Abstract
Lung cancer in young patients is an uncommon and understudied entity that harbors distinctive epidemiological, clinic-demographic, and genomic features. We carried out a systematic review of genomic profiling in young patients with lung cancer from 2010 to 2020 in the main electronic databases and selected 23 manuscripts. Lung cancer in young patients occurs more frequently in women with adenocarcinoma histology and at more advanced stages. Some studies report higher oncogenic genomic alteration in this population, with higher anaplastic lymphoma kinase rearrangements, a distinct profile of epidermal growth factor receptor mutations, and other novel genomic alterations. Although still uncommon, the implementation of next-generation sequencing (NGS) has shed some light on germline genomic alterations associated with lung cancer in young patients. Although outcomes when compared with the older population are conflicting, the overall prognosis is still poor in this subset of patients and efforts to find targetable genomic alterations should be made to improve survival.
Key words: genomic profiling, lung cancer, adolescents and young adults, non-small-cell lung cancer
Highlights
-
•
Lung cancer in adolescents and young adults (AYA) has specific features that may unveil a unique genomic background.
-
•
AYA harbor a higher percent of oncogenic genomic alterations, gene fusions, and a different profile of gene alterations.
-
•
Comprehensive genomic profiling is strongly recommended to offer the best treatment available.
-
•
In driver-negative non-small-cell lung cancer by DNA-based NGS assay, RNA-based NGS should be considered.
Introduction
Lung cancer is one of the most common malignancies worldwide. An estimated 2.1 million new cases of lung cancer (11.6% of all new tumors) and 1.8 million deaths (18.4%) were predicted for 2018.1 Although the mortality rate has declined in the last years, lung cancer is still the leading cause of cancer deaths.2 Lung cancer is not prevalent among adolescents and young adults (AYA), however, according to Cancer Statistics 2016 and the Surveillance, Epidemiology, and End Results (SEER) Program 18 database from 2009 to 2013, it is estimated that around 13 000 young adults will die from lung cancer in the United States each year.3 The aforementioned bad prognosis together with the distinct clinico-pathological features and different genomic alterations makes lung cancer in AYA an interesting field of research. In this review, we aim to summarize the published reports on genomic profiling in AYA with lung cancer [focusing on non-small-cell lung cancer (NSCLC)] in the last 10 years.
Materials and methods
Review design
This review was carried out according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.
Data sources and search strategy
We searched the electronic databases MEDLINE, Pubmed Central, Embase, and the Cochrane Library CENTRAL to identify all relevant records from January 2010 to May 2020. Our search strategy was restricted to manuscripts that included ‘lung adenocarcinoma’ or ‘lung malignancies’ or ‘lung cancer’ or ‘lung carcinomas’ and ‘young∗’ in the title.
Inclusion and exclusion criteria
We included manuscripts written in English. We included papers whose cut-off value for ‘young patient’ was age ≤50 years and that studied any germline or somatic genomic alteration. We excluded manuscripts focusing on other histologies than NSCLC. Case reports and reviews were excluded.
Study selection, data extraction, and quality assessment
The lead author (DV) carried out the first screening of the article. Another author (DM) validated the screening. DV and DM both assessed full-text articles. Discrepancies were discussed to reach a consensus. Two independent reviewers (JdC, OH) assessed study quality. The study was conducted in compliance with local ethics regulations. We included both clinical trials and observational studies.
Results
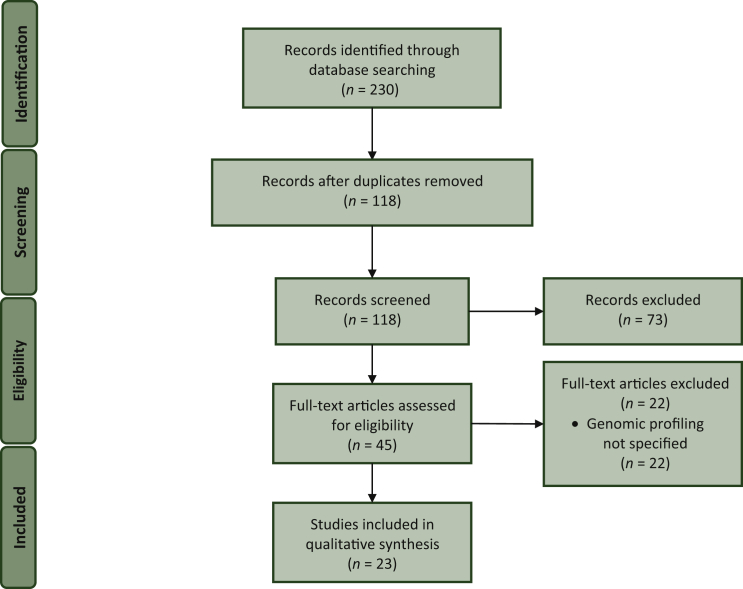
Within a multistep process, we screened 230 records for duplicates and eligibility, resulting in 66 publications undergoing abstract/full-text screening (Figure 1). We found 45 retrospective studies on AYAs with lung cancer.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48 Fourteen were reports from national databases. Our final selection for genomic profiling in AYA with lung cancer included 23 studies4,7, 8, 9,11,13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26,40,41,43,44 (Table 1).
Figure 1.
PRISMA flow diagram of the literature search.
Table 1.
Studies addressing the genomic profiling of young patients with lung cancer from 2010 to 2020
| Manuscript, location | Age cut-off (years) | Population | Technique | Oncogenic genomic alteration (%) | EGFR (%) | ALK (%) | ROS1 (%) | BRAF (%) | RET (%) | HER2 (%) | Other (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tian et al., 2020 West China Hospitala |
50 | NSCLC− ALK+. N = 101 Y = 52 |
NGS | – | – | EML4-ALK: 80.8 versus 79.6 EML4-ALK v1: 38.5 versus 14.3; P < 0.01 EML4-ALK v2: 1.9 versus 8.2 P = 0.19 EML4-ALK v3: 28.9 versus 40.8; P = 0.21 Non-EML4-ALK: 19 versus 21; P = 1.0 ALKm/TP53 mut: 34.5 versus 13.3; P = 0.07 |
– | – | – | – | – |
| Yang et al., 2019 Zhejiang Cancer Hospitala |
40 | Localized NSCLC N = 640 Y = 54 |
rtPCR | 76.7 versus 71.8; P = 0.49 | 48.8 versus 48 | 6.9 versus 3.3 | 6.9 versus 1.5 | 0 versus 1 | 9.3 versus 1 | 0 versus 2 | KRAS: 2 versus 5.6 PI3KCA: 2 versus 2 Fusion genes: 23.3 versus 5.9; P < 0.01 |
| Suidan et al., 2019 Davidoff Cancer Center, Rabin Medical Center, Israela |
50 | Lung cancer N = 186 Y = 62 |
rtPCR (24), NGS (78) |
63 versus 43 | 23 versus 18; P = 0.4 | 13 versus 2; P < 0.01 | – | – | – | – | Other genomic alterations (MET, KRAS, HER2, TP53, MYC, BRAF, BRCA1-2, APC): 27 versus 23; P = 0.45 |
| Pan et al., 2018 Jinling Hospital, China |
40 | NSCLC N = 272 LUAD N = 194 |
LUAD: rtPCR (EGFR) IHC (ALK) FISH (ROS1) |
– | 40 (29/73) | 34 (25/74) | 14 (1/7) | – | – | – | – |
| Hsu et al., 2016 National Taiwan Lung Cancer Registrya |
45 | Lung cancer N = 21536 Y = 1074 |
rtPCR, PCR, Sanger |
– | 52.5 versus 60.6; P < 0.01 | – | – | – | – | – | – |
| Wu et al., 2018 National Taiwan University Hospitala |
50 | Stage IV LUAD N = 872 Y = 142 |
rtPCR | 91 versus 84; P = 0.03 | 60 versus 67, P = 0.09 Uncommon mutations: 18 versus 9; P = 0.02 |
18.4 versus 5; P < 0.01a | 6 versus 1; P < 0.01 | 0.7 versus 1.6; P > 0.05 | 1.4 versus 2.1; P > 0.05 | 5 versus 2.7; P > 0.05 | KRAS: 0.7 versus 1.9; P > 0.05 PIK3CA: 0 versus 1.8 JAK2: 0 versus 0.6 |
| Wu et al., 2017 National Taiwan University Hospitala |
50 | LUAD N = 1039 Y = 161 |
rtPCR | – | 57.8 versus 66.1; P = 0.04 Uncommon mutations: 13.7 versus 8.4; P = 0.03 |
– | – | – | – | – | – |
| Hou et al., 2018 Hospital of Qingdao Universitya |
45 | Resected LUAD N = 177 Y = 87 |
NGS | 90.8 versus 85.6; P < 0.01 | 56.3 versus 52.2; P = 0.65 Concurrent TP53 mut: 81.6 versus 46.8; P < 0.001 |
16.1 versus 1.1; P < 0.01 | 0 versus 3.3; P = 0.24 | 3.4 versus 1.1; P = 0.36 | 1.1 versus 1.1; P = 1.0 | 13.8 versus 4.4; P = 0.03 | KRAS: 3.4 versus 18.9; P < 0.01 STK11: 1.1 versus 8.9; P = 0.03 MET: 0 versus 2.2; P = 0.49 TP53: 56.3 versus 48.9; P = 0.36 |
| Chen et al., 2019 First Affiliated Hospital, Zhejiang University |
35 | LUAD N = 89 |
NGS IHC for ALK and ROS1 |
67 | 21.3 | 16.9 | 1.1 | 3.4 | - | 24.7 | TP53: 9 PI3KCA: 1.1 |
| Chen et al., 2019 Affiliated Cancer Hospital of Zhengzhou Universitya |
40 | LUAD N = 1000 Y = 181 |
NGS | 84 versus 66; P < 0.01 | 34.3 versus 50.1 P < 0.01 Ex19del: 75.5 versus 40.2; P < 0.01 L858R: 16.1 versus 43.9; P < 0.01 G719X: 0 versus 4.6; P < 0.01 L861Q: 0 versus 2.4; P < 0.01 Ex20ins: 8.1 versus 1.2; P < 0.01 T790M: 0 versus 5.1; P < 0.01 |
37.6 versus 4.0; P < 0.01 | 1.7 versus 1.0; P = 0.25 | – | – | 2.2 versus 1.1; P = 0.23 | KRAS: 6.1 versus 10.4; P = 0.07 MET: 2.2 versus 0.2; P < 0.01 |
| Tanaka et al., 2017 Aichi Cancer Center Hospitala |
40 | LUAD N = 1746 Y = 81 |
NGS | 62 | 29.6 versus 45.4; P < 0.01 Ex19del: 75 versus 43; P < 0.01 L858R: 17 versus 48; P < 0.01 Ex20ins: 8 versus 1; P < 0.01 G719X: 0 versus 4; P = 0.32 L861X: 0 versus 1; P = 0.63 |
40.7 versus 4.2; P < 0.01 | 2.4 versus 0.001; P = 0.02 | 0 versus 0.4; P = 0.5 | 2.5 versus 0.01; P = 0.1 | 4.9 versus 1.1; P < 0.01 | KRAS: 2.2 versus 9.9; P: 0.02 |
| Sacher et al., 2016 Dana-Farber/Harvard Cancer Centera |
40 | NSCLC N = 2237 Y = 81 |
NGS, Sanger, PCR, FISH (ALK and ROS1) | 68 versus 52; P < 0.01 | 47 versus 40; P = 0.01 | 25 versus 1; P < 0.01 | 6 versus 1; P = 0.1 | 0 versus 3; P = 0.12 | – | 2 versus 3; P = 0.15 | KRAS: 9 versus 27; P < 0.01 |
| Yang et al., 2019 General Hospital of Chinesea |
36 | LUAD N = 44 Y = 20 |
Somatic and germline WES ALK IHC |
60 versus 58 | 35 versus 58.3; P = 0.12 | 25 versus 0; P = 0.50 | – | – | – | – | KRAS: 0 versus 8.3; P = 0.55 TP53: 35 versus 41.7; P = 0.88 FRG1: 40 versus 12.5; P = 0.08 KMT2: 50 versus 16.7; P = 0.04 N somatic mut/tumor: 92 versus 84; P: 0.42 |
| Kim et al., 2012 Inha University Hospital, Koreaa |
40 | NSCLC N = 1147 Y = 52 |
IHC for EGFR and ALK | 26 versus 31.9 | 22.6 versus 26.9; P = 0.60 | 3.2 versus 5.0; P = 0.86 | – | – | – | – | – |
| Wang et al., 2015 First Affiliated Hospital, Zhejiang University |
30 | Lung cancer N = 41 |
pyroPCR for EGFR FISH for ALK IHC for ROS1 |
68 | 22.7 | 27.2 | 11.7 | – | – | – | KRAS: 11.7 |
| Catania et al., 2015 Tumor Registry IEO |
40 | Lung cancer, N = 2847 Y = 100 |
rtPCR and FISH | 55.8 | 14.7 | 26.4 | – | – | – | 0 | PI3KA: 2.9 KRAS: 14.7 |
| Ye et al., 2014 Fudan University Shanghai Cancer Centera |
40 | Resected LUAD N = 123 Y = 36 |
rtPCR, FISH for ALK and RET |
69.5 versus 78.2; P = 0.39 | 52.8 versus 63.2; P = 0.13 Ex19: 73.7 versus 49.1 Ex20: 5.3 versus 3.6 Ex21: 21.0 versus 47.3 |
5.6 versus 4.6 | – | 0 versus 3.4 | 2.8 versus 1.1 | 0 versus 2.3 | KRAS: 8.3 versus 3.4 TP53: 72.2 versus 25.3; P < 0.01 LKB1: 11.1 versus 11.5; P = 0.95 |
| VandenBussche et al., 2014 Johns Hopkins Medical Institutions, Baltimore |
50 | NSCLC with molecular testing N = 53 |
pyroPCR and NGS FISH (ALK) |
52.8 | 20 | 11.6 | – | 0 | – | – | KRAS: 25.5 |
| Luo et al., 2018 West China Hospital |
45 | Never-smoking resected LUAD N = 36 |
WGS | 63.9 | 25 | 17 | – | 14 | 8 | – | MET: 14 Mean somatic mutation rate: 4.7 mut/MB. |
| He et al., 2020 Taipei Veterans General Hospitala |
40 | NSCLC N = 5051 Y = 168 |
rtPCR for EGFR IHC for ALK |
22.6 versus 16.2; P = 0.02 | 4.2 versus 0.5; P < 0.01 | – | – | – | – | – | |
| Hou et al., 2020 cBioPortal for Cancer Genomics databasea |
45 | LUAD N = 773 Y = 42 |
NGS | 59.6 versus 33.7; P < 0.01 | 30.9 versus 17.3; P < 0.01 | 9.5 versus 2.8; P = 0.01 | 4.7 versus 1.9; P = 0.05 | 2.3 versus 2.0; P = 0.05 | 9.5 versus 1.0; P < 0.01 | 0 versus 3.0; P = 0.72 | MET: 2.3 versus 5.6; P = 0.06 KRAS: 4.7 versus 32.2; P < 0.01 |
| Zhong et al., 2018 First Affiliated Hospital of Medical College, Zhejiang Universitya |
40 | Lung cancer N = 439 Y = 272 |
rtPCR | – | 41.3 versus 43.9; P = 0.76 EGFR mutation types: P < 0.01 Ex19del: 30.4 versus 14.0 L858R: 8.7 versus 21.1 Other types: 2.2 versus 8.8 Wild type: 58.7 versus 56.1 |
17.0 versus 4.1; P < 0.01 | 0 versus 2.1; P = 0.45 | – | – | – | – |
| Scarpino et al., 2016 Sant'Andrea Hospital of Romea |
50 | LUAD N = 789 Y = 78 |
RtPCR for EGFR FISH for ALK and ROS1 |
– | 12.8 versus 16 | 18 versus 6 | 8.6 versus 1 | – | – | – | – |
ALK, anaplastic lymphoma kinase; APC, adenomatous polyposis coli; BRAF, v-Raf murine sarcoma viral oncogene homolog B; BRCA1, breast cancer type 1; CT, chemotherapy; EGFR, epidermal growth factor receptor; Ex19del, exon 19 deletion; Ex20ins, exon 20 insertion; FISH, fluorescence in situ hybridization; HER2, human epidermal growth factor receptor 2; IEO, Instituto Europeo di Oncologia; IHC, immunohistochemistry; JAK2, Janus quinasa 2; KRAS, Kirsten rat sarcoma viral oncogene homolog; LKB1, Liver Kinase B1; LUAD, lung adenocarcinoma; MB, megabase; MET, mesenchymal epithelial transition receptor; MYC, MYC proto-oncogene; NGS, next-generation sequencing; NSCLC, non-small-cell lung cancer; PI3K, phosphatidylinositol-4,5-bisphosphate 3-kinase; RET, rearranged during transfection gene; ROS1, c-ROS oncogene 1; STK11, Serine/Threonine Kinase 11; TP53, tumor protein p53; WES, whole exome sequencing; WGS, whole genome sequencing; Y, young.
Comparative studies: young versus old patients.
Age cut-off value
The AYA patient is generally defined as an individual 15 to 39 years of age at the time of initial cancer diagnosis.49 However, the age cut-off in patients with lung cancer is still under debate and varies widely among studies. Of the 45 records included from 2010 to 2020, only 40% used 40 years as the cut-off value. Other reports chose a cut-off value between 30 and 50 years of age. The reason for these discrepancies may reflect that age is a continuum variable and differences in risk factors, clinico-pathological characteristics, and genetics may be observed across every range of ages. Even in patients between 15 to 39 years, some differences may be seen. For instance, Liu et al.34 found that patients aged ≤30 years compared with patients between 30 and 39 years had a higher proportion of adenocarcinoma, a lower proportion of large cell carcinoma, a higher proportion of stage I disease, and better lung-cancer-specific survival. Sacher et al.19 studied the frequency of targetable genomic alterations across small age groupings (5 years each) to look for an age cut-point where the likelihood of harboring a targetable genotype changes, finding a drop in the incidence of targetable genotypes above age 50 years.
Demographics and clinico-pathological characteristics
The median age at diagnosis of lung cancer is 71 years. However, 30% of patients are diagnosed at less than 65 years.3 Lung cancer is rare among AYAs, with an estimated incidence of fewer than 15 000 cases per year (0.2% of the total lung cancer diagnoses) in patients aged <35 years.1 Although it is the 18th incident tumor type among AYAs, it rises to the 11th position for the mortality rate. The incidence of lung cancer in AYAs seems to be decreasing over time. For example, in the USA, the incidence of NSCLC decreased in the young (aged <40 years) between 1975 and 2010 (0.9/100 000 in 1975 versus 0.4/100 000 in 2010 in females and 1.1/100 000 in 1975 versus 0.4/100 000 in 2010 in males),35 although these data must be interpreted cautiously.
Regarding sex, the historical patterns of higher incidence of lung cancer among men have reversed among non-Hispanic Whites and Hispanics. Among non-Hispanic Whites, the female-to-male incidence rate exceeded 1.0 in patients aged 30 to 49 years,50 and this change is not fully explained by sex differences in smoking behaviors. Most of the reports published to date found a higher percentage of young women with lung cancer.11,15,17,19,21,23,32,35,38,43,45
Historically, smoking prevalence was considerably higher among men than women. However, among Whites, sex differences have disappeared among people born during the 1960s and after.39 In patients with lung cancer, many studies that compare smoking history between young and old patients found significantly less exposure to tobacco in young lung cancer patients.4,7,11,14,15,17,19,21,23,24,28,37,40,43
Although most of the reports were carried out in Asia, some articles analyzed ethnic differences in the United States' population, showing a significantly higher percent of Asian and African-American patients among the younger patients.19,32,35,38,45
The histological pattern also seems to differ. NSCLC is still the most common type of lung cancer, but adenocarcinoma is even more common in younger patients.7,11,19,21,23,28,32,35, 36, 37, 38,40,43,45 Rich et al.36 divided patients with NSCLC extracted from the English National Lung Cancer Audit in six age groups from <39 to >80 years and showed a progressive decrease in the percent of adenocarcinoma from 48% in those aged <39 years to 31% in those aged >80 years.
Previous reports have shown that younger patients tend to present later, with more symptoms, and with tumors at a more advanced stage.51 This is probably due to the lack of suspicion, the denial of the patient, and the better performance status of young patients that could mask early symptoms. Data regarding the stage at presentation are conflicting; nevertheless, most of the studies15,17, 18, 19,21,23,32,35,38,40,43,45 show a significantly higher percentage of young patients presenting with metastatic disease.
Genomic profiling in AYA with lung cancer
The reduced involvement of tobacco and the higher percent of adenocarcinoma histology may unveil a different genomic background of lung cancer in AYAs. This is especially relevant in this era of personalized medicine and targeted therapy. An increasing number of reports are finding differences between old and young patients with lung adenocarcinoma (LUAD) (Table 1). However, although retrospective data in patients with Asian ethnicity are available, few studies conducted in other populations provide information about the genomic profiling of the tumors,8,19,23,25,44 thus challenging the generalization of the results. Another limitation of the interpretation of the results is that some of the studies have carried out molecular assessment only in patients with localized lung cancer.7,15,24,26
To date, some authors have reported interesting results regarding the percentage of oncogenic genomic alterations (OGA) in young LUAD patients compared with old patients. Four13,15,17,19 out of six comparative reports with more than two genomic alterations found that younger patients with LUAD had a significantly higher percent of OGA. Among these studies, Chen et al.17 had the largest cohort of young LUAD patients (N = 181) and showed that 84% of young Asian patients (aged <40 years) harbored a targetable genomic alteration, compared with 66% in the older counterpart. Sacher et al.19 recently published a retrospective study at the Dana-Farber Cancer Institute including 2237 patients of all races with NSCLC that had genotyping per standard protocol [including next-generation sequencing (NGS) in the past few years]. Among those patients that underwent testing for all five targetable genomic alterations at that time [epidermal growth factor receptor (EGFR), anaplastic lymphoma kinase (ALK), ROS1, human epidermal growth factor receptor 2 (HER2), and v-Raf murine sarcoma viral oncogene homolog B (BRAF)], an increased frequency of a targetable genotype was seen in patients aged 50 years or younger (68% versus 52%; P < 0.01). Other descriptive studies have found 52%-91% of OGA in young patients with AYA.9,16,18,22,23,25,26,38,41
Regarding EGFR mutations in young patients with LUAD, there have been some discrepancies. The PIONEER study52 was a prospective, epidemiological study of EGFR mutations in Asian patients with LUAD and did not find any significant correlation with age (P = 0.565). The prevalence of EGFR mutations varies widely between studies included in this systematic review, probably due to ethnicity and the retrospective nature of the studies. Only seven11,14,17, 18, 19,41,42 out of fourteen studies found statistically significant differences in the prevalence of EGFR mutations across age groups (see Table 1). However, four12,15,18,19 out of these seven studies found a significantly higher percentage of EGFR mutations among older patients with LUAD. One of the three studies that found significant differences in favor of young patients was the one carried out with a Caucasian population.20 Interestingly, the specific EGFR mutation may vary between young and old patients. At least three studies showed that EGFR Del19 is significantly more prevalent among young patients and L858R is more prevalent among older patients.17,18,43 Two studies by Wu et al.13,14 pointed out that although common mutations were less prevalent, uncommon mutations are more frequent among young patients (18% versus 9%; P = 0.02 and 13.7% versus 8.4%; P = 0.03, respectively). Chen et al.17 and Tanaka et al.18 found that insertions in exon 20 are more prevalent among young patients, with a prevalence of 8% versus 1% approximately in both studies. Additionally, Chen et al.17 found that de novo T790M was significantly more prevalent among older patients. Concurrent TP53 mutation is now considered a negative prognostic factor in patients with EGFR-mutated LUAD that receive EGFR-TKIs.53 A Chinese report found that concurrent TP53 mutation was more prevalent among young patients.15
Gene fusions such as ALK, ROS, and RET rearrangements, though uncommon, are among the most frequent gene alterations that can be targeted in lung cancer.54 It has been shown that young lung cancer patients harbor gene fusions more frequently than their older counterparts (23.3% versus 5.9% respectively; P < 0.01; in a study by Yang et al.7). Among them, ALK rearrangements are probably the most frequent gene fusions in the young population. Median age of patients with ALK-positive lung cancer included in phase III clinical trials varies between 50 and 60 years,55, 56, 57 while the estimated median age for unselected patients with lung cancer is approximately 70 years.58 In retrospective series included in this review, the prevalence of ALK rearrangement in AYAs is as much as 34% among the young Asian population.9 Studies in the US,19,25 Europe,23 and Latin America10 also show a prevalence of ALK rearrangements between 10% and 25% of AYAs with LUAD. Nine8,13,15,17, 18, 19,40,41,43 out of eleven studies that compared the prevalence of ALK rearrangements between young and old populations with NSCLC confirmed a higher, statistically significant percentage of ALK mutations in the younger group. In fact, in the study by Sacher et al.,19 ALK rearrangements were the only genetic alteration with a positive significant association with younger age in the multivariate analysis. Tian et al. analyzed the differences between young and old patients with ALK rearrangements.4 The most frequently occurring ALK fusion partner was EML4 (80.8% of young patients). Compared with the older patients, patients with early-onset disease were more likely to harbor EML4-ALK variant 1 (38.5% versus 14.3%; P = 0.007). Rare ALK fusions partners were also identified in the young cohort, including CHRNA7-ALK, TACR1-ALK, HIP1-ALK, DYSF-ALK, and ITGAV-ALK. It was also suggested that concomitant TP53 is more prevalent among young patients.
At least some of the published data also showed a higher frequency of ROS1 rearrangements in young patients with LUAD,7,13,18,19,44 although the prevalence of this alteration is lower than ALK (0%-6.9% in AYAs). Median age of patients with ROS-1 rearranged NSCLC in clinical trials varies between 53 and 55 years.59, 60, 61
Prevalence of RET alterations among young patients varies between 1.4% and 9.3%.8,14,16,19,25,27,41 Four comparative studies have addressed alterations in RET. Three of them did not find significant differences between age groups.14,16,19,41 Hou et al.41 analyzed data from patients with LUAD and NGS available from the cBioPortal for Cancer Genomics database and found that 9.5% of patients aged <45 years compared with 1% of older patients harbored RET fusions. In the LIBRETTO-001 trial, median age of patients with RET-fusion-positive NSCLC was 61 years.62
NTRK fusions are common in rare pediatric cancers. Among AYAs, median age varies from 48 to 58 years.63 However, the low frequency of this alteration limits the interpretation of the results. None of the studies included in our analysis were carried out on patients with NTRK fusions.
HER2 alterations might also be more frequent among AYAs, with two studies reporting significant differences when compared with older patients (Hou et al.15 described 13.8% of HER2 amplification in young patients compared with 4.4% in older patients). Conversely, six studies did not show differences among age groups according to HER2 status (see Table 1). Few studies have addressed alterations in MET, and results are inconclusive. In a report on 36 Asian, non-smoking LUAD patients <45 years old, the prevalence was 14%.26 Other reports showed a prevalence of 0% to 2% of MET alterations among young patients.15,17 Patients with MET-altered NSCLC included in clinical trials seem to be older than patients with other genotypes with approved targeted therapies (up to 74 years, depending on the trial and cohorts).64,65
On the contrary, BRAF and Kirsten rat sarcoma viral oncogene homolog (KRAS) mutations seem to be more frequent in the older populations, with at least three reports15,18,41 showing significant differences between old and young patients. Sacher et al.19 found a significant association between age and the frequency of KRAS mutation. Patients aged less than 40, 40-49, 50-59, 60-69, and more than 70 years old had a frequency of 9%, 13%, 26%, 34%, and 27%, respectively. In this study, BRAF and KRAS were independently associated with older age at diagnosis in the multivariate analysis (P < 0.01 and P = 0.04, respectively).
The recent implementation of NGS in clinical practice for genomic profiling may shed some light on new potential targets in young patients with LUAD, but at the moment there have been few reports with this technique. Luo et al.26 reported that in 36 never-smoking patients with LUAD younger than 45 years old, the mean somatic mutation rate was 4.7 per megabase (Mb), lower than that of LUAD from the TCGA cohort (8.87 per Mb). Patients aged older than 40 years and those with somatic TP53 mutation carried a higher somatic mutation rate than patients aged 40 years or younger (P = 0.035 and P < 0.01, respectively). They also identified several potential lung-cancer-associated gene mutations in young patients that were rarely reported (e.g. HOXA4 and MST1). Yang et al.20 reported in 20 patients with LUAD and ≤36 years a similar number of somatic mutations per tumor than their older counterparts. Mutations in FRG1 and KMT2C were associated with a younger age especially after correcting for tobacco smoking and sex. Hou et al.15 analyzed 71 surgically resected lung adenocarcinoma tissue samples from patients aged less than 45 years and compared them with LUAD tissue from older patients. A significant difference was seen between the younger and the older groups in a targeted genetic profile identified by NGS (90.8 versus 85.6; P < 0.001). Most importantly, 80.5% of younger patients harbored genetic alterations mapping to therapies approved by the National Comprehensive Cancer Network (NCCN) Guidelines (version 1.2018 updates, Non-Small-Cell Lung Cancer) compared with 54.4% (49/90) of older patients.
At this moment, several guidelines favor NGS rather than multiple single-gene tests as the preferred test to identify actionable oncogenic drivers.66 It is becoming clear that selecting the adequate NGS-based assay is important because each technique has its own caveats. DNA-based large-gene-panel NGS assay is probably the most widely available test for broad molecular sequencing of cancer patients. In fact, the FDA approved FoundationOne CDx (F1CDx) and MSK-IMPACT67, 68, 69 in 2017. However, DNA-based NGS assays may lead to false-negative gene fusion results due to inadequate coverage of long introns and the presence of intronic repetitive sequence elements also recurrently found across the genome. Furthermore, DNA-based NGS assays do not provide direct evidence of the mRNA expression of the rearrangement which may limit the interpretation of the rearrangements that seem noncanonical at the genomic DNA level.70,71 In an effort to assess the yield of RNA sequencing for targetable kinase fusions in LUAD, Benayed et al. carried out a targeted RNA sequencing in patients with no driver alteration found by DNA sequencing (MSK-IMPACT) and found that a previously undetected alteration was identified in 36 of the 254 cases (14%). This percentage of patients was even higher (31%) when selecting patients with low tumor mutational burden (TMB) (0-5 mutations/megabase). ROS1 was the most commonly detected fusion among patients with DNA-sequenced driver-negative tumors (10 out of 36 cases).70 This is especially relevant among AYAs, as they more frequently harbor tumors with targetable kinase fusions.
Germline genomic alterations in young patients with lung cancer
Patients with lung cancer have not been considered as having a high inherited predisposition to cancer. Familial aggregation varies widely across the studies (from 1% to 30%) and only one comparative study found significant differences between young and old patients.
Recently, some germline genomic alterations have been associated with lung cancer syndromes, including EGFRT790M, TP53, BRCA, HER2, YAP1, and CHECK2 among others,72 thus bringing back the question as to whether we should test for germline genetic alterations, especially in AYAs with lung cancer. NCCN and ESMO guidelines recommend performing germline analysis of EGFRT790M in patients with de novo somatic EGFRT790M mutation.67,69
Some of the manuscripts selected in our review provide some information. Yang et al.20 studied the prevalence of germline mutations in young LUAD, finding five germline mutations out of 20 patients; one of them showed a likely pathogenic variant (p.R141H) in TP53, which is known to cause Li–Fraumeni syndrome. The other mutations were two pathogenic variants in the ELAC2 gene (an endonuclease involved in the maturation of tRNA), which are not known to cause hereditary cancer syndromes, and two variants in the TGFBR gene, with conflicting evidence about its pathogenicity. Luo et al.26 studied 36 young LUAD patients with whole-genome sequencing and found that 78% of them carried at least one pathogenic mutation in 35 different genes. Six of these genes (BPIFB1, CHD4, PARP1, NUDT1, RAD52, and MFI2) were significantly enriched in comparison with a noncancer database. Mezquita et al.73 recently reported a cohort of 21 lung cancer patients with Li–Fraumeni syndrome and genomic profiling was carried out. They found that 90% of them had somatic oncogenic driver mutations, including EGFR mutations in 18 (exon 19 deletion in 12 cases, L858R in three cases, and G719A, exon 20 insertion, among others) and ROS1 fusion in one case. Additionally, a study carried out by Hu et al. in 2018 found that 1% of NSCLC (64/6220 patients) harbored a pathogenic BRCA1/2 mutation. The prevalence among patients with early-onset disease (aged <50 years) was significantly higher.74
The increasing availability of NGS in clinical practice may help to identify patients with suspicious somatic mutations, especially EGFRT790MF, BRCA, and TP53, that may eventually harbor a germline alteration. Case-by-case discussion on molecular tumor boards is recommended.
Treatment and prognosis in young patients with lung cancer
Some reports concluded that survival was similar between AYAs and older patients,8,17,21,23,24,51,75, 76, 77, 78 while others found that young patients performed better.28,34,35,38,40,45,79,80 One of the largest studies that used the National Cancer Database (NCDB) including 173 856 patients from 2003 to 2009 found that 5-year survival was significantly better for younger patients (P < 0.001), although this difference was more clinically significant in early stages. The absolute difference in 5-year overall survival (OS) at stages I and II (AJCC seventh-edition staging) between both groups was 25%, while at stage IV was only 2%.
Performing thorough genomic profiling can have an impact on the treatment of young patients. In a Middle-East single-center report that carried out NGS on half of the cohort of young patients, it was found that molecular testing affected treatment decisions more for younger patients (39% versus 22%; P = 0.002) and that significantly more patients finally received matched targeted therapy (48% versus 23% P = 0.001). Other reports did not find such a difference.17
Some reports have studied the efficacy of targeted therapy among AYAs with lung cancer. Tian et al.4 reported the efficacy of crizotinib in patients aged <50 versus 50 years or older and found that younger patients had a statistically significant longer progression-free survival (PFS) (17.5 months versus 9.0 months, respectively; P = 0.04). Other reports did not find any difference in survival in patients treated with EGFR tyrosine kinase inhibitors (TKIs) or ALK-TKIs therapy according to age.17,18 It is important to point out that young patients harboring driver genomic alterations may have concomitant genomic features that modify the response to treatment and prognosis, such as mutation in the TP53 gene.15 Hou et al.15 showed that concurrent TP53 mutation was significantly more prevalent among young patients with EGFR-mutated LUAD, and that conferred worse prognosis (PFS of 5.3 months in 10 young patients with concurrent EGFR/TP53 mutation). Tian et al. found that the median PFS of young patients harboring ALK rearrangements and concurrent TP53 mutations was only 6.2 months. Sacher et al. analyzed the survival of a cohort of 2237 patients with NSCLC from 2002 to 2014 according to the presence of a targetable genotype. In patients harboring a targetable genotype, the lowest median overall survival (mOS) was in patients younger than 40 years of age and those older than 70 years [21.4 months, 95% confidence interval (CI): 13.6-47.3 and 22.3 months, 95% CI: 16.9-28.6, respectively]. Those aged between 50 and 59 years had the longest OS (35.4 months, 95% CI: 29.6-41.4).
In young patients harboring oncogenic driver alterations, the use of immunotherapy remains controversial. Most of these tumors are associated with low tumor mutational burden, lack of CD8+ tumor-infiltrating lymphocytes, and develop in non-smokers, thus explaining the little efficacy of immunotherapy in this population. Furthermore, combining TKIs and immunotherapy has resulted in higher toxicity with no added benefit.81,82
Anyway, looking for a targetable genomic alteration is of special interest in the young population due to the higher probability of finding a targetable driver, the reduced toxicity of the treatment, and the potential better outcomes if matched treatment is started.
Final remarks and conclusions
The distinctive epidemiological and clinical characteristics of AYAs with lung cancer (predominantly in women, non-smokers with adenocarcinoma histology, and more advanced stages at diagnosis) unveil a unique genomic background that may tailor the treatment and ultimately improve outcomes. Young patients with lung cancer seem to harbor a higher percentage of OGA, higher prevalence of gene fusions, and a different profile of gene alteration. Comprehensive genomic profiling is strongly recommended to offer the best treatment available. In those patients with driver-negative NSCLC as determined by DNA-based NGS assay, an RNA-based NGS should be considered. Despite the recent advances in lung cancer, the prognosis is still dismal in this population and further studies are needed to better characterize and treat these patients.
Acknowledgments
Funding
None declared.
Disclosure
The authors have declared no conflicts of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA A Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 3.National Cancer Institute Surveillance, epidemiology and end results program. SEER stat fact sheets: lung and bronchus cancer. 2014. http://seer.cancer.gov/statfacts/html/lungb.html Available at:
- 4.Tian P., Liu Y., Zeng H. Unique molecular features and clinical outcomes in young patients with non-small cell lung cancer harboring ALK fusion genes. J Cancer Res Clin Oncol. 2020;146:935–944. doi: 10.1007/s00432-019-03116-6. [DOI] [PubMed] [Google Scholar]
- 5.Li F., He H., Qiu B. Clinicopathological characteristics and prognosis of lung cancer in young patients aged 30 years and younger. J Thorac Dis. 2019;11(10):4282–4291. doi: 10.21037/jtd.2019.09.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu B., Quan X., Xu C. Lung cancer in young adults aged 35 years or younger: a full-scale analysis and review. J Cancer. 2019;10(15):3553–3559. doi: 10.7150/jca.27490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang S., Song Z., Cheng G. Genomic alterations and survival in young patients aged under 40 years with completely resected non-small cell lung cancer. Ann Transl Med. 2019;7(7):140. doi: 10.21037/atm.2019.03.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suidan A.M., Roisman L., Belilovski Rozenblum A. Lung cancer in young patients: higher rate of driver mutations and brain involvement, but better survival. J Glob Oncol. 2019;5:1–8. doi: 10.1200/JGO.18.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan X., Lv T., Zhang F., Fan H., Liu H., Song Y. Frequent genomic alterations and better prognosis among young patients with non-small-cell lung cancer aged 40 years or younger. Clin Transl Oncol. 2018;20(9):1168–1174. doi: 10.1007/s12094-018-1838-z. [DOI] [PubMed] [Google Scholar]
- 10.Corrales-Rodríguez L., Arrieta O., Mas L. An international epidemiological analysis of young patients with non-small cell lung cancer (AduJov-CLICaP) Lung Cancer. 2017;113:30–36. doi: 10.1016/j.lungcan.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Hsu C.H., Tseng C.H., Chiang C.J. Characteristics of young lung cancer: analysis of Taiwan's nationwide lung cancer registry focusing on epidermal growth factor receptor mutation and smoking status. Oncotarget. 2016;7(29):46628–46635. doi: 10.18632/oncotarget.9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagashima O., Ohashi R., Yoshioka Y. High prevalence of gene abnormalities in young patients with lung cancer. J Thorac Dis. 2013;5(1):27–30. doi: 10.3978/j.issn.2072-1439.2012.12.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu S.G., Liu Y.N., Yu C.J., Yang J.C., Shih J.Y. Driver mutations of young lung adenocarcinoma patients with malignant pleural effusion. Genes Chromosomes Cancer. 2018;57(10):513–521. doi: 10.1002/gcc.22647. [DOI] [PubMed] [Google Scholar]
- 14.Wu S.G., Chang Y.L., Yu C.J., Yang P.C., Shih J.Y. Lung adenocarcinoma patients of young age have lower EGFR mutation rate and poorer efficacy of EGFR tyrosine kinase inhibitors. ERJ Open Res. 2017;3(3):00092–2016. doi: 10.1183/23120541.00092-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hou H., Zhu H., Zhao H. Comprehensive molecular characterization of young Chinese patients with lung adenocarcinoma identified a distinctive genetic profile. Oncologist. 2018;23(9):1008–1015. doi: 10.1634/theoncologist.2017-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z., Teng X., Zhang J. Molecular features of lung adenocarcinoma in young patients. BMC Cancer. 2019;19(1):777. doi: 10.1186/s12885-019-5978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L., Hu X., Wu H. Unique profiles of targetable genomic alterations and prognosis in young Chinese patients with lung adenocarcinoma. Pathol Res Pract. 2019;215(6):152407. doi: 10.1016/j.prp.2019.03.035. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka K., Hida T., Oya Y. Unique prevalence of oncogenic genetic alterations in young patients with lung adenocarcinoma. Cancer. 2017;123(10):1731–1740. doi: 10.1002/cncr.30539. [DOI] [PubMed] [Google Scholar]
- 19.Sacher A.G., Dahlberg S.E., Heng J., Mach S., Jänne P.A., Oxnard G.R. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2016;2(3):313–320. doi: 10.1001/jamaoncol.2015.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang B., Li J., Li F. Comprehensive analysis of age-related somatic mutation profiles in Chinese young lung adenocarcinoma patients. Cancer Med. 2019;8(4):1350–1358. doi: 10.1002/cam4.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim L., Kim K.H., Yoon Y.H. Clinicopathologic and molecular characteristics of lung adenocarcinoma arising in young patients. J Korean Med Sci. 2012;27(9):1027–1036. doi: 10.3346/jkms.2012.27.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Chen J., Ding W., Yan B., Gao Q., Zhou J. Clinical features and gene mutations of lung cancer patients 30 years of age or younger. PLoS One. 2015;10(9):e0136659. doi: 10.1371/journal.pone.0136659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catania C., Botteri E., Barberis M. Molecular features and clinical outcome of lung malignancies in very young people. Future Oncol. 2015;11(8):1211–1221. doi: 10.2217/fon.15.10. [DOI] [PubMed] [Google Scholar]
- 24.Ye T., Pan Y., Wang R. Analysis of the molecular and clinicopathologic features of surgically resected lung adenocarcinoma in patients under 40 years old. J Thorac Dis. 2014;6(10):1396–1402. doi: 10.3978/j.issn.2072-1439.2014.08.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VandenBussche C.J., Illei P.B., Lin M.T., Ettinger D.S., Maleki Z. Molecular alterations in non-small cell lung carcinomas of the young. Hum Pathol. 2014;45(12):2379–2387. doi: 10.1016/j.humpath.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Luo W., Tian P., Wang Y. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int J Cancer. 2018;143(7):1696–1705. doi: 10.1002/ijc.31542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galvez-Nino M., Ruiz R., Pinto J.A. Lung cancer in the young. Lung. 2020;198:195–200. doi: 10.1007/s00408-019-00294-5. [DOI] [PubMed] [Google Scholar]
- 28.Yoneyama R., Saji H., Kato Y. Clinicopathological characteristics and treatment strategies for young lung cancer patients. Ann Transl Med. 2019;7(5):100. doi: 10.21037/atm.2019.01.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia W., Wang A., Jin M. Young age increases risk for lymph node positivity but decreases risk for non-small cell lung cancer death. Cancer Manag Res. 2018;10:41–48. doi: 10.2147/CMAR.S152017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., Yang F., Li X. Characteristics, survival, and risk factors of Chinese young lung cancer patients: the experience from two institutions. Oncotarget. 2017;8(51):89236–89244. doi: 10.18632/oncotarget.19183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz I., Olsson H. Lung cancer in young women in southern Sweden: a descriptive study. Clin Respir J. 2018;12(4):1565–1571. doi: 10.1111/crj.12712. [DOI] [PubMed] [Google Scholar]
- 32.Arnold B.N., Thomas D.C., Rosen J.E. Lung cancer in the very young: treatment and survival in the National Cancer Data Base. J Thorac Oncol. 2016;11(7):1121–1131. doi: 10.1016/j.jtho.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 33.Gomes R., Dabó H., Queiroga H., Hespanhol V. Non-small cell lung cancer in young patients – a retrospective analysis of 10 years in a tertiary university hospital. Rev Port Pneumol. 2016;22(2):125–126. doi: 10.1016/j.rppnen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu M., Cai X., Yu W., Lv C., Fu X. Clinical significance of age at diagnosis among young non-small cell lung cancer patients under 40 years old: a population-based study. Oncotarget. 2015;6(42):44963–44970. doi: 10.18632/oncotarget.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas A., Chen Y., Yu T., Jakopovic M., Giaccone G. Trends and characteristics of young non-small cell lung cancer patients in the United States. Front Oncol. 2015;5:113. doi: 10.3389/fonc.2015.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rich A.L., Khakwani A., Free C.M. Non-small cell lung cancer in young adults: presentation and survival in the English National Lung Cancer Audit. QJM. 2015;108(11):891–897. doi: 10.1093/qjmed/hcv052. [DOI] [PubMed] [Google Scholar]
- 37.Jiang W., Kang Y., Shi G.Y. Comparisons of multiple characteristics between young and old lung cancer patients. Chin Med J (Engl) 2012;125(1):72–80. [PubMed] [Google Scholar]
- 38.Subramanian J., Morgensztern D., Goodgame B. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: a surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol. 2010;5(1):23–28. doi: 10.1097/JTO.0b013e3181c41e8d. [DOI] [PubMed] [Google Scholar]
- 39.Peng Y., Sun Y. Development and validation of nomograms for predicting overall and cancer-specific survival in young patients with non-small cell lung cancer. J Thorac Dis. 2020;12(4):1404–1416. doi: 10.21037/jtd.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He C.H., Shih J.F., Lai S.L., Chen Y.M. Non-small cell lung cancer in the very young: higher EGFR/ALK mutation proportion than the elder. J Chin Med Assoc. 2020;83(5):461–465. doi: 10.1097/JCMA.0000000000000311. [DOI] [PubMed] [Google Scholar]
- 41.Hou H., Zhang C., Qi X. Distinctive targetable genotypes of younger patients with lung adenocarcinoma: a cBioPortal for cancer genomics data base analysis. Cancer Biol Ther. 2020;21(1):26–33. doi: 10.1080/15384047.2019.1665392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bigay-Gamé L., Bota S., Greillier L. Characteristics of lung cancer in patients younger than 40 years: a prospective multicenter analysis in France. Oncology. 2018;95(6):337–343. doi: 10.1159/000489784. [DOI] [PubMed] [Google Scholar]
- 43.Zhong W., Zhao J., Huang K., Zhang J., Chen Z. Comparison of clinicopathological and molecular features between young and old patients with lung cancer. Int J Clin Exp Pathol. 2018;11(2):1031–1035. [PMC free article] [PubMed] [Google Scholar]
- 44.Scarpino S., Rampioni Vinciguerra G.L., Di Napoli A. High prevalence of ALK+/ROS1+ cases in pulmonary adenocarcinoma of adolescents and young adults. Lung Cancer. 2016;97:95–98. doi: 10.1016/j.lungcan.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 45.Lara M.S., Brunson A., Wun T. Predictors of survival for younger patients less than 50 years of age with non-small cell lung cancer (NSCLC): a California Cancer Registry analysis. Lung Cancer. 2014;85(2):264–269. doi: 10.1016/j.lungcan.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Duan L., You Q., Chen X. Outcome and prognosis for patients younger than thirty with primary lung cancer. Minerva Chir. 2013;68(2):175–182. [PubMed] [Google Scholar]
- 47.Hsu C.L., Chen K.Y., Shih J.Y. Advanced non-small cell lung cancer in patients aged 45 years or younger: outcomes and prognostic factors. BMC Cancer. 2012;12:241. doi: 10.1186/1471-2407-12-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang J., Chen S.F., Zhen Y. Multicenter analysis of lung cancer patients younger than 45 years in Shanghai. Cancer. 2010;116(15):3656–3662. doi: 10.1002/cncr.25100. [DOI] [PubMed] [Google Scholar]
- 49.Smith A.W., Seibel N.L., Lewis D.R. Next steps for adolescent and young adult oncology workshop: an update on progress and recommendations for the future. Cancer. 2016;122(7):988–999. doi: 10.1002/cncr.29870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jemal A., Miller K.D., Ma J. Higher lung cancer incidence in young women than young men in the United States. N Engl J Med. 2018;378(21):1999–2009. doi: 10.1056/NEJMoa1715907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skarin A.T., Herbst R.S., Leong T.L., Bailey A., Sugarbaker D. Lung cancer in patients under age 40. Lung Cancer. 2001;32(3):255–264. doi: 10.1016/s0169-5002(00)00233-6. [DOI] [PubMed] [Google Scholar]
- 52.Shi Y., Au J.S., Thongprasert S. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–162. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qin K., Hou H., Liang Y., Zhang X. Prognostic value of TP53 concurrent mutations for EGFR- TKIs and ALK-TKIs based targeted therapy in advanced non-small cell lung cancer: a meta-analysis. BMC Cancer. 2020;20(1):328. doi: 10.1186/s12885-020-06805-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bruno R., Fontanini G. Next generation sequencing for gene fusion analysis in lung cancer: a literature review. Diagnostics (Basel) 2020;10(8):521. doi: 10.3390/diagnostics10080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Solomon B.J., Mok T., Kim D.W. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371(23):2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 56.Camidge D.R., Kim H.R., Ahn M.J. Brigatinib versus crizotinib in ALK-positive non-small-cell lung cancer. N Engl J Med. 2018;379(21):2027–2039. doi: 10.1056/NEJMoa1810171. [DOI] [PubMed] [Google Scholar]
- 57.Peters S., Camidge D.R., Shaw A.T. Alectinib versus crizotinib in untreated ALK-positive non-small-cell lung cancer. N Engl J Med. 2017;377(9):829–838. doi: 10.1056/NEJMoa1704795. [DOI] [PubMed] [Google Scholar]
- 58.National Institutes of Health/National Cancer Institute SEER cancer stat facts: Lung and bronchus cancer. http://seer.cancer.gov/statfacts/html/lungb.html Available at:
- 59.Shaw A.T., Riely G.J., Bang Y.J. Crizotinib in ROS1-rearranged advanced non-small-cell lung cancer (NSCLC): updated results, including overall survival, from PROFILE 1001. Ann Oncol. 2019;30(7):1121–1126. doi: 10.1093/annonc/mdz131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shaw A.T., Ou S.H., Bang Y.J. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371(21):1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drilon A., Siena S., Dziadziuszko R. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol. 2020;21(2):261–270. doi: 10.1016/S1470-2045(19)30690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Drilon A., Oxnard G.R., Tan D.S.W. Efficacy of selpercatinib in RET fusion-positive non-small-cell lung cancer. N Engl J Med. 2020;383(9):813–824. doi: 10.1056/NEJMoa2005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Drilon A., Laetsch T.W., Kummar S. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378(8):731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paik P.K., Felip E., Veillon R. Tepotinib in non-small-cell lung cancer with MET Exon 14 skipping mutations. N Engl J Med. 2020;383(10):931–943. doi: 10.1056/NEJMoa2004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wolf J., Seto T., Han J.Y. Capmatinib in MET Exon 14-mutated or MET-amplified non-small-cell lung cancer. N Engl J Med. 2020;383(10):944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 66.Colomer R., Mondejar R., Romero-Laorden N., Alfranca A., Sanchez-Madrid F., Quintela-Fandino M. When should we order a next generation sequencing test in a patient with cancer? EClinicalMedicine. 2020;25:100487. doi: 10.1016/j.eclinm.2020.100487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Planchard D., Popat S., Kerr K. Metastatic non-small cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29(suppl 4):iv192–iv237. doi: 10.1093/annonc/mdy275. [DOI] [PubMed] [Google Scholar]
- 68.Maconachie R., Mercer T., Navani N., McVeigh G., Guideline Committee Lung cancer: diagnosis and management: summary of updated NICE guidance. BMJ. 2019;364:l1049. doi: 10.1136/bmj.l1049. [DOI] [PubMed] [Google Scholar]
- 69.National Comprehensive Cancer Network Non-Small Cell Lung Cancer (Version 1.2021-November 25, 2020) https://www.nccn.org/professionals/physician_gls/pdf/nscl_blocks.pdf Available at: Accessed December 10, 2020.
- 70.Benayed R., Offin M., Mullaney K. High yield of RNA sequencing for targetable kinase fusions in lung adenocarcinomas with no mitogenic driver alteration detected by DNA sequencing and low tumor mutation burden. Clin Cancer Res. 2019;25(15):4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies K.D., Aisner D.L. Wake up and smell the fusions: single-modality molecular testing misses drivers. Clin Cancer Res. 2019;25(15):4586–4588. doi: 10.1158/1078-0432.CCR-19-1361. [DOI] [PubMed] [Google Scholar]
- 72.de Alencar V.T.L., Formiga M.N., de Lima V.C.C. Inherited lung cancer: a review. Ecancermedicalscience. 2020;14:1008. doi: 10.3332/ecancer.2020.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mezquita L., Jové M., Nadal E. High prevalence of somatic oncogenic driver alterations in patients with NSCLC and Li–Fraumeni syndrome. J Thorac Oncol. 2020;15(7):1232–1239. doi: 10.1016/j.jtho.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 74.Hu X., Yang D., Li Y. Prevalence and clinical significance of pathogenic germline BRCA1/2 mutations in Chinese non-small cell lung cancer patients. Cancer Biol Med. 2019;16(3):556–564. doi: 10.20892/j.issn.2095-3941.2018.0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanagiri T., Sugio K., Mizukami M. Postoperative prognosis in patients with non-small cell lung cancer according to the method of initial detection. J Thorac Oncol. 2007;2(10):907–911. doi: 10.1097/JTO.0b013e318156079c. [DOI] [PubMed] [Google Scholar]
- 76.Gadgeel S.M., Ramalingam S., Cummings G. Lung cancer in patients <50 years of age: the experience of an academic multidisciplinary program. Chest. 1999;115(5):1232–1236. doi: 10.1378/chest.115.5.1232. [DOI] [PubMed] [Google Scholar]
- 77.Maruyama R., Yoshino I., Yohena T. Lung cancer in patients younger than 40 years of age. J Surg Oncol. 2001;77(3):208–212. doi: 10.1002/jso.1096. [DOI] [PubMed] [Google Scholar]
- 78.Mauri D., Pentheroudakis G., Bafaloukos D. Non-small cell lung cancer in the young: a retrospective analysis of diagnosis, management and outcome data. Anticancer Res. 2006;26(4B):3175–3181. [PubMed] [Google Scholar]
- 79.Nugent W.C., Edney M.T., Hammerness P.G., Dain B.J., Maurer L.H., Rigas J.R. Non-small cell lung cancer at the extremes of age: impact on diagnosis and treatment. Ann Thorac Surg. 1997;63(1):193–197. doi: 10.1016/s0003-4975(96)00745-x. [DOI] [PubMed] [Google Scholar]
- 80.Kuo C.W., Chen Y.M., Chao J.Y., Tsai C.M., Perng R.P. Non-small cell lung cancer in very young and very old patients. Chest. 2000;117(2):354–357. doi: 10.1378/chest.117.2.354. [DOI] [PubMed] [Google Scholar]
- 81.Calles A., Riess J.W., Brahmer J.R. Checkpoint blockade in lung cancer with driver mutation: choose the road wisely. Am Soc Clin Oncol Educ Book. 2020;40:372–384. doi: 10.1200/EDBK_280795. [DOI] [PubMed] [Google Scholar]
- 82.Mazieres J., Drilon A., Lusque A. Immune checkpoint inhibitors for patients with advanced lung cancer and oncogenic driver alterations: results from the IMMUNOTARGET registry. Ann Oncol. 2019;30:1321–1328. doi: 10.1093/annonc/mdz167. [DOI] [PMC free article] [PubMed] [Google Scholar]