Highlights
-
•
CircANKIB1 was upregulated in OS tissues and cells.
-
•
CircANKIB1 downregulation suppressed the progression of OS by upregulating miR-217.
-
•
MiR-217 exerted its anti-cancer roles in OS cells via targeting PAX3.
-
•
CircANKIB1 modulated PAX3 expression through sponging miR-217.
Keywords: Osteosarcoma, circANKIB1, miR-217, PAX3
Abstract
Background
Circular RNAs (circRNAs) have been discovered to exert essential roles in human cancers, including osteosarcoma (OS). The aim of this study was to investigate the exact roles and regulatory mechanism of circRNA ankyrin repeat and IBR domain containing 1 (circANKIB1) in OS.
Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to measure the expression levels of circANKIB1, microRNA-217 (miR-217) and paired box 3 (PAX3). Cell proliferation was assessed by colony formation assay. Cell cycle distribution and apoptosis rate were determined by flow cytometry analysis. Wound healing assay and transwell assay were employed to evaluate cell migration and invasion abilities. Western blot assay was used to analyze the protein levels of PAX3, E-cadherin and Vimentin. Targeting relationship between miR-217 and circANKIB1 or PAX3 was predicted by Circular RNA Interactome or TargetScan and demonstrated by dual-luciferase reporter assay. The mice xenograft model was established to confirm the role of circANKIB1 in vivo.
Results
CircANKIB1 and PAX3 were high-expressed, whereas miR-217 was low-expressed in OS tissues and cells. Knockdown of circANKIB1 inhibited the progression of OS by reducing cell proliferation, migration, invasion, and tumor growth (in vivo), and inducing apoptosis. MiR-217 was a direct target of circANKIB1, and its inhibition reversed the inhibitory effect of circANKIB1 knockdown on the progression of OS cells. Moreover, PAX3 was a direct target of miR-217, and miR-217 exerted the anti-tumor role in OS cells by targeting PAX3. Furthermore, circANKIB1 positively regulated PAX3 expression by sponging miR-217.
Conclusion
Knockdown of circANKIB1 suppressed OS progression by upregulating miR-217 and downregulating PAX3, which might provide a novel insight into the pathogenesis of OS.
1. Introduction
Osteosarcoma (OS) is one of the most common primary bone tumors in children and adolescents [1]. OS is highly invasive and easy to metastasis, leading to poor prognosis, high mortality and disability [2]. Despite some significant improvements have been made in the treatment of OS, the patients with advanced stage and metastasis show a lower survival rate [3], [4]. Hence, it is important to understand the molecular mechanisms of OS initiation, development and metastasis.
With the rapid development of sequencing technology in recent years, circular RNAs (circRNAs) are identified as a new type of non-coding RNAs (ncRNAs), and characterized by covalently closed-loop structures without 5′-cap and 3′-end poly A tail [5]. CircRNAs have higher tolerance to exonuclease digestion and also have higher stability than linear RNAs [6], [7]. Currently, many reports have revealed that circRNAs can serve as tumor suppressors or oncogenes in a variety of cancers, including OS [8], [9]. For example, circRNA hsa_circ_0001564, hsa_circ_0002052, circNASP, etc, have been identified to play pivotal roles in regulating OS progression [10], [11], [12]. CircRNA ankyrin repeat and IBR domain containing 1 (ANKIB1) (circANKIB1; circRNA ID: hsa_circ_0009112, position: chr7:91972337-91981956) is a product of ANKIB1 mRNA splicing, which is a downregulated circRNA in OS tissues [13]. Nevertheless, the exact roles and regulatory mechanism of circANKIB1 in OS have not been studied deeply in vivo or in vitro.
In comparison to circRNAs, microRNAs (miRNAs), a group of short ncRNAs, usually bind to 3′untranslated regions (3′UTR) of target mRNAs to suppress the expression of target genes [14]. Mechanically, circRNAs can function as competing endogenous RNAs (ceRNAs; also known as miRNA sponges) by competitively binding to miRNA response elements, thus inducing release of genes that are targeted by specific miRNAs, and forming a functional circRNA-miRNA-mRNA network [15]. A previous report suggested that miR-217 was lowly expressed in OS tissues and cells, and played a tumor-suppressive role in OS [16]. Besides, paired box 3 (PAX3) has been reported to promote the progression of OS [17]. Through biological analysis, we found that circANKIB1 could specifically bind to miR-217, and miR-217 also had binding sites with PAX3. So, we focused on whether circANKIB1 served as a sponge for miR-217 to modulate PAX3 level in OS progression.
In this research, we examined the expression of circANKIB1, miR-217 and PAX3 in OS tissue specimens and cell lines. Moreover, we investigated their functions by performing a series of experiments and also explored the potential regulatory mechanism among them in the progression of OS. We aimed to provide a novel insight into the diagnosis and treatment of OS.
2. Materials and methods
2.1. Specimen collection
OS tissue specimens (n = 35) and adjacent normal bone tissue specimens (n = 35) were collected during the surgery at General Hospital of Ningxia Medical University, and preserved at −80 °C until RNA or protein extraction. Informed consent was obtained from patients. These subjects did not receive any treatment before surgery. This research had been approved by the Research Ethics Committee of General Hospital of Ningxia Medical University.
2.2. Cell culture and transfection
OS cell lines (U2OS and HOS) and fetal osteoblastic cell line (hFOB) were obtained from BeNa Culture Collection (Beijing, China). These cells were grown in Dulbecco’s modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS, Invitrogen), and maintained in a 5% CO2 incubator at 37 °C.
The small interfering RNA (siRNA) against circANKIB1 (si-circANKIB1), circANKIB1-overexpressing plasmid (oe-circANKIB1), miR-217 mimic, miR-217 inhibitor, pcDNA-PAX3 overexpression plasmid (pcDNA-PAX3) and their negative controls (si-NC, oe-NC, miR-NC mimic, miR-NC inhibitor, pcDNA-NC) were obtained from Genechem (Shanghai, China). Transient transfection was carried out in our research by using Lipofectamine 3000 Reagent (Invitrogen).
2.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA in tissue specimens and cell lines was extracted with TRIzol reagent (Invitrogen). Subsequently, extracted RNA was reverse transcribed into complementary DNA (cDNA) using the Primescript RT Reagent (TaKaRa, Kusatsu, Japan) for analysis of circANKIB1, ANKIB1 and PAX3, or using microRNA First-Strand cDNA Synthesis Kit (Sangon Biotech, Shanghai, China) for detection of miR-217. After that, qRT-PCR reactions were performed using the SYBR Green PCR Kit (TaKaRa) on CFX96 Touch Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). In this study, the following primers were used for qRT-PCR: circANKIB1 (Forward, 5′-GGACACCTCTTCTGCTGACT-3′; Reverse, 5′-GAACACCTGATCGTTGGCAG-3′), ANKIB1 (Forward, 5′-GGAAAAGGACACCTCTTCTGCTG-3′; Reverse, 5′-CTCGTAGGCTTCACTAACTCCC-3′), miR-217 (Forward, 5′-TTGAGGTTGCTTCAGTGA-3′; Reverse, 5′-GGAGTAGATGATGGTTAGC-3′), PAX3 (Forward, 5′-TGATCGGAACACTGTGCCCTC-3′; Reverse, 5′-GCTTTCAACCATCTCATTCCGG-3′), β-Actin (Forward, 5′-GACCTCTATGCCAACACAGT-3′; Reverse, 5′-AGTACTTGCGCTCAGGAGGAG-3′), U6 (Forward, 5′-GTGCGTGTCGTGGAGTCG-3′; Reverse, 5′-AACGCTTCACGAATTTGCGT-3′). The expression levels of circANKIB1, ANKIB1, PAX3, and miR-217 were evaluated by 2-ΔΔCt method, followed by normalizing to β-Actin or U6. All experiments are performed for three times.
2.4. Actinomycin D and RNase R treatment
Actinomycin D (2 mg/mL) or dimethyl sulfoxide solution (DMSO; as the negative control, Sigma-Aldrich, St. Louis, MO, USA) was placed into cell culture medium to block transcription. To validate the circular characteristic of circANKIB1, total RNA (2 μg) was incubated by RNase R (3 U/μg, Epicentre Technologies, Madison, WI, USA) for 0.5 h at 37 °C. Following treatment with Actinomycin D and RNase R, the abundance of circANKIB1 and ANKIB1 was tested using qRT-PCR analysis. All experiments are performed for three times.
2.5. Colony formation assay
Following transfection for 48 h, U2OS and HOS cells were inoculated in a 6-well plate and then cultured for 14 days. After that, these colonies were fixed with paraformaldehyde (4%, Beyotime, Jiangsu, China) for 0.5 h, and stained with crystal violet (0.1%, Sigma-Aldrich) for 2 h. The colonies (one colony more than 50 cells) were photographed and analyzed. All experiments are performed for three times.
2.6. Flow cytometry
For cell cycle determination, U2OS and HOS cells were collected by centrifugation and fixed with ice-cold 75% ethanol (Beyotime) for 12 h at −20 °C. Next, these cells were collected and stained with propidium iodide (PI; 25 μg/mL, Sangon Biotech) and 50 µg/mL RNase A (Sangon Biotech) in phosphate-buffered saline (PBS; Beyotime) for 15 min. Subsequently, flow cytometer (Partec AG, Arlesheim, Switzerland) was applied for detecting cell cycle distribution. Cell apoptosis assay was conducted using the Annexin V-fluorescein isothiocyanate (FITC)/PI apoptosis detection kit (KeyGen Biotech, Nanjing, China). U2OS and HOS cells were harvested following transfection for 48 h, and then stained with Annexin V-FITC and PI in the darkness for 15 min, followed by the detection of apoptotic cells using flow cytometry. All experiments are performed for three times.
2.7. Wound healing assay
Wound healing assay was utilized for detecting migration ability of U2OS and HOS cells. In short, U2OS and HOS cells were inoculated in 12-well plates. At ~ 80% confluence, we used a pipette tip (200 µL) to scrape the center of the well and washed with PBS. Images were captured using a microscope (Leica, Wetzlar, Germany) at indicated time with × 40 magnification. The distance of cell migration was examined using ImageJ software. All experiments are performed for three times.
2.8. Transwell assay
Transwell chamber (8 μm pore size, Costar, Corning, NY, USA) without (for migration assay) or with (for invasion assay) pre-coated Matrigel (BD Biosciences, Franklin, NJ, USA) was employed to detect cell migration or invasion abilities. In short, transfected cells suspended in serum-free medium (DMEM, 200 µL) were seeded into the top chamber. Meanwhile, DMEM with 10% FBS was placed into the bottom chamber as a chemoattractant. After 48 h of incubation, cells remaining on the top membrane were erased by cotton swabs. At the same time, the cells on the bottom surface were fixed with paraformaldehyde (4%, Beyotime) for 20 min and stained by crystal violet solution (0.1%, Beyotime) for 30 min, and photographed under a microscope (Leica) with a magnification of × 100. All experiments are performed for three times.
2.9. Western blot assay
Clinical tissues and cultured cells were lysed using RIPA lysis buffer (Solarbio, Beijing, China) for extracting total protein. Next, the protein samples were denatured by heating at 100 °C for 3–5 min, and then quantified using a BCA protein assay kit (Beyotime). After that, protein samples (about 30 μg/lane) were resolved and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE; Beyotime), and then blotted onto nitrocellulose membranes (Invitrogen). After being incubated with 5% non-fat milk (Sangon Biotech), these membranes were subsequently incubated for 12 h at 4 °C with primary antibody against E-cadherin (ab15148, 1:500, Abcam, Cambridge, MA, USA), Vimentin (ab137321, 1:1000, Abcam), PAX3 (ab180754, 1:1000, Abcam) or β-Actin (1:5000, ab227387, Abcam). Subsequently, these membranes were continuously probed with goat anti-rabbit IgG H&L (HRP) (ab205718, 1:4000, Abcam). At last, the blots were visualized by enhanced chemiluminescence reagent (Tanon, Shanghai, China). All experiments are performed for three times.
2.10. Bioinformatics prediction and dual-luciferase reporter assay
The potential binding sites for miR-217 in circANKIB1 or PAX3 were predicted by Circular RNA Interatome or TargetScan. Wild-type (WT) or mutant (mut; the binding site of miR-217 was mutated) circANKIB1 or PAX3 3′UTR was cloned into pmirGlO luciferase reporter vector (Promega, Madison, WI, USA), named as WT-circANKIB1, MUT-circANKIB1, WT-PAX3, and MUT-PAX3. Each of the above-mentioned plasmids was respectively transfected into U2OS and HOS cells along with miR-NC mimic or miR-217 mimic. After 48 h of transfection, dual-luciferase reporter assay system (Promega) was employed for examining the luciferase activity in the cells lysates. All experiments are performed for three times.
2.11. Tumor formation assay in vivo
The animal experiments obtained the approval from the Animal Care and Use Committee of General Hospital of Ningxia Medical University. BALB/c nude mice (female 5–6 weeks old, weighing 20–25 g, Huafukang, Beijing, China) were used for the in vivo tumor formation assay. Lentivirus-mediated shRNA targeting circANKIB1 (sh-circANKIB1) and its negative control (sh-NC) were provided by RiboBio (Guangzhou, China). HOS cells transiently transfected with sh-NC (as control) or sh-circANKIB1 were subcutaneously injected into nude mice (5 mice/group). Every week, we detected tumor volume by calipers and calculated by the formula: volume = length × width2/2. After 4 weeks, the mice were sacrificed, tumor specimens were weighed and collected to detect the abundance of circANKIB1, miR-217 and PAX3.
2.12. Statistical analysis
GraphPad Prism software (GraphPad Software, La Jolla, CA, USA) was used for the general statistical analysis. All data from at least 3 independent experiments were shown as the mean ± standard deviation (SD). The Student’s t-test was utilized to analyze 2-group differences, multiple comparisons were analyzed by one-way analysis of variance (ANOVA). The correlations among circANKIB1, miR-217 and PAX3 were analyzed with Pearson’s correlation coefficient. P < 0.05 was considered statistically significant.
3. Results
3.1. The level of circANKIB1 was increased in OS tissues and cells
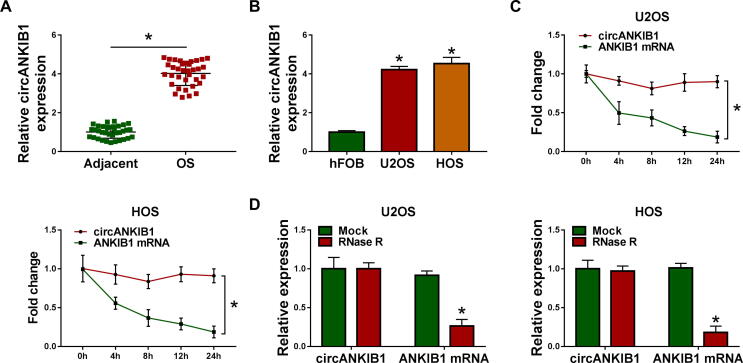
Firstly, the expression of circANKIB1 was detected in OS tissues and cells by qRT-PCR. As presented in Fig. 1A, circANKIB1 level was increased in OS tissues compared with adjacent normal tissues. Likewise, circANKIB1 expression was upregulated in U2OS and HOS cells compared to hFOB cells (Fig. 1B). Next, we explored the characteristics of circANKIB1 in OS cells. The results from Actinomycin D assay showed that the half-life of circANKIB1 transcript exceeded 24 h, while that of the ANKIB1 mRNA displayed about 4 h (Fig. 1C), suggesting that circANKIB1 transcript was more stable than the linear ANKIB1 mRNA transcript in U2OS and HOS cells. Moreover, circANKIB1 was resistant to RNase R digestion (Fig. 1D), implying that circANKIB1 had a loop structure. Our results revealed that circANKIB1 expression was enhanced in OS and had a stable closed-loop structure.
Fig. 1.
CircANKIB1 was overexpressed in OS tissues and cells. (A) The expression of circANKIB1 was detected by qRT-PCR in OS tissues (n = 35) and adjacent normal tissues (n = 35). (B) The expression of circANKIB1 was measured by qRT-PCR in OS cells (U2OS and HOS) and hFOB cells. (C and D) The expression levels of circANKIB1 and ANKIB1 mRNA were determined after treatment with Actinomycin D and RNase R by qRT-PCR in U2OS and HOS cells. *P < 0.05.
3.2. Knockdown of circANKIB1 inhibited cell growth, migration and invasion, and induced apoptosis in OS cells
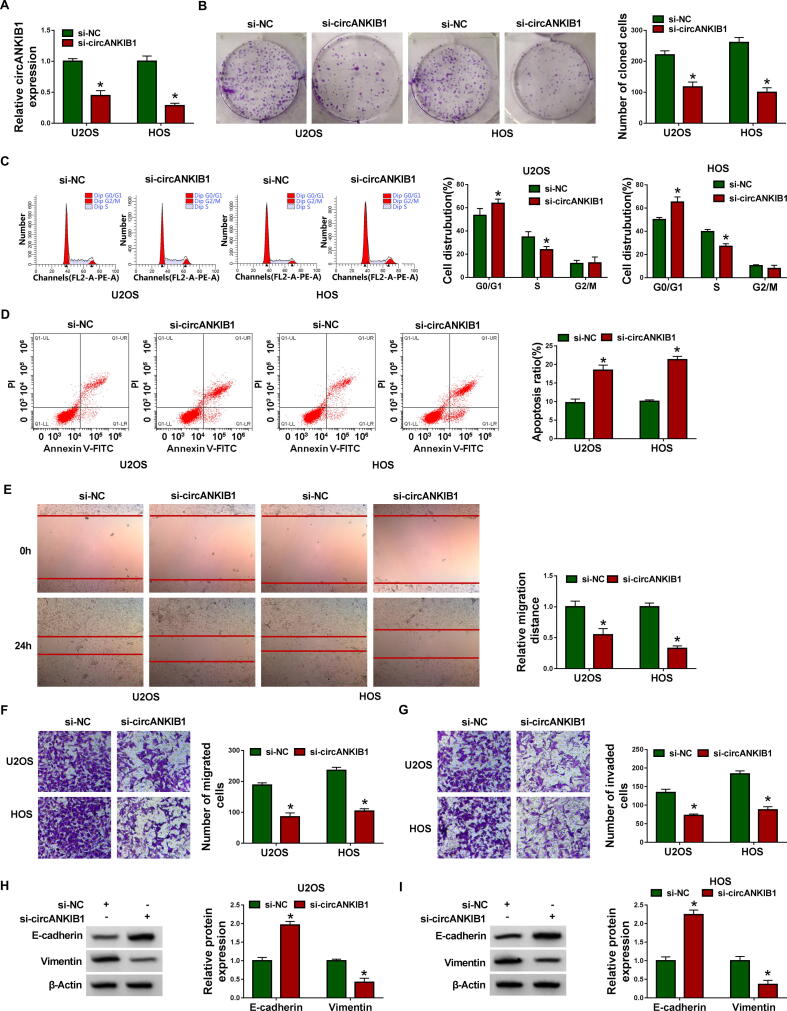
To investigate the functional role of circANKIB1 in OS, U2OS and HOS cells were transfected with si-NC or si-circANKIB1. We performed qRT-PCR assay to confirm the transfection efficacy of si-circANKIB1. Results showed that circANKIB1 expression was significantly reduced after transfection with si-circANKIB1 (Fig. 2A). Colony formation assay indicated that knockdown of circANKIB1 decreased the number of cloned cells (Fig. 2B). To further assess whether circANKIB1 could influence the cell cycle distribution and apoptosis, the flow cytometry analysis was applied. We found that more cells were distributed in the G0/G1 phase after knockdown of circANKIB1, and fewer cells were distributed in S phase (Fig. 2C), indicating that the cell cycle was arrested at the G0/G1 phase to inhibit cell growth. Moreover, circANKIB1 knockdown induced U2OS and HOS cell apoptosis (Fig. 2D). Wound healing and transwell assays indicated that circANKIB1 downregulation inhibited U2OS and HOS cell migration and invasion (Fig. 2E-2G). Epithelial-mesenchymal transition (EMT) is a key step in cancer metastasis [18]. Thus, EMT-related proteins were analyzed in OS cells. Western blot assay showed that the protein level of E-cadherin (an epithelial marker) was increased and the protein expression of Vimentin (a mesenchymal marker) was decreased after knocking down the circANKIB1 (Fig. 2H and 2I). These data suggested that circANKIB1 knockdown could inhibit the progression of OS in vitro.
Fig. 2.
CircANKIB1 knockdown inhibited cell proliferation, migration and invasion, and accelerated apoptosis in OS cells. U2OS and HOS cells were transfected with si-NC or si-circANKIB1. (A) Knockdown efficiency of circANKIB1 was determined by qRT-PCR. (B) Colony formation assay was performed to examine the number of colonies. (C and D) Flow cytometry analysis was used to detect cell cycle distribution and apoptosis rate. (E) Wound healing assay was utilized to assess cell migration ability (×40). (F and G) Transwell assay was employed to evaluate cell migration and invasion abilities (×100). (H and I) Western blot assay was carried out to measure the expression levels of E-cadherin and Vimentin. *P < 0.05.
3.3. CircANKIB1 served as a molecular sponge for miR-217 in OS cells
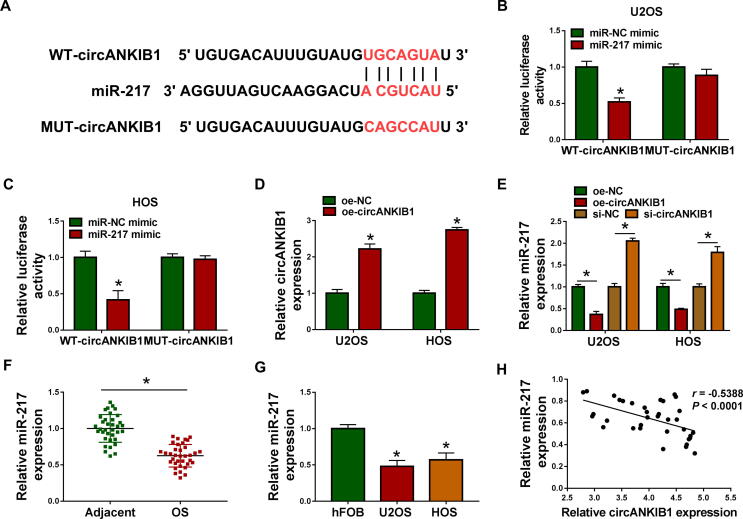
Presently, circRNAs are widely considered as miRNA sponges and may be involved in cancer occurrence and development. Therefore, Circular RNA Interactome was used to predict the potential target miRNAs of circANKIB1. We found that there were binding sites between circANKIB1 and miR-217, suggesting that miR-217 might be a target of circANKIB1 (Fig. 3A). To confirm whether circANKIB1 could directly bind to miR-217, dual-luciferase reporter assay was performed. The results displayed that the luciferase activity of WT-circANKIB1 was obviously decreased after transfection with miR-217 mimic in U2OS and HOS cells, whereas the luciferase activity of MUT-circANKIB1 was unaffected after transfection with miR-217 mimic (Fig. 3B and 3C). Next, overexpression efficiency of circANKIB1 was determined by qRT-PCR in U2OS and HOS cells transfected with oe-circANKIB1 or oe-NC. Results showed that the expression of circANKIB1 was obviously increased after transfection with oe-circANKIB1 (Fig. 3D). Moreover, miR-217 expression was reduced by overexpression of circANKIB1 and enhanced by knockdown of circANKIB1 (Fig. 3E). Next, it was found that miR-217 expression was decreased in OS tissues and cells compared with their controls (Fig. 3F and 3G). Furthermore, we observed that circANKIB1 expression was negatively correlated with miR-217 level in OS tissues (P < 0.0001, r = -0.5388) (Fig. 3H). Overall, circANKIB1 could bind with miR-217.
Fig. 3.
CircANKIB1 acted as a sponge of miR-217 in OS cells. (A) The predicted binding sites for circANKIB1 and miR-217 were presented. (B and C) The luciferase activity in U2OS and HOS cells co-transfected with WT-circANKIB1 or MUT-circANKIB1 and miR-217 mimic or miR-NC mimic was measured by dual-luciferase reporter assay. (D) The expression of circANKIB1 was determined by qRT-PCR in U2OS and HOS cells transfected with oe-NC or oe-circANKIB1. (E) The expression of miR-217 was examined by qRT-PCR in U2OS and HOS cells transfected with oe-NC, oe-circANKIB1, si-NC, or si-circANKIB1. (F and G) The abundance of miR-217 was examined by qRT-PCR in OS tissues, adjacent normal tissues, OS cells (U2OS and HOS), and hFOB cells. (H) The correlation between miR-217 and circANKIB1 expression was analyzed in OS tissues. *P < 0.05.
3.4. CircANKIB1 knockdown inhibited the progression of OS cells by upregulating miR-217
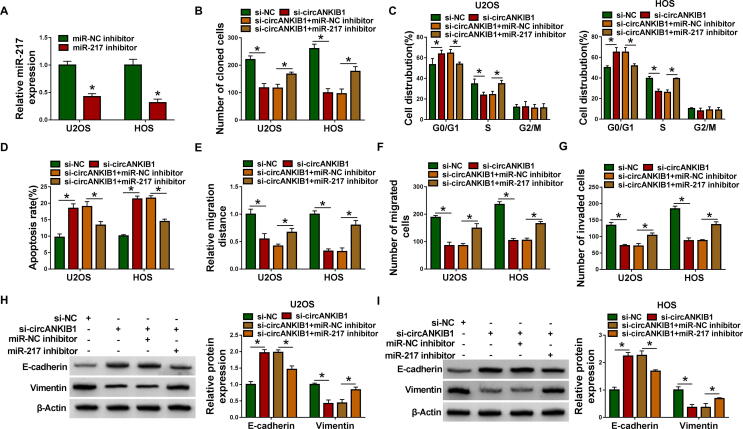
Inhibition efficiency of miR-217 was examined by qRT-PCR in OS cells transfected with miR-NC inhibitor or miR-217 inhibitor. The results indicated that miR-217 was successfully knocked down in U2OS and HOS cells (Fig. 4A). To explore whether circANKIB1 exerted its biological functions by sponging miR-217 in OS cells, U2OS and HOS cells were transfected with si-NC, si-circANKIB1, si-circANKIB1 + miR-NC inhibitor, or si-circANKIB1 + miR-217 inhibitor. We found that the inhibitory effect of si-circANKIB1 on colony formation was reversed by knockdown of miR-217 (Fig. 4B). Furthermore, downregulation of miR-217 abolished si-circANKIB1-induced promotion of G0/G1 phase cells and reduction of S phase cells (Fig. 4C). Moreover, circANKIB1 silence-mediated pro-apoptosis, anti-migration and anti-invasion effects were abated by silencing miR-217 (Fig. 4D-4G). Besides, the effects of circANKIB1 knockdown on increase of E-cadherin expression and decrease of Vimentin expression were reversed by downregulating miR-217 (Fig. 4H and 4I). These data illustrated that circANKIB1 influenced OS cell proliferation, cell cycle, apoptosis, and metastasis by sponging miR-217.
Fig. 4.
Silence of circANKIB1 suppressed the progression of OS cells by sponging miR-217. (A) The knockdown efficiency of miR-217 was examined by qRT-PCR in U2OS and HOS cells transfected with miR-NC inhibitor or miR-217 inhibitor. (B-I) U2OS and HOS cells were transfected with si-NC, si-circANKIB1, si-circANKIB1 + miR-NC inhibitor, or si-circANKIB1 + miR-217 inhibitor. (B) Colony formation assay was used to examine colony formation ability. (C and D) Cell cycle distribution and cell apoptosis rate were determined by flow cytometry analysis. (E-G) Cell migration and invasion were measured by wound healing assay and transwell assay. (H and I) Western blot assay was performed to test the protein levels of E-cadherin and Vimentin. *P < 0.05.
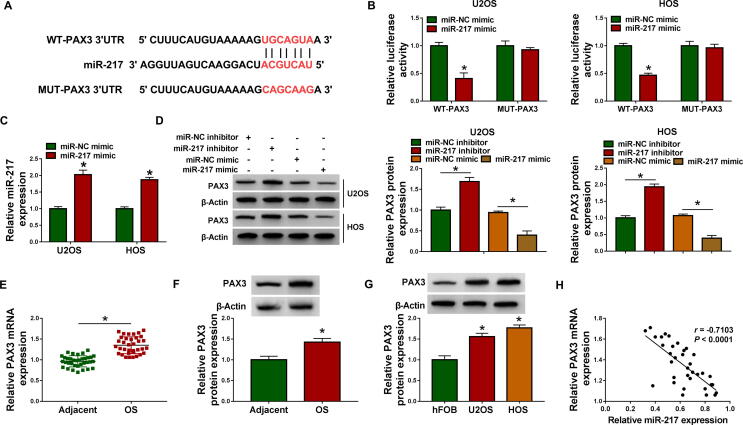
3.5. PAX3 was a direct target of miR-217 in OS cells
To further investigate the downstream mRNAs of circANKIB1/miR-217, TargetScan was used to search for the possible target genes of miR-217. Results showed that PAX3 had binding sequence with miR-217 (Fig. 5A). Next, we performed the dual-luciferase reporter assay to verify whether miR-217 could directly target bind to PAX3 3′UTR through the targeting sites. The results showed that miR-217 overexpression decreased the luciferase activity of WT-PAX3, but did not alter the luciferase activity of MUT-PAX3 in U2OS and HOS cells (Fig. 5B). The results of qRT-PCR displayed that miR-217 expression was overexpressed after transfection with miR-217 mimic (Fig. 5C). Western blot assay suggested that the protein expression of PAX3 was increased by inhibiting miR-217 and decreased by overexpressing miR-217 (Fig. 5D). Moreover, we observed that PAX3 mRNA expression and protein expression were both increased in OS tissues in contrast to adjacent normal tissues (Fig. 5E and 5F). Similarly, the protein expression of PAX3 was also upregulated in OS cells (U2OS and HOS) relative to hFOB cells (Fig. 5G). In addition, we observed a negative correlation between PAX3 mRNA expression and miR-217 expression in OS tissues (P < 0.0001, r = -0.7103) (Fig. 5H). To sum up, miR-217 directly targeted PAX3 and negatively regulated PAX3 expression.
Fig. 5.
PAX3 was a target gene of miR-217 in OS cells. (A) PAX3 had binding sites for miR-217. (B) Relative luciferase activity was measured in U2OS and HOS cells co-transfected with WT-PAX3 or MUT-PAX3 and miR-217 mimic or miR-NC mimic. (C) Overexpression efficiency of miR-217 was determined by qRT-PCR in U2OS and HOS cells transfected with miR-NC mimic or miR-217 mimic. (D) The protein expression of PAX3 was tested by western blot analysis in U2OS and HOS cells transfected with miR-NC inhibitor, miR-217 inhibitor, miR-NC mimic, or miR-217 mimic. (E and F) PAX3 mRNA and protein expression were detected in OS tissues and adjacent normal tissues by qRT-PCR and western blot analyses, respectively. (G) Western blot sassy was conducted to measure the protein expression of PAX3 in OS cells (U2OS and HOS) and hFOB cells. (H) The correlation between PAX3 mRNA expression and miR-217 expression was analyzed in OS tissues. *P < 0.05.
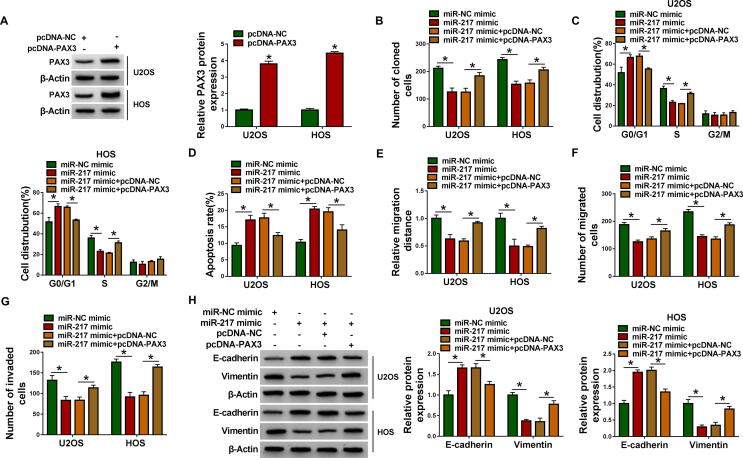
3.6. MiR-217 upregulation suppressed the progression of OS cell by targeting PAX3
To further explore whether miR-217 function was regulated by PAX3, rescue assays were performed. Western blot assay indicated that transfection of pcDNA-PAX3 greatly increased the protein expression of PAX3 in U2OS and HOS cells (Fig. 6A). Colony formation assay and flow cytometry analysis suggested that overexpression of miR-217 reduced the number of cloned cells and arrested cells at G0/G1 phase, which was reversed by upregulating PAX3 (Fig. 6B and 6C). Moreover, enforced expression of miR-217 promoted apoptosis and inhibited cell migration and invasion, while these effects were abated by overexpression of PAX3 (Fig. 6D-6G). Furthermore, transfection of miR-217 mimic increased the protein abundance of E-cadherin and decreased the protein expression of Vimentin, which was abolished by co-transfection of pcDNA-PAX3 in U2OS and HOS cells (Fig. 6H). In summary, miR-217 exerted the anti-cancer role by targeting PAX3 in OS cells.
Fig. 6.
MiR-217 exerted the anti-tumor role in OS cells by targeting PAX3. (A) Overexpression efficiency of PAX3 was determined by western blot assay in U2OS and HOS cells transfected with pcDNA-NC or pcDNA-PAX3. (B-H) U2OS and HOS cells were transfected with miR-NC mimic, miR-217 mimic, miR-217 mimic + pcDNA-NC, or miR-217 mimic + pcDNA-PAX3. (B) Cell proliferation was evaluated by colony formation assay. (C and D) Cell cycle distribution and apoptosis rate were examined by flow cytometry analysis. (E-G) Wound healing assay and transwell assay were used to assess cell migration and invasion abilities. (H) The protein levels of E-cadherin and Vimentin were measured by western blot assay. *P < 0.05.
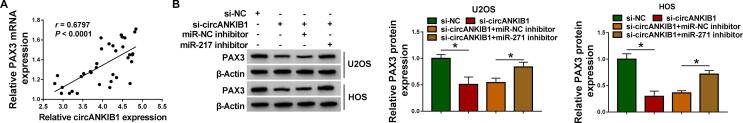
3.7. CircANKIB1 modulated PAX3 expression by sponging miR-217
Next, we further explored the relationships among circANKIB1, miR-217 and PAX3. We found that PAX3 mRNA level was positively correlated with circANKIB1 expression in OS tissues (P < 0.0001, r = 0.6797) (Fig. 7A). Western blot assay indicated that knockdown of circANKIB1 inhibited the protein level of PAX3, which was rescued by inhibiting miR-217 in U2OS and HOS cells (Fig. 7B). All findings indicated that circANKIB1 positively regulated PAX3 expression via targeting miR-217.
Fig. 7.
CircANKIB1 regulated PAX3 expression by acting as a sponge of miR-217 in OS cells. (A) The correlation between PAX3 mRNA expression and circANKIB1 expression was analyzed in OS tissues. (B) Western blot assay was performed to analyze the protein expression of PAX3 in U2OS and HOS cells transfected with si-NC, si-circANKIB1, si-circANKIB1 + miR-NC inhibitor, or si-circANKIB1 + miR-217 inhibitor. *P < 0.05.
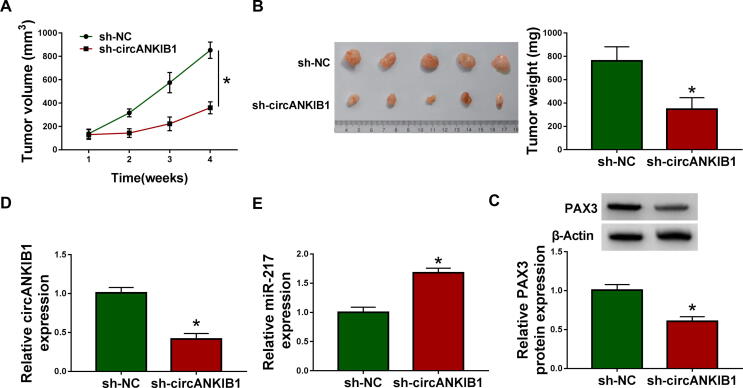
3.8. CircANKIB1 interference inhibited tumor growth by increasing miR-217 and inhibiting PAX3 in vivo
Given that circANKIB1 suppressed cell growth in vitro, we further evaluated the effect of circANKIB1 on tumor growth in vivo. HOS cells transfected with sh-circANKIB1 or sh-NC were subcutaneously injected into female nude mice. We found that knockdown of circANKIB1 reduced tumor volume and weight (Fig. 8A and 8B). Moreover, circANKIB1 silence inhibited the expression of circANKIB1 and increased the expression of miR-217 in tumor tissues (Fig. 8C and 8D). Western blot analysis showed that circANKIB1 downregulation reduced the protein expression of PAX3 in tumor tissues (Fig. 8E). Hence, in vivo experiments proved that circANKIB1 silence inhibited tumor growth by regulating miR-217 and PAX3 expression.
Fig. 8.
CircANKIB1 downregulation suppressed tumor growth via regulating miR-217 and PAX3 in vivo. Sh-NC or sh-circANKIB1-transfected HOS cells were subcutaneously injected into BALB/c nude mice to establish mice xenograft model. (A) Tumor volume was analyzed every week in mice. (B) Tumor weight was calculated after injection for 4 weeks. (C and D) The expression levels of circANKIB1 and miR-217 in tumor tissues were detected by qRT-PCR. (E) The protein expression of PAX3 in tumor tissues was measured by western blot assay. *P < 0.05.
4. Discussion
OS is one of the most common malignant bone tumors. Recently, emerging evidence has indicated that circRNAs have important functions in the progression and development of cancers [19], [20]. But, the roles of circRNAs in OS are largely unclear. In our research, we confirmed that circANKIB1 was overexpressed in OS, and knockdown of circANKIB1 repressed the progression of OS by regulating miR-217/PAX3 axis.
CircRNAs are initially considered as the “junk by-products” of pre-mRNA splicing [21]. However, multiple studies have demonstrated that circRNAs have critical regulatory roles in various pathological environments [22]. Moreover, increasing evidence has demonstrated that circRNAs are abundant in eukaryotic cells and recognized as significant prognostic biomarkers for tumors due to their abundance and stability [23], [24]. CircANKIB1 has been reported to be overexpressed in OS [25]. Moreover, Du et al. revealed that interference of circANKIB1 restrained cell growth and invasion, and facilitated apoptosis by regulating miR-19b on SOCS3/STAT3 pathway in OS cells [13]. In accordance with these findings, we found that circANKIB1 was overexpressed in OS tissue samples and cell lines. Silence of circANKIB1 in OS cells could repress cell growth and metastasis, and induce apoptosis. These results suggested that circANKIB1 served as an oncogene in OS.
It is widely accepted that circRNAs can serve as miRNA sponges to interact with miRNAs and affect the expression of target genes [26]. Bioinformatics analysis showed that circANKIB1 might bind to miR-217, which was demonstrated by performing dual-luciferase reporter assay. Some studies revealed that miR-217 acted as an anti-oncogene in some tumors, such as hepatocellular carcinoma [27], ovarian cancer [28] and lung cancer [29]. MiR-217 also served as a tumor-suppressive miRNA in OS. For instance, He et al. proved that miR-217 was lowly expressed in OS tissue specimens and cell lines, and upregulation of miR-217 could suppress OS cell growth, migration, and invasion via targeting SIRT1 [16]. Moreover, Shen et al. stated that inhibition of miR-217 markedly facilitated cell proliferation and metastasis of OS cells through targeting WASF3 [30]. Consistent with these results, we uncovered that miR-217 level was declined in OS tissue samples and cell lines. Furthermore, rescue experiments proved that downregulation of miR-217 reversed the si-circANKIB1-mediated inhibition of cell growth and metastasis, and promotion of apoptosis in OS cell. Our findings strongly revealed that circANKIB1 exerted its functions in OS cells via sponging miR-217.
Increasing evidence has shown that miRNAs can exert biological functions through inhibiting their target genes in many diseases [31]. Next, we searched for the miR-217 downstream target genes by TargetScan. PAX3 was identified as a downstream target gene of miR-217. PAX3 has been suggested to play pivotal roles in diverse tumors, including OS [32], [33]. For example, Fujii et al. showed that PAX3 downregulation repressed OS cell growth and induced cell cycle arrest at the G0/G1 phase via modulating p21 expression [17]. Moreover, Zhan et al. demonstrated that PAX3 abundance was enhanced in OS tissue specimens, and PAX3 upregulation abated the anti-tumor roles of miR-206 overexpression in OS cells [34]. In line with these findings, we observed that the abundance of PAX3 was increased in OS tissue specimens and cell lines. Furthermore, rescue assays demonstrated that enforced expression of PAX3 could partly abolish the effects of miR-217 overexpression on reduction of cell growth and metastasis, and enhancement of apoptosis in OS cells. Besides, circANKIB1 positively regulated PAX3 expression by sponging miR-217 in OS cells. Constant with in vitro results, knockdown of circANKIB1 also repressed tumor growth in vivo by upregulating miR-217 and downregulating PAX3. Taken together, our research demonstrated that circANKIB1 sponged miR-217 to promote the progression of OS via regulating PAX3 expression.
In conclusion, circANKIB1 and PAX3 were upregulated and miR-217 was downregulated in OS tissues and cells. Moreover, circANKIB1 silence repressed the progression of OS by upregulating miR-217 and downregulating PAX3. The circANKIB1/miR-217/PAX3 axis might provide a new sight for OS treatment.
5. Declarations
5.1. Ethics approval and consent to participate
The present study was approved by the ethical review committee of General Hospital of Ningxia Medical University. Written informed consent was obtained from all enrolled patients.
Consent for publication
Patients agree to participate in this work
Availability of data and materials
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Funding
This work was supported by Chengdu Municipal Health Commission Project (No:2019107).
CRediT authorship contribution statement
Xi Zhu: Validation, Investigation, Writing - original draft, Writing - review & editing. Changhao Liu: Conceptualization, Methodology, Validation, Investigation, Writing - original draft, Writing - review & editing. Jiandang Shi: Conceptualization, Methodology, Writing - original draft, Writing - review & editing, Project administration. Zhanwen Zhou: Formal analysis, Data curation. Suoli Chen: Formal analysis, Data curation, Software. Sayed Abdulla Jami: Formal analysis, Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
References
- 1.Simpson E., Brown H.L. Understanding osteosarcomas. Jaapa. 2018;31:15–19. doi: 10.1097/01.JAA.0000541477.24116.8d. [DOI] [PubMed] [Google Scholar]
- 2.Harrison D.J., Geller D.S., Gill J.D., Lewis V.O., Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert. Rev. Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 3.Messerschmitt P.J., Garcia R.M., Abdul-Karim F.W., Greenfield E.M., Getty P.J. Osteosarcoma. J. Am. Acad. Orthop. Surg. 2009;17:515–527. doi: 10.5435/00124635-200908000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Bielack S.S., Kempf-Bielack B., Delling G., Exner G.U., Flege S., Helmke K., Kotz R., Salzer-Kuntschik M., Werner M., Winkelmann W., Zoubek A., Jürgens H., Winkler K. Prognostic factors in high-grade osteosarcoma of the extremities or trunk: an analysis of 1,702 patients treated on neoadjuvant cooperative osteosarcoma study group protocols. J. Clin. Oncol. 2002;20:776–790. doi: 10.1200/JCO.2002.20.3.776. [DOI] [PubMed] [Google Scholar]
- 5.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H.D., Jiang L.H., Sun D.W., Hou J.C., Ji Z.L. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 9.Patop I.L., Kadener S. circRNAs in Cancer. Curr. Opin. Genet. Dev. 2018;48:121–127. doi: 10.1016/j.gde.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song Y.Z., Li J.F. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem. Biophys. Res. Commun. 2018;495:2369–2375. doi: 10.1016/j.bbrc.2017.12.050. [DOI] [PubMed] [Google Scholar]
- 11.Wu Z., Shi W., Jiang C. Overexpressing circular RNA hsa_circ_0002052 impairs osteosarcoma progression via inhibiting Wnt/β-catenin pathway by regulating miR-1205/APC2 axis. Biochem. Biophys. Res. Commun. 2018;502:465–471. doi: 10.1016/j.bbrc.2018.05.184. [DOI] [PubMed] [Google Scholar]
- 12.Huang L., Chen M., Pan J., Yu W. Circular RNA circNASP modulates the malignant behaviors in osteosarcoma via miR-1253/FOXF1 pathway. Biochem. Biophys. Res. Commun. 2018;500:511–517. doi: 10.1016/j.bbrc.2018.04.131. [DOI] [PubMed] [Google Scholar]
- 13.Du Y.X., Guo L.X., Pan H.S., Liang Y.M., Li X. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion. Hum. Cell. 2020;33:252–260. doi: 10.1007/s13577-019-00298-6. [DOI] [PubMed] [Google Scholar]
- 14.Ardekani A.M., Naeini M.M. The role of microRNAs in human diseases. Avicenna J. Med. Biotechnol. 2010;2:161. [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 16.He S., Wang Z., Tang H., Dong J., Qu Y., Lv J. MiR-217 Inhibits Proliferation, Migration, and Invasion by Targeting SIRT1 in Osteosarcoma. Cancer Biother. Radiopharm. 2019;34:264–270. doi: 10.1089/cbr.2017.2394. [DOI] [PubMed] [Google Scholar]
- 17.Fujii R., Osaka E., Sato K., Tokuhashi Y. MiR-1 Suppresses Proliferation of Osteosarcoma Cells by Up-regulating p21 via PAX3. Cancer Genomics Proteomics. 2019;16:71–79. doi: 10.21873/cgp.20113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudo-Saito C., Shirako H., Takeuchi T., Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 19.Haque S., Harries L.W. Circular RNAs (circRNAs) in Health and Disease. Genes (Basel). 2017;8 doi: 10.3390/genes8120353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou R., Wu Y., Wang W., Su W., Liu Y., Wang Y., Fan C., Li X., Li G., Li Y., Xiong W., Zeng Z. Circular RNAs (circRNAs) in cancer. Cancer Lett. 2018;425:134–142. doi: 10.1016/j.canlet.2018.03.035. [DOI] [PubMed] [Google Scholar]
- 21.Liu J., Liu T., Wang X., He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kristensen L.S., Andersen M.S., Stagsted L.V.W., Ebbesen K.K., Hansen T.B., Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 23.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., Loewer A., Ziebold U., Landthaler M., Kocks C., le Noble F., Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 25.Kun-Peng Z., Chun-Lin Z., Jian-Ping H., Lei Z. A novel circulating hsa_circ_0081001 act as a potential biomarker for diagnosis and prognosis of osteosarcoma. Int. J. Biol. Sci. 2018;14:1513–1520. doi: 10.7150/ijbs.27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulcheski F.R., Christoff A.P., Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J. Biotechnol. 2016;238:42–51. doi: 10.1016/j.jbiotec.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Su J., Wang Q., Liu Y., Zhong M. miR-217 inhibits invasion of hepatocellular carcinoma cells through direct suppression of E2F3. Mol. Cell. Biochem. 2014;392:289–296. doi: 10.1007/s11010-014-2039-x. [DOI] [PubMed] [Google Scholar]
- 28.Li J., Li D., Zhang W. Tumor suppressor role of miR-217 in human epithelial ovarian cancer by targeting IGF1R. Oncol. Rep. 2016;35:1671–1679. doi: 10.3892/or.2015.4498. [DOI] [PubMed] [Google Scholar]
- 29.Guo J., Feng Z., Huang Z., Wang H., Lu W. MicroRNA-217 functions as a tumour suppressor gene and correlates with cell resistance to cisplatin in lung cancer. Mol. Cells. 2014;37:664–671. doi: 10.14348/molcells.2014.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen L., Wang P., Yang J., Li X. MicroRNA-217 regulates WASF3 expression and suppresses tumor growth and metastasis in osteosarcoma. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0109138. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Felekkis K., Touvana E., Stefanou C., Deltas C. microRNAs: a newly described class of encoded molecules that play a role in health and disease. Hippokratia. 2010;14:236. [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L., Xia L., Zhao L., Chen Z., Shang X., Xin J., Liu M., Guo X., Wu K., Pan Y., Fan D. Activation of PAX3-MET pathways due to miR-206 loss promotes gastric cancer metastasis. Carcinogenesis. 2015;36:390–399. doi: 10.1093/carcin/bgv009. [DOI] [PubMed] [Google Scholar]
- 33.Xu G., Fang P., Chen K., Xu Q., Song Z., Ouyang Z. MicroRNA-362-3p Targets PAX3 to Inhibit the Development of Glioma through Mediating Wnt/β-Catenin Pathway. NeuroImmunoModulation. 2019;26:119–128. doi: 10.1159/000499766. [DOI] [PubMed] [Google Scholar]
- 34.Zhan F.B., Zhang X.W., Feng S.L., Cheng J., Zhang Y., Li B., Xie L.Z., Deng Q.R. MicroRNA-206 Reduces Osteosarcoma Cell Malignancy In Vitro by Targeting the PAX3-MET Axis. Yonsei Med. J. 2019;60:163–173. doi: 10.3349/ymj.2019.60.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.