Key Points
Question
What is the in vivo RAS dependency of BRAF alterations in cancer samples?
Findings
In this cross-sectional study of genomic data analysis with more than 11 9000 cancer samples, it was found that current BRAF alteration classification does not accurately represent in vivo RAS dependency for non-V600 alterations.
Meaning
Results of this study suggest a need to revisit the classification of BRAF alterations, which has important clinical implications because personalized treatment strategies could be developed for RAS-dependent BRAF variant cancers based on different mechanisms of RAS activation.
Abstract
Importance
Understanding RAS dependency and mechanisms of RAS activation in non-V600 BRAF variant cancers has important clinical implications. This is the first study to date to systematically assess RAS dependency of BRAF alterations with real-world cancer genomic databases.
Objective
To evaluate RAS dependency of individual BRAF alterations through alteration coexistence analysis using cancer genomic databases.
Design and Setting
A cross-sectional data analysis of 119 538 nonredundant cancer samples using cancer genomics databases including GENIE (Genomics Evidence Neoplasia Information Exchange) and databases in cBioPortal including TCGA (The Cancer Genome Atlas) (accessed March 24, 2020), in addition to 2745 cancer samples from Mayo Clinic Genomics Laboratory (January 1, 2015, to July 1, 2020). Frequencies and odds ratios of coexisting alterations of RAS (KRAS, NRAS and HRAS) and RAS regulatory genes (NF1, PTPN11 and CBL) were calculated for individual BRAF alterations, and compared according to the current BRAF alteration classification; cancer type specificity of coexisting alterations of RAS or RAS regulatory genes was also evaluated.
Main Outcomes and Measures
Primary outcome measurement is enrichment of RAS (KRAS, NRAS and HRAS) alterations in BRAF variant cancers. Secondary outcome measurement is enrichment of RAS regulatory gene (NF1, PTPN11, and CBL) in BRAF variant cancers.
Results
A total of 2745 cancer samples from 2708 patients (female/male ratio: 1.0) tested by Mayo Clinic Genomics Laboratory and 119 538 patients (female/male ratio: 1.1) from GENIE and cBioPortal database were included in the study. In 119 538 nonredundant cancer samples, class 1 BRAF alterations and BRAF fusions were found to be mutually exclusive to alterations of RAS or RAS regulatory genes (odds ratio range 0.03-0.13 and 0.03-0.73 respectively), confirming their RAS independency. Both class 2 and class 3 BRAF alterations show variable and overlapping levels of enriched RAS alterations (odds ratio range: 0.03-5.9 and 0.63-2.52 respectively), suggesting heterogeneity in RAS dependency and a need to revisit BRAF alteration classification. For RAS-dependent BRAF alterations, the coexisting alterations also involve RAS regulatory genes by enrichment analysis (for example, S467L shows an odds ratio of 8.26 for NF1, 9.87 for PTPN11, and 15.23 for CBL) and occur in a variety of cancer types with some coalterations showing cancer type specificity (for example, HRAS variations account for 46.7% of all coexisting RAS alterations in BRAF variant bladder cancers, but 0% in non–small cell lung cancers). Variant-level assessment shows that BRAF alterations involving the same codon may differ in RAS dependency. In addition, RAS dependency of previously unclassified BRAF alterations could be assessed.
Conclusions and Relevance
Current BRAF alteration classification based on in vitro assays does not accurately predict RAS dependency in vivo for non-V600 BRAF alterations. RAS-dependent BRAF variant cancers with different mechanisms of RAS activation suggest the need for different treatment strategies.
This cross-sectional study assesses existing BRAF alteration classifications and the spectrum of coexisting alterations of RAS pathway genes and cancer types in which they occur using nonredundant cancer samples from cancer genomics databases.
Introduction
BRAF alterations have been classified into class 1, 2, and 3 based on dimer formation, enzyme activity, and RAS dependence from in vitro experiments.1,2 Different classes of BRAF alterations differ in mechanism of action and require different treatment strategies.3,4,5,6 Class 3 alterations are different from class 1 (V600) and class 2 (non-V600 activating alterations) in that they are RAS-dependent with impaired kinase activity.1,2 Class 2 BRAF alterations were defined as RAS independent, and as activating BRAF alterations they are expected to be mutually exclusive to RAS alterations similar to class 1 BRAF alterations. However, some class 2 BRAF alterations were reported to coexist with RAS alterations.7,8 In contrast, class 3 BRAF variants, defined as RAS dependent, can be found without coexisting RAS alterations.7,8 Variation coexistence analysis using cancer genomics database provides evidence for functional interactions between activated RAS and BRAF alterations, and thus can support in vivo RAS dependency or independency. However, such analysis requires a large cancer genomics database, and most published clinical studies are limited in size to systematically assess RAS dependency with real-world clinical data.
Herein with 119 538 nonredundant cancer samples in GENIE (Genomics Evidence Neoplasia Information Exchange)5 and cancer alteration databases in cBioPortal including TCGA (The Cancer Genome Atlas) (accessed March 24, 2020), in addition to 2745 cancer samples sequenced at Mayo Clinic Genomics Laboratory (January 1, 2015, to July 1, 2020), we systematically assessed RAS dependence of BRAF alterations by examining their coexisting alterations of RAS and other RAS regulatory genes including NF1, PTPN11, and CBL. We aim to evaluate existing BRAF alteration classifications and understand the spectrum of coexisting alterations of RAS pathway genes and cancer types in which they occur. In addition, leveraging the size of the database, variant level assessment of RAS dependency can be performed for some previously unclassified BRAF alterations.
Methods
Targeted next generation sequencing data from 2745 cancer samples (lung cancer, colorectal cancer, and melanoma) that have undergone clinical testing at Mayo Clinic Genomics Laboratory from January 1, 2015, to July 1, 2020, were collected with ACE (Advanced Cohort Explorer). This study was approved by the institutional review board at Mayo Clinic Rochester. A waiver was granted by the institutional review board at Mayo Clinic Rochester because the study is a retrospective data analysis with deidentified data. In addition, targeted next generation sequencing data from 79 720 samples from 75 191 patients in GENIE database (v.7.0, public release: January 2020, access date: March 24, 2020), and 176 nonduplicate studies available in cBioPortal (access date: March 24, 2020)9,10 including 46 697 samples from 44 347 patients were also collected. Both GENIE and cBioPortal databases are publicly available to the entire scientific community. We removed duplicated patients and only kept their earliest record, resulting in a total number of 119 538 non-Mayo samples. We removed duplicate records and kept only the earliest record, resulting in a total number of 119 538 non-Mayo samples.
Statistical Analysis
To analyze the coexistence pattern of genes of interest (KRAS, NRAS, HRAS, NF1, PTPN11, CBL), distributions of samples based on BRAF alteration status (BRAF altered and BRAF unaltered) and alteration status of genes of interest were summarized in 2 × 2 contingency tables. The SNVs and indels, fusions (for BRAF), and copy number alterations (RAS amplification and NF1 loss) were included in the analysis. Variants with less than 2 Catalogue of Somatic Mutations in Cancer counts were excluded as passenger alterations. Odds ratios (ORs) and frequencies of coalterations were calculated for individual BRAF alterations. BRAF alteration classification was based on previous studies.1,2 The χ2 test was used with the 2 × 2 contingency tables to test whether alterations coexist or are mutually exclusive to each other. The 95% CIs of ORs were calculated as exp(ln(OR)±1.96*SE(ln(OR))). Standard error SE(ln(OR)) was calculated as [(1/A) + (1/B) + (1/C) + (1/D)]1/2, where A, B, C, D are 4 numbers in the 2 × 2 contingency table. For contingency table with a value of 0, we applied Haldane-Anscombe correction by adding 0.05 to each cell and then calculated OR and its CI. P value was calculated through χ2 test for each contingency table. A P < .05 was considered statistically significant. This study followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline for cross-sectional studies as applicable.
Results
A total of 2745 cancer samples from 2708 patients (female/male ratio: 1.0) tested by Mayo Clinic Genomics Laboratory and 119 538 patients (female/male ratio: 1.1) from GENIE and cBioPortal database were included in the study. Targeted gene alteration data from 2708 patients tested by Mayo Clinic Genomics Laboratory showed that both class 3 and class 2 BRAF alterations have higher frequencies of coexisting RAS alterations than class 1 (10 of 38 [26.3%] for class 3, 4 of 22 [18.1%] for class 2, and 1 of 304 [0.3%] for class 1; P < .001 by χ2 for both class 3 vs class 1 and class 2 vs class 1). The Table lists the BRAF alterations that coexist with RAS alterations among the Mayo Clinic tested patient cohort. Shown by large genomic database analysis using GENIE and cancer alteration databases in cBioPortal, 5767 cases harbored BRAF alterations, 3761 of which belong to class 1, 529 class 2, 651 class 3, and 282 BRAF fusions, with the remaining unclassifiable. Among them, 138 (3.6%) cases with class 1 BRAF alterations, 107 (20.2%) class 2, 212 (32.6%) class 3, and 8 (2.8%) cases with BRAF fusions harbor alterations in RAS or RAS regulatory genes. As shown in Figure 1, both BRAF class 1 alterations and fusions are mutually exclusive to coexisting RAS alterations (P < .001 by χ2 for both; odds ratios range from 0.003-0.13 and 003-0.73, respectively) (eTable in the Supplement),2,11 but class 2 and class 3 alterations show heterogeneous and overlapping levels of ORs for coexisting RAS alterations (odds ratio range: 0.03-5.9 and 0.63-2.52 respectively) (Figure 1A). Some class 2 alterations show high ORs of RAS coalteration supporting their RAS dependence (eg, p.E586K [OR, 3.8; 95% CI, 1.6-9.2] and p.F595L [OR, 3.5; 95% CI, 1.5-8.4]). Some class 3 BRAF alterations (such as N581S) show lower OR (0.63) of coexisting RAS alterations than many class 2 alterations (Figure 1A-B and the eTable in the Supplement).
Table. BRAF-RAS Coalteration Cases From Mayo Clinic Cohort.
| Case | Diagnosis | BRAF alteration | RAS alteration | BRAF alteration class |
|---|---|---|---|---|
| 1 | Metastatic colorectal adenocarcinoma | p.V600E | KRAS p.G12V | 1 |
| 2 | Lung adenocarcinoma | p.L597Q | NRAS p.Q61R | 2 |
| 3 | Histiocytic sarcoma | p.K601N | NRAS p.Q61K | 2 |
| 4 | Melanoma | p.Leu597Ser | NRAS p.G13R | 2 |
| 5 | Colorectal adenocarcinoma | p.Gly469Val | KRAS p.G12S | 2 |
| 6 | Invasive high-grade urothelial carcinoma | p.D594G | KRAS p.G12V | 3 |
| 7 | Lung adenocarcinoma | p.N581I | KRAS p.G12S | 3 |
| 8 | Metastatic lung adenocarcinoma | p.D594H | KRAS p.G12V | 3 |
| 9 | Metastatic melanoma | p.G466E | NRAS p.A146V | 3 |
| 10 | Metastatic melanoma | p.G466V | NRAS p.A146V | 3 |
| 11 | Soft tissue, melanoma | p.S467L | NRAS p.Q61R | 3 |
| 12 | Metastatic melanoma | p.S467L | NRAS p.Q61R | 3 |
| 13 | Metastatic melanoma | p.G466R | NRAS p.Q61H | 3 |
| 14 | Metastatic melanoma | p.G596C | NRAS p.Q61L | 3 |
| 15 | Melanoma | p.D594E | NRAS p.Q61H | 3 |
| 16 | Lung adenocarcinoma | p.G596V | KRAS p.G13C | NA |
Abbreviation: NA, not applicable.
Figure 1. Summary of Coexisting Alterations of RAS and RAS Regulatory Genes for Individual BRAF Alterations.
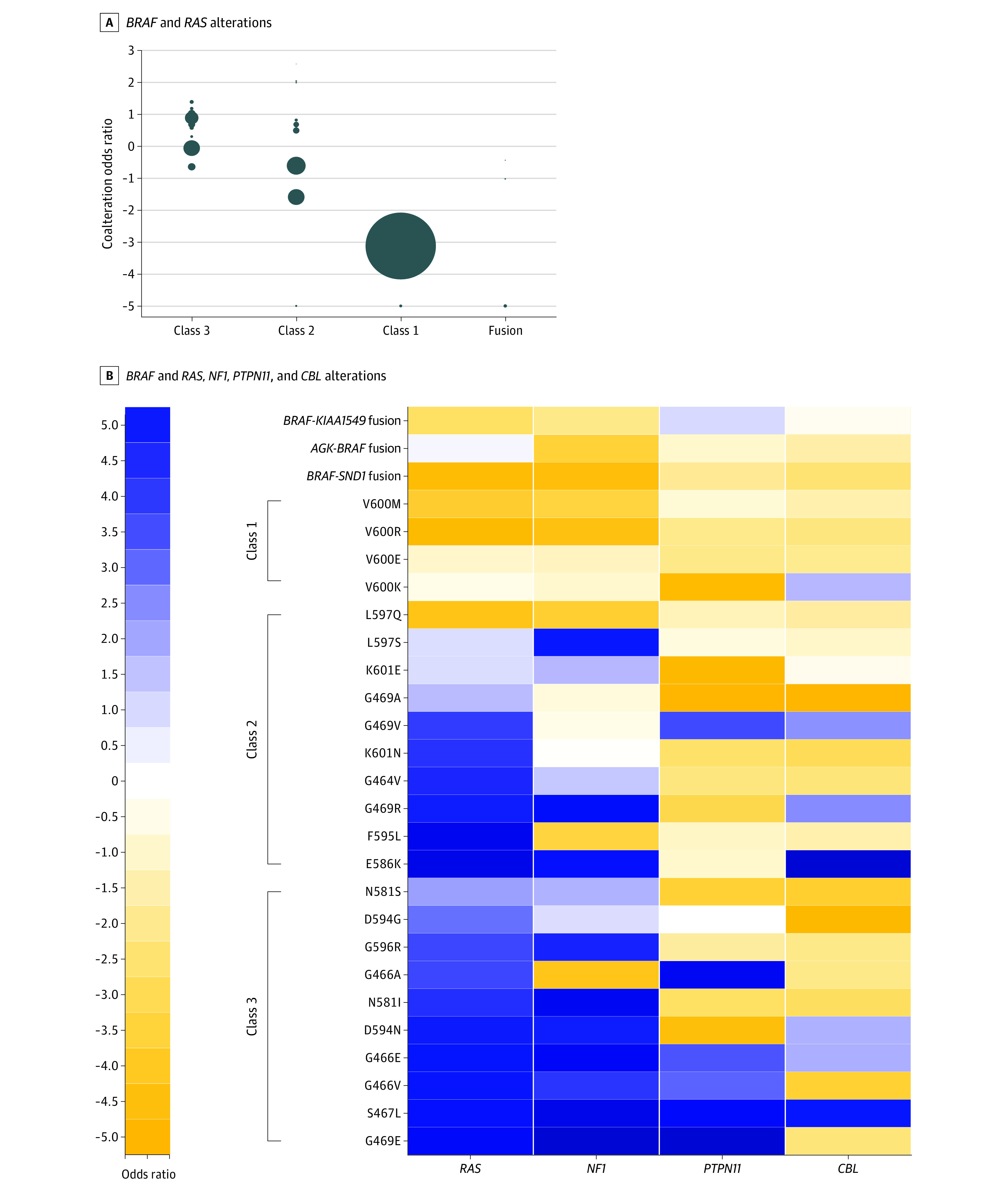
A, Coalteration associations between BRAF alterations and RAS (KRAS, NRAS, and HRAS) alterations. Individual dots represent different alterations in the class. Size of dots represents numbers of patients with this alteration. B, Coalteration associations between BRAF alterations and RAS, NF1, PTPN11, and CBL alterations. Colors represent log2 odds ratio, and positive values were normalized to a −5 to 0 range, and negative values were normalized to a 0 to 5 range.
For BRAF alterations showing RAS dependency, coalterations of NF1, PTPN11 and CBL also exist (Figure 1B), supporting a variety of mechanisms of RAS activation in these tumors. With the variant level assessment, our study suggests alterations involving the same codon may have different RAS dependency. For example, p.G469E shows much higher frequency of coalterations of RAS or RAS regulatory genes than p.G469A (20 of 34 [58.8%] vs 21 of 162 [13.0%]; P < .001 by χ2) (Figure 1B). In addition, RAS dependency manifested as enriched alterations of RAS or RAS regulatory genes is a common phenomenon in various types of tumors (Figure 2), not limited to melanoma as previously suggested.2 Relative cancer type-specific coalterations exist. While KRAS is the overall dominant gene that is coaltered, HRAS contributed to most coalteration cases in bladder cancer, and NF1 coalteration preferably occurs in melanoma and endometrial cancer. In addition, our study assessed RAS dependency of previously unclassified alterations. For example, p.E501K (OR, 4.3; 95% CI, 1.9-10.0), p.S363F (OR, 3.4; 95% CI, 1.0-12.0), p.R354Q (OR, 3.2; 95% CI, 1.1-9.9), and p.E26D (OR, 2.5; 95% CI, 1.2-5.1) show enriched coexisting RAS or RAS regulatory genes (eTable in the Supplement), suggesting that they are RAS dependent.
Figure 2. Cancer Type Distributions of Coexisting Mutations of RAS or RAS Regulatory Genes for BRAF Mutations.
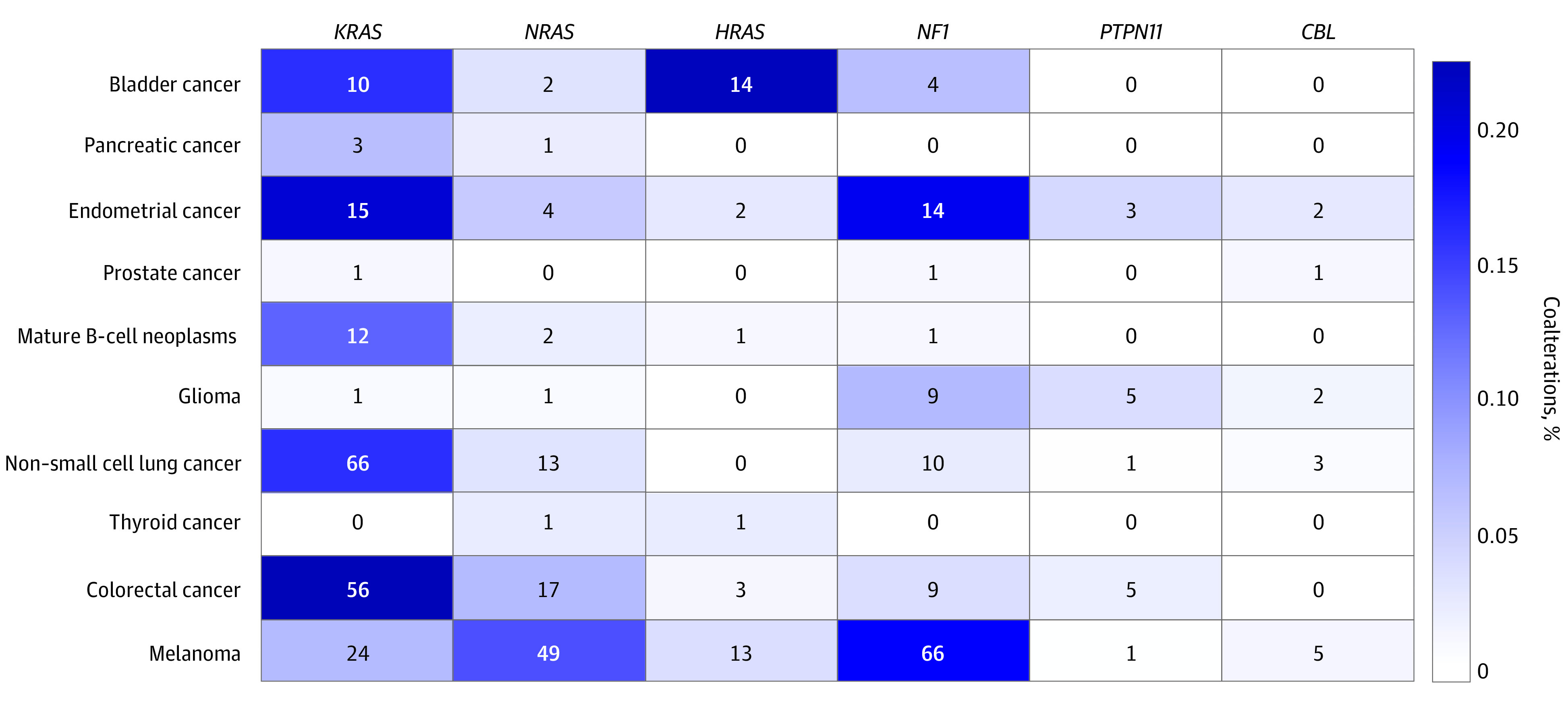
Numbers represent sample counts. Colors reflect percentage of coalterations among all BRAF alterations within the cancer type.
Discussion
Findings of this cross-sectional study suggest RAS independency in class 1 BRAF (V600) alterations and that enriched coexisting RAS alterations for some class 3 BRAF alterations support RAS dependency. In addition, our study presents an extended spectrum of coexisting alterations of RAS regulatory genes including NF1, PTPN11, and CBL, indicating different mechanisms of activating RAS for RAS-dependent BRAF variant tumors. This study also shows heterogeneity in RAS dependency in class 2 and 3 alterations, which cannot be explained by current BRAF alteration classification. Classification of BRAF alterations was mostly achieved by in vitro assays and RAS dependency established with mouse embryonal fibroblasts established in literature.12,13 This study suggests that they may not accurately predict in vivo function. Thus, revisiting the classification of BRAF alterations is warranted. In addition, our study shows that coexisting alterations of RAS or RAS regulatory gene are common phenomena in various types of tumors, more broadly than previous recognized.2 Moreover, the variant level assessment can be used to evaluate previously unclassified alterations and is important in deciphering its clinical significance and enabling precision medicine.
Our study has notable clinical implications. First, class 2 but not class 3 BRAF alterations may respond to MEK inhibitors like trametinib.3,4,5,6 Our study showed that some class 2 BRAF alterations are actually RAS dependent and may behave similarly to class 3 alterations and not respond to MEK inhibitors. Second, RAS dependency suggests targeting RAS or signaling upstream may be effective, and the presence of coexisting alterations may affect treatment selection. For those without coexisting RAS or NF1 alterations, upstream signaling including EGFR,2 tyrosine kinase receptor,14 and SHP23 may be targeted. PTPN11 and CBL are potentially targetable if their alterations are present. If RAS or NF1 coalterations are present, however, targeting downstream signaling including RAF (such as sorafnib)15 may be more effective. In addition, our study also suggests that when a BRAF alteration with RAS dependency is identified, reflex testing should be performed to assess alteration status of RAS and RAS regulatory genes, which may lead to more precise assessment of therapeutic vulnerabilities and strategies. These study findings suggest that large cancer genomics data analysis provides evidence-based gene alteration interpretation to assist in clinical decision-making in the context of precision medicine.
Limitations
This study has several limitations. First, clonal architecture of the cancer samples could not be fully assessed with the available single-time point genomic data; in theory, the coexisting alterations could be present in different tumor subclones, though a previous study showed that concomitant BRAF and RAS alterations were mostly present in the same tumor cell populations.16 Second, some rare non-V600 BRAF alterations are underrepresented in the genomic databases; thus their RAS dependency cannot be reliably assessed with the approach in the study. In addition, for analysis of individual BRAF alterations, multiple-testing-inflated false discoveries could not be fully excluded. Third, this is a cross-sectional data mining study, and future laboratory and clinical research are needed to confirm the findings in the study.
Conclusions
These findings suggest that there are different mechanisms of RAS activation for RAS-dependent BRAF alterations in a wide variety of cancer types, including alterations of RAS genes and RAS regulatory genes. In addition, current BRAF alteration classification based on in vitro assays does not accurately predict RAS dependency in vivo for non-V600 BRAF alterations.
eTable. Frequencies and Odds Ratios of Coexisting Mutations of KRAS, NRAS, HRAS, NF1, PTPN11 and CBL for all BRAF Mutations With ≥5 Count
References
- 1.Dankner M, Rose AAN, Rajkumar S, Siegel PM, Watson IR. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene. 2018;37(24):3183-3199. doi: 10.1038/s41388-018-0171-x [DOI] [PubMed] [Google Scholar]
- 2.Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. . Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017;548(7666):234-238. doi: 10.1038/nature23291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bracht JWP, Karachaliou N, Bivona T, et al. . BRAF mutations classes I, II, and III in NSCLC patients included in the SLLIP trial: the need for a new pre-clinical treatment rationale. Cancers (Basel). 2019;11(9):E1381. doi: 10.3390/cancers11091381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowyer SE, Rao AD, Lyle M, et al. . Activity of trametinib in K601E and L597Q BRAF mutation-positive metastatic melanoma. Melanoma Res. 2014;24(5):504-508. doi: 10.1097/CMR.0000000000000099 [DOI] [PubMed] [Google Scholar]
- 5.Posch C, Cholewa BD, Vujic I, et al. . Combined inhibition of MEK and Plk1 has synergistic antitumor activity in NRAS mutant melanoma. J Invest Dermatol. 2015;135(10):2475-2483. doi: 10.1038/jid.2015.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richtig G, Aigelsreiter A, Kashofer K, et al. . Two case reports of rare BRAF mutations in Exon 11 and Exon 15 with discussion of potential treatment options. Case Rep Oncol. 2016;9(3):543-546. doi: 10.1159/000449125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dagogo-Jack I, Martinez P, Yeap BY, et al. . Impact of BRAF mutation class on disease characteristics and clinical outcomes in BRAF-mutant lung cancer. Clin Cancer Res. 2019;25(1):158-165. doi: 10.1158/1078-0432.CCR-18-2062 [DOI] [PubMed] [Google Scholar]
- 8.Lokhandwala PM, Tseng LH, Rodriguez E, et al. . Clinical mutational profiling and categorization of BRAF mutations in melanomas using next generation sequencing. BMC Cancer. 2019;19(1):665. doi: 10.1186/s12885-019-5864-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cerami E, Gao J, Dogrusoz U, et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401-404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao J, Aksoy BA, Dogrusoz U, et al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirripa M, Biason P, Lonardi S, et al. . Class 1, 2, and 3 BRAF-mutated metastatic colorectal cancer: a detailed clinical, pathologic, and molecular characterization. Clin Cancer Res. 2019;25(13):3954-3961. doi: 10.1158/1078-0432.CCR-19-0311 [DOI] [PubMed] [Google Scholar]
- 12.Yao Z, Torres NM, Tao A, et al. . BRAF mutants evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell. 2015;28(3):370-383. doi: 10.1016/j.ccell.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan PT, Garnett MJ, Roe SM, et al. ; Cancer Genome Project . Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855-867. doi: 10.1016/S0092-8674(04)00215-6 [DOI] [PubMed] [Google Scholar]
- 14.Sen B, Peng S, Tang X, et al. . Kinase-impaired BRAF mutations in lung cancer confer sensitivity to dasatinib. Sci Transl Med. 2012;4(136):136ra70. doi: 10.1126/scitranslmed.3003513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smalley KS, Xiao M, Villanueva J, et al. . CRAF inhibition induces apoptosis in melanoma cells with non-V600E BRAF mutations. Oncogene. 2009;28(1):85-94. doi: 10.1038/onc.2008.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng G, Tseng LH, Chen G, et al. . Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer. 2015;15:779. doi: 10.1186/s12885-015-1811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Frequencies and Odds Ratios of Coexisting Mutations of KRAS, NRAS, HRAS, NF1, PTPN11 and CBL for all BRAF Mutations With ≥5 Count


