. J Infect Dis 179(Suppl 1):S155–S163. doi: 10.1086/514299. [DOI] [PubMed] [Google Scholar]
- 111.Caron A, Bourgarel M, Cappelle J, Liégeois F, De Nys HM, Roger F. 2018. Ebola virus maintenance: if not (only) bats, what else? Viruses 10:549. doi: 10.3390/v10100549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hranac CR, Marshall JC, Monadjem A, Hayman DTS. 2019. Predicting Ebola virus disease risk and the role of African bat birthing. Epidemics 29:100366. doi: 10.1016/j.epidem.2019.100366. [DOI] [PubMed] [Google Scholar]
- 113.Jayme SI, Field HE, de Jong C, Olival KJ, Marsh G, Tagtag AM, Hughes T, Bucad AC, Barr J, Azul RR, Retes LM, Foord A, Yu M, Cruz MS, Santos IJ, Lim TMS, Benigno CC, Epstein JH, Wang L-F, Daszak P, Newman SH. 2015. Molecular evidence of Ebola Reston virus infection in Philippine bats. Virol J 12:107. doi: 10.1186/s12985-015-0331-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goldstein T, Anthony SJ, Gbakima A, Bird BH, Bangura J, Tremeau-Bravard A, Belaganahalli MN, Wells HL, Dhanota JK, Liang E, Grodus M, Jangra RK, DeJesus VA, Lasso G, Smith BR, Jambai A, Kamara BO, Kamara S, Bangura W, Monagin C, Shapira S, Johnson CK, Saylors K, Rubin EM, Chandran K, Lipkin WI, Mazet JAK. 2018. The discovery of Bombali virus adds further support for bats as hosts of ebolaviruses. Nat Microbiol 3:1084–1089. doi: 10.1038/s41564-018-0227-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Forbes KM, Webala PW, Jääskeläinen AJ, Abdurahman S, Ogola J, Masika MM, Kivistö I, Alburkat H, Plyusnin I, Levanov L, Korhonen EM, Huhtamo E, Mwaengo D, Smura T, Mirazimi A, Anzala O, Vapalahti O, Sironen T. 2019. Bombali virus in Mops condylurus bat, Kenya. Emerg Infect Dis 25:955–957. doi: 10.3201/eid2505.181666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez J-P, Swanepoel R. 2005. Fruit bats as reservoirs of Ebola virus. Nature 438:575–576. doi: 10.1038/438575a. [DOI] [PubMed] [Google Scholar]
- 117.Kock RA, Begovoeva M, Ansumana R, Suluku R. 2019. Searching for the source of Ebola: the elusive factors driving its spillover into humans during the West African outbreak of 2013–2016. Rev Sci Tech 38:113–117. doi: 10.20506/rst.38.1.2946. [DOI] [PubMed] [Google Scholar]
- 118.Leendertz SAJ, Gogarten JF, Düx A, Calvignac-Spencer S, Leendertz FH. 2016. Assessing the evidence supporting fruit bats as the primary reservoirs for Ebola viruses. Ecohealth 13:18–25. doi: 10.1007/s10393-015-1053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Anthony NM, Atteke C, Bruford MW, Dallmeier F, Freedman A, Hardy O, Ibrahim B, Jeffery KJ, Johnson M, Lahm SA, Lepengue N, Lowenstein JH, Maisels F, Mboumba J-F, Mickala P, Morgan K, Ntie S, Smith TB, Sullivan JP, Verheyen E, Gonder MK. 2015. Evolution and conservation of central African biodiversity: priorities for future research and education in the Congo Basin and Gulf of Guinea. Biotropica 47:6–17. doi: 10.1111/btp.12188. [DOI] [Google Scholar]
- 120.Paknia O, Rajaei Sh H, Koch A. 2015. Lack of well-maintained natural history collections and taxonomists in megadiverse developing countries hampers global biodiversity exploration. Org Divers Evol 15:619–629. doi: 10.1007/s13127-015-0202-1. [DOI] [Google Scholar]
- 121.Fisher MA, Vinson JE, Gittleman JL, Drake JM. 2018. The description and number of undiscovered mammal species. Ecol Evol 8:3628–3635. doi: 10.1002/ece3.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Demos TC, Webala PW, Lutz HL, Kerbis Peterhans JC, Goodman SM, Cortés‐Delgado N, Bartonjo M, Patterson BD. 2020. Multilocus phylogeny of a cryptic radiation of Afrotropical long‐fingered bats (Chiroptera, Miniopteridae). Zool Scr 49:1–13. doi: 10.1111/zsc.12388. [DOI] [Google Scholar]
- 123.Kupferschmidt K 24 January 2019. This bat species may be the source of the Ebola epidemic that killed more than 11,000 people in West Africa. Science Magazine. [Google Scholar]
- 124.Amman BR, Carroll SA, Reed ZD, Sealy TK, Balinandi S, Swanepoel R, Kemp A, Erickson BR, Comer JA, Campbell S, Cannon DL, Khristova ML, Atimnedi P, Paddock CD, Crockett RJK, Flietstra TD, Warfield KL, Unfer R, Katongole-Mbidde E, Downing R, Tappero JW, Zaki SR, Rollin PE, Ksiazek TG, Nichol ST, Towner JS. 2012. Seasonal pulses of Marburg virus circulation in juvenile Rousettus aegyptiacus bats coincide with periods of increased risk of human infection. PLoS Pathog 8:e1002877. doi: 10.1371/journal.ppat.1002877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schuh AJ, Amman BR, Jones MEB, Sealy TK, Uebelhoer LS, Spengler JR, Martin BE, Coleman-McCray JAD, Nichol ST, Towner JS. 2017. Modelling filovirus maintenance in nature by experimental transmission of Marburg virus between Egyptian rousette bats. Nat Commun 8:14446. doi: 10.1038/ncomms14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Biek R, Walsh PD, Leroy EM, Real LA. 2006. Recent common ancestry of Ebola Zaire virus found in a bat reservoir. PLoS Pathog 2:e90. doi: 10.1371/journal.ppat.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olivero J, Fa JE, Farfán MÁ, Márquez AL, Real R, Juste FJ, Leendertz SA, Nasi R. 2020. Human activities link fruit bat presence to Ebola virus disease outbreaks. Mamm Rev 50:1–10. doi: 10.1111/mam.12173. [DOI] [Google Scholar]
- 128.Olson SH, Reed P, Cameron KN, Ssebide BJ, Johnson CK, Morse SS, Karesh WB, Mazet JAK, Joly DO. 2012. Dead or alive: animal sampling during Ebola hemorrhagic fever outbreaks in humans. Emerg Health Threats J 5:913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kuisma E, Olson SH, Cameron KN, Reed PE, Karesh WB, Ondzie AI, Akongo M-J, Kaba SD, Fischer RJ, Seifert SN, Muñoz-Fontela C, Becker-Ziaja B, Escudero-Pérez B, Goma-Nkoua C, Munster VJ, Mombouli J-V. 2019. Long-term wildlife mortality surveillance in northern Congo: a model for the detection of Ebola virus disease epizootics. Philos Trans R Soc Lond B Biol Sci 374:20180339. doi: 10.1098/rstb.2018.0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.ACR. 2019. African Chiroptera Report 2019, Appendix 2a. AfricanBats NPC, Pretoria, South Africa. [Google Scholar]
- 131.Dunnum JL, McLean BS, Dowler RC, Systematic Collections Committee of the American Society of Mammalogists . 2018. Mammal collections of the Western Hemisphere: a survey and directory of collections. J Mammal 99:1307–1322. doi: 10.1093/jmammal/gyy151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hutterer R, Kryštufek B. 2020. A history of mammal research in Europe, p 1–22. In Hackländer K, Zachos FE (ed), Handbook of the mammals of Europe. Springer International Publishing, Cham, Switzerland. [Google Scholar]
- 133.Young CCW, Olival KJ. 2016. Optimizing viral discovery in bats. PLoS One 11:e0149237. doi: 10.1371/journal.pone.0149237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hope AG, Sandercock BK, Malaney JL. 2018. Collection of scientific specimens: benefits for biodiversity sciences and limited impacts on communities of small mammals. Bioscience 68:35–42. doi: 10.1093/biosci/bix141. [DOI] [Google Scholar]
- 135.Sikes RS, Animal Care and Use Committee of the American Society of Mammalogists . 2016. 2016 Guidelines of the American Society of Mammalogists for the use of wild mammals in research and education. J Mammal 97:663–688. doi: 10.1093/jmammal/gyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Phillips CD, Dunnum JL, Dowler RC, Bradley LC, Garner HJ, MacDonald KA, Lim BK, Revelez MA, Campbell ML, Lutz HL, Garza NO, Cook JA, Bradley RD, Alvarez-Castañeda ST, Bradley JE, Bradley RD, Carraway LN, Carrera-E JP, Conroy CJ, Coyner BS, Demboski JR, Dick CW, Dowler RC, Doyle K, Dunnum JL, Esselstyn JA, Gutiérrez E, Hanson JD, Holahan PM, Holmes T, Iudica CA, Leite RN, Lee TE, Lim BK, Malaney JL, McLean BS, McLaren SB, Moncrief ND, Olson L, Ordóñez-Garza N, Phillips CD, Revelez MA, Rickart EA, Rogers DS, Thompson CW, Upham NS, Velazco PM, Systematic Collections Committee of the American Society of Mammalogists . 2019. Curatorial guidelines and standards of the American Society of Mammalogists for collections of genetic resources. J Mammal 100:1690–1694. doi: 10.1093/jmammal/gyz111. [DOI] [Google Scholar]
- 137.Simmons JE 2014. Fluid preservation: a comprehensive reference. Rowman & Littlefield, Lanham, MD. [Google Scholar]
- 138.Ruane S, Austin CC. 2017. Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Mol Ecol Resour 17:1003–1008. doi: 10.1111/1755-0998.12655. [DOI] [PubMed] [Google Scholar]
- 139.Code of Federal Regulations. 2013. Title 42. Public Health. Section 71.53. Requirements for importers of nonhuman primates. [Google Scholar]
- 140.Bradley RD, Bradley LC, Garner HJ, Baker RJ. 2014. Assessing the value of natural history collections and addressing issues regarding long-term growth and care. Bioscience 64:1150–1158. doi: 10.1093/biosci/biu166. [DOI] [Google Scholar]
- 141.Simmons JE 2006. Things great and small: collections management policies. American Association of Museums, Washington, DC. [Google Scholar]
- 142.Nelson G, Sweeney P, Gilbert E. 2018. Use of globally unique identifiers (GUIDs) to link herbarium specimen records to physical specimens. Appl Plant Sci 6:e1027. doi: 10.1002/aps3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zimkus BM, Ford LS. 2014. Genetic resource collections associated with natural history museums: a survey and analysis to establish a benchmark of standards, p 9–44. In Applequist WL, Campbell LM (ed), DNA banking for the 21st century. Missouri Botanical Garden, St. Louis, MO. [Google Scholar]
- 144.Zimkus BM, Ford LS. 2014. Best practices for genetic resources associated with natural history collections: recommendations for practical implementation. Collection Forum 28:77–112. doi: 10.14351/0831-4985-28.1.77. [DOI] [Google Scholar]
- 145.Kraft CS, Kortepeter MG, Gordon B, Sauer LM, Shenoy ES, Eiras DP, Larson L, Garland JA, Mehta AK, Barrett K, Price CS, Croyle C, West LR, Noren B, Kline S, Arguinchona C, Arguinchona H, Grein JD, Connally C, McLellan S, Risi GF, Uyeki TM, Davey RT, Jr, Schweinle JE, Schwedhelm MM, Harvey M, Hunt RC, Kratochvil CJ. 2019. The special pathogens research network: enabling research readiness. Health Secur 17:35–45. doi: 10.1089/hs.2018.0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Robertson T, Döring M, Guralnick R, Bloom D, Wieczorek J, Braak K, Otegui J, Russell L, Desmet P. 2014. The GBIF integrated publishing toolkit: facilitating the efficient publishing of biodiversity data on the internet. PLoS One 9:e102623. doi: 10.1371/journal.pone.0102623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Miller SE, Barrow LN, Ehlman SM, Goodheart JA, Greiman SE, Lutz HL, Misiewicz TM, Smith SM, Tan M, Thawley CJ, Cook JA, Light JE. 2020. Building natural history collections for the twenty-first century and beyond. Bioscience 70:674–687. doi: 10.1093/biosci/biaa069. [DOI] [Google Scholar]
- 148.Bradley RD, Bradley LC, Honeycutt RL, MacDonald KA, Amarilla-Stevens HN, Stevens RD. 2020. Nomenclatural, curatorial, and archival best practices for symbiotypes and other type materials in natural history collections. Occas Pap Mus Tex Tech Univ 366:1–20. [Google Scholar]
- 149.Courtot M, Cherubin L, Faulconbridge A, Vaughan D, Green M, Richardson D, Harrison P, Whetzel PL, Parkinson H, Burdett T. 2019. BioSamples database: an updated sample metadata hub. Nucleic Acids Res 47:D1172–D1178. doi: 10.1093/nar/gky1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.El Zowalaty ME, Järhult JD. 2020. From SARS to COVID-19: a previously unknown SARS-related coronavirus (SARS-CoV-2) of pandemic potential infecting humans − call for a One Health approach. One Health 9:100124. doi: 10.1016/j.onehlt.2020.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gryseels S, Watts TD, Kabongo Mpolesha J-M, Larsen BB, Lemey P, Muyembe-Tamfum J-J, Teuwen DE, Worobey M. 2020. A near full-length HIV-1 genome from 1966 recovered from formalin-fixed paraffin-embedded tissue. Proc Natl Acad Sci U S A 117:12222–12229. doi: 10.1073/pnas.1913682117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schindel DE, Cook JA. 2018. The next generation of natural history collections. PLoS Biol 16:e2006125. doi: 10.1371/journal.pbio.2006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Malaney JL, Cook JA. 2018. A perfect storm for mammalogy: declining sample availability in a period of rapid environmental degradation. J Mammal 99:773–788. doi: 10.1093/jmammal/gyy082. [DOI] [Google Scholar]
- 154.Kading RC, Kingston T. 2020. Common ground: the foundation of interdisciplinary research on bat disease emergence. PLoS Biol 18:e3000947. doi: 10.1371/journal.pbio.3000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tesh RB, Wilson ML, Salas R, De Manzione NM, Tovar D, Ksiazek TG, Peters CJ. 1993. Field studies on the epidemiology of Venezuelan hemorrhagic fever: implication of the cotton rat Sigmodon alstoni as the probable rodent reservoir. Am J Trop Med Hyg 49:227–235. doi: 10.4269/ajtmh.1993.49.227. [DOI] [PubMed] [Google Scholar]
- 156.Fulhorst CF, Milazzo ML, Carroll DS, Charrel RN, Bradley RD. 2002. Natural host relationships and genetic diversity of Whitewater Arroyo virus in southern Texas. Am J Trop Med Hyg 67:114–118. doi: 10.4269/ajtmh.2002.67.114. [DOI] [PubMed] [Google Scholar]
- 157.Milazzo ML, Barragán-Gomez A, Hanson JD, Estrada-Franco JG, Arellano E, González-Cózatl FX, Fernández-Salas I, Ramirez-Aguilar F, Rogers DS, Bradley RD, Fulhorst CF. 2010. Antibodies to Tacaribe serocomplex viruses (family Arenaviridae, genus Arenavirus) in cricetid rodents from New Mexico, Texas, and Mexico. Vector Borne Zoonotic Dis 10:629–637. doi: 10.1089/vbz.2009.0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bates M 1944. The Saimiri monkey as an experimental host for the virus of Yellow Fever. Am J Trop Med Hyg 1944:83–89. doi: 10.4269/ajtmh.1944.s1-24.83. [DOI] [Google Scholar]
- 159.Davis NC 1930. The transmission of yellow fever: experiments with the “woolly monkey” (Lagotrix lagotricha Humboldt), the “spider monkey” (Ateleus ater F. Cuvier), and the “squirrel monkey” (Saimiri scireus Linnaeus). J Exp Med 51:703–720. doi: 10.1084/jem.51.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Davis NC 1931. The transmission of yellow fever: further experiments with monkeys of the New World. Am J Trop Med Hyg 1931:113–125. doi: 10.4269/ajtmh.1931.s1-11.113. [DOI] [Google Scholar]
- 161.Kumm HW 1932. Yellow fever transmission experiments with South American bats. Ann Trop Med Parasitol 26:207–213. doi: 10.1080/00034983.1932.11684716. [DOI] [Google Scholar]
- 162.Lloyd W, Penna HA. 1933. Yellow fever virus encephalitis in South American monkeys. Am J Trop Med Hyg 1933:243–264. doi: 10.4269/ajtmh.1933.s1-13.243. [DOI] [Google Scholar]
- 163.Williams SC 1994. Nationalism and public health: the convergence of Rockefeller Foundation technique and Brazilian Federal authority during the time of Yellow Fever, 1925–1930, p 23–51. In Cueto M (ed), Missionaries of science: the Rockefeller Foundation and Latin America. Indiana University Press, Bloomington, IN. [Google Scholar]
- 164.Yahnke CJ, Meserve PL, Ksiazek TG, Mills JN. 2001. Patterns of infection with Laguna Negra virus in wild populations of Calomys laucha in the central Paraguayan chaco. Am J Trop Med Hyg 65:768–776. doi: 10.4269/ajtmh.2001.65.768. [DOI] [PubMed] [Google Scholar]
- 165.Mills JN, Johnson JM, Ksiazek TG, Ellis BA, Rollin PE, Yates TL, Mann MO, Johnson MR, Campbell ML, Miyashiro J, Patrick M, Zyzak M, Lavender D, Novak MG, Schmidt K, Peters CJ, Childs JE. 1998. A survey of hantavirus antibody in small-mammal populations in selected United States National Parks. Am J Trop Med Hyg 58:525–532. doi: 10.4269/ajtmh.1998.58.525. [DOI] [PubMed] [Google Scholar]
- 166.Malmstrom CM, Shu R, Linton EW, Newton LA, Cook MA. 2007. Barley yellow dwarf viruses (BYDVs) preserved in herbarium specimens illuminate historical disease ecology of invasive and native grasses. J Ecol 95:1153–1166. doi: 10.1111/j.1365-2745.2007.01307.x. [DOI] [Google Scholar]
- 167.Tiee MS, Harrigan RJ, Thomassen HA, Smith TB. 2018. Ghosts of infections past: using archival samples to understand a century of monkeypox virus prevalence among host communities across space and time. R Soc Open Sci 5:171089. doi: 10.1098/rsos.171089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Doty JB, Malekani JM, Kalemba LN, Stanley WT, Monroe BP, Nakazawa YU, Mauldin MR, Bakambana TL, Liyandja Dja Liyandja T, Braden ZH, Wallace RM, Malekani DV, McCollum AM, Gallardo-Romero N, Kondas A, Peterson AT, Osorio JE, Rocke TE, Karem KL, Emerson GL, Carroll DS. 2017. Assessing monkeypox virus prevalence in small mammals at the human–animal interface in the Democratic Republic of the Congo. Viruses 9:283. doi: 10.3390/v9100283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Traub R, Wisseman CL, Jr, Ahmad N. 1967. The occurrence of scrub typhus infection in unusual habitats in West Pakistan. Trans R Soc Trop Med Hyg 61:23–57. doi: 10.1016/0035-9203(67)90052-1. [DOI] [PubMed] [Google Scholar]
- 170.Hromada M, Klimovičová M. 2015. From dusty collections to descriptions of new species–birds in Sarisske Museum Bardejov as valuable source for investigating mite biodiversity. Folia Oecologica 7:98–108. [Google Scholar]
- 171.Light JE, Hafner MS. 2007. Cophylogeny and disparate rates of evolution in sympatric lineages of chewing lice on pocket gophers. Mol Phylogenet Evol 45:997–1013. doi: 10.1016/j.ympev.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 172.Hoogstraal H 1967. Ticks in relation to human diseases caused by Rickettsia species. Annu Rev Entomol 12:377–420. doi: 10.1146/annurev.en.12.010167.002113. [DOI] [PubMed] [Google Scholar]
- 173.Hoogstraal H 1966. Ticks in relation to human diseases caused by viruses. Annu Rev Entomol 11:261–308. doi: 10.1146/annurev.en.11.010166.001401. [DOI] [PubMed] [Google Scholar]
- 174.Jiménez FA, Patterson BD. 2012. A new species of Pterygodermatites (Nematoda: Rictulariidae) from the Incan shrew opossum, Lestoros inca. J Parasitol 98:604–607. doi: 10.1645/GE-3014.1. [DOI] [PubMed] [Google Scholar]
- 175.Asakawa M, Nicolas V. 2003. A new host and locality records of a spirurid nematode species, Protospirura muricola, from Gabonese wild murids. Biogeography 5:67–70. [Google Scholar]
- 176.Hughes MR, Negovetich NJ, Mayes BC, Dowler RC. 2018. Prevalence and intensity of the sinus roundworm (Skrjabingylus chitwoodorum) in rabies-negative skunks of Texas, USA. J Wildl Dis 54:85–94. doi: 10.7589/2017-02-023. [DOI] [PubMed] [Google Scholar]
- 177.Van Steenberge M, Pariselle A, Huyse T, Volckaert FAM, Snoeks J, Vanhove MPM. 2015. Morphology, molecules, and monogenean parasites: an example of an integrative approach to cichlid biodiversity. PLoS One 10:e0124474. doi: 10.1371/journal.pone.0124474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Emery AM, Allan FE, Rabone ME, Rollinson D. 2012. Schistosomiasis collection at NHM (SCAN). Parasit Vectors 5:185. doi: 10.1186/1756-3305-5-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Wyatt KB, Campos PF, Gilbert MTP, Kolokotronis S-O, Hynes WH, DeSalle R, Ball SJ, Daszak P, MacPhee RDE, Greenwood AD. 2008. Historical mammal extinction on Christmas Island (Indian Ocean) correlates with introduced infectious disease. PLoS One 3:e3602. doi: 10.1371/journal.pone.0003602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hershkovitz P 1983. Two new species of night monkeys, genus Aotus (Cebidae, platyrrhini): a preliminary report on Aotus taxonomy. Am J Primatol 4:209–243. doi: 10.1002/ajp.1350040302. [DOI] [PubMed] [Google Scholar]
- 181.Fooden J 1994. Malaria in macaques. Int J Primatol 15:573–596. doi: 10.1007/BF02735972. [DOI] [Google Scholar]
- 182.Lutz HL, Patterson BD, Kerbis Peterhans JC, Stanley WT, Webala PW, Gnoske TP, Hackett SJ, Stanhope MJ. 2016. Diverse sampling of East African haemosporidians reveals chiropteran origin of malaria parasites in primates and rodents. Mol Phylogenet Evol 99:7–15. doi: 10.1016/j.ympev.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 183.Lutz HL, Hochachka WM, Engel JI, Bell JA, Tkach VV, Bates JM, Hackett SJ, Weckstein JD. 2015. Parasite prevalence corresponds to host life history in a diverse assemblage of afrotropical birds and haemosporidian parasites. PLoS One 10:e0121254. doi: 10.1371/journal.pone.0121254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schaer J, Perkins SL, Ejotre I, Vodzak ME, Matuschewski K, Reeder DM. 2017. Epauletted fruit bats display exceptionally high infections with a Hepatocystis species complex in South Sudan. Sci Rep 7:6928. doi: 10.1038/s41598-017-07093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schaer J, Reeder DM, Vodzak ME, Olival KJ, Weber N, Mayer F, Matuschewski K, Perkins SL. 2015. Nycteria parasites of Afrotropical insectivorous bats. Int J Parasitol 45:375–384. doi: 10.1016/j.ijpara.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 186.Dietrich M, Wilkinson DA, Soarimalala V, Goodman SM, Dellagi K, Tortosa P. 2014. Diversification of an emerging pathogen in a biodiversity hotspot: Leptospira in endemic small mammals of Madagascar. Mol Ecol 23:2783–2796. doi: 10.1111/mec.12777. [DOI] [PubMed] [Google Scholar]
- 187.Lagadec E, Gomard Y, Guernier V, Dietrich M, Pascalis H, Temmam S, Ramasindrazana B, Goodman SM, Tortosa P, Dellagi K. 2012. Pathogenic Leptospira spp. in bats, Madagascar and Union of the Comoros. Emerg Infect Dis 18:1696–1698. doi: 10.3201/eid1810.111898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Rothschild BM, Martin LD, Lev G, Bercovier H, Bar-Gal GK, Greenblatt C, Donoghue H, Spigelman M, Brittain D. 2001. Mycobacterium tuberculosis complex DNA from an extinct bison dated 17,000 years before the present. Clin Infect Dis 33:305–311. doi: 10.1086/321886. [DOI] [PubMed] [Google Scholar]
- 189.Harbeck M, Seifert L, Hänsch S, Wagner DM, Birdsell D, Parise KL, Wiechmann I, Grupe G, Thomas A, Keim P, Zöller L, Bramanti B, Riehm JM, Scholz HC. 2013. Yersinia pestis DNA from skeletal remains from the 6th century AD reveals insights into Justinianic plague. PLoS Pathog 9:e1003349. doi: 10.1371/journal.ppat.1003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Martin MD, Cappellini E, Samaniego JA, Zepeda ML, Campos PF, Seguin-Orlando A, Wales N, Orlando L, Ho SYW, Dietrich FS, Mieczkowski PA, Heitman J, Willerslev E, Krogh A, Ristaino JB, Gilbert MTP. 2013. Reconstructing genome evolution in historic samples of the Irish potato famine pathogen. Nat Commun 4:2172. doi: 10.1038/ncomms3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ristaino JB, Groves CT, Parra GR. 2001. PCR amplification of the Irish potato famine pathogen from historic specimens. Nature 411:695–697. doi: 10.1038/35079606. [DOI] [PubMed] [Google Scholar]
- 192.Ristaino JB 2002. Tracking historic migrations of the Irish potato famine pathogen, Phytophthora infestans. Microbes Infect 4:1369–1377. doi: 10.1016/s1286-4579(02)00010-2. [DOI] [PubMed] [Google Scholar]
- 193.Campana MG, Kurata NP, Foster JT, Helgen LE, Reeder DM, Fleischer RC, Helgen KM. 2017. White-nose syndrome fungus in a 1918 bat specimen from France. Emerg Infect Dis 23:1611–1612. doi: 10.3201/eid2309.170875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Oghenekaro AO, Miettinen O, Omorusi VI, Evueh GA, Farid MA, Gazis R, Asiegbu FO. 2014. Molecular phylogeny of Rigidoporus microporus isolates associated with white rot disease of rubber trees (Hevea brasiliensis). Fungal Biol 118:495–506. doi: 10.1016/j.funbio.2014.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Microbiology sampling methods and museums—a human survey. Download Text S1, DOCX file, 0.03 MB (32.4KB, docx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Microbiology sampling methods and museums—a human survey. Download Data Set S1, XLSX file, 0.03 MB (132.8KB, xlsx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Natural history collections with previous experience and/or interest in voucher-based host-pathogen studies. Abbreviations of natural history collections are as follows: AMNH, American Museum of Natural History; CTUA, Colección Teriológica, Universidad de Antioquia; FMNH, Field Museum of Natural History; GNM, Gothenburg Natural History Museum; LMUSP, Laboratório de Mamíferos da Escola Superior de Agricultura “Luiz de Queiroz,” Universidade de São Paulo; MECN, Museo del Instituto Nacional de Biodiversidad, Ecuador; MN, Museu Nacional, Universidade Federal do Rio de Janeiro; MNHN, Muséum national d’Histoire naturelle; MPEG, Museu Paraense Emilio Goeldi; MSB, Museum of Southwestern Biology, University of New Mexico; NHM, Natural History Museum, London; OMNH, Sam Noble Museum of Natural History, University of Oklahoma; PSUNHM, Princess Maha Chakri Sirindhorn Natural History Museum, Prince of Songkla University; QCAZ, Museo de Zoología de la Pontificia Universidad Católica del Ecuador; RMCA, Royal Museum for Central Africa; TTU, Natural Science Research Laboratory, Texas Tech University; UACH, Universidad Austral de Chile; UMMZ, University of Michigan Museum of Zoology. Download Table S1, DOCX file, 0.02 MB (17.2KB, docx) .
Copyright © 2021 Thompson et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.