Abstract
Colorectal carcinoma is one of the leading causes of cancer-related deaths and has a high tendency for metastasis, which makes it a priority to find novel methods to diagnose and treat colorectal carcinoma at a very early stage. We studied the role of the regulator of G-protein signaling (RGS) family of proteins RGS17 in colorectal carcinoma growth and metastasis. We found that RGS17 was upregulated in both clinical colorectal carcinoma tissues and cultured colorectal carcinoma cells. Knockdown of RGS17 by specific siRNA decreased the cell proliferation rate, whereas overexpression of RGS17 with expression plasmid increased the rate in cultured cells. Consistently, a mouse model for colorectal carcinoma also showed that depletion of RGS17 significantly inhibited tumor growth in vivo. Moreover, a Transwell assay showed that RGS17 promoted the ability of colorectal carcinoma cells to migrate and invade. These data suggest that RGS17 is overexpressed in colorectal carcinoma and promotes cell proliferation, migration, and invasion.
Key words: RGS17, Colorectal carcinoma, Cell growth, Migration, Invasion
INTRODUCTION
Colorectal carcinoma is one of the most frequently diagnosed malignant tumors around the world, with high morbidity and mortality in both males and females1–3. Current therapeutic strategies against colorectal carcinoma are basically dependent on curative surgery and postoperative adjuvant chemotherapy. The survival of patients has indeed improved slowly but steadily during the past decades3–5. However, because of postsurgical recurrence as well as fatal distant metastasis, only limited improvement has been made with respect to the prognosis of patients, despite the progress in its treatment. Therefore, it is a great urgency to search for valuable biomarkers for the sake of the determination of suitable interventional strategies and the improvement of patient outcomes.
The regulator of G-protein signaling (RGS) family members traditionally regulate a subset of signaling events, working as GTPase accelerating proteins (GAPs) for suppressing Gα-coupled GPCRs by accelerating the intrinsic hydrolysis rate of G proteins and terminating GPCR signaling much faster6–8. In recent studies, it has also been demonstrated that RGS proteins are involved in a variety of cancers. Rangel et al. testified that RGS1 was overexpressed in melanoma and associated with decreased relapse-free survival9. RGS6 was also shown to have significance in regulating bladder cancer tumorigenesis10. Wu et al.11 and Tso and colleagues12,13 reported that RGS19 could stimulate cell proliferation by arresting the cell cycle and increasing the Akt signaling pathway. These findings suggested the crucial roles for RGS proteins in human oncogenesis.
RGS17, also named RGSZ2, is a member of the RZ subfamily of RGS proteins, which is mainly characterized by the RGS domain (a cysteine-rich N terminus) and its ability to regulate the Gαz signaling pathway14. RGS17 was shown by Larminie et al.15 to express almost exclusively in the brain. Nevertheless, recent studies suggest that RGS17 is overexpressed in lung and prostate cancers and associated with tumor cell proliferation16–18. In addition, RGS17 exerted a proto-oncogenic role through interacting with the AMP–PKA–CREB signaling pathway17. However, the exact role of RGS17 in colorectal carcinoma has not been determined.
In this study, we aimed to investigate the role of RGS17 in human colorectal carcinoma growth and metastasis. To this end, specific siRNA against RGS17 and its expression plasmid were used to modulate the expression of RGS17. mRNA and protein levels of RGS17 were determined in human clinical colorectal carcinoma tissues and in cultured human colorectal carcinoma cell lines. Furthermore, whether RGS17 had any effect on tumor cell proliferation and metastasis is also discussed.
MATERIALS AND METHODS
Human Samples
A total of 25 cases of colorectal carcinoma patients who were admitted to Renmin Hospital of Wuhan University were collected. Matched adjacent noncancerous tissues were also obtained. All patients gave their full consent to participate in this study, and a written consent form was obtained from each patient.
Cell Culture and Antibodies
Human colorectal carcinoma cell lines HCT116 and COLO205 as well as HT-29 and the control cells 293T were purchased from the Cell Bank of the Chinese Academy of Sciences (CAS; Shanghai, P.R. China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Los Angeles, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco). Primary antibodies against RGS17 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and secondary antibodies were commercially purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Total RNA Extraction and cDNA Synthesis
The total RNAs from both human tissues and cultured colorectal carcinoma cells were extracted with TRIzol reagent (TaKaRa, Shiga, Japan) according to the manufacturer’s recommendations. The quality and concentration were measured by collecting the absorbance with Nanodrop 2000 (Thermo Scientific, Waltham, MA, USA) at 260 nm. First-strand cDNAs were reverse transcribed with the PrimeScript RT Master Mix Perfect Real Time (TaKaRa).
Relative Real-Time PCR
All real-time polymerase chain reactions (RT-PCRs) were developed in an ABI PRISM 7900 Real-Time System with the SYBR Premix Ex Taq Kit (TaKaRa). Briefly, the protocol was as follows: initial denaturation step at 95°C for 2 min, 35 repetitions of the three-step cycling program consisting of 30 s at 95°C (denaturation), 1 min at 53°C (primers annealing), and 30 s at 72°C (elongation), and the final extension step for 10 min at 72°C. Amplified DNAs were determined by 1% (w/v) agarose gels (Bio-Rad, Hercules, CA, USA). The primers of the RT-PCR assay are shown in Table 1, and housekeeping gene GAPDH was included as the internal control. All quantitative data were normalized to GAPDH.
Table 1.
Primers Used in RT-PCR
| Genes | Gene ID | Primer Nucleotide Sequences |
|---|---|---|
| RGS17 | 26575 | Forward: 5′-CAGAGGCCCAACAACACCTG-3′ |
| Reverse: 5′-TGTGGGTCTTCCCGCATTTT-3′ | ||
| GAPDH | 2597 | Forward: 5′-GTGGACATCCGCAAAGAC-3′ |
| Reverse: 5′-AAAGGGTGTAACGCAACTA-3′ |
Western Blot Analysis
Total proteins from both clinical tissues and cultured cells were extracted. For human tissue, each sample from colorectal carcinoma tissue and the adjacent noncancerous tissue was cut into pieces and incubated with the lysis buffer with protease inhibitor. After ultracentrifugation, the supernatant was collected for subsequent immunoblot analysis. For in vitro assays, cells were allowed to grow until 95% confluence. Cells were then washed twice with PBS and lysed with a general lysis buffer to generate the total protein lysate. An equal amount of 50 μg of whole protein was loaded into each lane in a 12% SDS-PAGE gel. GAPDH was synchronously detected as a loading control. Immunoreactivity was determined with enhanced chemoluminescence autoradiography (Thermo Scientific, Pittsburgh, PA, USA), and each experiment was repeated at least three times.
siRNA Interference and Plasmid Transfection
For knockdown of RGS17, specific siRNA was designed and chemically synthesized by Santa Cruz Biotechnology. The RGS17 expression plasmid, which was cloned into pLIC-SGC1 vector, was purchased from Addgene (Cambridge, MA, USA). The transfection assay was preceded with Lipofectamine 2000 transfection reagent (Life Technologies, New York, NY, USA) according to the manufacturer’s instructions. Six hours after transfection, the medium was replaced with fresh DMEM with 10% FBS.
Cell Proliferation Assay
Colorectal carcinoma cells HCT116 were seeded into 96-well plates (3,000 cells/well) and allowed to grow overnight. Cells were then transfected with 10 μM of siRGS17 or RGS17 expression plasmid, followed by incubation in DMEM for another 72 h. Cell viabilities were determined for a consecutive 5 days with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution. MTT solution (2 mg/ml) was added into each well and monitored each day. After cells were incubated for another 4 h at 37°C, medium was discarded, and 200 μl of DMSO was added. The plate was then shaken for 5 min, and the optical density was determined at 570 nm.
Cell Transwell Assay
HCT116 cells were cultured in 24-well plates and transfected with specific RGS17 siRNA or plasmid. Forty-eight hours after transfection, cells were harvested in serum-free DMEM as single-cell suspension, and 150 μl of cell suspension (3 × 104 cells) was seeded into the upper chamber (Corning, Corning, NY, USA). The lower chamber was filled with 600 μl of DMEM supplemented with 10% FBS. For the invasion assay, the chamber was coated with Matrigel 6 h before seeding cells into the chamber. After incubating at 37°C for 12 h, the cells were fixed with ice-cold methanol for 20 min and stained with 0.1% crystal violet for 5 min. The images were acquired under a microscope at 200% crystal violet for 5 min. Images were captured under a microscope (magnification: 200×; Nikon Corp., Tokyo, Japan).
Wound Healing Assay
Colorectal carcinoma cells HCT116 were seeded into six-well plates and transfected with either siRGS17 or RGS17 expression plasmid. Forty-eight hours after transfection, a 10-μl sterile pipette tip was used to scrape a cross in the center of each well. After scratching, cells were rinsed with PBS three times and replaced with serum-free medium immediately. Cells were allowed to migrate for another 24 h when scratches were observed and photographed for each group. Light microscopic images were captured with a Nikon microscope. Each assay was repeated at least three times in triplicate.
Mouse Xenograft Model of Human Colorectal Carcinoma
All animal experimental procedures were reviewed and approved by the Institution Animal Care and Use Committee of the Wuhan University. A total of 20 female athymic nude mice (6 weeks old) were randomly divided into two groups and housed in 12-h light/dark cages and received water and food freely. Prior to injection, HCT116 cells that had RGS17 stably removed were constructed. HCT116 cells without RGS17 ablation were also constructed as a control. HCT116 cells (5 × 106 cells/ml) with or without RGS17 ablation were then suspended in PBS with 20% Matrigel and later injected subcutaneously into the right flank of each mouse. Tumor dimensions were measured every 5 days, and the tumor volume for each mouse was calculated as previously described. Forty days after inoculation, mice were sacrificed, and the dissected tumors were weighed and measured. All efforts were made to minimize suffering.
Statistical Analysis
The results were all exhibited as means ± standard deviation (SD). Statistical analysis was carried out with the Student’s t-test. A value of p < 0.05 was considered significant. All experiments were repeated at least three times unless otherwise noted.
RESULTS
RGS17 Is Overexpressed in Human Colorectal Carcinoma Tissues and in Cultured Colorectal Carcinoma Cell Lines
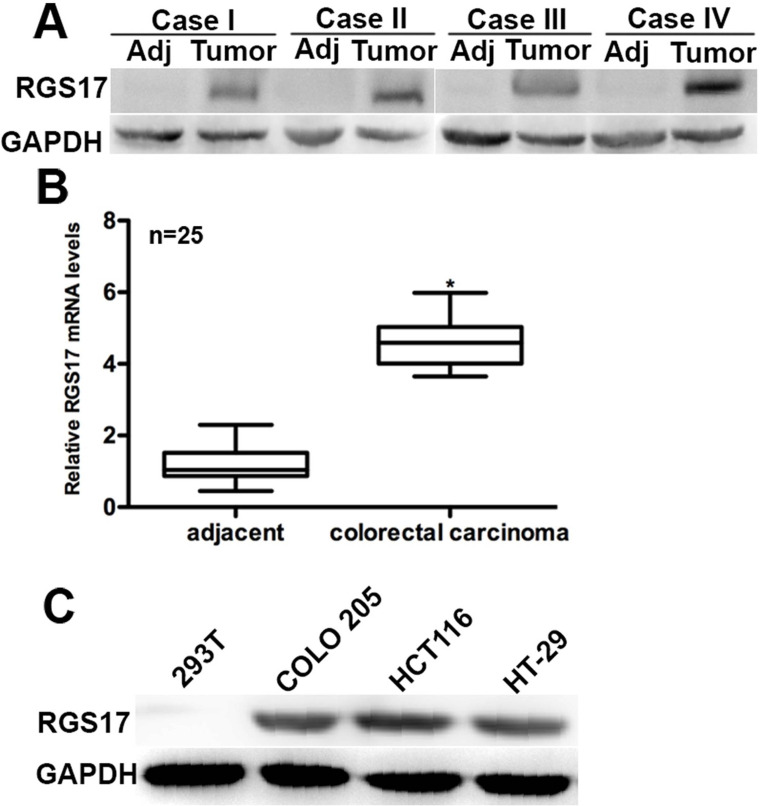
To affirm the role of RGS17 in colorectal carcinoma, we initially examined the expression of RGS17 in 25 cases of clinical human colorectal carcinoma tissues and their adjacent noncancerous tissues. Western blot analysis showed that the protein level of RGS17 was significantly increased in colorectal carcinoma tissues compared with the adjacent normal tissues. In the four representative cases, expression of RGS17 was rarely detected in the adjacent tissues, whereas it was remarkably detected in the matched cancer tissues (Fig. 1A). The RT-PCR assay also showed that the relative mRNA level of RGS17 in the clinical colorectal carcinoma tissues was approximately fivefold that in the adjacent tissues (n = 25) (Fig. 1B). COLO205, HCT116, and HT-29 cells are three colorectal carcinoma cell lines that are widely used in the literature. 293T cells were used as a noncancerous control cell line. Protein levels of RGS17 in the three colorectal carcinoma cell lines were markedly higher than those in 293T cells (Fig. 1C). These data showed that RGS17 is highly expressed in both human colorectal carcinoma tissues and in cultured colorectal carcinoma cells.
Figure 1.
RGS17 is overexpressed in human colorectal carcinoma tissues and cultured cells. (A) In the four representative cases, Western blot analysis showed that RGS17 was highly expressed in cancer tissues, whereas its expression was rarely detectable in the adjacent noncancerous tissues. (B) Real-time polymerase chain reaction (RT-PCR) analysis of 25 clinical colorectal carcinoma cases showed that the mRNA level of RGS17 was significantly different between colorectal carcinoma and adjacent tissues. *p < 0.05 colorectal carcinoma versus adjacent. (C) In all colorectal carcinoma cell lines, RGS17 was highly expressed compared with 293T control cells. Data were obtained from three independent experiments.
Transfection Efficiency of Specific siRNA Against RGS17 (siRGS17) and RGS17 Expression Plasmid
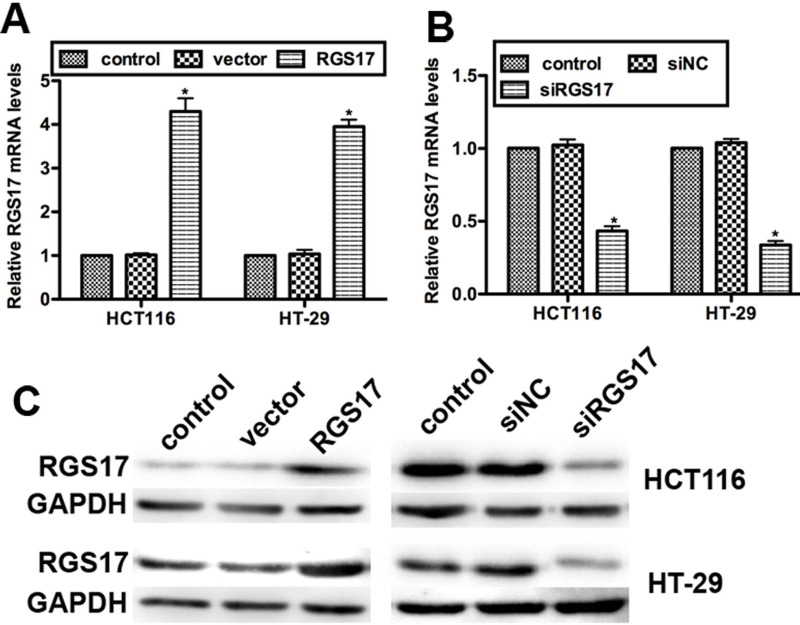
To further examine the role of RGS17 in colorectal carcinoma progression, we transfected two colorectal carcinoma cell lines, HCT116 and HT-29, with specific siRNA against RGS17 (named siRGS17) or RGS17 expression plasmid. RT-PCR data showed that the relative mRNA level of RGS17 in RGS17-transfected HCT116 cells was increased by up to 4.2-fold compared with the control, while it was raised to 3.8-fold in HT-29 cells transfected with RGS17 expression plasmid (Fig. 2A). On the contrary, the mRNA level of RGS17 was decreased by approximately 50% in HCT116 cells and by 45% in HT-29 cells, respectively, when the siRGS17 was transfected (Fig. 2B). Consistently, Western blot analysis showed that the protein level of RGS17 was markedly increased by transfection with the RGS17 plasmid, while it was notably suppressed by the transfection of siRGS17 in both cell lines (Fig. 2C). These results confirmed the highly effective transfection and the efficiency of specific siRGS17 and RGS17 expression plasmid with respect to modulation of RGS17 expression.
Figure 2.
Transfection efficiency of siRGS17 and RGS17 expression plasmid. (A) Relative mRNA levels of RGS17 when RGS17 expression plasmid was transfected in HCT116 and HT-29 cell lines. (B) Relative mRNA levels of RGS17 when siRGS17 was transfected in HCT116 and HT-29 cell lines. siNC, negative control siRNA; vector means pLIC-SGC1. *p < 0.05 versus control. (C) The protein levels of RGS17 were determined in colorectal carcinoma cell lines HCT116 and HT-29, which were transfected with RGS17 expression plasmid or siRGS17. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was included as an internal control.
RGS17 Increases Cell Proliferation Ability in Both Colorectal Carcinoma Cell Lines
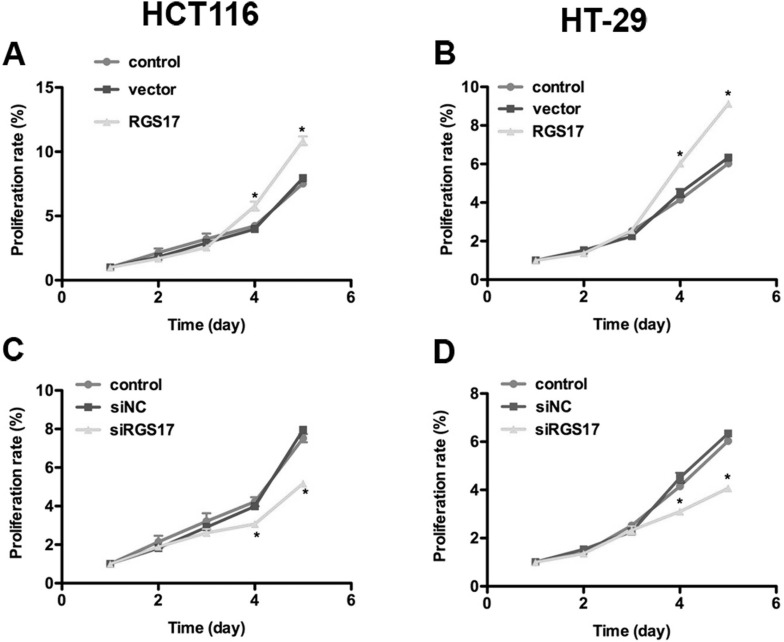
RGS17 functioned as an activator of cell proliferation in lung cancer18. Therefore, we examined the effect of RGS17 modulation on colorectal carcinoma cell proliferation. An MTT assay showed that cell proliferative rates were not significantly different in the first 3 days among different groups, though a minimal difference could still be observed. However, overexpression of RGS17 increased the cell proliferation rate by 19% in HCT116 cells and 33.3% in HT-29 cells on the fourth day. It further increased cell proliferation by 25% in HCT116 cells and 37.2% in HT-29 cells on the fifth day (Fig. 3A and B). On the contrary, the proliferation rate of siRGS17-transfected HCT116 cells was significantly lowered and decreased by 25% relative to the HCT116 control cells on the fourth day. The decrease in the proliferative rate was even more significant by the fifth day when cell proliferation was inhibited by approximately 37.5% in HCT116 cells (Fig. 3C). Similar results were observed in HT-29 cells that were significantly inhibited from proliferation after transfection with siRGS17 (Fig. 3D). These data suggest that RGS17 could promote cell proliferation in colorectal carcinoma.
Figure 3.
RGS17 promotes cell proliferation in both colorectal carcinoma cell lines. Viability of HCT116 cells and HT-29 cells was monitored for 5 consecutive days by MTT assay. HCT116 cells and HT-29 cells were transfected with RGS17 expression plasmid or siRGS17 prior to MTT assay. It was shown that (A) the proliferation rate was increased by 19% on the fourth day and 25% on the fifth day when RGS17 plasmid was transfected into HCT116 cells. (B) The proliferation rate was increased by 33.3% on the fourth day and 37.2% on the fifth day when RGS17 plasmid was transfected into HT-29 cells. (C) The proliferation rate was decreased by 25% on the fourth day and 37.5% on the fifth day when siRGS17 was transfected into HCT116 cells. (D) The proliferation rate was decreased by 25% on the fourth day and 33.3% on the fifth day when siRGS17 was transfected into HT-29 cells. *p < 0.05 versus control.
RGS17 Promotes Cell Migration and Invasion in Colorectal Carcinoma
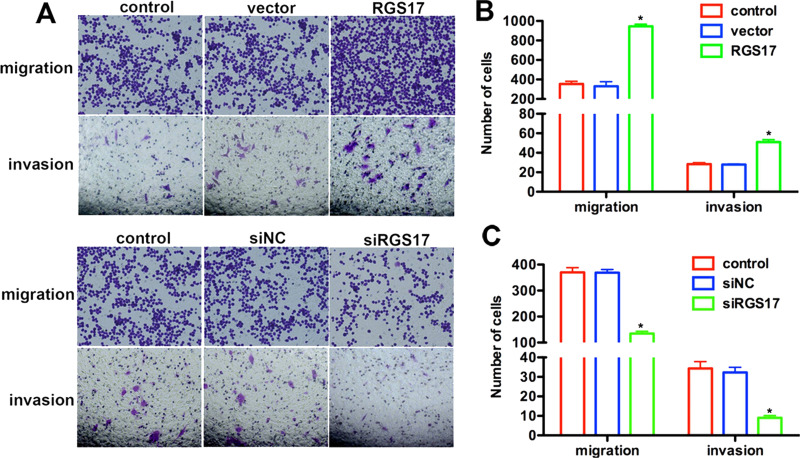
Colorectal carcinoma has a high tendency to spread. Consequently, we wanted to confirm whether RGS17 played any role in this process. A Transwell assay, together with the following crystal violet staining, showed that the number of transmigrated cells was markedly different between the experimental and control groups (Fig. 4A). Counting the transmigrated cells further showed that, compared with the corresponding control group, the number of transmigrated cells that attached to the permeable membrane was significantly increased, up to threefold, in the RGS-treated group and decreased by almost 60% when siRGS17 was transfected in HCT116 cells (Fig. 4B and C). For the invasion assay, HCT116 cells with RGS17 overexpression exhibited a higher invasive ability, while cells with TRIM59 knockdown showed a lower ability to invade through the Matrigel and migrate through the pores (Fig. 4B and C). These data support the notion of RGS17 as a contributor to cell migration and invasion in colorectal carcinoma.
Figure 4.
RGS17 promotes cell migration and invasion in colorectal carcinoma. (A) Transwell assay was performed in HCT116 cells with RGS17 overexpression or knockdown. Cells were allowed to migrate or invade for 12 h after plating. Cells treated with RGS17 plasmid showed a higher ability to migrate and invade, whereas siRGS17-transfected cells exhibited lower migration and invasive abilities. (B, C) Quantification of transmigrated cells in the Transwell assay. *p < 0.05 versus control.
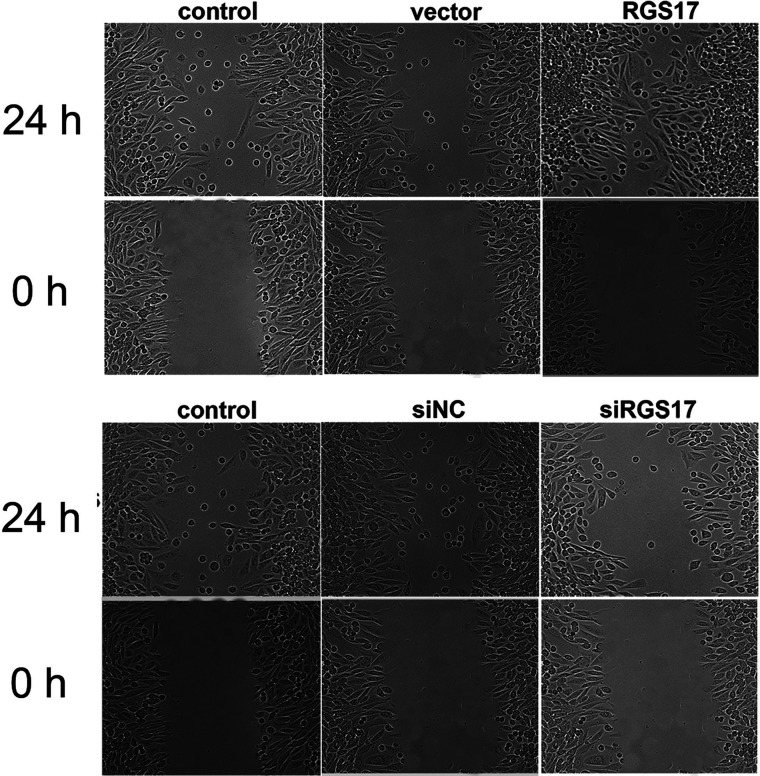
The wound healing assay is a classical and commonly used method for studying cell migration in vitro. It was observed that cells transfected with RGS17 expression plasmid actively recovered the wound 24 h after the artificial scratch, while the control cells showed minimal activity in recovering the wound. In contrast, recovery activity in RGS17-depleted HCT116 cells was significantly blocked at 24 h after artificial scratching (Fig. 5). These observations further confirm that RGS17 promotes cell migration in colorectal carcinoma cells.
Figure 5.
Effects of RGS17 on the wound healing processes of colorectal carcinoma cell HCT116. HCT116 cells were transfected with siRGS17 or RGS17 expression plasmid 24 h prior to scratching with a sterile pipette. Migration was observed under a microscope at a 200× magnification.
Knockdown of RGS17 Inhibits Tumor Growth in a Mouse Model of Colorectal Carcinoma
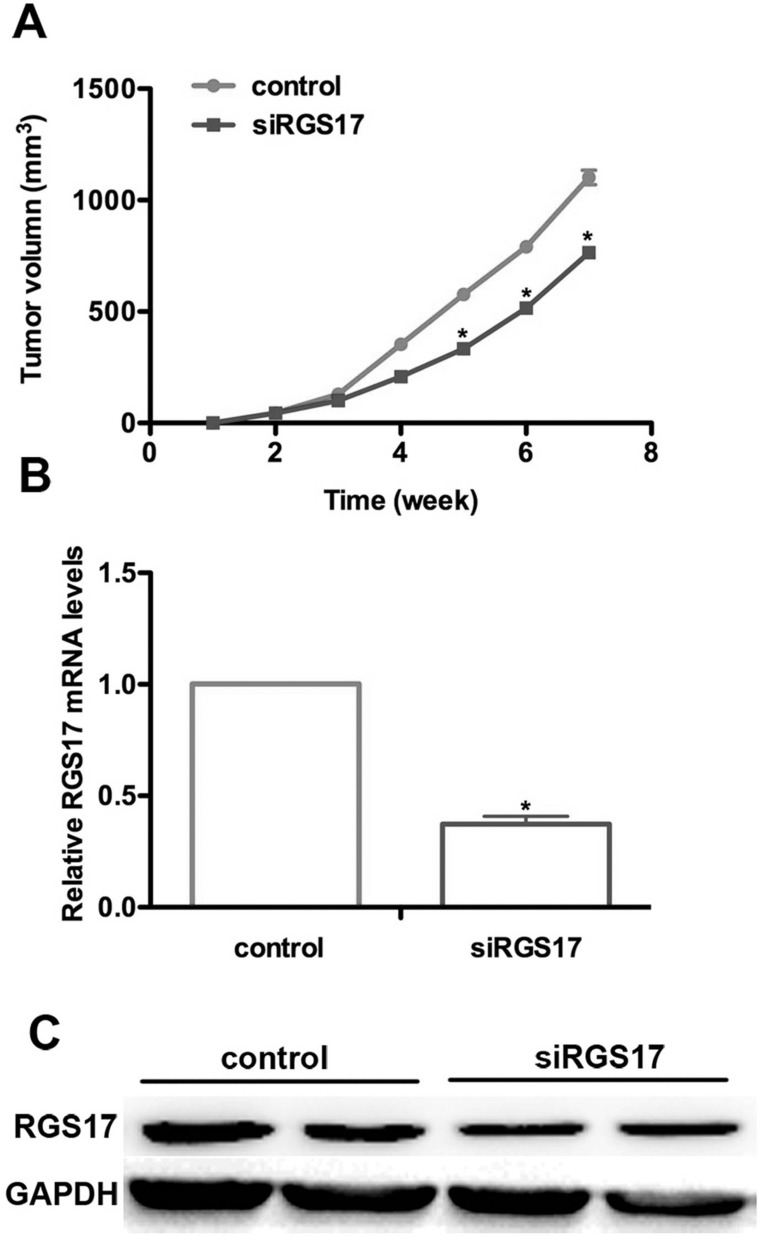
Moreover, we established a nude mouse model for colorectal carcinoma. Mice from both groups were injected with HCT116 cells with and without RGS17 stable depletion. One week after inoculation, all mice were observed to bear tumors. Periodic measurement of tumor size showed that they were enlarging as time went on. More importantly, mice in the control group showed a larger tumor volume compared with the ones with RGS17 ablation. At the fourth week, the average tumor size of mice from the RGS17-depleted group was significantly smaller than that in the control (Fig. 6A). We then examined the mRNA and protein levels of both groups. There was a significant difference between the siRGS17-treated and control groups (Fig. 6B). The mRNA level of RGS17 in the siRGS17-treated group was decreased by approximately 60% compared with the control group. This was also observed from the protein level in Western blot assays (Fig. 6C). These data strongly suggest that the knockdown of RGS17 could notably decrease the ability for tumor growth.
Figure 6.
Knockdown of RGS17 inhibits tumor growth in a mouse model of colorectal carcinoma. (A) A nude mouse model for colorectal carcinoma was established, and the tumor volume was measured every 5 days. Generally, mice in the control group exhibited a larger tumor size compared with the siRGS17-treated ones beginning the fourth week after inoculation. The difference in tumor size between the two groups became more significant as time passed. (B) A qRT-PCR assay showed a lower RGS17 mRNA level in the siRGS17-treated group compared with the control group. *p < 0.05, siRGS17 versus control. (C) Western blot assay showed that the protein level in siRGS17-transfected mice was significantly decreased. GAPDH was included as an internal control.
DISCUSSION
Colorectal carcinoma has been defined as a widespread malignancy all over the world, with approximately 1,400,000 new cases diagnosed in 2012 alone19,20. Although remarkable progress has been made in the last two decades, surgery remains effective, but mainly in localized colorectal carcinoma. For patients suffering from metastatic colorectal carcinoma, only 10%–25% are eligible for surgical resection, and the 5-year survival rate is limited to 30%–40%21–23. Therefore, finding new diagnostic and prognostic molecular markers and a schematic understanding of the development of colorectal carcinoma remain to be uncovered as soon as possible.
In this study, our data suggest that RGS17 is highly expressed in clinical human colorectal carcinoma tissues and in colorectal carcinoma cell lines at both the mRNA and protein levels. After knockdown of RGS17 with specific siRNA in HCT116 and HT-29 cells, an MTT assay showed that the cell proliferation rate was notably restricted. Furthermore, overexpression of RGS17 promoted, whereas knockdown of RGS17 inhibited, cell migration and invasion in both colorectal carcinoma cell lines. An in vivo mouse xenograft model of colorectal carcinoma revealed that knockdown of RGS17 significantly inhibited tumor growth. Furthermore, the migration and invasion abilities of colorectal carcinoma cells were regulated by RGS17, as illuminated by the Transwell and wound healing assays. Taken together, it could be concluded that RGS17 is a key mediator of colorectal carcinoma progression based on the strong evidence. However, our study represented only a basic investigation of the roles of RGS17 in colorectal carcinoma. The underlying mechanisms remain obscure. More details are required to further reveal the critical role of RGS17 in colorectal carcinoma tumorigenesis.
In previous research, RGS17 was traditionally considered to be an RGS, which regulates the Gαi/o and Gαq signaling pathways and has no activity against Gαs signaling14,24–26. Functionally, emerging studies have focused on the role of RGS17 in tumorigenesis. RGS17 was initially considered to be exclusively expressed in the brain. However, recent studies showed that RGS17 was also overexpressed in lung and prostate cancers and associated with familial lung cancer and prostate cancer cell proliferation process, in which the AMP–PKA–CREB signaling pathway was involved16,17. These findings suggest that RGS17 may be aberrantly expressed during tumorigenesis and associated with oncogenic activities, as evidenced by our study in colorectal carcinoma. Of note, several studies also showed that knockdown of RGS17 is significant in developing chemoresistance in ovarian cancer, and lysophosphatidic acid (LPA) is involved in this process16,27,28. However, whether a similar mechanism is involved in the regulation of colorectal carcinoma by RGS17 remains to be solved. More work needs to be done to provide clinical and fundamental researchers with a more detailed understanding of the biological activities of RGS17 in human tumorigenesis.
In conclusion, RGS17 is overexpressed in clinical colorectal carcinoma tissues and cultured cells. Knockdown of RGS17 inhibits, whereas overexpression of RGS17 increases, aggressive activities of colorectal carcinoma cells, including proliferation, migration, and invasion. This is the first report, to the authors’ knowledge, to investigate the critical roles of RGS17 in colorectal carcinoma progression.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Ferlay J, Parkin DM, Steliarova-Foucher E. Estimates of cancer incidence and mortality in Europe in 2008. Eur J Cancer 2010;46(4):765–81. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127(12):2893–917. [DOI] [PubMed] [Google Scholar]
- 3. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global Cancer Statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 4. Guo P, Huang ZL, Yu P, Li K. Trends in cancer mortality in China: An update. Ann Oncol. 2012;23(10):2755–62. [DOI] [PubMed] [Google Scholar]
- 5. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383(9927):1490–502. [DOI] [PubMed] [Google Scholar]
- 6. Dohlman HG, Thorner J. RGS proteins and signaling by heterotrimeric G proteins. J Biol Chem. 1997;272(7):3871–4. [DOI] [PubMed] [Google Scholar]
- 7. Druey KM, Blumer KJ, Kang VH, Kehrl JH. Inhibition of C-protein-mediated MAP kinase activation by a new mammalian gene family. Nature 1996;379(6567):742–6. [DOI] [PubMed] [Google Scholar]
- 8. Koelle MR, Horvitz HR. EGL-10 regulates G protein signaling in the C-elegans nervous system and shares a conserved domain with many mammalian proteins. Cell 1996;84(1):115–25. [DOI] [PubMed] [Google Scholar]
- 9. Rangel J, Nosrati M, Leong SPL, Haqq C, Miller JR, Sagebiel RW, Kashani-Sabet M. Novel role for RGS1 in melanoma progression. Am J Surg Pathol. 2008;32(8):1207–12. [DOI] [PubMed] [Google Scholar]
- 10. Berman DM, Wang YF, Liu ZY, Dong Q, Burke LA, Liotta LA, Fisher R, Wu XF. A functional polymorphism in RGS6 modulates the risk of bladder cancer. Cancer Res. 2004;64(18):6820–6. [DOI] [PubMed] [Google Scholar]
- 11. Wu T, Li YY, Huang DL, Han F, Zhang YY, Zhang DW, Han JH. Regulator of G-protein signaling 19 (RGS19) and its partner G alpha-inhibiting activity polypeptide 3 (GNAI3) are required for zVAD-induced autophagy and cell death in L929 cells. PLos One 2014;9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang YC, Tong Y, Tso PH, Wong YH. Regulator of G protein signaling 19 suppresses Ras-induced neoplastic transformation and tumorigenesis. Cancer Lett. 2013;339(1):33–41. [DOI] [PubMed] [Google Scholar]
- 13. Tso PH, Yung LY, Wang YC, Wong YH. RGS19 stimulates cell proliferation by deregulating cell cycle control and enhancing Akt signaling. Cancer Lett. 2011;309(2):199–208. [DOI] [PubMed] [Google Scholar]
- 14. Mao H, Zhao QS, Daigle M, Ghahremani MH, Chidiac P, Albert PR. RGS17/RGSZ2, a novel regulator of G(i/o), G(z), and G(q) signaling. J Biol Chem. 2004;279(25):26314–22. [DOI] [PubMed] [Google Scholar]
- 15. Larminie C, Murdock P, Walhin JP, Duckworth M, Blumer KJ, Scheideler MA, Garnier M. Selective expression of regulators of G-protein signaling (RGS) in the human central nervous system. Mol Brain Res. 2004;122(1):24–34. [DOI] [PubMed] [Google Scholar]
- 16. You M, Wang D, Liu P, Vikis H, James M, Lu Y, Wang Y, Wang M, Chen Q, Jia D, Liu Y, Wen W, Yang P, Sun Z, Pinney SM, Zheng W, Shu XO, Long J, Gao YT, Xiang YB, Chow WH, Rothman N, Petersen GM, de Andrade M, Wu Y, Cunningham JM, Wiest JS, Fain PR, Schwartz AG, Girard L, Gazdar A, Gaba C, Rothschild H, Mandal D, Coons T, Lee J, Kupert E, Seminara D, Minna J, Bailey-Wilson JE, Amos CI, Anderson MW. Fine mapping of chromosome 6q23-25 region in familial lung cancer families reveals RGS17 as a likely candidate gene. Clin Cancer Res. 2009;15(8):2666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. James MA, Lu Y, Liu Y, Vikis HG, You M. RGS17, an overexpressed gene in human lung and prostate cancer, induces tumor cell proliferation through the cyclic AMP-PKA-CREB pathway. Cancer Res. 2009;69(5):2108–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodle CR, Mackie DI, Roman DL. RGS17: An emerging therapeutic target for lung and prostate cancers. Future Med Chem. 2013;5(9):995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim JL, Cho KH, Park EC, Cho WH. A single measure of cancer burden combining incidence with mortality rates for worldwide application. Asian Pac J Cancer Prev. 2014;15(1):433–9. [DOI] [PubMed] [Google Scholar]
- 20. Ferro A, Peleteiro B, Malvezzi M, Bosetti C, Bertuccio P, Levi F, Negri E, La Vecchia C, Lunet N. Worldwide trends in gastric cancer mortality (1980-2011), with predictions to 2015, and incidence by subtype. Eur J Cancer 2014;50(7):1330–44. [DOI] [PubMed] [Google Scholar]
- 21. Tomlinson JS, Jarnagin WR, DeMatteo RP, Fong Y, Kornprat P, Gonen M, Kemeny N, Brennan MF, Blumgart LH, D’Angelica M. Actual 10-year survival after resection of colorectal liver metastases defines cure. J Clin Oncol 2007;25(29):4575–80. [DOI] [PubMed] [Google Scholar]
- 22. Fong YM, Cohen AM, Fortner JG, Enker WE, Turnbull AD, Coit DG, Marrero AM, Prasad M, Blumgart LH, Brennan MF. Liver resection for colorectal metastases. J Clin Oncol. 1997;15(3):938–46. [DOI] [PubMed] [Google Scholar]
- 23. Dale PS, Howard RJ, Henderson JM, Bolton JS, Stain SC, Abdalla EK. Recurrence and outcomes following hepatic resection, radiofrequency ablation, and combined resection/ablation for colorectal liver metastases. Ann Surg. 2004;239(6):825–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Doupnik CA, Xu T, Shinaman JM. Profile of RGS expression in single rat atrial myocytes. Biochim Biophys Acta 2001;1522(2):97–107. [DOI] [PubMed] [Google Scholar]
- 25. Zielinski T, Kimple AJ, Hutsell SQ, Koeff MD, Siderovski DP, Lowery RG. Two G alpha(i1) rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screening 2009;14(10):1195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-G alpha protein-protein interaction by CCG-4986. Mol Pharmacol. 2010;78(3):360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nature Rev. 2003;3(8):582–91. [DOI] [PubMed] [Google Scholar]
- 28. Hooks SB, Callihan P, Altman MK, Hurst JH, Ali MW, Murph MM. Regulators of G-Protein signaling RGS10 and RGS17 regulate chemoresistance in ovarian cancer cells. Mol Cancer 2010;9:289. [DOI] [PMC free article] [PubMed] [Google Scholar]