Abstract
This study is an integrated analysis of the transcriptome profile microRNA (miRNA) and its experimentally validated mRNA targets differentially expressed in the tumorigenic stem-like fraction of oral squamous cell carcinoma (OSCC). We had previously reported the coexistence of multiple drug-resistant tumorigenic fractions, termed side population (SP1, SP2, and MP2), and a nontumorigenic fraction, termed main population (MP1), in oral cancer. These fractions displayed a self-renewal, regenerative potential and expressed known stemness-related cell surface markers despite functional differences. Flow cytometrically sorted pure fractions of SP1 and MP1 cells were subjected to differential expression analysis of both mRNAs and miRNAs. A significant upregulation of genes associated with inflammation, cell survival, cell proliferation, drug transporters, and antiapoptotic pathways, in addition to enhanced transcriptome reprogramming mediated by DNA–histone binding proteins and pattern recognition receptor-mediated signaling, was found to play a crucial role in the transformation of the nontumorigenic MP1 fraction to the tumorigenic SP1 fraction. We also identified several differentially expressed miRNAs that specifically target genes distinctive of tumorigenic SP1 fraction. miRNA-mediated downregulation of stemness-associated markers CD44 and CD147 and upregulation of CD151 may also account for the emergence and persistence of multiple tumorigenic stem cell fractions with varying degrees of malignancy. The phenotypic switch of cancer cells to stem-like OSCC cells mediated by transcriptomal regulation is effectual in addressing biological tumor heterogeneity and subsequent therapeutic resistance leading to a minimal residual disease (MRD) condition in oral cancer. A detailed study of the interplay of miRNAs, mRNA, and the cellular phases involved in the gradual transition of nontumorigenic cancer cells to tumorigenic stem-like cells in solid tumors would enable detection and development of a treatment regimen that targets and successfully eliminates multiple, drug-resistant fractions of cancer cells.
Key words: Cancer stem cells (CSCs), Oral cancer, Side population (SP), Tumorigenic, MicroRNAs (miRNAs)
INTRODUCTION
Ranked as the 11th most common cancer in the world1, oral cancer has one of the lowest (∼50%) overall 5-year survival rates2. The accumulation of random errors or somatic mutations resulting in altered cellular integrity over subsequent cell divisions in normal stem cells often leads to multiple altered stem-like cells, otherwise termed cancer stem cells (CSCs)3. Previous studies from our laboratory have demonstrated that multiple fractions of interconverting oral cancer cells with distinct tumorigenic potential, drug resistance, and malignant transformation coexist within an apparently homogenous pool of cancer cells4. This initiated the “cancer stem cell shift hypothesis,” experimentally proven in a select oral squamous cell carcinoma (OSCC) cell line, SCC172, and an in vitro model to explore the role of oral epithelial stem cells in tumorigenesis4,5. The presence of distinct yet interconverting phenotypes with cells of one phenotype generating progeny resembling another phenotype and vice versa has recently been discovered in breast cancer patient-derived circulating tumor cells (CTCs) and was found to contribute to progression and acquisition of drug resistance6. A parallel study also supported the plasticity of oral CSCs and the transient shift between proliferative epithelial and metastatic mesenchymal CSC subpopulations causing heterogeneity and therapeutic resistance in oral cancer7. Equivalent interconverting tumorigenic stem cell populations have also been identified in breast cancer8.
We identified four phenotypically similar cell fractions with regard to CD147 expression (CD147+ cells) that differed in functional assays [cell cycle stage, aldehyde dehydrogenase (ALDH) expression, clonogenic, and self-renewal potentials] and were isolated based on the expression and lack of drug transporter protein ATP-binding cassette subfamily G member 2 (ABCG2), termed as side population 1/2 (SP1/SP2) and main population 1/2 (MP1/MP2), respectively4. SP1 fraction was the least tumorigenic in comparison to SP2/MP2, whereas the MP1 fraction failed to initiate tumors in NOD/SCID mice4. SP1 and MP1 fractions also displayed a profound regenerative capacity and changeover in culture4. We hypothesize that temporary silencing of genes normally expressed in SP1 could transform it to SP2/MP2 and MP1 fractions and also revert back to SP1 depending on cues from the microenvironment. A lucid comprehension of how each of these fractions actually differs in functionality and phenotype could only be established at the molecular level by identifying genes that are predominantly expressed in the tumorigenic fraction (SP1) in comparison to the nontumorigenic fraction (MP1). To investigate the molecular mechanisms that regulate this shift between cellular fractions leading to differential tumorigenic and malignant state amidst coexisting oral cancer cells, we performed a preliminary comparative analysis of mRNA and microRNA (miRNA) expression profile in tumorigenic SP1 versus nontumorigenic MP1 fraction of SCC172 OSCC cell line.
MATERIALS AND METHODS
Side Population (SP) Analysis
Maintenance of the SCC172 cell line and SP analysis was based on the previously described method4. Sorted SP and MP fractions (see supplemental Fig. S1A, available at http://rgcb.res.in/OncologyResearch.php) were resorted to ensure 100% sort purity using flow cytometer and microscopic visualizing before performing RNA isolation (see supplemental Fig. S1B, available at http://rgcb.res.in/OncologyResearch.php).
Microarray Analysis
Three batches of 6 × 105 SP1 and non-SP (MP1) cells were flow cytometrically sorted as biological replicates and stored in RNA Later (Ambion Inc., Austin, TX, USA). Total RNA was isolated using Qiagen’s All Prep DNA/RNA Mini Kit (Cat. No. 80204), and Human Genome U133 Plus 2.0 Arraychip (Affymetrix, Santa Clara, CA, USA) was used following the manufacturer’s instructions. Biotin-labeled targets (cRNA) were prepared from 100 ng of total RNA (two SP1 and three MP1 samples) using modified MessageAmp™-based protocols (Ambion Inc.). The standard Affymetrix protocol was followed for labeled cRNA, array hybridization, and washing. The hybridization step was carried out in an Affymetrix Model 640 hybridization oven at 45°C for 16 h and stained on an Affymetrix FS450 Fluidics station. Affymetrix GeneChip Scanner 3000 7G was used to scan the arrays. Affymetrix Statistical Algorithm MAS 5.0 (GCOS v1.3) algorithm was used to generate a summary of the image signal data, detection cells with all arrays scaled to 500, and gene annotations on the array. Statistical differential analysis was performed using the one-way ANOVA model to test the null hypothesis followed by a two-sample t-test for every gene and multiplicity correction to control the false discovery rate (FDR) at 0.05.
Real-Time PCR Validation of Analysis
RNA was isolated from FACS-sorted cells isolated independently from the samples used for microarray analysis using Qiagen’s All Prep DNA/RNA Mini Kit (Cat. No. 80204). High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) was used for the synthesis of cDNA from total RNA using a random primer scheme. Total RNA was reverse transcribed in a total reaction volume of 20 μl. The reaction mix consisted of 10 μl of total RNA sample (from SP1 and MP1 fractions) and 10 μl of master mix (10× RT buffer, 25× dNTP mix of 10 mM concentration, 10× RT random primers, reverse transcriptase enzyme, RNase inhibitor, and RNase free water). Thermal cycler conditions for the preparation of cDNA were as follows: step 1—25°C for 10 min; step 2—37°C for 120 min; and step 3—85°C for 5 min and at 4°C∞. TaqMan Gene Expression Assays/primer (FAM) sets selected were all inventoried from Applied Biosystems. Ten nanograms of total RNA equivalent cDNA (assuming 100% cDNA synthesis efficiency) product was used in a volume of 15 μl for each PCR well. The reaction mix for real-time amplification consisted of cDNA, TaqMan gene expression master mix (Applied Biosystems), and individual TaqMan mRNA assays. β-2-Microglobulin (β2M) was used as an endogenous control. No-template (water) reaction mixtures were included as negative controls. All amplifications were performed in triplicate on a validated ABI 7900 real-time thermocycler. Thermal cycler conditions for the qRT-PCR were as follows: step 1 (repeated 1 cycle each)—50°C for 2 min and 95°C for 10 min; step 2 (repeated 40 cycles)—95°C for 15 s and 60°C for 1 min.
Differential miRNA Expression Profiling of SP Cells
Approximately 1 × 106 of SP1 and non-SP (MP1) cells were sorted using flow cytometer, and total RNA was isolated using mirVana miRNA Isolation Kit (Applied Biosystems) following the manufacturer’s protocol. cDNA synthesis was done with TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems) following the manufacturer’s protocol. The SP1 fraction was set as the sample, and MP1 was the control from which differences in the sample were deduced. Input RNA (500 ng) was used for TaqMan Low Density Array Human MicroRNA panel. RNU44 and RNU48 were used as endogenous controls. The array was run on 7900 Real-Time PCR System (Applied Biosystems).
MicroRNA Target Identification
The miRNA–mRNA interactions (miRNA targets) were evaluated using DIANA-Tarbase, the database that hosts experimentally proven miRNA–mRNA interactions curated from published research articles (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=tarbase/index). This ensures the selection of a set of high-confidence miRNAs and their targets for further analysis. The miRNAs for which experimentally validated targets not yet reported in this database were excluded from further analysis. The mRNA targets of the upregulated miRNAs obtained from DIANA-Tarbase were matched with the downregulated mRNA list from our microarray study and vice versa.
In Silico Analysis of Transcriptional Profile of SP Cells
The mRNAs differentially regulated in SP1 fractions compared to the MP1 fractions were subjected to KEGG pathway enrichment analysis using the ClueGO/CluePedia (version v2.2.6) in Cytoscape. The experimentally validated target mRNAs of upregulated miRNAs were correlated with the downregulated mRNAs identified in our microarray study and vice versa. The common targets were also subjected to KEGG pathway enrichment analysis using ClueGO/CluePedia in Cytoscape to identify the significantly perturbed signaling pathways.
Statistics and Software
Student’s t-test and one-way ANOVAs were used for statistical comparisons where appropriate. Software used for microarray analysis was Partek Genomic Solutions 6.2 (Partek Inc., St. Charles, MO, USA), Matlab 7 (Mathworks Inc., Natick, MA, USA), and GCOS 1.3 (Affymetrix Inc., Foster City, CA, USA). Real-time PCR data were analyzed by AB’s SDS software. TaqMan miRNA profiling data were analyzed using SDS v2.2 software. Error bars on all graphs represent the mean ± SEM.
Accession Number
Microarray data can be found in Gene Expression Omnibus (accession GSE36111) (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hxurlwaacikocbc&acc=GSE36111).
RESULTS
Microarray Analysis and Real-Time Validation of Genes in SP1
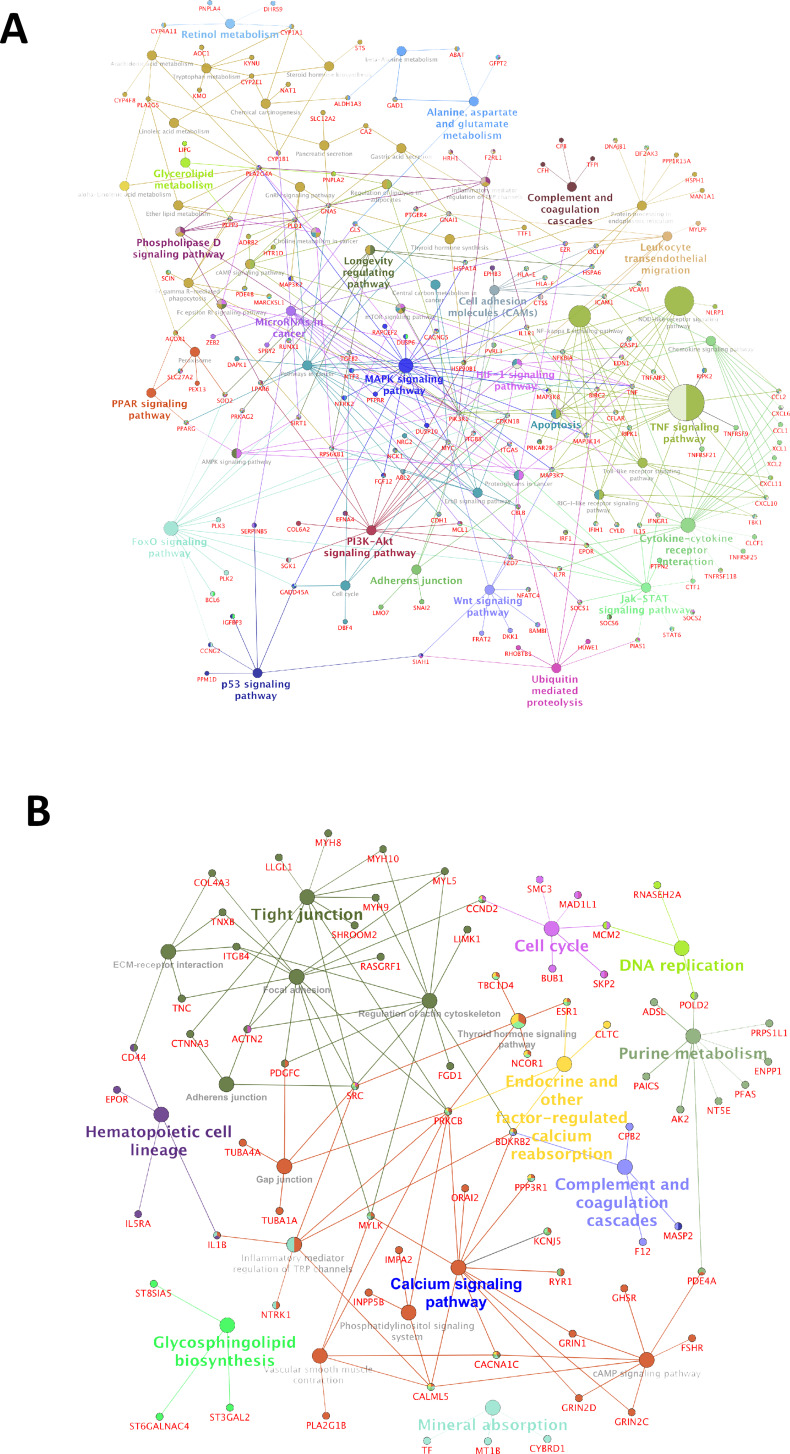
Analysis of the gene expression microarray data (deposited in GEO under accession No. GSE36111) resulted in the identification of 688 mRNAs to be upregulated and 362 to be downregulated in the tumorigenic SP1 fraction compared to the nontumorigenic MP1 fraction (≥2-fold, p < 0.05) of SCC172 OSCC cells (see supplemental Excel Table S1, available at http://rgcb.res.in/OncologyResearch.php). The majority of genes upregulated in SP1 belonged to proinflammatory pathways as mitogen-activated protein kinase (MAPK), tumor necrosis factor (TNF), transcriptional misregulation in cancer, Janus kinase/signal transducers and activators of transcription (JAK/STAT), Forkhead box protein O (FoxO), nuclear factor-κB (NF-κB), nucleotide-binding oligomerization domain-like (NOD-like) receptor, and retinoid-inducible gene 1-like (RIG-I-like) receptor signaling pathways also linked to cell proliferation, differentiation, migration, apoptosis, and tumorigenesis (Fig. 1A).
Figure 1.
Network mapping of differentially expressed genes in side population 1 (SP1). (A) Schematic representation of pathways and genes upregulated in the tumorigenic SP fraction in comparison to the nontumorigenic main population (MP) fraction. (B) Functional classification of downregulated genes and their interactants specifically expressed in SP1 fraction in comparison to MP1. All major pathways have been represented in colored circles (also see supplementary Excel sheet S1 available at http://rgcb.res.in/OncologyResearch.php).
We also validated a subset of upregulated genes with higher fold values in the microarray experiment using qRT-PCR and their relative expression against an endogenous control, β2M. These genes pertaining to drug transporters (ABCG2 and ABCA5), chemokines (CCL2 and CXCL10), and regulation of cell adhesion (VCAM1 and MMP13) correlated well with the microarray result (see supplemental Fig. S2, available at http://rgcb.res.in/OncologyResearch.php).
Differential MicroRNA Profile in SP1 and MP1 Fractions
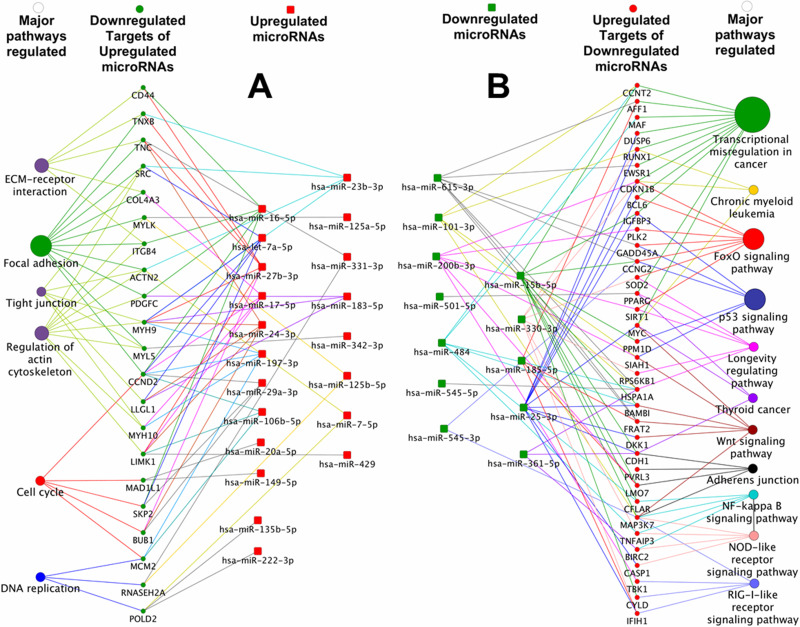
To add onto the molecular characterization of SP1 and MP1 fractions, a second line of regulatory network mediated by small RNA molecules termed miRNAs was investigated across these two fractions (see supplemental Excel Table S2, available at http://rgcb.res.in/OncologyResearch.php). From a total of 369 miRNAs analyzed by TaqMan real-time PCR array, we identified 103 upregulated and 14 downregulated miRNAs in the SP1 fraction to be differentially expressed by a fold change of 1.5 and a value of p < 0.05 in comparison to the MP1 fraction. Correlation analysis of targets of miRNAs (experimentally proven) and the corresponding mRNAs (microarray data) resulted in the identification of 199 downregulated mRNA targets of 87 upregulated miRNAs (Fig. 2A) and 205 upregulated mRNA gene targets for the corresponding 13 downregulated miRNAs related to our microarray study (Fig. 2B). These miRNA targets included transcriptional regulators such as RUNX1, MYC, and BCL6 (targets of hsa-miR-25-3p), and CCNT2 and CD151 (targets of hsa-miR-15-5p), which are examples of multiple modes of regulation of gene expression at the transcriptome level in the SP1 fraction.
Figure 2.
MicroRNA–mRNA target interaction and pathways perturbed. (A) Functional grouping and network mapping of interactants of upregulated microRNAs in SP1 fraction and its corresponding downregulated targets from mRNA array list of upregulated genes (green color represents downregulated mRNAs in SP1; functional clustering of targeted downregulated mRNAs in SP1; red color represents upregulated microRNAs in SP1). (B) Similarly, downregulated microRNAs in SP1 were correlated with upregulated mRNA targets in SP1 (red color represents functional clustering of upregulated mRNAs in SP1; green color represents targeted downregulated microRNAs in SP1) (also see supplementary Excel sheet S2 available at http://rgcb.res.in/OncologyResearch.php).
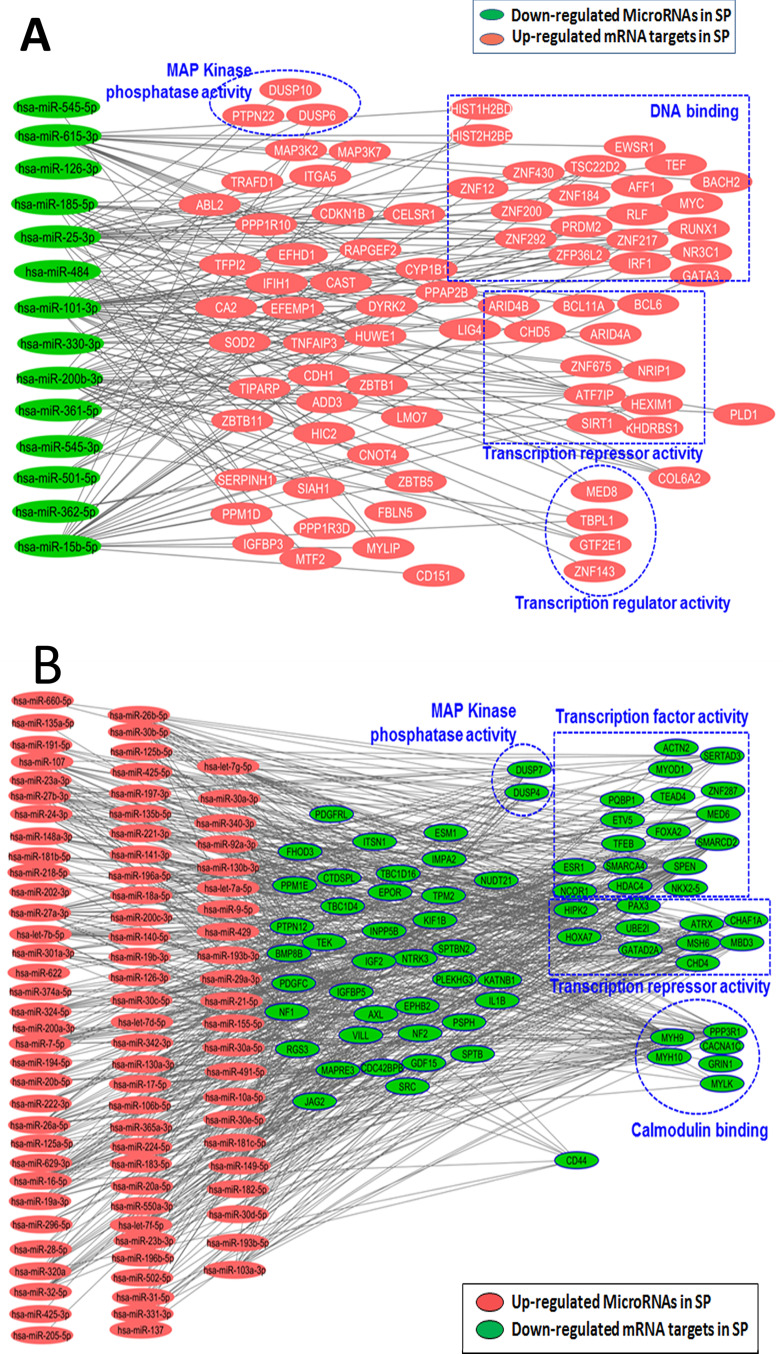
Similarly, multiple clusters of upregulated miRNAs had a few downregulated mRNA interactants (41 miRNAs targeting SPEN and 20 miRNAs targeting NCOR1) (Fig. 3A) annotated as DNA-binding proteins capable of epigenetic reprogramming, transcriptional repressor activity, and remarkably reduced expression of CD44, a CSC marker targeted by 10 miRNAs (see supplemental Excel Table S2, available at http://rgcb.res.in/OncologyResearch.php) (Fig. 3B). This also correlated with our published immunofluorescence analysis, wherein tumorigenic SP1 fraction displayed CD147+/EGFR+/CD44−/lo/CD44v6+ phenotypic expression4. Experimental validation of the targets of these miRs in other OSCC cell lines and patient-derived tumor samples might establish tumorigenic fraction (SP1)-specific miRNA signature and reveal a clear picture of how these miRNAs regulate gene expression, thereby facilitating tumor stem cell maintenance and survival.
Figure 3.
Significance of transcriptome reprogramming in cancer. (A) Functional grouping and network mapping of interactants of downregulated microRNAs in SP1 fraction and its corresponding targets from mRNA array list of upregulated genes (green color represents downregulated microRNAs in SP1; red color represents functional clustering of targeted upregulated mRNAs in SP1). (B) Similarly upregulated microRNAs in SP1 were correlated with downregulated mRNA targets in SP1 (red color represents functional clustering of upregulated microRNAs in SP1; green color represents targeted downregulated mRNAs in SP1) (also see supplementary Excel sheet S2 available at http://rgcb.res.in/OncologyResearch.php).
DISCUSSION
Similar to normal stem cells, CSCs and the CSC progeny have been reported to display significant phenotypic and functional heterogeneity9. An understanding of the interaction between tumor-initiating cells (TICs) and their differentiated progeny could lead to effective treatment of hierarchically organized cancers that can avert the development of tumor cell variants capable of seeding new tumors and distant metastases10. Even though all tumor cells appear similar in phenotype, reprogramming of transcriptome and subsequent epigenetic alterations result in multiple fractions with varying degrees of differentiation and metastatic and tumorigenic potential11. Uninterrupted diagnoses of the TIC fraction that can be obtained by minimally invasive procedures from patients have major limitations due to the poor reliability of stem cell markers and important sampling biases10. Several studies have proven the presence of a distinct metastatic clone amidst multiple nonmetastatic clones of tumor tissue12. The large multitude of assays currently employed fail to address the heterogenic and dynamic nature of CSCs, the pivotal mechanisms that govern this shift of cancer cells through a hierarchically organized structure featuring fractions of highly differentiated subpopulations of cells as well as cancer stem and progenitor cells13.
Relating to our CSC shift hypothesis5, we had earlier proposed that TICs evade therapy by undergoing a temporary shift between four states, depending on varying expressions of drug transporter proteins and cell cycle status: a dormant/chemoresistant phase (SP2 cells), proliferative/chemoresistant phase (MP2 cells), dormant/chemosensitive (SP1 cells), and finally the bulk of cancer cells categorized as proliferative/chemosensitive (MP1 cells)4,5. We also analyzed the concept of biological heterogeneity displayed by these four fractions: SP1, SP2, MP1, and MP24. Characterization studies revealed that four fractions subsisted not as distinct clones but merely showed a transient shift from one cell type to another4, probably mediated by transcriptomal regulation at the miRNA level. SP1 cells displayed a higher potential for self-renewal and plasticity to regenerate MP1, SP2, and MP24. Each of these fractions was also capable of repopulating the other fractions, indicative of cellular heterogeneity. A series of transitional events were brought about by regulation of protein expression by differentially expressed mRNAs, which are, in turn, controlled by miRNAs. Even though we analyzed the population as four distinct fractions, ultimately it was the same CSC undergoing a transient progressive shift to attain malignancy at all stages of the disease. Discovery of regulatory molecules overriding this transient shift of stem cell types would enable the development of a patient-oriented therapeutic regimen. This is a preliminary study connecting miRNAs and stem-like, drug-resistant cells in oral cancer.
Microarray analysis of SP1 and MP1 was performed and compared to exemplify the signaling molecules that play a major role in addressing properties of resistance, self-renewal, and tumor formation. Inflammatory pathways and interactants were found to be significantly upregulated in SP1 fraction. Inflammation has been proposed to mediate all phenomena such as initiation and promotion of tumors, malignant transformation, angiogenesis, and metastasis in which cytokines are prominent players14. An inflammatory microenvironment can enhance tumor initiation by increasing mutation rates and triggering the production of cytokines and growth factors, which can stimulate stem cell expansion or confer a stem cell-like phenotype14. The pattern recognition receptors were categorized into four different groups, Toll-like receptors (TLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), and RIG-1-like receptors (RLRs), which play a role in the development of inflammation-associated carcinogenesis15. An upregulation of cytokines and receptors mediates TLR activation and phosphorylation of MAPKs, ultimately leading to the activation of NF-κB and production of TNF-α and interleukins (ILs) with an overall outcome of tumorigenesis through MAPK and NF-κB signaling cascade16. An upregulated expression of these signaling pathways, specifically in the SP1 fraction, imparted the potentials for tumor initiation and chemoresistance through expression of drug transporter proteins.
Several stemness and malignancy-associated cell surface markers such as ALDH1A3, CD151, CEACAM 21, ABCG2, MUC16, and MMP13 were also found to be upregulated in the SP1 fraction. In contrast to several reports, as reviewed by Ghuwalewala et al.17, suggesting that CD44high cells are tumorigenic, we found that our tumorigenic SP1 fraction displayed CD44low, ALDHhigh, and CD151high expression levels. The tetraspanin CD151 has a proven regulatory role in posttranslational modifications of interactants, namely migration and invasion leading to metastasis, and also in the promotion of tumor neovascularization18,19. As previously reported in studies of head and neck squamous cell carcinoma (HNSCC) CSCs, high levels of ALDH may regulate the oxidative stress response caused by radiation and chemotherapy, thus enhancing the survival and tumorigenic potential of CSCs20. Antiapoptotic signaling molecules (e.g., NFKB1, BCL6, caspases) and drug-resistant transporters ABCG2 and ABCA5 were also strongly expressed in the SP1 fraction, enhancing the properties of chemoresistance and survival advantage to this particular fraction. HOXA7, an inducer of keratinocyte differentiation, was significantly downregulated in SP1, indicating a more primordial fraction than MP1. SP1 fraction had exhibited the highest proliferation rate in vitro followed by MP1, MP2, and SP2 fractions with the least proliferation potential4. Upregulated expression of the polymorphic P450 (CYP) enzyme superfamily CYP1A1 (12-fold) and ABCG2 (8-fold) in the SP fraction confirms the presence of an efficient machinery to break down and efflux drugs from within a fraction of cells, whereas MP cells are eradicated by treatment. An overexpression of transmembrane glycoprotein VCAM1 (CD106) and MMP13 in CD151+ SP1 cells apparently simplified the transition to a more malignant counterpart (SP2/MP2 fractions) that displayed higher tumorigenic and metastatic potentials in addition to chemoresistance4.
Notably, we identified over 131 genes involved in the regulation of gene expression [transcription factors (TFs) and their positive/negative regulators] and a significant number of components of the chromatin reorganization complexes to be differentially regulated across both fractions. Analysis of the TF network using STRING21 has enriched a set of nine upregulated TFs (RUNX1, CCNT2, PPARG, AFF1, MYC, EWSR1, BCL6, CDKN1B, and MAF) that were annotated to be deregulated in cancer and five downregulated transcriptional regulators (NCOR1, SPEN, MBD3, GATAD2A, and HDAC4) enriched for components of the transcriptional repressor complex. Among the upregulated TFs, RUNX1 has been reported to mark basal stem cells of human oral epithelium and also has a major role in tumor initiation and maintenance of stem cells22. The orientation of the transcriptional network, including expression of stemness and metastasis-specific markers in SP1, implies a significant regulation at the gene level responsible for the observed increase in the oncogenic potential between two fractions derived from the same parental cell line.
The set of major downregulated genes were pertaining to calcium signaling, focal adhesion, regulation of actin cytoskeleton, tight junction signaling pathways, and ECM–receptor interactions (Fig. 1B). An altered calcium signaling pathway in two fractions of the same oral cancer cell line may be a function of tumorigenic remodeling, also evidenced in a tumorigenic breast cancer cell line in comparison to a nontumorigenic cell line23. The transcriptional profile of SP1 highlighted an upregulated gene expression of drug transporters, cell proliferation, and survival signals, enzymes responsible for drug metabolism and antiapoptosis, cell adhesion molecules, and metalloproteases facilitating migration and metastasis.
The variable regeneration of SP1 from MP1 and MP1 from SP1 cells4 indicated the presence of an additional genre of transcriptional regulation, probably at the miRNA level. miRNAs are small RNA molecules that can regulate protein expression and operate highly complex regulatory networks by targeting numerous mRNAs that are associated with tumor initiation, development, and progression by either translational inhibition or mRNA degradation24. This could also lead to transient gain or loss of stemness and malignancy-associated features in cancer cells. Hence, we initiated a complete transcriptome profiling to define tumor-initiating SP1 fraction from its nontumorigenic counterpart MP1.
Among all miRs analyzed, miR-134 was found to be upregulated by the maximum fold (2.6) in the SP1 fraction. High expression of miR-134 has been found to play a role in promoting EMT and cell survival in lung cancer cells25. Among the four downregulated miRs with a fold change below −0.5, miR-15b scores high. This is a homolog of mir-15a, which negatively regulates an antiapoptotic gene, BCL2, often overexpressed in many types of human cancers and plays a role in the development of multidrug resistance (MDR), in part, by modulation of apoptosis26. Downregulation of miR-15b in SP1 cells compared to MP1 confirms the apoptotic resistance property of the SP fraction. Vascular endothelial growth factor (VEGF), which has a significant role in tumor angiogenesis by promoting endothelial cell division, proliferation, and migration, is also targeted by antiangiogenic miRNAs miR-15b and miR-200b26,27. Combined downregulation of miR-15b and miR-200b indicates an upregulation of antiapoptotic and angiogenic factors that transformed the SP1 fraction into a more malignant state, as we had previously observed in the SP2 fraction4. Histological analysis of a xenograft tumor generated from the enriched SP2 fraction revealed a moderately differentiated OSCC, with blood vessel-like structures and a combination of both epithelial and mesenchymal cells (partial EMT)4. miRNA-based posttranscriptional regulatory measures could be decisive in this temporary transition from MP1 to more malignant SP1, SP2, and MP2 fractions.
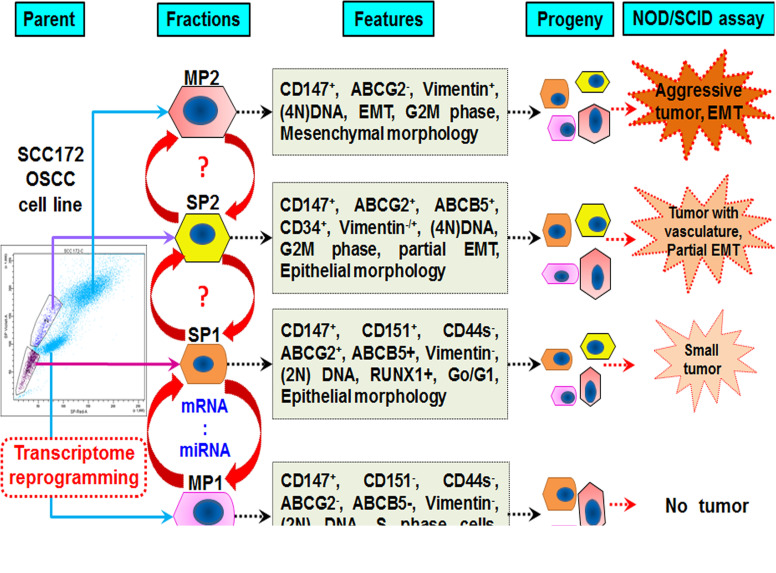
Biological heterogeneity in oral dysplasia is marked by the coexistence of four fractions. This transient CSC shift model originates with MP1 ↔ SP1 ↔ SP2 ↔ MP2 and eventually reverts back to SP1 to reenter the cycle of events (Fig. 4). Several oncogenic events in SP1 cells generate more SP2, which then transforms into MP2 cells with loss of drug transporters, adhesion markers, and epithelial markers. MP2 undergoes complete transition into mesenchymal cells to facilitate invasion and metastasis or may even revert to the SP1 phenotype by molecular reprogramming and reiterates tumorigenesis or persists as dormant, minimal residual disease (MRD). An upregulated coexpression of miRNAs acts as a second level of regulation that could inhibit translation of target genes. The mode of interaction wherein miRNAs balance the expression of genes through either transcriptional degradation or translational repression may not be accounted herewith. Even though certain miRNAs are not yet reported in oral cancer, a detailed study is strongly suggested as it can identify novel therapeutic targets.
Figure 4.
Biological heterogeneity in oral dysplasia is marked by coexistence of four fractions.
ACKNOWLEDGMENTS
The OSCC cell line UPCI:SCC172 was a gift from Professor Susanne M. Gollin, Ph.D., at the University of Pittsburgh. We gratefully acknowledge the expertise of Rajesh Krishna (Flow Cytometer Facility, NII) for excellent assistance in the FACS experiments. This study was supported by a research grant from the Department of Biotechnology, Government of India (BT/PR/7793/MED/14/1112/2006). The funding body has no role in the design of the study, nor the collection, analysis, or interpretation of the data, or in the writing of the manuscript. The study was approved by the Institutional Review Board of RGCB. Since there were no human subjects involved, consent to participate is not applicable. Author contributions: V.R. (methodology, validation, investigation, and writing); R.R., A.M.P., and R.G. (software, formal analysis, visualization, and data curation); V.R. and R.R. (writing, review, and editing); M.R.P. and T.R.S.K. (conceptualization, methodology, supervision, project administration, writing, editing, fund acquisition, and resources). All authors read and approved the manuscript for publication.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Sankaranarayanan R, Ramadas K, Amarasinghe H, Subramanian S, Johnson N. Oral cancer: Prevention, early detection, and treatment. In: Gelband H, Jha P, Sankaranarayanan R, Horton S, editors. Cancer: Disease control priorities, 3rd ed., Vol. 3 Washington (DC): The International Bank for Reconstruction and Development/The World Bank; 2015. Chapter 5. [PubMed] [Google Scholar]
- 2. National Cancer Institute. Surveillance, epidemiology, and end results program. SEER Stat fact sheets: Oral cavity and pharynx cancer [Internet]. Available from https://seer.cancer.gov/statfacts/html/oralcav.html
- 3. Tomasetti C, Vogelstein B. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science 2015;347(6217):78–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Richard V, Sebastian P, Nair MG, Nair SN, Malieckal TT, Santhosh Kumar TR, Pillai MR. Multiple drug resistant, tumorigenic stem-like cells in oral cancer. Cancer Lett. 2013;338(2):300–16. [DOI] [PubMed] [Google Scholar]
- 5. Richard V, Pillai MR. The stem cell code in oral epithelial tumorigenesis: ‘The cancer stem cell shift hypothesis’. Biochim Biophys Acta 2010;1806(2):146–62. [DOI] [PubMed] [Google Scholar]
- 6. Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, Yu M, Sundaresan TK, Licausi JA, Desai R, O’Keefe RM, Ebright RY, Boukhali M, Sil S, Onozato ML, Iafrate AJ, Kapur R, Sgroi D, Ting DT, Toner M, Ramaswamy S, Haas W, Maheswaran S, Haber DA. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature 2016;537:102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Biddle A, Gammon L, Liang X, Costea DE, Mackenzie IC. Phenotypic plasticity determines cancer stem cell therapeutic resistance in oral squamous cell carcinoma. EBioMedicine 2016,4:138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, Hong S, Adams A, D’Angelo R, Ginestier C, Charafe-Jauffret E, Clouthier SG, Birnbaum D, Wong ST, Zhan M, Chang JC, Wicha MS. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports 2014;2:78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Werner B, Scott JG, Sottoriva A, Anderson ARA, Traulsen A, Altrock PM. The cancer stem cell fraction in hierarchically organized tumors can be estimated using mathematical modeling and patient-specific treatment trajectories. Cancer Res. 2016;76:1705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aceto N, Toner M, Maheswaran S, Haber DA. En route to metastasis: Circulating tumor cell clusters and epithelial-to-mesenchymal transition. Trends Cancer 2015;1(1):44–52. [DOI] [PubMed] [Google Scholar]
- 13. Akrap N, Andersson D, Bom E, Gregersson P, Ståhlberg A, Landberg G. Identification of distinct breast cancer stem cell populations based on single-cell analyses of functionally enriched stem and progenitor pools. Stem Cell Reports 2016;6:121–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010;140:805–20. [DOI] [PubMed] [Google Scholar]
- 16. Manzoor Z, Koo JE, Koh Y-S. Mitogen-activated protein kinase signaling in inflammation-related carcinogenesis. J Bacteriol Virol. 2014;44:297–304. [Google Scholar]
- 17. Ghuwalewala S, Ghatak D, Das P, Dey S, Sarkar S, Alam N, Panda CK, Roychoudhury S. CD44(high)CD24(low) molecular signature determines the cancer stem cell and EMT phenotype in oral squamous cell carcinoma. Stem Cell Res. 2016;16:405–17. [DOI] [PubMed] [Google Scholar]
- 18. Sadej R, Grudowska A, Turczyk L, Kordek R, Romanska HM. CD151 in cancer progression and metastasis: A complex scenario. Lab Invest. 2014;94(1):41–51. [DOI] [PubMed] [Google Scholar]
- 19. Kumari S, Devi G, Badana A, Dasari VR, Malla RR. CD151—A striking marker for cancer therapy. Biomark Cancer 2015;7:7–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kurth I, Hein L, Mäbert K, Peitzsch C, Koi L, Cojoc M, Kunz-Schughart L, Baumann M, Dubrovska A. Cancer stem cell related markers of radioresistance in head and neck squamous cell carcinoma. Oncotarget 2015;6:34494–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scheitz CJF, Lee TS, McDermitt DJ, Tumbar T. Defining a tissue stem cell-driven Runx1/Stat3 signalling axis in epithelial cancer. EMBO J. 2012;31(21):4124–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stewart TA, Yapa KTDS, Monteith GR. Altered calcium signaling in cancer cells. Biochim Biophys Acta 2015;1848:2502–11. [DOI] [PubMed] [Google Scholar]
- 24. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010;466:835–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kitamura K, Seike M, Okano T, Matsuda K, Miyanaga A, Mizutani H, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-134/487b/655 cluster regulates TGF-β–induced epithelial–mesenchymal transition and drug resistance to gefitinib by targeting MAGI2 in lung adenocarcinoma cells. Mol Cancer Ther. 2014;13:444–53. [DOI] [PubMed] [Google Scholar]
- 26. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005;102:13944–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choi Y-C, Yoon S, Jeong Y, Yoon J, Baek K. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol Cells 2011;32:77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]