Abstract
Gastric cancer (GC) is one of the most common cancers in the world. The cathepsin F (CTSF) gene has recently been found to participate in the progression of several types of cancer. However, the clinical characteristics and function of CTSF in GC as well as its molecular mechanisms are not clear. Six GC cell lines and 44 paired adjacent noncancerous and GC tissue samples were used to assess CTSF expression by quantitative polymerase chain reaction (qPCR). We used lentivirus-mediated small hairpin RNA (Lenti-shRNA) against CTSF to knock down the expression of CTSF in GC cells. Western blot and qPCR were used to analyze the mRNA and related protein expression. The biological phenotypes of gastric cells were examined by cell proliferation and apoptosis assays. Microarray-based mRNA expression profile screening was also performed to evaluate the potential molecular pathways in which CTSF may be involved. The CTSF mRNA level was associated with tumor differentiation, depth of tumor invasion, and lymph node metastasis. Downregulation of CTSF expression efficiently inhibited apoptosis and promoted the proliferation of GC cells. Moreover, a total of 1,117 upregulated mRNAs and 1,143 downregulated mRNAs were identified as differentially expressed genes (DEGs). Further analysis identified the involvement of these mRNAs in cancer-related pathways and various other biological processes. Nine DEGs in cancer-related pathways and three downstream genes in the apoptosis pathway were validated by Western blot, which was mainly in agreement with the microarray data. To our knowledge, this is the first report investigating the effect of CTSF on the growth and apoptosis in GC cells and its clinical significance. The CTSF gene may function as a tumor suppressor in GC and may be a potential therapeutic target in the treatment of GC.
Key words: Gastric cancer (GC), Cathepsin F (CTSF), Proliferation, Apoptosis, Microarray
INTRODUCTION
Gastric cancer (GC) is a significant health issue worldwide and is the second most frequent cause of cancer-associated mortality1. The tumorigenesis and progression of GC is a multifactorial and multistage process, and thus an increased understanding of gene expression changes during carcinogenesis, particularly the identification of novel biomarkers for carcinoma diagnosis and novel therapeutic targets, will improve the diagnosis, treatment, and prevention of GC.
Cathepsins are a papain family of cysteine proteinases that represent a major component of the lysosomal proteolytic system. In general, cathepsins contain a signal sequence, followed by a propeptide, and then a catalytically active mature region2. Cathepsin F (CTSF), also known as CATSF/CLN13, is located on 11q133. The CTSF proregion is very long and is unique within the papain family of cysteine proteases in that it contains an additional N-terminal segment predicted to share structural similarities with cysteine protease inhibitors in the cystatin superfamily. This cystatin-like domain contains some of the elements known to be important for inhibitory activity4. The CTSF gene, ubiquitously expressed, encodes a predicted protein of 484 amino acids that contains a 19-residue signal peptide. CTSF also contains five potential N-glycosylation sites, and it may be targeted to the endosomal/lysosomal compartment via the mannose 6-phosphate receptor pathway5. High levels of CTSF were detected in cervical cancer6 in contrast to the low levels observed in different pediatric brain tumors7. The different expression of CTSF determines its distinct effects in human malignancies.
CTSF transcripts were also found in several cancer cell lines, suggesting that this enzyme could be involved in degradative processes during tumor progression4. Recently, Yang et al.8 found that CTSF could serve as a novel serum biomarker, which is also a noninvasive diagnostic index for GC and may potentially be used as a predictor of overall GC survival rate. Until now, only a small amount of data have been available on CTSF, and the biological role of CTSF in the progress of GC is not thoroughly known.
In the present study, we investigated the effects of downregulated CTSF expression on the proliferation and apoptosis of GC cells and explored the underlying molecular mechanisms by mRNA expression profile screening.
MATERIALS AND METHODS
Cell Culture
Six human GC cell lines (SGC7901, BGC823, MGC803, HGC27, AGS, and MKN45) and one immortalized normal gastric cell line (GES1) were obtained from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, P.R. China). These cells were cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco BRL) and 1% penicillin–streptomycin (10,000 U/ml penicillin and 10,000 μg/ml streptomycin; SolarBio, Beijing) and incubated at 37°C in a humidified 5% CO2 atmosphere.
Tissue Samples
Human GC samples and nonmalignant gastric tissues, which were at least 5 cm away from the tumor samples, were collected from 44 patients who underwent a gastrectomy at the Department of Surgery, Shengjing Hospital, China Medical University (Shenyang, P.R. China) between January 2015 and March 2016. All GC cases were pathologically confirmed.
Lentiviral Production and Stable Cell Lines
CTSF Lenti-shRNA and negative control Lenti-shRNA (NC-shRNA) were purchased from Shanghai GeneChem Company (Shanghai, P.R. China). CTSF stable silencing cell lines were selected with puromycin and identified by Western blot in SGC7901, BGC823, and MKN45 cells.
RNA Extraction and Quantitative Polymerase Chain Reaction (qPCR)
The total RNA was extracted from GC cells, human GC tissues, and the adjacent noncancerous tissues from the same patients. Extraction was performed using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions and reversely transcribed into cDNA using an Expand Reverse Transcriptase Kit (Takara, Dalian, P.R. China). The expression of CTSF mRNA was detected using qPCR with the following program: 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s. The reaction mixture contained 10 μl of SYBR Green (Takara), 0.4 μl of each primer, 2 μl of cDNA, and 7.2 μl of diethylpyrocarbonate (DEPC)-treated water. Real-time PCR was performed according to the protocol of SYBR Premix Ex Taq II Kit. Specific primer pairs in the experiment are listed in Table 1. Gene expression levels were calculated relative to the housekeeping gene GAPDH.
Table 1.
Primers Used in the Present Study
| Name | Primer Sequence | Distribution | Annealing Temperature (°C) | Product Size (bp) | Extension Time (s) |
|---|---|---|---|---|---|
| CTSF | F: 5′-CCCAAAACCATCCAGACAA-3′ | NM_003793.3545-704 | 55.27 | 160 | 40 |
| R: 5′-CGGGCTTCTTCCTTTGACT-3′ | 57.37 | ||||
| GAPDH | F: 5′-TGACTTCAACAGCGACACCCA-3′ | NM_002046.4273-393 | 62.18 | 121 | 40 |
| R: 5′-CACCCTGTTGCTGTAGCCAAA-3′ | 61.09 |
Western Blot
Total protein was extracted from cells in a lysate buffer: 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5% Nonidet P40, 0.5% sodium deoxycholate, and phenylmethylsulfonyl fluoride (all from Beyotime Institute of Biotechnology, Shanghai, P.R. China). Each sample (60 μg) was electrophoresed in 10% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA) by a BG-blotMiMi transfer machine (Baygene, Beijing, P.R. China). After blocking with 5% nonfat milk for 2 h at room temperature and then incubating with primary antibodies for CTSF (1:500 dilution; Proteintech, Wuhan, Hubei, P.R. China), MDM2 (1:500 dilution; Abcam, Cambridge, MA, USA), RALB (1:200 dilution; Abcam), PCNA (1:2,000 dilution; CST), Smad2 (1:300 dilution; Abcam), FAS (1:1,000 dilution; Abcam), CDK4 (1:1,000 dilution; CST), CDK6 (1:500 dilution; Abcam), BAX (1:1,000 dilution; Abcam), survivin (1:500 dilution; Abcam), Bid (1:1,000 dilution; CST), Bcl-2 (1:500 dilution; Abcam), C-IAP1 (1:1,000 dilution; CST), or GAPDH (1:2,000 dilution; Santa Cruz Biotechnology, Santa Cruz, CA, USA) overnight at 4°C. The next day, after incubation with a 1:5,000 dilution of secondary antibodies (Santa Cruz Biotechnology) at 37°C for 2 h and after washing, the immunoreactive protein bands were visualized using an electrochemiluminescence (ECL) detection kit (Pierce Biotechnology, Rockford, IL, USA). The ratio between the optical density of the protein of interest and GAPDH was calculated as the relative content of the protein detected. Each experiment was repeated three times.
Cell Counting Kit-8 (CCK-8) Assay
The cell proliferative ability was evaluated by a CCK-8 assay (C0037; Beyotime Institute of Biotechnology). The cells were seeded into 96-well plates (1.5 × 103/well). Following culture, CCK-8 solution (10 μl/100 μl medium) was added to each well, and the cells were incubated for 3 h at 37°C. The absorbance was measured at 450 nm using a Synergy2 Multi-Mode Microplate Reader (BioTek, Winooski, VT, USA). The GC cells cultured in RPMI-1640 affected with NC-shRNA and without any infection were used as the controls. The assay was conducted in five replicate wells for each sample, and three parallel experiments were performed.
Apoptosis Analysis by Flow Cytometry (FCM)
To detect the apoptotic variation induced by the effect of CTSF knockdown, the Annexin-V–PE/7-AAD double-labeling kit (KeyGEN, Nanjing, P.R. China) was used. Briefly, cells (1–2 × 105 cells/well) were collected and washed with cold PBS twice. Following the manufacturer’s instructions, apoptotic cells were analyzed via FCM. Finally, cells were analyzed using a FACSCalibur (BD Biosciences, San Jose, CA, USA). A proportion of cells were divided into four groups as viable cells (PE−/7-AAD−), necrotic cells (PE−/7-AAD+), early apoptosis (PE+/7-AAD−), and late apoptosis (PE+/7-AAD+).
RNA Isolation and GeneChip Hybridization
Total RNA from SGC7901-Lenti-shRNA against CTSF cell lines and negative control SGC7901-Lenti-shRNA cell lines was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA purity was determined using an ultraviolet and visible spectrophotometer (UNIC, Shanghai, P.R. China). Samples with RNA purity were used for GeneChip hybridization and scanning. Labeling, hybridization, and staining of these samples were performed according to the Eukaryotic Target Preparation protocol in the Affymetrix Technical Manual (701021 rev. 4; Affymetrix, Santa Clara, CA, USA). In summary, 1 μg of purified total RNA was used in the synthesis of cDNA, and the cDNA was purified using the GeneChip Sample Cleanup Module (Affymetrix). The purified cDNA was amplified to produce biotin-labeled cRNA using the GeneChip 3′IVT Express Kit (T7; Enzo Life Sciences, Farmingdale, NY, USA). Labeled cRNA was fragmented and hybridized to GeneChip Human Gene 2.0 ST according to the manufacturer’s protocol (Affymetrix). The hybridized gene chips were washed and stained for antibody amplification. The gene chips were then scanned using the Gene array Scanner 3000 7G (Affymetrix).
Data Preprocessing and Screening of DEGs
GeneChip® Operating Software (Affymetrix) was used to gather a signal value. Normalization algorithms were used to adjust sample signals by minimizing the effects of variation caused by nonbiological factors. Quantile normalization was performed by the robust multiarray average9 algorithm with application of the Affy package in R statistical software program10. Gene expression values of samples were log2 transformed and median centered for further analysis. If multiple probes corresponded to the same gene, the mean value was calculated as the expression value of this gene. The Limma package11 in R language was used to screen DEGs. The DEGs with fold change (FC) ≥1.0 and a value of p < 0.05 were considered to be significant.
Pathway Analysis
Genes showing a onefold and a value of p < 0.05 in expression changes (microarray; Table S1 available at https://pan.baidu.com/s/1dFeOpcX) were analyzed using Ingenuity Pathway Analysis (IPA).
Statistical Analysis
Statistical analysis was performed with the GraphPad Prism software package (v. 5.0; San Diego, CA, USA) or SPSS 22.0 software (Chicago, IL, USA), and p < 0.05 was considered to be statistically significant.
RESULTS
CTSF Expression Is Downregulated in GC Cells and Tissues
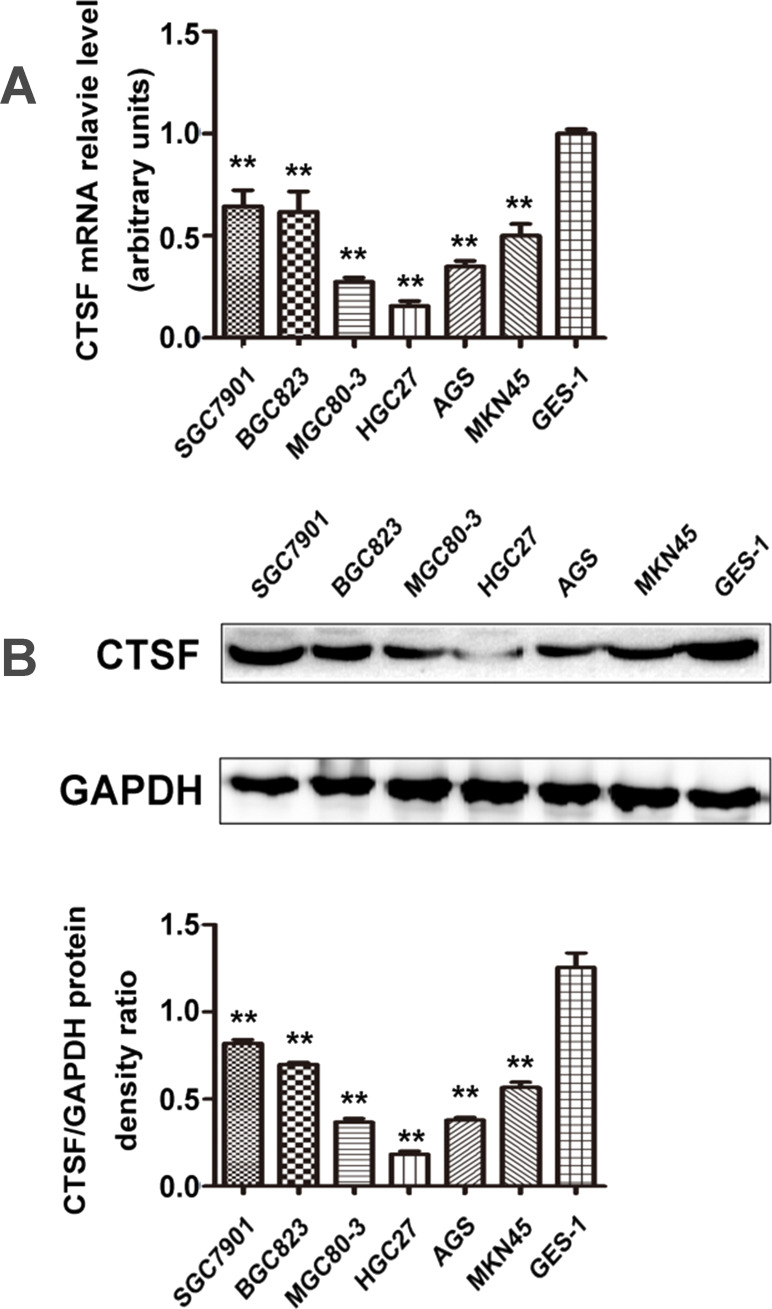
To investigate the CTSF transcription profile in GC cells, a qPCR was employed. With primers specific to the CTSF gene, qPCR results revealed that transcription of CTSF was found at different levels in various GC cell lines. The mRNA expression of CTSF was substantially downregulated in SGC7901 (0.645 ± 0.077), BGC823 (0.618 ± 0.100), MGC803 (0.275 ± 0.020), HGC27 (0.156 ± 0.023), AGS (0.348 ± 0.0208), and MKN45 (0.501 ± 0.055) cells compared with the normal mucosa line GES1 (onefold as the control; p < 0.01) (Fig. 1A). Western blot was then performed to assess the level of CTSF protein expression in these cells. The result was also remarkably downregulated in six GC cells compared with GES1 (p < 0.01) (Fig. 1B). Among the GC cells, CTSF protein expression was found at a comparatively higher level in SGC7901, BGC823, and MKN45 cells and was lowest in HGC 27.
Figure 1.
Expression of the cathepsin F (CTSF) gene in gastric carcinoma cell lines and a gastric epithelial cell line. (A) CTSF mRNA was detectable at different levels in gastric carcinoma cell lines (SGC7901, BGC823, MKN45, HGC27, AGS, and MGC803) and a gastric epithelial cell line (GES1). (B) Cell lysate was loaded and probed with anti-CTSF antibody (60 kDa) with GAPDH (36 kDa) as an internal control. CTSF protein expression was comparatively higher in SGC7901, BGC823, and MKN45 cells than in other gastric cancer (GC) cell lines (**p < 0.01). Results are representative of three different experiments, and data are expressed as mean ± standard deviation.
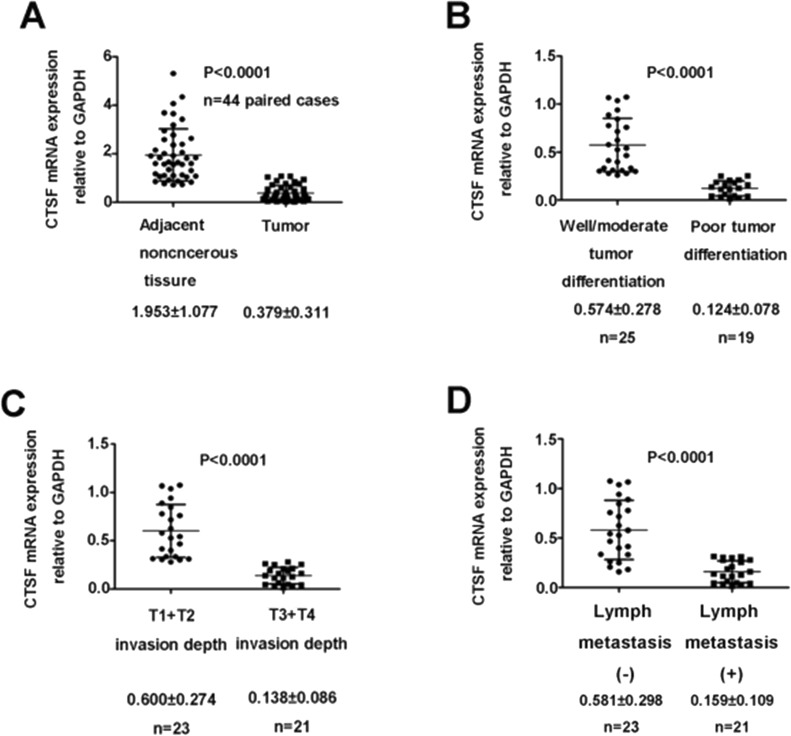
CTSF mRNA expression was also analyzed in 44 paired GC specimens and adjacent normal tissues by qPCR. It was identified that the expression of CTSF was significantly lower in the GC tissues than in their adjacent normal tissues (1.953 ± 1.077 vs. 0.379 ± 0.311; p < 0.0001) (Fig. 2A, Table 2). Furthermore, the correlation between CTSF mRNA expression and the clinicopathological factors of GC was examined. The CTSF mRNA expression level was associated with tumor differentiation, depth of tumor invasion, and lymph node metastasis (Fig. 2B–D, Table 2).
Figure 2.
Quantitative polymerase chain reaction (qPCR) analysis of CTSF mRNA expression in human GC tissues. (A) CTSF expression in adjacent noncancerous tissues and gastric cancer tissues. (B) CTSF mRNA expression level in patients with good/moderate tumor differentiation and those in tumors with poor tumor differentiation. (C) Association between increased invasion depth and downregulation of CTSF mRNA. (D) CTSF mRNA expression level in patients without lymph node metastasis and those in tumors with lymph node metastasis.
Table 2.
Correlation Between the Clinicopathological Features and Cathepsin F (CTSF) mRNA Expression in 44 Gastric Cancer Patients
| Variable | Patients (n) | CTSF mRNA Expression Relative to GAPDH | p Value |
|---|---|---|---|
| Normal | 44 | 1.953 ± 1.077 | <0.01 |
| Tumor | 44 | 0.379 ± 0.311 | |
| Age (years) | |||
| <65 | 24 | 0.349 ± 0.323 | 0.483 |
| ≥65 | 20 | 0.419 ± 0.297 | |
| Gender | |||
| Male | 30 | 0.290 ± 0.278 | 0.258 |
| Female | 14 | 0.571 ± 0.299 | |
| Tumor differentiation | |||
| Well/moderate | 25 | 0.574 ± 0.278 | <0.01 |
| Poor | 19 | 0.124 ± 0.078 | |
| Invasion depth | |||
| T1 + T2 | 23 | 0.600 ± 0.274 | <0.01 |
| T3 + T4 | 21 | 0.137 ± 0.086 | |
| Tumor location | |||
| Upper + middle | 18 | 0.477 ± 0.351 | 0.949 |
| Lower | 26 | 0.312 ± 0.265 | |
| Size (cm) | |||
| <3 | 16 | 0.435 ± 0.251 | 0.149 |
| ≥3 | 28 | 0.348 ± 0.340 | |
| Lymph node metastasis | |||
| No | 23 | 0.580 ± 0.298 | <0.01 |
| Yes | 21 | 0.159 ± 0.109 |
Knockdown of CTSF Significantly Promotes the Proliferation and Suppresses Apoptosis of Gastric Cancer Cells
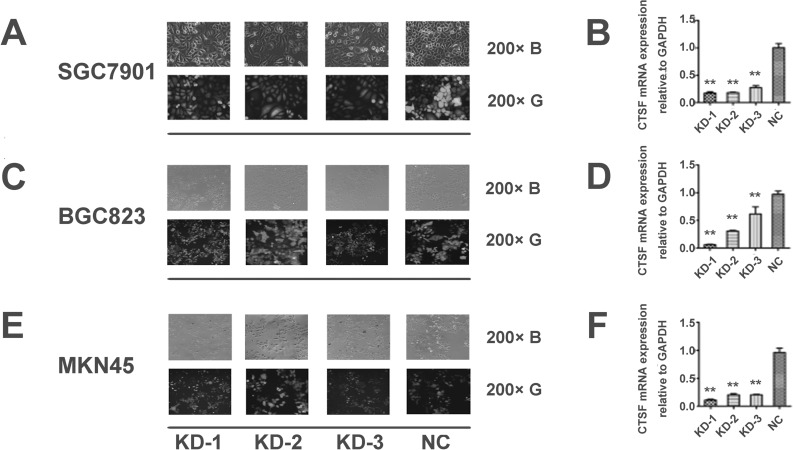
To investigate the effects of CTSF suppression on malignant growth potential, we introduced Lenti-shRNA targeting CTSF into SGC7901, BGC823, and MKN45 cells (Fig. 3A, C, and E). Using qPCR, we detected that the efficiency of CTSF knockdown in the Lenti-shRNA1 (KD-1), Lenti-shRNA2 (KD-2), and Lenti-shRNA3 (KD-3) groups was, respectively, 0.827 ± 0.026, 0.817 ± 0.009, and 0.728 ± 0.038 in SGC7901; 0.940 ± 0.008, 0.692 ± 0.011, and 0.384 ± 0.129 in BGC823; and 0.889 ± 0.015, 0.795 ± 0.019, and 0.797 ± 0.008 in MNK45, compared with the group infected with negative control lentivirus (NC; onefold as the control; p < 0.01) (Fig. 3B, D, and F). Compared with the NC group, the CTSF expression was notably decreased by KD-1 and moderately decreased by the other two shRNAs (KD-2 and KD-3) (Fig. 3B, D, and F). Thus, KD-1 was chosen for further experiments.
Figure 3.
Transfection efficiency tested by GFP-Lenti-shRNA under a fluorescence microscope and knockdown efficiency examined by qPCR. (A, C, E) Fluorescent particles within the cells indicated that Lenti-shRNA was successfully transfected into GC cells. (B, D, F) qPCR results showed that KD-1 has a higher efficiency of silencing CTSF expression than the other two shRNAs (KD-2 and KD-3). The ratio of CTSF knockdown was compared in the three groups (KD-1, KD-2, and KD-3). Each bar indicates the mean silencing rate ± standard deviation per group. All experiments were done three times, and data are presented as mean ± SD. **p < 0.01. (B) Visualization of the cells under common light. (G) Visualization of the cells under GFP (green fluorescent protein) fluorescence light. Lenti-shRNA, lentivirus-mediated small hairpin RNA; KD-1, Lenti-shRNA1; KD-2, Lenti-shRNA2; KD-3, Lenti-shRNA3. Magnification: ×200.
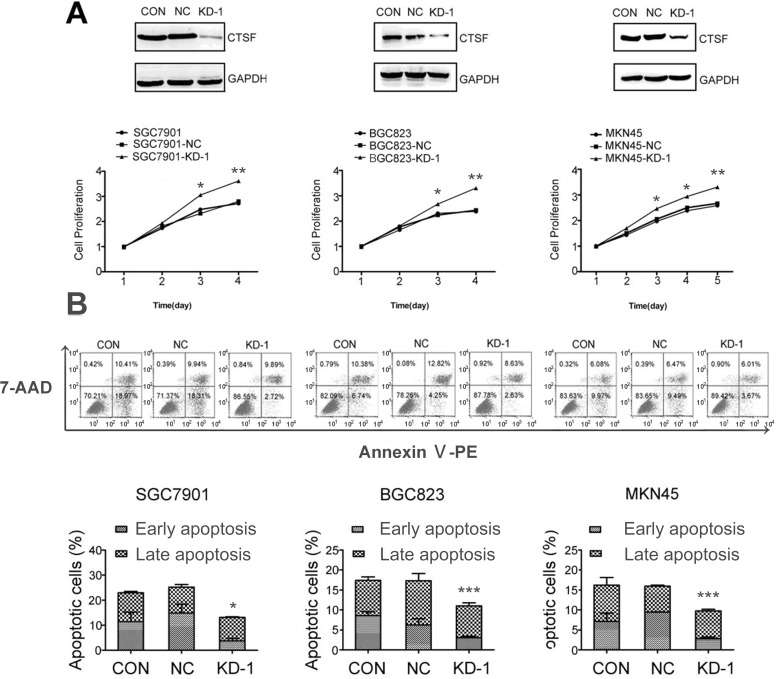
We also identified that the protein expression of CTSF was remarkably decreased in the KD-1 group, compared with the NC and control groups without any infection (CON). To evaluate the potential effects of transfection with KD-1 on the growth of SGC7901, BGC823, and MKN45 cells, growth curves were examined by a CCK-8 assay. The results showed that GC-KD-1 cells grew much more quickly than the other two control cells. Depletion of CTSF promoted growth of GC cells in a time-dependent manner, and the statistical analysis showed a significant difference on the third day (Fig. 4A).
Figure 4.
Knockdown of CTSF increases proliferation and inhibits the apoptosis of GC cells. (A) Growth curves for the indicated GC cells (CON, NC, and KD-1) were evaluated using a cell counting kit-8 assay. (B) The ratio of apoptosis in the cells of the indicated GC cells (CON, NC, and KD-1) was detected by flow cytometry. The ratio of apoptosis was compared in the three groups. Each bar indicates the mean apoptosis rate ± standard deviation per group. All experiments were done three times, and data are presented as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
We also examined whether the level of cell apoptosis was altered in CTSF-depleted GC cells. A flow-based annexin V assay was used to determine the effects of Lenti-shRNA1-mediated CTSF silencing on cell apoptosis. The apoptotic rates of the CON, NC, and KD-1 groups were 12.60 ± 0.31%, 12.24 ± 0.73%, and 7.85 ± 0.64% in SGC7901; 9.85 ± 0.61%, 9.63 ± 0.90%, and 6.63 ± 0.26% in BGC823; 8.94 ± 0.20%, 7.79 ± 0.61%, and 6.08 ± 0.07% in MNK45, respectively. The apoptotic rate of the KD-1 group compared with other control groups showed a statistical significance (p < 0.05). No significant difference was observed among the other two control groups (p > 0.05). The results indicated that the extent of apoptosis is much lower in the GC-KD-1 group than in the control cells groups, which is in agreement with the proliferation measurement using the CCK-8 approach (Fig. 4B).
CTSF Knockdown Induces Major Gene Expression Changes in Gastric Cancer Cells
To investigate the distinct downstream differentiation observed in GC cells with CTSF knockdown in more detail, we sequenced the whole transcriptome of both the KD-1 and NC groups in SGC7901 cells and compared their gene expression profiles. The microarray analysis showed that the DEGs were identified by the FC in filtering (FC ≥ 1 showed statistical significance). The results showed that 1,117 mRNAs were upregulated, whereas 1,143 mRNAs were downregulated (Table S1 available at https://pan.baidu.com/s/1dFeOpcX). The CTSF expression of the sh-CTSF SGC7901 group was decreased 4.24-fold.
Between the two cell groups, we detected 20 genes that mediate regulation of the PI3K/Akt signaling pathway, apoptosis, and transcriptional misregulation in cancer, including NUPR1, ATF4, SERP1, RPUSD4, GDF15, and MZT1, via up- and downregulation. To address how the DEGs in these cells corresponded to changes in biological functions, we analyzed all up- and downregulated DEGs using the IPA software. The top 10 most significantly altered canonical pathways in sh-CTSF SGC7901 cells included p53 signaling, PI3K/AKT signaling, hereditary breast cancer signaling, and molecular mechanisms of cancer. The top 10 most significantly altered disease and function pathways involved cancer, organismal injury and abnormalities, cellular growth and proliferation, cell death and survival, and gastrointestinal disease.
Validation of Downstream Target Genes in SGC7901-Silenced CTSF Cell Lines
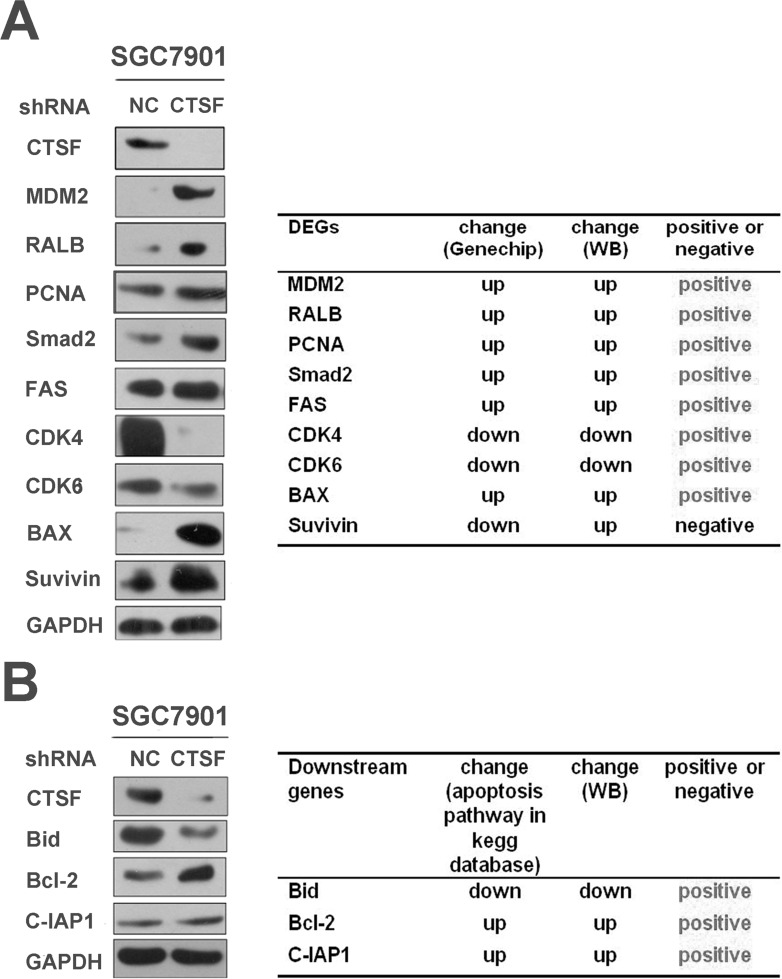
Using Western blot, the present study showed that the expression of MDM2, RALB, PCNA, smad2, FAS, and survivin was significantly increased in the SGC7901-KD-1 group compared with the SGC7901-NC group. Decreased expression of CDK4, CDK6, and Bax in the SGC7901-KD-1 group was also confirmed by Western blot. The results indicated that CTSF may be involved in cellular proliferation, apoptosis, and other processes in GC cells. The relative change in expression was mainly consistent with the microarray data (Fig. 5A).
Figure 5.
Western blot validation of downstream genes indicated by the microarray and KEGG pathway data in SGC7901 cell lines. (A) Expression change of the KD-1 group versus the NC group was verified by calculating Western blot results. The result showed that the change in gene expression by Western blot was mainly consistent with the microarray data. (B) Western blot analyses of Bid, Bcl-2, and C-IAP1 protein levels, which are apoptosis-related genes in the KEGG pathway database, between the two groups. GAPDH was used as the loading control.
Western blot assay confirmed the reduced expression of Bid and increased expression of Bcl-2 and C-IAP1 in the SGC7901-KD-1 group compared with the SGC7901-NC group at the protein level in order to affirm the underlying molecular mechanisms on cellular apoptosis in the KEGG pathway database (Fig. 5B).
DISCUSSION
Differential CTSF expression in certain malignant tumors suggests that this gene plays a distinct role in human malignancies6,7,12. We found that CTSF expression was significantly reduced in GC cells and tissues compared with normal mucosal cells and adjacent noncancerous tissues in the stomach. CTSF mRNA expression in GC tissues was related to tumor differentiation, invasion depth, and lymph node metastasis. This is in agreement with the result that differential CTSF expression in GC cells may be related to cellular differentiation. CTSF mRNA expression in the human moderately differentiated GC cell line SGC7901 was higher than in the human poorly differentiated GC cell line HGC27, which indicates that the degree of malignancy of GC is inversely proportional to the expression of CTSF.
Apoptosis is a process of programmed cell death that occurs in multicellular organisms. Biochemical events lead to characteristic morphological changes and death13. These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, chromosomal DNA fragmentation, and global mRNA decay14. Growth disorders can occur at the cellular level, and these consequently underpin much of the subsequent course in cancer, in which a group of cells display uncontrolled growth and division beyond the normal limits15. To investigate further tumor-suppressive functions of CTSF, SGC7901, BGC823, and MKN45 cells were treated with CTSF-Lenti-shRNA to knock down CTSF expression. Cell growth was markedly enhanced, and the level of cellular apoptosis was reduced simultaneously. This provides further evidence that CTSF may function as a tumor suppressor in GC.
To illuminate the fact that CTSF has a critical role in possible cellular processes required for proliferative development, we analyzed microarray data by IPA and identified DEGs affected by CTSF knockdown. Interestingly, consensus genes that were differentially regulated following CTSF alteration were significantly linked to ATM signaling, p53 signaling, transcriptional regulation, and other cancer-related cellular processes. We then used IPA to further analyze the disease and function pathways altered in our models. Some genes that showed altered expression form part of cancer, cellular growth, and proliferation, as well as cell death and survival pathways. These pathways have important roles in GC processes, but a deeper mechanistic analysis is required to achieve more conclusions from these data.
The protein expression levels of nine differentially expressed mRNAs (MDM2, RALB, PCNA, smad2, FAS, CDK4, CDK6, Bax, and survivin) in the cancer-related pathways were verified by Western blot analysis. The relative change in protein expression was mainly consistent with the microarray data. Several human tumor types have been shown to have an increased level of murine double minute (MDM2), as an oncogene, including soft tissue sarcomas and osteosarcomas as well as breast tumors16. MDM2 antagonizes p53 but may also carry out p53-independent functions. Removing MDM2 can arrest cell proliferation17. Ras-related protein (RalB) has been associated with the progression of several cancers, including bladder and prostate cancers18. RalB, involved in cellular apoptosis and motility, promotes tumor invasion and metastasis. Consequently, inhibition of RalB inhibits further progression of cancer19. Proliferating cell nuclear antigen (PCNA) is a DNA clamp that acts as a processivity factor and is essential for replication. PCNA protein, which is ubiquitinated, is involved in the RAD6-dependent DNA repair pathway. PCNA is also a potential therapeutic target in cancer therapy20. Mothers against decapentaplegic homolog 2 (SMAD2) mediates the signal of transforming growth factor-β (TGF-β) and thus regulates multiple cellular processes, such as cell proliferation, apoptosis, and differentiation21. This protein is recruited to the TGF-β receptors through its interaction with the SMAD anchor for receptor activation (SARA) protein22. Fatty acid synthase (FAS) has been investigated as a possible oncogene, which is upregulated in breast cancer23. It is also an indicator of poor prognosis and acts as a chemotherapeutic target24. Cyclin-dependent kinase inhibitor 2A (p16) acts as a tumor suppressor protein, which is a cyclin-dependent kinase (CDK) inhibitor that slows down the cell cycle by prohibiting progression from the G1 phase to the S phase25. Normally, CDK4/6 binds cyclin D and forms an active protein complex that phosphorylates retinoblastoma protein (pRB)26,27. Bcl-2-associated X protein (Bax) is regulated by the tumor suppressor p53 and is involved in p53-mediated apoptosis. Drugs that activate Bax act as anticancer treatments by inducing apoptosis in cancer cells28. Survivin protein functions to inhibit caspase activation, thereby leading to negative regulation of apoptosis or programmed cell death. Overexpression of survivin, which is regarded as an oncogene, decreases cellular apoptosis and increases in tumor growth29–31. The Western blot results indicated that MDM2, RALB, PCNA, smad2, FAS, and survivin were upregulated; on the contrary, CDK4, CDK6, and Bax were downregulated in the KD-1 group compared with the NC group in SGC7901. As a result, we have reason to believe that silencing CTSF can promote cellular growth and inhibit apoptosis in GC-KD-1 cells due to the potent roles of these DEGs, which were validated by Western blot analysis.
By analyzing the KEGG pathway database, we found that CTSF enhanced Bid and reduced Bcl-2 and C-IAPs in the cellular apoptosis pathway. The expression of Bid is upregulated by the tumor suppressor p53, and Bid has been shown to be involved in p53-mediated apoptosis32. The antiapoptotic Bcl-2 can bind Bid and inhibit Bid’s ability to activate Bax. As a result, the antiapoptotic Bcl-2 proteins may inhibit apoptosis by sequestering Bid, leading to reduced Bax activation. Simultaneous overexpression of Bcl-2 and the proto-oncogene Myc may produce aggressive B-cell malignancies including lymphoma33. C-IAP1 is important for the activation of MAPK signaling, which is involved in directing cellular processes that regulate cell functions including proliferation, gene expression, differentiation, cell survival, and apoptosis34. Therefore, CTSF may be useful for cellular growth and apoptosis in many malignant diseases. By Western blot analysis, the present study also indicated that Bcl-2 and C-IAPs were upregulated and Bid was downregulated in the KD-1 group compared with the NC group in SGC7901. However, further research on whether CTSF could also be a prognostic factor for GC patients remains to be explored.
In summary, we believe our study to be the first to report on the tumor suppressor functions of CTSF and that the silencing of CTSF after transfection with Lenti-shRNA promotes GC progression. We also identified the differentially expressed downstream genes in CTSF that downexpressed SGC7901 GC cells by microarray analysis, which provides clues for further experiments on the molecular mechanisms of CTSF in tumorigenesis. The present study provides new targets for prognosis and pharmacological intervention in human GC. Therefore, further studies, in vitro and in vivo, are required to determine the precise mechanism of CTSF in the progression of GC.
ACKNOWLEDGMENTS
This study was supported by Liaoning Science and Technology Grants (2014021010 and 2014021024). We thank Ci Lu (Ph.D.) and Xin-Lu Liu (Ph.D.) for their valuable comments and excellent technical assistance.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. de Martel C, Forman D, Plummer M. Gastric cancer: Epidemiology and risk factors. Gastroenterol Clin North Am. 2013;42(2):219–40. [DOI] [PubMed] [Google Scholar]
- 2. Wex T, Levy B, Wex H, Bromme D. Human cathepsins F and W: A new subgroup of cathepsins. Biochem Biophys Res Commun. 1999;259(2):401–7. [DOI] [PubMed] [Google Scholar]
- 3. Wang B, Shi GP, Yao PM, Li Z, Chapman HA, Bromme D. Human cathepsin F. Molecular cloning, functional expression, tissue localization, and enzymatic characterization. J Biol Chem. 1998;273(48):32000–8. [DOI] [PubMed] [Google Scholar]
- 4. Santamaria I, Velasco G, Pendas AM, Paz A, Lopez-Otin C. Molecular cloning and structural and functional characterization of human cathepsin F, a new cysteine proteinase of the papain family with a long propeptide domain. J Biol Chem. 1999;274(20):13800–9. [DOI] [PubMed] [Google Scholar]
- 5. Nagler DK, Sulea T, Menard R. Full-length cDNA of human cathepsin F predicts the presence of a cystatin domain at the N-terminus of the cysteine protease zymogen. Biochem Biophys Res Commun. 1999;257(2):313–8. [DOI] [PubMed] [Google Scholar]
- 6. Vazquez-Ortiz G, Pina-Sanchez P, Vazquez K, Duenas A, Taja L, Mendoza P, Garcia JA, Salcedo M. Overexpression of cathepsin F, matrix metalloproteinases 11 and 12 in cervical cancer. BMC Cancer 2005;5:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Rosa M, Sanfilippo C, Libra M, Musumeci G, Malaguarnera L. Different pediatric brain tumors are associated with different gene expression profiling. Acta Histochem. 2015;117(4–5):477–85. [DOI] [PubMed] [Google Scholar]
- 8. Yang L, Wang J, Li J, Zhang H, Guo S, Yan M, Zhu Z, Lan B, Ding Y, Xu M, Li W, Gu X, Qi C, Zhu H, Shao Z, Liu B, Tao SC. Identification of serum biomarkers for gastric cancer diagnosis using a human proteome microarray. Mol Cell Proteomics 2016;15(2):614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19(2):185–93. [DOI] [PubMed] [Google Scholar]
- 11. Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- 12. Oliveras-Ferraros C, Vazquez-Martin A, Cuyas E, Corominas-Faja B, Rodriguez-Gallego E, Fernandez-Arroyo S, Martin-Castillo B, Joven J, Menendez JA. Acquired resistance to metformin in breast cancer cells triggers transcriptome reprogramming toward a degradome-related metastatic stem-like profile. Cell Cycle 2014;13(7):1132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi YJ, Choi YK, Lee KM, Cho SG, Kang SY, Ko SG. SH003 induces apoptosis of DU145 prostate cancer cells by inhibiting ERK-involved pathway. BMC Complement Altern Med. 2016;16(1):507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yu X, Zhou X, Fu C, Wang Q, Nie T, Zou F, Guo R, Liu H, Zhang B, Dai M. Celastrol induces apoptosis of human osteosarcoma cells via the mitochondrial apoptotic pathway. Oncol Rep. 2015;34(3):1129–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai X, Dong M, Yu H, Xie Y, Yu Y, Cao Y, Kong Z, Zhou B, Xu Y, Yang T, Li K. Knockdown of TCTN1 strongly decreases growth of human colon cancer cells. Med Sci Monit. 2017;23:452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wade M, Wong ET, Tang M, Stommel JM, Wahl GM. Hdmx modulates the outcome of p53 activation in human tumor cells. J Biol Chem. 2006;281(44):33036–44. [DOI] [PubMed] [Google Scholar]
- 17. Wienken M, Dickmanns A, Nemajerova A, Kramer D, Najafova Z, Weiss M, Karpiuk O, Kassem M, Zhang Y, Lozano G, Johnsen SA, Moll UM, Zhang X, Dobbelstein M. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol Cell 2016;61(1):68–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kashatus DF. Ral GTPases in tumorigenesis: Emerging from the shadows. Exp Cell Res. 2013;319(15):2337–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tecleab A, Zhang X, Sebti SM. Ral GTPase down-regulation stabilizes and reactivates p53 to inhibit malignant transformation. J Biol Chem. 2014;289(45):31296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang SC. PCNA: A silent housekeeper or a potential therapeutic target? Trends Pharmacol Sci. 2014;35(4):178–86. [DOI] [PubMed] [Google Scholar]
- 21. Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, Thomsen GH, Wrana JL, Attisano L. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell 1996;86(4):543–52. [DOI] [PubMed] [Google Scholar]
- 22. Hocevar BA, Smine A, Xu XX, Howe PH. The adaptor molecule disabled-2 links the transforming growth factor beta receptors to the Smad pathway. EMBO J. 2001;20(11):2789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hunt DA, Lane HM, Zygmont ME, Dervan PA, Hennigar RA. MRNA stability and overexpression of fatty acid synthase in human breast cancer cell lines. Anticancer Res. 2007;27(1A):27–34. [PubMed] [Google Scholar]
- 24. Gansler TS, Hardman W 3rd, Hunt DA, Schaffel S, Hennigar RA. Increased expression of fatty acid synthase (OA-519) in ovarian neoplasms predicts shorter survival. Hum Pathol. 1997;28(6):686–92. [DOI] [PubMed] [Google Scholar]
- 25. Nobori T, Miura K, Wu DJ, Lois A, Takabayashi K, Carson DA. Deletions of the cyclin-dependent kinase-4 inhibitor gene in multiple human cancers. Nature 1994;368(6473):753–6. [DOI] [PubMed] [Google Scholar]
- 26. Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature 1993;366(6456):704–7. [DOI] [PubMed] [Google Scholar]
- 27. Khor GH, Froemming GR, Zain RB, Abraham MT, Omar E, Tan SK, Tan AC, Vincent-Chong VK, Thong KL. DNA methylation profiling revealed promoter hypermethylation-induced silencing of p16, DDAH2 and DUSP1 in primary oral squamous cell carcinoma. Int J Med Sci. 2013;10(12):1727–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Westphal D, Kluck RM, Dewson G. Building blocks of the apoptotic pore: How Bax and Bak are activated and oligomerize during apoptosis. Cell Death Differ. 2014;21(2):196–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–21. [DOI] [PubMed] [Google Scholar]
- 30. Tamm I, Wang Y, Sausville E, Scudiero DA, Vigna N, Oltersdorf T, Reed JC. IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer Res. 1998;58(23):5315–20. [PubMed] [Google Scholar]
- 31. De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell 2012;21(5):655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4(11):842–9. [DOI] [PubMed] [Google Scholar]
- 33. Ortiz JL, Cortijo J, Sanz C, De Diego A, Esplugues J, Morcillo E. Cooling-induced contraction of trachea isolated from normal and sensitized guinea-pigs. Naunyn Schmiedebergs Arch Pharmacol. 1991;343(4):418–26. [DOI] [PubMed] [Google Scholar]
- 34. Pearson G, Robinson F, Beers Gibson T, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: Regulation and physiological functions. Endocr Rev. 2001;22(2):153–83. [DOI] [PubMed] [Google Scholar]