Abstract
Dysregulated microRNA (miRNA) expression is involved in the occurrence and development of colorectal cancer (CRC) through the regulation of various important physiological events. Hence, miRNAs may be used as effective targets for CRC treatment; however, this hypothesis warrants further investigation. miRNA-511 (miR-511) plays vital roles in the progression of different tumor types. However, the expression, exact role, and the mechanisms underlying the regulation of colorectal carcinogenesis and progression by miR-511 remain poorly understood. This study presents that miR-511 expression was decreased in CRC tissues and cell lines compared with that in adjacent nonneoplastic tissues and normal human colon epithelium cell lines, respectively. The enforced expression of miR-511 in CRC cells significantly reduced cell proliferation and invasion. Hepatoma-derived growth factor (HDGF) was mechanically validated as a direct target of miR-511 in CRC. Furthermore, miR-511 was negatively associated with HDGF in CRC tissues. The restored HDGF expression can abrogate the tumor-suppressive roles of miR-511 in CRC cells. More importantly, miR-511 overexpression suppressed the PI3K/AKT signaling pathway in CRC. These results suggest that miR-511 can potentially serve as a therapeutic target for the therapy of patients with CRC.
Key words: Colorectal cancer (CRC), MicroRNA-511, Hepatoma-derived growth factor, Proliferation, Invasion
INTRODUCTION
Colorectal cancer (CRC) is the third most prevalent cancer and the fourth leading cause of cancer-associated deaths worldwide1. Approximately 1.2 million novel diagnosed cases and 600,000 deaths are reported annually2. Numerous risk factors, including advanced age, hereditary components, obesity, drinking, smoking, high consumption of red meat, and a lack of physical exercise, are involved in the formation and progression of CRC3,4. Despite considerable improvement in treatments in the past few decades, the therapeutic outcome for patients with CRC remains unsatisfactory, especially for those with recurrence and metastasis5. Approximately 30%–50% of patients with CRC undergo recurrence and metastasis6. The 5-year overall survival rate for patients with CRC is 90%; however, this rate is greatly reduced to less than 5% for those with metastasis or recurrence7. Therefore, understanding the molecular mechanisms underlying CRC occurrence and progression is essential to explore novel therapeutic methods and improve the prognosis.
MicroRNAs (miRNAs) are endogenous and noncoding small RNAs with a length of approximately 19–24 nucleotides8. miRNAs have been validated as novel gene regulators by directly binding to their 3′-untranslated regions (3′-UTRs) in a sequence-specific way and consequently induces translational inhibition or mRNA degradation9. Bioinformatics studies have shown that miRNAs may modulate almost two thirds of all human protein-coding genes and are therefore involved in the regulation of various cellular processes, including embryonic development, cell differentiation, transformation, proliferation, apoptosis, invasion, epithelial-to-mesenchymal transition, and metastasis10,11. More than half of miRNAs are located at the fragile sites and genomic regions associated with cancer, suggesting that miRNAs may play a crucial role in tumorigenesis and tumor development12. The dysregulation of miRNAs (either downregulation or upregulation) has been reported in various types of malignant tumors, including CRC13. miRNAs may function as either tumor suppressor miRNAs or oncomiRNAs primarily based on the biological functions of their target genes14. Therefore, further investigation on the biological roles of miRNAs in CRC may be beneficial in developing efficient therapeutic targets for patients with CRC.
miR-511 plays vital roles in the progression of different tumor types15–17. However, the expression, exact role, and the mechanisms underlying the regulation of colorectal carcinogenesis and progression by miR-511 remain poorly understood. Our study aims to analyze miR-511 expression and investigates its biological roles and associated molecular mechanism in CRC.
MATERIALS AND METHODS
Patients and Tissue Specimens
This study was approved by the Ethics Committee of The Seventh People’s Hospital of Shanghai University of TCM (Shanghai, P.R. China). Written informed consent was also obtained from all patients with CRC before the collection of tissue samples. Thirty-six paired CRC tissues and adjacent nonneoplastic tissues were obtained from patients who underwent surgery at The Seventh People’s Hospital of Shanghai University of TCM. None of these patients had undergone radiation therapy or chemotherapy before surgical resection. All tissue specimens were immediately frozen in liquid nitrogen and stored in a refrigerator at −80°C.
Cell Culture
Four CRC cell lines (HCT116, HT29, SW480, and SW620) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). A normal human colon epithelium cell line (FHC) was obtained from the American Type Culture Collection (Manassas, VA, USA). All cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 mg/ml streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). All cells were grown at 37°C in a humidified incubator with 5% CO2.
miRNA Mimics and Plasmid Transfection
miR-511 mimics and negative control miRNA mimics (miR-NC) were acquired from Guangzhou RiboBio Co., Ltd. (Guangzhou, P.R. China). Hepatoma-derived growth factor (HDGF) overexpression plasmid pcDNA3.1-HDGF and empty plasmid pcDNA3.1 were chemically synthesized by Shanghai GenePharma Co. Ltd. (Shanghai, P.R. China). Cells were transfected with miRNA mimics or plasmid using Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA) following the manufacturer’s protocol. The medium was replaced with fresh DMEM supplemented with 10% FBS after 6 h of transfection.
Total RNA Extraction and Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA of the tissue specimens and cells was isolated using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The quality and quantity of the total RNA were determined using a Nanodrop® ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc., Pittsburgh, PA, USA). For miRNA expression, the total RNA was reversibly transcribed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). The complementary DNA (cDNA) was subjected to qPCR with the TaqMan MicroRNA PCR Kit (Applied Biosystems). cDNA was synthesized from the total RNA using a PrimeScript RT Reagent kit (Takara Bio, Dalian, P.R. China) to quantify the HDGF mRNA level. Subsequent qPCR was performed using a SYBR Premix Ex Taq™ kit (Takara Bio). U6 snRNA and glyceraldehyde 3 phosphate dehydrogenase (GAPDH) were selected as the endogenous control for miR-511 and HDGF mRNA, respectively. Data were analyzed using the 2−ΔΔCq method18.
3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
MTT assay was conducted to investigate cell proliferation. Transfected cells were collected 24 h after transfection and plated in 96-well plates with a density of 3 × 103 cells per well. These cells were allowed to grow at 37°C in a humidified incubator with 5% CO2 for 0, 24, 48, and 72 h. At every time point, 20 μl of MTT solution (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA) was added into each well and incubated at 37°C for 4 h. Afterward, the medium was removed, and 100 μl of dimethyl sulfoxide (DMSO; Sigma-Aldrich) was added to dissolve the formazan crystals. Finally, the absorbance of the suspension at a wavelength of 490 nm was determined using a Victor 3 Multi-Label Microplate Reader (PerkinElmer, Inc., Waltham, MA, USA).
Matrigel Invasion Assay
Transwell 24-well chambers (Corning Incorporated, Corning, NY, USA) coated with Matrigel matrix (BD Biosciences, Franklin Lakes, NJ, USA) were employed to evaluate the cell invasion capacity. In brief, 5 × 104 transfected cells in 200 μl of FBS-free DMEM were seeded into the upper chambers. The lower chambers were filled with 500 μl of DMEM containing 20% FBS. Following incubation at 37°C in a humidified incubator with 5% CO2 for 24 h, the remaining cells on the upper surface of the chambers were removed gently using cotton wool. The invasive cells were fixed with 100% methanol and stained with 0.1% crystal violet. The stained cells were photographed and counted under an IX71 inverted microscope (Olympus Corporation, Tokyo, Japan) with five random fields per chamber.
Bioinformatics Analysis and Luciferase Reporter Assay
Bioinformatics analysis was conducted with TargetScan (http://www.targetscan.org) and miRBase (http://www.mirbase.org/) to predict the potential target of miR-511. A highly conserved binding site with a perfect match to the seed region of miR-511 was found in the 3′-UTR of HDGF.
The luciferase reporter plasmids containing the wild-type (Wt) or mutant (Mut) sequences of miR-511 in the 3′-UTR of HDGF were synthesized by GenePharma Co. Ltd. and named as pGL3-HDGF-3′-UTR Wt and pGL3-HDGF-3′-UTR Mut, respectively. These cells were plated into 24-well plates with a density of 1.5 × 105 cells per well. miR-511 mimics or miR-NC, together with pGL3-HDGF-3′-UTR Wt or pGL3-HDGF-3′-UTR Mut, was transfected into the cells using Lipofectamine 2000 according to the manufacturer’s instructions. The cells were incubated at 37°C with 5% CO2 for 48 h. Afterward, luciferase activity was detected using a dual-luciferase assay system (Promega, Madison, WI, USA) according to the manufacturer’s protocol. Renilla luciferase activity served as the normalization for the luciferase activity.
Western Blot Analysis
Total protein was extracted from the tissues and cells using RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, P.R. China). A BCA assay kit was used to detect the total protein concentration. A specific amount of total protein was electrophoresed by 10% sodium dodecyl sulfate-polyacrylamide gel and transferred into polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA). After blocking with 5% skimmed milk for 1 h, the membranes were incubated with the primary antibodies overnight at 4°C, washed with Tris-based saline-Tween 20 three times, and incubated with corresponding horseradish peroxidase-conjugated secondary antibody (sc-2005; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Enhanced chemiluminescence (ECL) prime Western blot substrate (GE, Amersham, UK) was used to visualize the protein bands. The primary antibodies used in this study were acquired from Santa Cruz Biotechnology and included mouse anti-human HDGF antibody (sc-271344), mouse anti-human phosphotidylinositide 3 kinase (PI3K) antibody (sc-376412), mouse anti-human phosphorylated (p)-PI3K antibody (sc-56938), mouse anti-human p-AKT antibody (sc-271966), mouse anti-human AKT antibody (sc-81434), and mouse anti-human β-actin antibody (sc-69879). The densities of the protein bands were quantified using ImageJ 1.49 (National Institutes of Health, Bethesda, MD, USA).
Statistical Analysis
Results were presented as mean ± standard deviation (SD) and compared with Student’s t-test or one-way analysis of variance (ANOVA) test. Student–Newman–Keuls test was used as a post hoc test following ANOVA. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
miR-511 Is Downregulated in CRC Tissues and Cell Lines
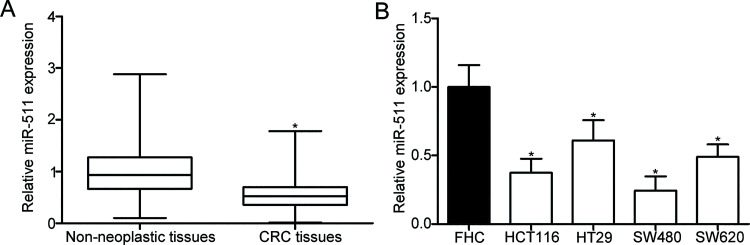
To explore whether miR-511 is involved in CRC formation and progression, we first measured the miR-511 expression in 36 paired CRC tissues and adjacent nonneoplastic tissues using RT-qPCR. The results showed that the expression level of miR-511 was decreased in the CRC tissues compared with that in the adjacent nonneoplastic tissues (p < 0.05) (Fig. 1A). Consistent with the results observed in CRC tissues, miR-511 expression was downregulated in all four detected CRC cell lines (HCT116, HT29, SW480, and SW620) compared with that in a normal human colon epithelium cell line (FHC) (p < 0.05) (Fig. 1B). These observations suggest that miR-511 may play key roles in the initiation and progression of CRC.
Figure 1.
Expression level of microRNA (miR)-511 in the colorectal cancer (CRC) tissues and cell lines. (A) miR-511 expression in 36 paired CRC tissues and adjacent nonneoplastic tissues was determined using reverse transcription quantitative polymerase chain reaction (RT-qPCR). *p < 0.05 versus nonneoplastic tissues. (B) Expression levels of miR-511 in four CRC cell lines (HCT116, HT29, SW480, and SW620) and a normal human colon epithelium cell line (FHC) were also examined by RT-qPCR. *p < 0.05 versus FHC.
miR-511 Prohibits Cell Proliferation and Invasion of CRC
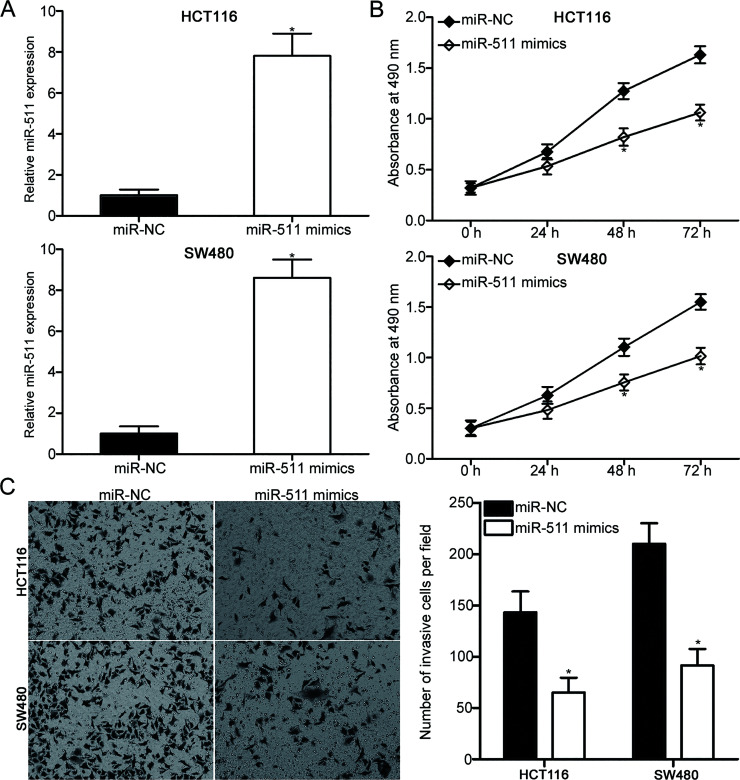
Based on the observation that miR-511 was lowly expressed in CRC, we examined whether miR-511 plays tumor-suppressing roles in CRC. Among the four cell lines, the HCT116 and SW480 cells with relatively low miR-511 expression were transfected with miR-511 mimics to increase its endogenous expression and confirm the previous hypothesis. Following transfection, RT-qPCR analysis revealed that miR-511 was evidently upregulated in the HCT116 and SW480 cells after transfection with the miR-511 mimics (p < 0.05) (Fig. 2A). The effect of miR-511 overexpression on CRC cell proliferation was determined using MTT assay. As shown in Figure 2B, the restored expression of miR-511 prohibited the proliferation of HCT116 and SW480 cells (p < 0.05). We also evaluated the effect of miR-511 on the invasion ability of CRC cells. Matrigel invasion assay results revealed that the enhanced expression of miR-511 attenuated the invasion capacities of HCT116 and SW480 cells compared with those of cells transfected with miR-NC (p < 0.05) (Fig. 2C). These findings suggest that miR-511 may serve tumor-suppressive roles in CRC.
Figure 2.
Restored expression of miR-511 suppresses the proliferative and invasion capacities of the HCT116 and SW480 cells. (A) Relative miR-511 expression in the HCT116 and SW480 cells transfected with the miR-511 mimics or miR-NC was detected using RT-qPCR. *p < 0.05 versus miR-NC. (B) MTT assay was utilized to evaluate the proliferation of the HCT116 and SW480 cells transfected with miR-511 mimics or miR-NC. *p < 0.05 versus miR-NC. (C) Matrigel invasion assay was used to assess the effect of the miR-511 overexpression on the invasion ability of the HCT116 and SW480 cells. *p < 0.05 versus miR-NC.
miR-511 Inhibits HDGF Expression by Interacting With the Binding Site in its 3′-UTR
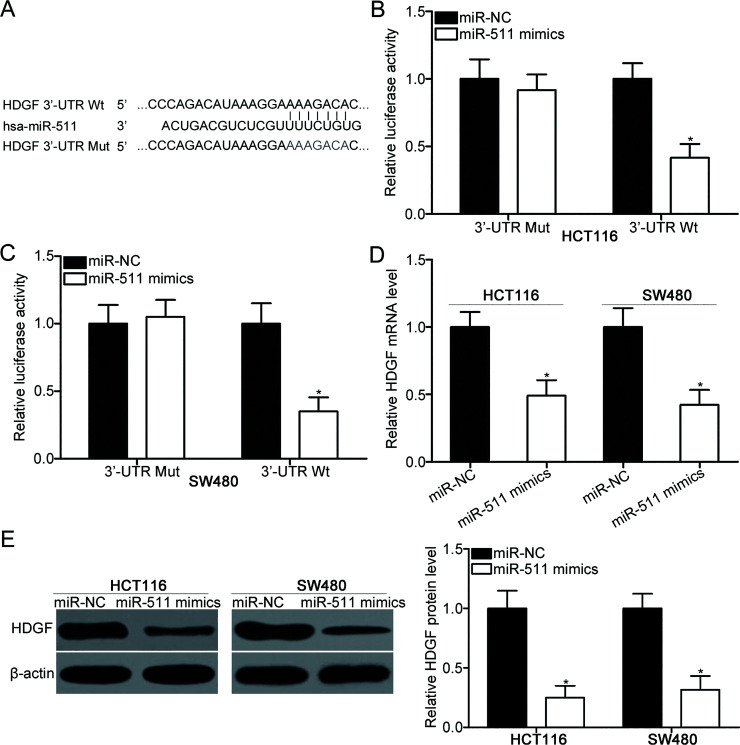
To elucidate the mechanisms responsible for the tumor-suppressing roles of miR-511 in CRC, we conducted bioinformatics analysis to predict the potential targets of miR-511. HDGF, which was overexpressed in CRC and involved in CRC occurrence and development19–21, was selected because its 3′-UTR contains a highly conserved binding site (Fig. 3A). To confirm this possibility, we performed luciferase reporter assays, wherein the HCT116 and SW480 cells were cotransfected with luciferase reporter plasmid and miR-511 mimics or miR-NC. As illustrated in Figure 3B and C, the upregulation of miR-511 significantly reduced the luciferase activities of Wt HDGF 3′-UTR plasmid in the HCT116 and SW480 cells (p < 0.05). However, the luciferase activity of the Mut HDGF 3′-UTR plasmid was not affected, suggesting that miR-511 can directly interact with the 3′-UTR of HDGF. RT-qPCR and Western blot analyses were carried out to investigate whether miR-511 could regulate the endogenous HDGF expression in CRC. The results indicated that the enforced expression of miR-511 suppressed the HDGF expression in HCT116 and SW480 cells at both mRNA (p < 0.05) (Fig. 3D) and protein (p < 0.05) (Fig. 3E) levels. Thus, HDGF is a direct target of miR-511 in CRC.
Figure 3.
Hematoma-derived growth factor (HDGF) is a direct target of miR-511 in CRC. (A) Wild-type (Wt) and mutant (Mut) miR-511 binding sequences in the 3′-untranslated region (3′-UTR) of HDGF. (B, C) Dual-luciferase assay system was used to measure the luciferase activity in the HCT116 and SW480 cells cotransfected with miR-511 mimics or miR-NC and pGL3-HDGF-3′-UTR Wt or pGL3-HDGF-3′-UTR Mut. *p < 0.05 versus miR-NC. (D, E) The HDGF expression at both mRNA and protein levels was decreased in the miR-511 mimics transfected into the HCT116 and SW480 cells as revealed by RT-qPCR and Western blot analyses, respectively. *p < 0.05 versus miR-NC.
HDGF Is Upregulated in CRC Tissues and Negatively Correlated With miR-511
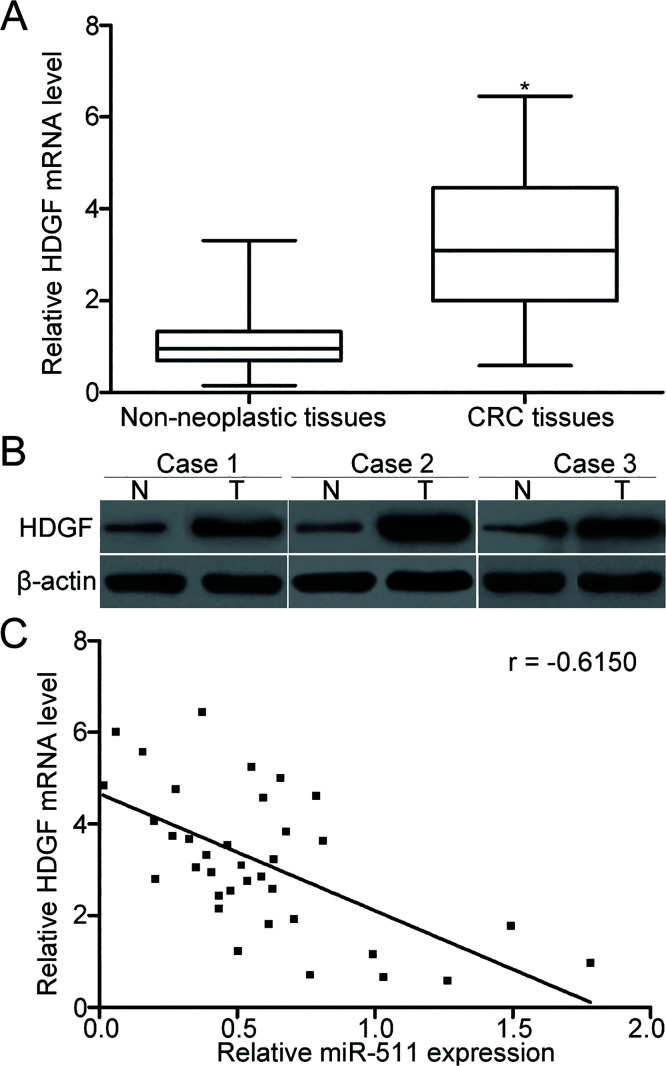
To evaluate the association between miR-511 and HDGF in CRC, we detected the mRNA and protein expression levels of HDGF in CRC tissues and adjacent nonneoplastic tissues. Results of the RT-qPCR and Western blot analyses showed that both the mRNA (p < 0.05) (Fig. 4A) and protein (Fig. 4B) levels of HDGF were overexpressed in CRC tissues compared with those in adjacent nonneoplastic tissues. In addition, the HDGF mRNA was negatively associated with miR-511 levels in the CRC tissues based on Spearman’s correlation analysis (r = −0.6150, p < 0.001) (Fig. 4C). These results support that HDGF is a direct target of miR-511 in CRC.
Figure 4.
High expression of HDGF was inversely correlated with miR-511 level in the CRC tissues. (A, B) HDGF mRNA and protein expression levels were determined by RT-qPCR and Western blot analysis in CRC tissues and adjacent nonneoplastic tissues, respectively. *p < 0.05 versus nonneoplastic tissues. (C) Spearman’s correlation analysis was performed to analyze the association between the HDGF mRNA and miR-511 levels in CRC tissues. r = −0.6150, p < 0.001.
HDGF Mediates the Tumor-Suppressive Roles of miR-511 in CRC
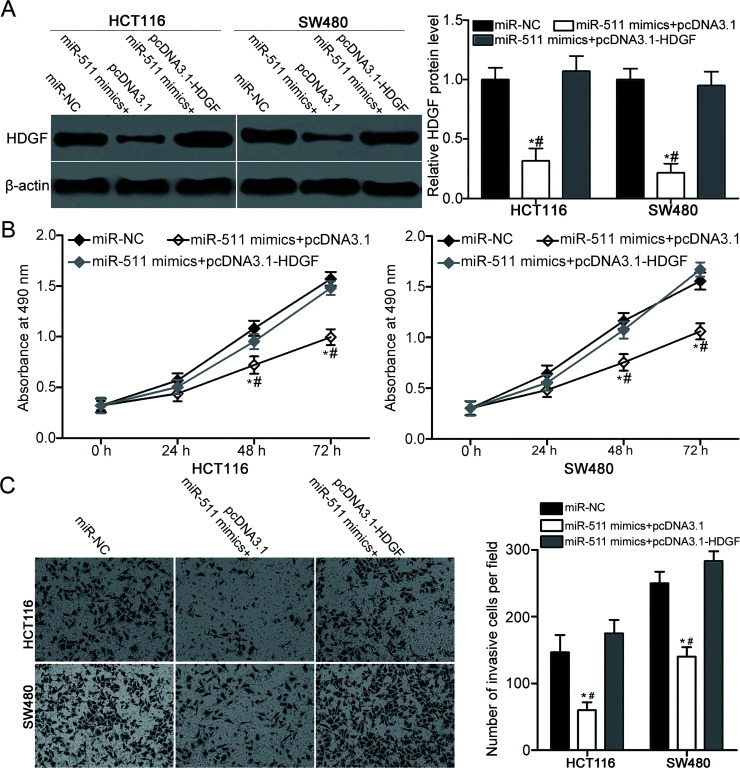
We performed a rescue experiment to further validate whether the inhibitory effects of miR-511 overexpression on CRC cell proliferation and invasion are mediated by HDGF. miR-511 mimics together with pcDNA3.1 or pcDNA3.1-HDGF were transfected into the HCT116 and SW480 cells. Following the transfection, Western blot analysis revealed that the decreased HDGF protein expression in the HCT116 and SW480 cells caused by the miR-511 overexpression was restored by the cotransfection with pcDNA3.1-HDGF (p < 0.05) (Fig. 5A). Subsequent MTT and Matrigel invasion assays revealed that the restored HDGF expression can recover the inhibitory effects of miR-511 mimics on the proliferation (p < 0.05) (Fig. 5B) and invasion (p < 0.05) (Fig. 5C) of HCT116 and SW480 cells. Our data indicate that miR-511 may impede CRC progression, to some extent, by inhibiting HDGF expression.
Figure 5.
Restored HDGF expression rescues the suppressive effects of the miR-511 overexpression on CRC cell proliferation and invasion. The HCT116 and SW480 cells were cotransfected with miR-511 mimics and pcDNA3.1 or pcDNA3.1-HDGF. (A) HDGF protein expression was detected by Western blot analysis. *p < 0.05 versus miR-NC. #p < 0.05 versus miR-511 mimics + pcDNA3.1-HDGF. (B, C) Proliferation and invasion of the indicated cells were determined using MTT assay and Matrigel invasion assay, respectively. *p < 0.05 versus miR-NC. #p < 0.05 versus miR-511 mimics + pcDNA3.1-HDGF.
miR-511 Suppresses the PI3K/AKT Signaling Pathway in CRC
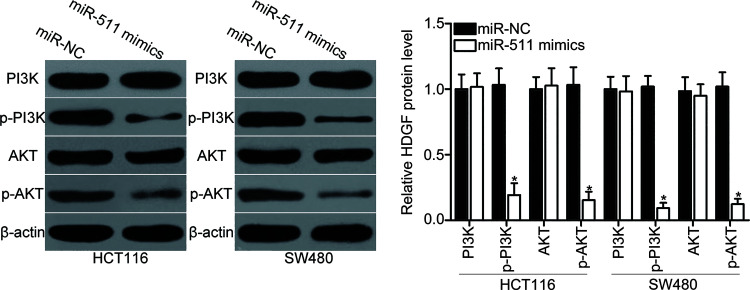
HDGF was previously reported to participate in the regulation of the PI3K/AKT signaling pathway22. Therefore, we explored whether miR-511 could suppress the PI3K/AKT signaling pathway in CRC. Western blot analysis was performed to detect PI3K, p-PI3K, AKT, and p-AKT expression levels in HCT116 and SW480 cells after transfection with miR-511 mimics or miR-NC. The results showed that upregulation of miR-511 reduced the p-PI3K and p-AKT protein levels in HCT116 and SW480 cells (p < 0.05) (Fig. 6). However, the total expression levels of PI3K and AKT were unaffected. These results suggest that miR-511 may suppress the PI3K/AKT signaling pathway in CRC.
Figure 6.
miR-511 inhibits the activation of phosphotidylinositide 3 kinase (PI3K)/AKT signaling pathway in CRC. HCT116 and SW480 cells were transfected with miR-511 mimics or miR-NC. Following transfection for 72 h, Western blot analysis was conducted to detect PI3K, p-PI3K, AKT, and p-AKT protein levels. *p < 0.05 versus miR-NC.
DISCUSSION
Dysregulated miRNA expression is involved in the occurrence and development of CRC through the regulation of various important physiological events23,24. Hence, miRNAs may be used as effective targets for CRC treatment; however, this hypothesis warrants further investigation. In this study, we found that miR-511 was underexpressed in CRC tissues and cell lines. The ectopic expression of miR-511 inhibited cell proliferation and invasion in CRC. In addition, HDGF was identified as a direct target of miR-511 in CRC. Furthermore, miR-511 overexpression suppressed the PI3K/AKT pathway in CRC. This study suggests that miR-511 may be a potential therapeutic target for CRC.
miR-511 is involved in the progression of several types of human malignancy. For example, miR-511 was downregulated in gastric cancer tissues and cell lines. The upregulation of miR-511 attenuated cell proliferation, colony formation ability, and cell cycle progression in gastric cancer by inhibiting the activation of the PI3K/AKT and wingless-related integration site (Wnt)/β-catenin signaling pathways through tripartite motif-containing 24 (TRIM24) suppression15. Zhang et al. reported that the expression level of miR-511 was reduced in lung adenocarcinoma tissues compared with that in paracarcinoma tissues. The restored expression of miR-511 induced tribbles pseudokinase 2 (TRIB2) inhibition to suppress cell proliferation and promoted apoptosis of lung adenocarcinoma in vitro16. In addition, miR-511 overexpression prohibited cell proliferation, increased apoptosis, and induced cell cycle arrest at the G1–S transition in radioresistant lung adenocarcinoma by directly targeting B-cell lymphoma 2 X-associated protein (BAX)17. However, miR-511 was overexpressed in hepatocellular carcinoma. The resumed expression of miR-511 promoted cell proliferation of hepatocellular carcinoma via the regulation of B-cell translocation gene 1 protein (BTG1)25. These conflicting studies implied that the expression pattern and biological roles of miR-511 in human malignancy have tissue specificity.
Identifying CRC-related miR-511 and its target genes is vital for the development of promising therapeutic targets. In this study, HDGF was found as a novel direct target of miR-511 in CRC. First, bioinformatics analysis indicated a highly conserved binding site in the 3′-UTR of HDGF with a perfect match to the seed region of miR-511. Second, luciferase reporter assays revealed that the 3′-UTR of HDGF can be directly targeted by miR-511. Third, miR-511 reduced the HDGF expression at both the mRNA and protein levels in CRC as determined by RT-qPCR and Western blot analysis. Fourth, the high expression of HDGF was inversely correlated with the low miR-511 expression in CRC tissues. Finally, the restored HDGF expression rescued the tumor-suppressive functions in CRC, which was induced by miR-511 overexpression. These results implied that miR-511 exhibits important functions in CRC carcinogenesis and progression, to some extent, through HDGF regulation.
HDGF, which is located on chromosome 1q21–q23, is a heparin-binding growth factor that was first purified from a culture medium conditioned with the hepatoma cell line HuH726. Substantial studies have reported that HDGF was overexpressed in various types of human cancer and played important roles in carcinogenesis and cancer progression27–29. For instance, HDGF was upregulated in hepatocellular carcinoma and was associated with tumor differentiation. Hepatocellular carcinoma patients with high HDGF levels had poorer disease-free and overall survival than those with low HDGF expression30. The inhibition of HDGF repressed cell proliferation, metastasis, and apoptosis in vitro and reduced tumor growth of hepatocellular carcinoma in vivo31–33. HDGF was also highly expressed in CRC tissues and cell lines. A high HDGF expression level is an independent prognostic factor for the reduced overall survival in patients with CRC19. HDGF knockdown reduced CRC cell growth and metastasis in vitro and in vivo and increased cell apoptosis rate in vitro19–21. Hence, targeting HDGF may be an effective therapeutic target for the treatment of CRC.
In conclusion, this study provided evidence that miR-511 may be involved in colorectal carcinogenesis and progression partially by directly targeting HDGF and regulating the PI3K/AKT signaling pathway. Modulating the miR-511/HDGF axis is a potential strategy for the treatment of patients with CRC.
ACKNOWLEDGMENTS
The study was supported by grants from the Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxk2017-06), the Key Disciplines Group Construction Project of Pudong Health Bureau of Shanghai (PWZxq2014-12), Natural Science Foundation of China (No. 81571718), Science and Technology Development Fund of Shanghai Pudong New Area (PKJ2016-Y19), and Talents Training Program of The Seventh People’s Hospital of Shanghai University of TCM (Grant No. XX2017-06).
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. [DOI] [PubMed] [Google Scholar]
- 2. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 3. Andrews L. Dietary flavonoids for the prevention of colorectal cancer. Clin J Oncol Nurs. 2013;17:671–2. [DOI] [PubMed] [Google Scholar]
- 4. Altobelli E, Lattanzi A, Paduano R, Varassi G, di Orio F. Colorectal cancer prevention in Europe: Burden of disease and status of screening programs. Prev Med. 2014;62:132–41. [DOI] [PubMed] [Google Scholar]
- 5. Moghimi-Dehkordi B, Safaee A. An overview of colorectal cancer survival rates and prognosis in Asia. World J Gastrointest Oncol. 2012;4:71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheer A, Auer RA. Surveillance after curative resection of colorectal cancer. Clin Colon Rectal Surg. 2009;22:242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cartwright TH. Treatment decisions after diagnosis of metastatic colorectal cancer. Clin Colorectal Cancer 2012;11:155–66. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116:281–97. [DOI] [PubMed] [Google Scholar]
- 9. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: The implications for cancer research. Nat Rev Cancer 2010;10:389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–87. [DOI] [PubMed] [Google Scholar]
- 12. Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA 2004;101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stiegelbauer V, Perakis S, Deutsch A, Ling H, Gerger A, Pichler M. MicroRNAs as novel predictive biomarkers and therapeutic targets in colorectal cancer. World J Gastroenterol. 2014;20:11727–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esquela-Kerscher A, Slack FJ. Oncomirs—MicroRNAs with a role in cancer. Nat Rev Cancer 2006;6:259–69. [DOI] [PubMed] [Google Scholar]
- 15. Fang Z, Zhang L, Liao Q, Wang Y, Yu F, Feng M, Xiang X, Xiong J. Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer. J Exp Clin Cancer Res. 2017;36:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C, Chi YL, Wang PY, Wang YQ, Zhang YX, Deng J, Lv CJ, Xie SY. miR-511 and miR-1297 inhibit human lung adenocarcinoma cell proliferation by targeting oncogene TRIB2. PLoS One 2012;7:e46090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang HH, Pang M, Dong W, Xin JX, Li YJ, Zhang ZC, Yu L, Wang PY, Li BS, Xie SY. miR-511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX. Oncol Rep. 2014;31:1473–9. [DOI] [PubMed] [Google Scholar]
- 18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 19. Lian J, Tang J, Shi H, Li H, Zhen T, Xie W, Zhang F, Yang Y, Han A. Positive feedback loop of hepatoma-derived growth factor and beta-catenin promotes carcinogenesis of colorectal cancer. Oncotarget 2015;6:29357–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liao F, Dong W, Fan L. Apoptosis of human colorectal carcinoma cells is induced by blocking hepatoma-derived growth factor. Med Oncol. 2010;27:1219–26. [DOI] [PubMed] [Google Scholar]
- 21. Sun B, Gu X, Chen Z, Xiang J. MiR-610 inhibits cell proliferation and invasion in colorectal cancer by repressing hepatoma-derived growth factor. Am J Cancer Res. 2015;5:3635–44. [PMC free article] [PubMed] [Google Scholar]
- 22. Kung ML, Tsai HE, Hu TH, Kuo HM, Liu LF, Chen SC, Lin PR, Ma YL, Wang EM, Liu GS, Liu JK, Tai MH. Hepatoma-derived growth factor stimulates podosome rosettes formation in NIH/3T3 cells through the activation of phosphatidylinositol 3-kinase/Akt pathway. Biochem Biophys Res Commun. 2012;425:169–76. [DOI] [PubMed] [Google Scholar]
- 23. Wang J, Li H, Wang Y, Wang L, Yan X, Zhang D, Ma X, Du Y, Liu X, Yang Y. MicroRNA-552 enhances metastatic capacity of colorectal cancer cells by targeting a disintegrin and metalloprotease 28. Oncotarget 2016;7:70194–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y, Uzair Ur R, Guo Y, Liang H, Cheng R, Yang F, Hong Y, Zhao C, Liu M, Yu M, Zhou X, Yin K, Chen J, Zhang J, Zhang CY, Zhi F, Chen X. miR-181b functions as an oncomiR in colorectal cancer by targeting PDCD4. Protein Cell 2016;7:722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang SQ, Yang Z, Cai XL, Zhao M, Sun MM, Li J, Feng GX, Feng JY, Ye LH, Niu JQ, Zhang XD. miR-511 promotes the proliferation of human hepatoma cells by targeting the 3’UTR of B cell translocation gene 1 (BTG1) mRNA. Acta Pharmacol Sin. 2017;38:1161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang JS, Chao CC, Su TL, Yeh SH, Chen DS, Chen CT, Chen PJ, Jou YS. Diverse cellular transformation capability of overexpressed genes in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2004;315:950–8. [DOI] [PubMed] [Google Scholar]
- 27. Giri K, Pabelick CM, Mukherjee P, Prakash YS. Hepatoma derived growth factor (HDGF) dynamics in ovarian cancer cells. Apoptosis 2016;21:329–39. [DOI] [PubMed] [Google Scholar]
- 28. Enomoto H, Nakamura H, Liu W, Nishiguchi S. Hepatoma-derived growth factor: Its possible involvement in the progression of hepatocellular carcinoma. Int J Mol Sci. 2015;16:14086–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai CC, Huang SC, Tai MH, Chien CC, Huang CC, Hsu YC. Hepatoma-derived growth factor upregulation is correlated with prognostic factors of early-stage cervical adenocarcinoma. Int J Mol Sci. 2014;15:21492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yoshida K, Tomita Y, Okuda Y, Yamamoto S, Enomoto H, Uyama H, Ito H, Hoshida Y, Aozasa K, Nagano H, Sakon M, Kawase I, Monden M, Nakamura H. Hepatoma-derived growth factor is a novel prognostic factor for hepatocellular carcinoma. Ann Surg Oncol. 2006;13:159–67. [DOI] [PubMed] [Google Scholar]
- 31. Tsang TY, Tang WY, Tsang WP, Co NN, Kong SK, Kwok TT. Mechanistic study on growth suppression and apoptosis induction by targeting hepatoma-derived growth factor in human hepatocellular carcinoma HepG2 cells. Cell Physiol Biochem. 2009;24:253–62. [DOI] [PubMed] [Google Scholar]
- 32. Enomoto H, Nakamura H, Liu W, Iwata Y, Nishikawa H, Takata R, Yoh K, Hasegawa K, Ishii A, Takashima T, Sakai Y, Aizawa N, Ikeda N, Iijima H, Nishiguchi S. Down-regulation of HDGF inhibits the growth of hepatocellular carcinoma cells in vitro and in vivo. Anticancer Res. 2015;35:6475–9. [PubMed] [Google Scholar]
- 33. Yang GY, Zhang AQ, Wang J, Li CH, Wang XQ, Pan K, Zhou C, Dong JH. Hepatoma-derived growth factor promotes growth and metastasis of hepatocellular carcinoma cells. Cell Biochem Funct. 2016;34:274–285. [DOI] [PubMed] [Google Scholar]