Abstract
This work aims to elucidate the effects and the potential underlying mechanisms of microRNA-216a-3p (miR-216a-3p) on the proliferation, migration, and invasion of gastric cancer (GC) cells. In this study, we revealed that the expression of miR-216a-3p was significantly elevated in GC tissues and cell lines. The different expression level of miR-216a-3p was firmly correlated with clinicopathological characteristics of GC patients. We next demonstrated that upregulation of miR-216a-3p could dramatically promote the ability of proliferation, migration, and invasion of GC cells using a series of experiments, whereas downregulation essentially inhibited these properties. Additionally, through bioinformatics analysis and biological approaches, we confirmed that runt-related transcription factor 1 (RUNX1) was a direct target of miR-216a-3p, and overexpression of RUNX1 could reverse the potential effect of miR-216a-3p on GC cells. Furthermore, mechanistic investigation using Western blot analysis showed that downregulation of RUNX1 by miR-216a-3p could stimulate the activation of NF-κB signaling pathway. In summary, this work proved that miR-216a-3p can promote GC cell proliferation, migration, and invasion via targeting RUNX1 and activating the NF-κB signaling pathway. Therefore, miR-216a-3p/RUNX1 could be a possible molecular target for innovative therapeutic agents against GC.
Key words: Gastric cancer (GC), miR-216a-3p, RUNX1, NF-κB signaling pathway, Proliferation, Migration and invasion
INTRODUCTION
Gastric cancer (GC) is the most fatal type of gastrointestinal tract malignancy and the second leading cause of cancer-related deaths worldwide1. Each year, almost 990,000 people are diagnosed with GC worldwide, of whom approximately 738,000 die from this disease2. The explicit cause of GC is still uncertain, but most of the risk is correlated with race, Helicobacter pylori, intake of briny and smoked food, obesity, cigarette smoking, and statins3. Though a great deal of development has been made, including surgery, simultaneous radio- and chemotherapy, molecular targeted therapy and biotherapy, GC still remains a problematic malignancy with invariable appearance of tumor recurrence and 5-year survival rates of 10%–30%4. Therefore, a further investigation of the pathogenesis and molecular mechanisms that participate in GC progression is essential to achieve early diagnosis as well as positive responses to therapy.
MicroRNAs (miRNAs, miRs) are a family of naturally existing, small, noncoding RNAs, which can suppress translation or modulate transcription of target genes through binding to 3′-UTRs5–7. Undoubtedly, miRNAs are involved in many biological processes, including regulation of metabolism, stress resistance, immunity, cellular differentiation, proliferation, and apoptosis8–10. Increasing evidence demonstrates that miRNA deregulation can be correlated with the development of numerous human cancers because they can act as tumor suppressors or oncogenes by directly modulating well-known oncogenes or tumor-suppressive genes11,12. These findings thus provide a reliable rationale for the practicality of miRNAs in the detection, diagnosis, prognosis, and possible therapeutic targets of human cancer.
A considerable number of studies have clarified the ability of some miRNAs to act as tumor promoters (miR-21, miR-30e, miR-196b, miR-183, miR-1288, and miR-143) or suppressors (miR-433, miR-206, miR-133, miR-219-5p, and Let-7g) in GC13,14. miR-216a (ENSG00000207798), a typical multifunctional miRNA, lies in the genomic region of human chromosome 2p16.1, which has been found to be aberrantly expressed in several human malignancies and identified as participating in tumorigenesis. Xia et al.15 reported that miR-216a was found to cause epithelial–mesenchymal transition (EMT) and adjust the drug resistance in liver cancer via activating the PI3K/Akt and TGF-β pathways. Also, the study by Chen et al.16 discovered that miR-216a was apparently upregulated in liver cancer tissues and stimulated transcriptionally by the androgen pathway, which would be an innovative mechanism for the role that the androgen pathway played in early hepatocarcinogenesis. Recently, Liu et al.17 showed for the first time that miR-216a was greatly increased in ovarian cancer tissues as well as cell lines, and supported the metastasis and EMT by repressing the PTEN/Akt signaling pathway. Contrarily, other reports indicated that miR-216a was a tumor suppressor in pancreatic carcinoma18, lung carcinoma19, and glioma20. These findings thus indicate that miR-216a may play dual roles in tumorigenicity, determined by tissue type and specific targets. However, how miR-216a-3p is involved in the development and progression of GC has been largely unexplained.
RUNX1, also known as acute myeloid leukemia 1 (AML1) protein, is a transcription factor that consists of 453 amino acids and modulates the differentiation of hematopoietic stem cells into functional blood cells21. As a transcription factor, RUNX1 collaborates with another protein named core-binding factor β (CBFβ), composing one version of a complex acknowledged as core-binding factor (CBF), which binds to the core section, 5′-PYGPYGGT-3′, of many enhancers and promoters. In fact, the RUNX1 gene is frequently mutated in sporadic myeloid and lymphoid leukemia via point mutation22, translocation23, or amplification24, suggesting its role in carcinogenesis of blood cancers. Contrarily, RUNX1 was also regarded as a tumor suppressor in other solid cancer types including GC25–27, with lower expression in corresponding tumor tissues. However, the molecular mechanisms underlying the low expression of RUNX1 in GC are still largely unknown. The NF-κB signaling pathway is known to play a key role in the development and progression of cancers28–30; however, the functional analysis of miR-216 and the NF-κB signaling pathway has not been well documented.
In the present study, we examined the expression of miR-216a-3p in GC tissues and cell lines (GES-1, MKN-45, and HGC-27) and also investigated its significance in the progression, migration, and invasion of GC cells. As a result, our findings demonstrated that miR-216a-3p might act as a tumor promoter in GC by targeting RUNX1 and activating the NF-κB signaling pathway, suggesting important roles of miR-216a-3p in GC pathogenesis and indicating its potential utilization for the treatment of GC.
MATERIALS AND METHODS
Patients and Tissue Specimens
One hundred forty samples of tumor tissues and adjacent tissues were obtained from patients going through tumor resections between 2013 and 2016 in Zhejiang Provincial People’s Hospital. All samples were collected, and operations were performed after obtaining informed consent. The median age of the patients was 68 years (43–79 years). All patients had a definite histologic diagnosis of GC according to the American Joint Committee on Cancer. The Institute Research Medical Ethics Committee of Zhejiang Provincial People’s Hospital granted approval for this study. Resected tissues were directly frozen in liquid nitrogen after resection from the body and were stored at −80°C until further operation, or were fixed in 4% paraformaldehyde for paraffin embedding.
Cell Culture and Transfection
The gastric adenocarcinoma cell lines (AGS, MKN-45, and HGC-27) and normal gastric epithelial cell line (GES-1) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured in RPMI-1640 medium (Hyclone, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin (Life Technologies, Grand Island, NY, USA) at 37°C in a 5% CO2 incubator. For transfection, cells were allocated in a six-well culture plate at a density of 2 × 105 cells per well and cultured at 37°C. After the cells had grown to about 70% confluence, either the miR-216a-3p mimic or inhibitor (Ribobio, Guangzhou, P.R. China) or both with the RUNX1-overexpressing plasmids were transfected into cells for 48 h using Lipofectamine 3000 (Invitrogen, Shanghai, P.R. China), depending on the manufacturer’s instructions. Nontargeting scrambled oligonucleotide (Ribobio) was used as a negative control for the transfection. Finally, the transfection efficiency of the cells was determined by qRT-PCR).
Immunoblotting
After 48 h of transfection, protein extracts were quantified using the BCA kit (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and heated for 10 min at 100°C. Total proteins (30 μg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and then transferred to nitrocellulose membranes (Merk Millipore, Germany), which was subsequently blocked by incubating with 5% (w/v) nonfat dry milk in TBST (Tris-buffered saline with Tween 20) for 1 h at room temperature. Membranes were then incubated with specific primary antibodies [anti-cyclin D1, anti-matrix metalloproteinase 2 (MMP2), anti-RUNX1/AML1, anti-NF-κB p65, anti-IKB α (Abcam, Shanghai, P.R. China), anti-Bcl-2 (BioWorld Technology, Edina, MN, USA), anti-MMP9 (GeneTex, Irvine, CA, USA), and anti-β-actin (HuaAn Biotechnology Co., Ltd., Hangzhou, P.R. China)] overnight at 4°C. Membranes were subsequently washed with TBST and incubated with horseradish peroxidase-conjugated secondary antibodies in dilution solution for 1 h at room temperature. Finally, the blots were washed and then detected using ChemiDoc™ MP Imaging System (Bio-Rad, Hercules, CA, USA) with a SuperEnhanced Chemiluminescence Detection Kit (Applygen, Beijing, P.R. China). Densitometric measurements were subsequently performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
qRT-PCR
At 48 h posttransfection, total RNA was removed from tissues and cell lines using TRIzol reagent (Invitrogen), and cDNA was synthesized utilizing a TaqMan Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). The quantification of miRNA levels was determined with Stem-Loop RT-PCR assay utilizing the miRNA Q-PCR Detection Kit (ABI, Abilene, TX, USA). All data were normalized to the expression of U6 small nuclear RNA. For mRNA analysis, the expression of RUNX1 was detected with SYBR® Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). GAPDH mRNA levels were utilized for normalization. The primers for miR-216a-3p, RUNX1, GAPDH, and U6 were purchased from GeneCopoeia (Guangzhou, P.R. China). The cycling programmer was conducted in a total volume of 20 μl using the following procedure: preliminary denaturation at 95°C for 10 min, followed by 39 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and elongation at 72°C for 60 s, followed by a final elongation step at 95°C for 10 min. qPCR was operated in triplicate, involving no-template controls, using the Bio-Rad CFX-96 Real-time PCR System and iQ SYBR Green Supermix (Bio-Rad). Relative gene expression of miRNA or mRNA was quantified via the 2−ΔΔCt method.
Cell Proliferation Assay
Cell viability was determined using the 3-(4,5-dimethylthiazol-2-yl)-5- (3-carboxymethonyphenol)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay. In brief, GC cells transfected with either the miR-216a-3p mimic or inhibitor, or both with the RUNX1 overexpression plasmid were plated into 96-well plates at a density of 1 × 104 cells per well and cultured for 1, 2, 3, or 4 days at 37°C. Then the cells were incubated with 20 μl/well MTS Reagent (Promega, Madison, WI, USA) at 37°C for another 4 h. Finally, the plate was shaken briefly on a shaker for 1 min and then read at 490 nm using a microplate reader (Beckman Coulter, Brea, CA, USA). Background absorbance was first deducted using a set of wells including medium only. The experiment was duplicated three times, and the mean OD values obtained were utilized to generate cell growth curves.
Plate Colony Formation Assay
On the plate colony formation assay, cells were seeded onto a 60-mm dish at a density of 500 cells/well after transfection as indicated above. Approximately 2 weeks later, colonies were fixed with 4% paraformaldehyde for 10 min and stained for 10 min with 2.5% crystal violet. Then representative images were obtained, and the mean number of colonies was counted. All experiments were performed in triplicate.
Transwell Invasion Assay
Matrigel-coated chambers (Millipore, Billerica, MA, USA) were used to evaluate GC cell invasion abilities depending on the manufacturer’s instructions. Briefly, at 48 h posttransfection, HGC-27 cells were suspended in serum-free medium at a density of 5 × 104/ml, and 200 μl of this cell suspension was plated into the top chamber of the Matrigel-coated Transwells. RPMI-1640 medium with 20% FBS was augmented into the lower chamber as a cell chemoattractant. After incubation for 24 h at 37°C, the cells on the upper side of the membrane were detached with a cotton swab, while cells on the lower surface were fixed with 95% alcohol and stained with 0.5% crystal violet. The number of invaded cells was counted in five fields (magnification: 200×) for triplicate membranes under a Nikon standard fluorescence microscope (Ti-U; Nikon, Japan) with NIS-Elements BR 4.50.00 imaging software.
Wound Healing Assay
The property of migration was evaluated utilizing a scratch wound healing assay. Transfected HGC-27 cells were cultured in a six-well plate and grown to confluence overnight. The linear wounds were then scratched with a 200-μl pipette tip. Cells were next washed to clear away the debris using PBS and incubated at 37°C for 24 h. The wound width after scratching was determined at 0 and 24 h in three fields for triplicate experiments.
Luciferase Assay
The wild-type (Wt) and mutant (Mut) 3′-UTR of the human RUNX1 gene, which contains a miR-216a-3p binding site, was PCR amplified and inserted downstream of the firefly luciferase open reading frame in the pGL3 vector (Promega, San Luis, CA, USA). The Mut construct of RUNX1 3′-UTR was created by a QuickChange Mutagenesis Kit (Agilent, Palo Alto, CA, USA). HGC-27 cells were seeded on a 24-well plate and then cotransfected with firefly luciferase reporter vectors, the Wt Renilla luciferase reporter vector, and the miR-216a-3p mimic or negative control using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. After 48 h, luciferase activity was determined using a dual-luciferase reporter assay system (Promega, Madison, WI, USA), in which Renilla luciferase activity served as an internal reference. The firefly luciferase activities were normalized to Renilla luciferase activity and expressed as the percentage of activity relative to that of cells that were transfected with negative controls. Experiments were independently repeated three times.
Immunohistochemical Detection
Immunohistochemical staining was performed with the Vectastain Elite ABC Kit (Burlingame, CA, USA). Briefly, all GC tissues were dealt with the following procedures: 4% paraformaldehyde fixation at room temperature, paraffin embedding, section (4-μm thickness), deparaffinization in xylene with series concentration gradients, then rehydration, 3% hydrogen peroxide incubation for 25 min, antigen retrieval at 100°C by citrate buffer solution, 10% goat serum (Invitrogen, Grand Island, NY, USA) blocking, followed by mouse anti-RUNX1 antibody incubation (1:100; Abcam) at 4°C overnight, biotinylated anti-mouse secondary antibody incubation (1:100; BioWorld) at 37°C for 20 min, and then incubation of streptavidin–horseradish peroxidase complex. Finally, the slides were visualized in diaminobenzidine solution (Sigma-Aldrich, St. Louis, MO, USA). All sections were separately assessed by two certified pathologists who were uninformed of the patients’ clinical pathology and other information. Quantitative analysis for percentage of RUNX1+ cells was conducted (four random fields, five sections for each classification, two magnifications: 200× and 400×).
Apoptosis Assay
Cell apoptosis was detected by Annexin-V–FITC/PI Kit (Beyotime, Jiangsu, P.R. China) according to the manufacturer’s instructions. Briefly, cells were transfected as indicated above. At 48 h posttransfection, cells separated by trypsinization were harvested at a density of 1 × 106 cells/ml. The collected cells were washed twice with cold PBS, then resuspended in 300 μl of 1× binding buffer, and next stained with 5 μl of annexin V-FITC and 10 μl of PI. Following short incubation for 10 min in the dark at room temperature, the fluorescence of cells was analyzed by the NovoCyte Flow Cytometer (ACEA Biosciences Inc., San Diego, CA, USA) utilizing the built-in software (NovoExpress).
Statistical Analysis
Statistical analysis was achieved with IBM SPSS Statistics software version 20 (SPSS Inc., Chicago, IL, USA). The t-test or one-way analysis of variance (ANOVA) was utilized to analyze the statistical significance between different groups. Overall survival was appraised using the Kaplan–Meier method as well as the log-rank test. Correlation between RUNX1 expression and clinicopathological features was considered by utilizing the Pearson’s chi-squared test. The relationship between miR-216a-3p and RUNX1 expressions was detected with a two-tailed Pearson’s correlation analysis. All experiments were repeated three times, and data are represented as mean ± standard deviation (SD). A value of p < 0.05 was considered statistically significant.
RESULTS
miR-216a-3p Is Significantly Elevated in GC Tissues and Cell Lines
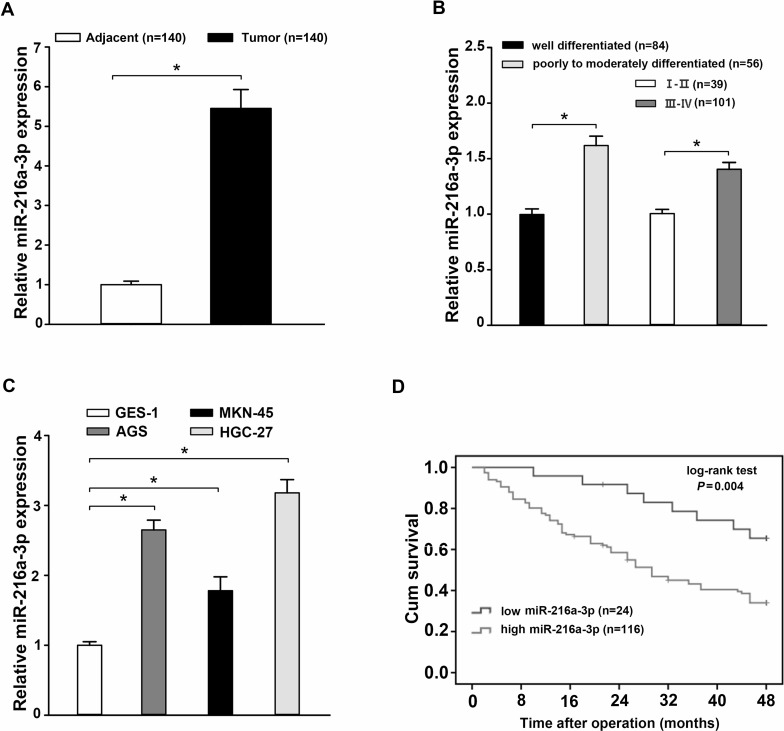
The expression of miR-216a-3p in 140 pairs of GC samples and paired paracancerous tissues was analyzed by qRT-PCR. The relative expression of miR-216a-3p was obviously elevated in GC samples compared to paired paracancerous tissues (p < 0.05) (Fig. 1A). In addition, a significant difference in miR-216a-3p expression was observed between the well-differentiated group and the poorly to moderately differentiated group or between the phases I–II and phases III–IV groups (p < 0.05) (Fig. 1B). According to the expression level of miR-216a-3p, 140 cases of GC patients were divided into either the miR-216a-3p high-expression group or the miR-216a-3p low-expression group, and the relationship between miR-216a-3p levels and clinicopathological factors was analyzed (Table 1). As for the overall survival curve, patients in the miR-216a-3p low-expression group had a significantly poorer prognosis than those in the miR-216a-3p high-expression group (p = 0.004) (Fig. 1C). We also detected the expression of miR-216a-3p in three GC cell lines (AGS, MKN-45, and HGC-27) compared to normal human gastric epithelium cell line GES-1 using qRT-PCR. In the AGS, MKN-45, and HGC-27 cell lines, the expression of miR-216a-3p was significantly upregulated compared with that in GES-1 cells (p < 0.05) (Fig. 1D). In addition, HGC-27 cells had the highest miR-216a-3p expression level. Taken together, these data suggest that miR-216a-3p might be involved in the development of human GC.
Figure 1.
miR-216a-3p expression is elevated in human gastric cancer (GC) tissues and cell lines and is associated with patient survival. (A) The expression of miR-216a-3p in 140 cases of GC tissues was higher than in matched paracancerous tissues as determined using qRT-PCR. (B) Comparison of miR-216a-3p expression between the well-differentiated group and the poorly to moderately differentiated group, as well as between the phases I–II and phases III–IV groups. (C) The expression of miR-216a-3p was increased in three GC cell lines (AGS, MKN-45, and HGC-27) compared to the normal human gastric epithelial cell line GES-1, detected using qRT-PCR. (D) Kaplan–Meier overall survival curve revealed the prognosis of GC patients was worse in those with a high expression of miR-216a-3p (p = 0.004). Three independent experiments were conducted in duplicate. Data are presented as mean ± standard deviation. U6 was used as the internal control. *p < 0.05.
Table 1.
Relationship Between miR-216a-3p Expression and Clinicopathological Factors in 140 Primary Gastric Cancer Tissues
| Clinicopathological Features | Low Expression (n = 24) | High Expression (n = 116) | p Value |
|---|---|---|---|
| Gender | 0.888 | ||
| Male | 11 | 55 | |
| Female | 13 | 61 | |
| Age (years) | 0.589 | ||
| ≤65 | 12 | 51 | |
| >65 | 12 | 65 | |
| Tumor location | 0.662 | ||
| Distal third | 14 | 62 | |
| Middle or proximal | 10 | 54 | |
| Tumor size (cm) | 0.002‡ | ||
| ≥4 | 7 | 74 | |
| <4 | 17 | 42 | |
| Histologic grade | 0.013† | ||
| Well | 15 | 41 | |
| Poor to moderate | 9 | 75 | |
| Lauren classification | 0.615 | ||
| Intestinal | 13 | 60 | |
| Diffuse | 15 | 56 | |
| Invasion | 0.000§ | ||
| T1 + T2 | 14 | 22 | |
| T3 + T4 | 10 | 94 | |
| Lymph node metastasis | 0.000§ | ||
| Negative | 16 | 14 | |
| Positive | 8 | 102 | |
| Distant metastasis | 0.098 | ||
| Negative | 15 | 51 | |
| Positive | 9 | 65 | |
| Clinical stages | 0.000§ | ||
| I + II | 19 | 35 | |
| III + IV | 5 | 81 | |
| Prognosis* | 0.001‡ | ||
| Survival group | 16 | 37 | |
| Death group | 8 | 79 |
Survival outcomes at 48 months postoperation.
p < 0.05;
p < 0.01;
p < 0.001.
miR-216a-3p Promotes Proliferation, Migration, and Invasion of GC Cells
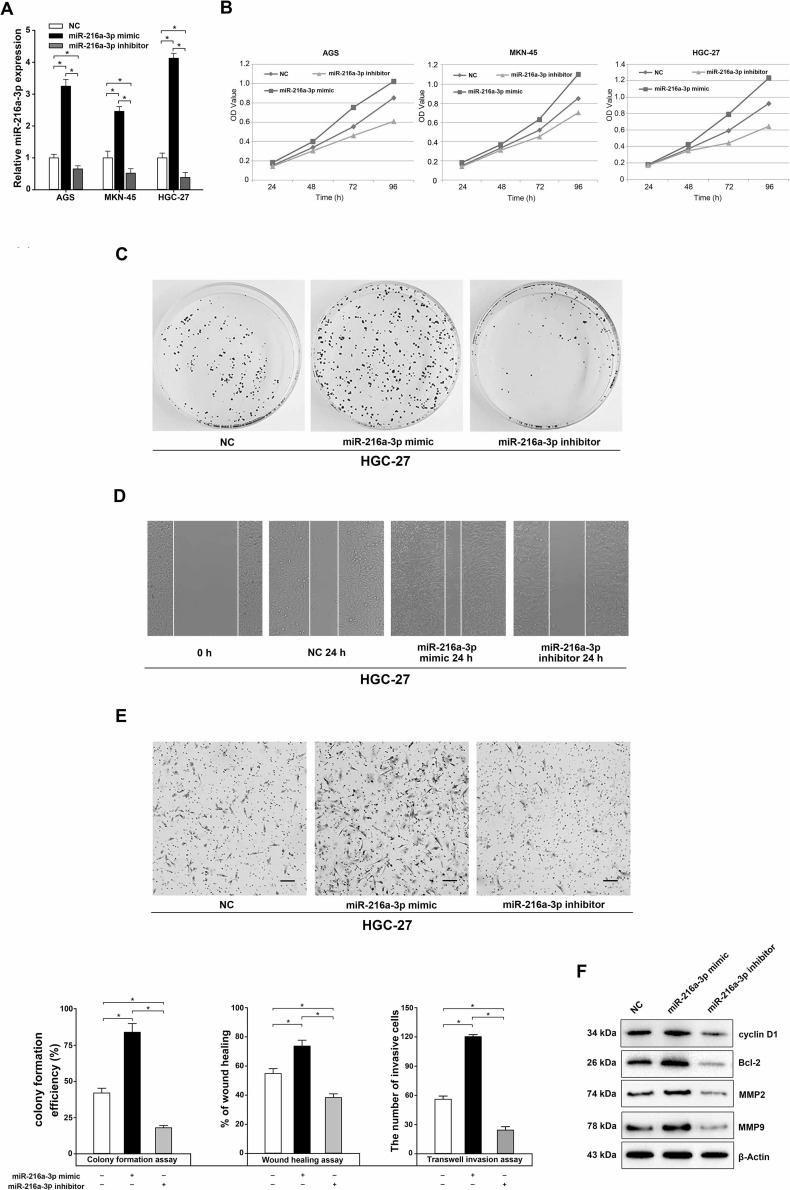
To identify the functional importance of miR-216a-3p in the proliferation, migration, and invasion of GC cells, we first assessed the transfection efficiency of the miR-216a-3p mimic or the miR-216a-3p inhibitor by qRT-PCR at 48 h posttransfection. The results showed that the intracellular expression of miR-216a-3p was significantly upregulated in the miR-216a-3p mimic group, while it was obviously downregulated after miR-216a-3p inhibitor transfection (p < 0.05) (Fig. 2A). As for cell viability, the OD value (representing cell viability) of the cells transfected with the miR-216a-3p mimic was apparently higher than that of cells transfected with negative control, while the miR-216a-3p inhibitor had an opposite effect (Fig. 2B). Similar results were found for HGC-27 cells in the plate colony formation assay (p < 0.05) (Fig. 2C). The wound healing assay showed that at 24 h posttransfection cells in the miR-216a-3p mimic group had enhanced wound healing ability compared with those in either the miR-216a-3p inhibitor or the negative control group (p < 0.05) (Fig. 2D). Using Transwell invasion assay, we found that the number of invading cells in the miR-216a-3p mimic group was remarkably increased (120.33 ± 2.027) compared to the negative control group (56.82 ± 3.317), while that of the miR-216a-3p inhibitor group was significantly decreased (24.38 ± 2.465) (p < 0.05) (Fig. 2E). Finally, Western blot analysis showed that the protein levels of cyclin D1, Bcl-2, MMP2, and MMP9 were significantly upregulated in the miR-216a-3p mimic-transfected cells, while downregulated in the miR-216a-3p inhibitor-transfected cells (Fig. 2F). These results suggest the striking role that miR-216a-3p plays in promoting the proliferation, migration, and invasion of GC cells.
Figure 2.
Effects of miR-216a-3p on the proliferation, migration, and invasion of human GC cells. (A) The miR-216a-3p expression was measured by qRT-PCR based on whole-cell lysate from negative control (NC), miR-216a-3p mimic-, or inhibitor-transfected AGS, MKN-45, and HGC-27 cells. (B) MTS assay of proliferation in AGS, MKN-45, and HGC-27 cells following a time course of transfection with NC, miR-216a-3p mimic, or miR-216a-3p inhibitor. (C–E) The effects of miR-216a-3p on the plate colony formation efficiency (C), migration (D), and invasion (E) of human HGC-27 cells were assessed. The rate of wound healing was calculated with the following formula: (0-h width of wound − 24-h width of wound)/(0-h width of wound). (F) Effects of miR-216a-3p on cyclin D1, Bcl-2, matrix metalloproteinase 2 (MMP2), and MMP9 expression were measured by Western blot. All experiments were performed in triplicate. U6 or β-actin was used as the internal control. *p < 0.05, one-way analysis of variance (ANOVA).
RUNX1 Is a Potential Target of miR-216a-3p in GC Cells
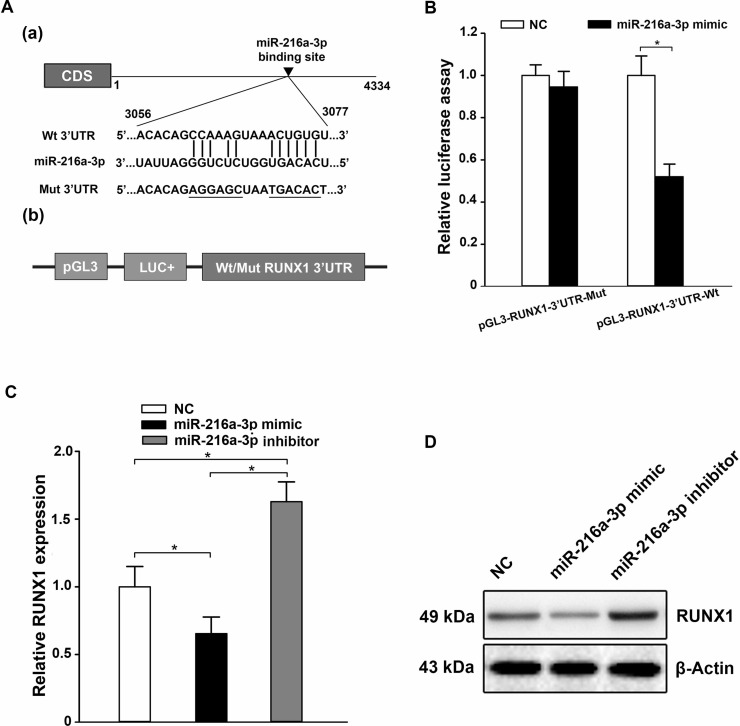
Bioinformatics analysis predicted the 3′-UTR of RUNX1 to comprise a theoretical binding site for miR-216a-3p (Fig. 3A). To test whether RUNX1 was a functional target of miR-216a-3p, a dual-luciferase reporter system was applied. Briefly, HGC-27 cells were transfected with a Wt or Mut pGL3-RUNX1-3′-UTR luciferase reporter plasmid (Fig. 3A), and then cotransfected with the miR-216a-3p mimic or negative control scrambled oligonucleotide. The intensity of fluorescence after miR-216a-3p mimic cotransfection was reduced markedly compared with NC, when the reporter gene included the Wt RUNX1 3′-UTR in HGC-27; however, there was no obvious distinction in luciferase activity when the 3′-UTR of RUNX1 was mutated (p < 0.05) (Fig. 3B). Thus, the luciferase assay proved that miR-216a-3p could directly target RUNX1. Then qRT-PCR and Western blot analysis were used to further determine whether miR-216a-3p could regulate RUNX1 at both the mRNA and protein levels, respectively. In HGC-27 cells, the ectopic expression of miR-216a-3p decreased the endogenous expression of RUNX1 compared with NC, while the level of RUNX1 expression was upregulated by the miR-216a-3p inhibitor (p < 0.05) (Fig. 3C and D). Thus, these data suggest that RUNX1 is a direct functional target of miR-216a-3p.
Figure 3.
miR-216a-3p negatively regulates runt-related transcription factor 1 (RUNX1) by binding to the 3′-UTR. (A) Bioinformatics prediction of the miR-216a-3p binding site in the 3′-UTR of the human RUNX1 mRNA (a) and diagram of the firefly luciferase reporter plasmids with the wild-type or mutant RUNX1 3′-UTR (b). (B) The firefly luciferase reporter plasmid was cotransfected into HGC-27 cells with the miR-216a-3p mimic or negative control scrambled oligonucleotide. The luciferase activity of the cells was determined at 48 h posttransfection, and the values were normalized to the negative control values. (C, D) The expression of RUNX1 in HGC-27 was obviously decreased when transfected with the miR-216a-3p mimic, while it was upregulated by the miR-216a-3p inhibitor, as elucidated by qRT-PCR (C) and Western blot (D). Three independent experiments were conducted in duplicate. U6 or GAPDH was used as the internal control in qRT-PCR and β-actin in the Western blot analysis. All the data are shown as mean ± standard deviation. *p < 0.05.
RUNX1 Is Downregulated and Inversely Associated With the Expression of miR-216a-3p in GC Tissues and Cell Lines
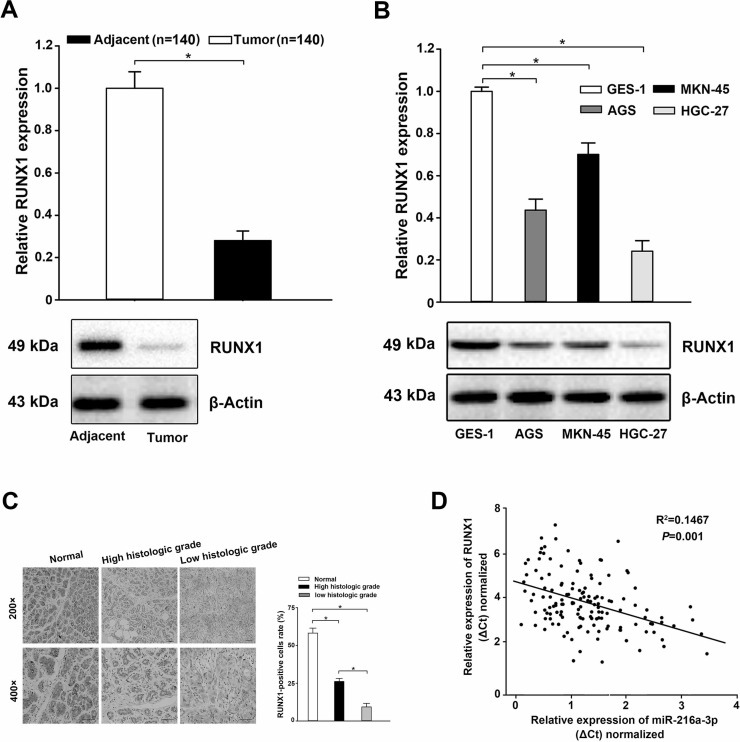
To further clarify the correlation between the expression of miR-216a-3p and that of RUNX1, we first adopted qRT-PCR and Western blot to determine RUNX1 expression both at the tissue and cell levels. The results showed that the expression levels of RUNX1 mRNA and protein in GC tissues and cell lines were lower compared to those in the noncancerous tissues and the normal human gastric epithelial cell line GES-1 (p < 0.05) (Fig. 4A and B). Furthermore, the results of immunohistochemistry (IHC) staining displayed that the expression of RUNX1 was decreased in comparison to that in the noncancerous tissues and correlated significantly with the histologic grade of GC (Fig. 4C). Finally, a further correlation analysis between the expression of miR-216a-3p and RUNX1 in the same GC specimen was conducted. We found a conspicuous inverse correlation between miR-216a-3p expression and RUNX1 transcript levels (Pearson’s correlation coefficient analysis, R 2 = 0.1467, p = 0.001) (Fig. 4D). These results were consistent with the results shown in Figure 1, supporting a potential role for RUNX1 as a target of miR-216a-3p, which might be involved in the development of human GC.
Figure 4.
Decreased expression of RUNX1 in GC tissues and cell lines. (A, B) The expression of RUNX1 was significantly decreased in GC tissues (A) and cell lines (B) compared to that in the noncancerous tissues and normal human gastric epithelial cell line GES-1, detected using qRT-PCR and Western blot. (C) Weak RUNX1 staining in cancer tissues with corresponding strong RUNX1 staining in adjacent normal gastric tissues, and lower protein expression was detected in low histologic grade tumors in comparison with high histologic grade tumors. Scale bars: 500 μm. (D) Negative correlation between the expression of miR-216a-3p and RUNX1 in 140 GC tissues (Pearson’s correlation coefficient analysis, R 2 = 0.1467, p = 0.001). Three independent experiments were executed in duplicate. U6 or GAPDH was used as the internal control for qRT-PCR and β-Actin for Western blot analysis. All the data are presented as mean ± standard deviation. *p < 0.05.
Overexpression of RUNX1 Impaired miR-216a-3p-Induced Promotion of Proliferation, Migration, and Invasion in GC Cells
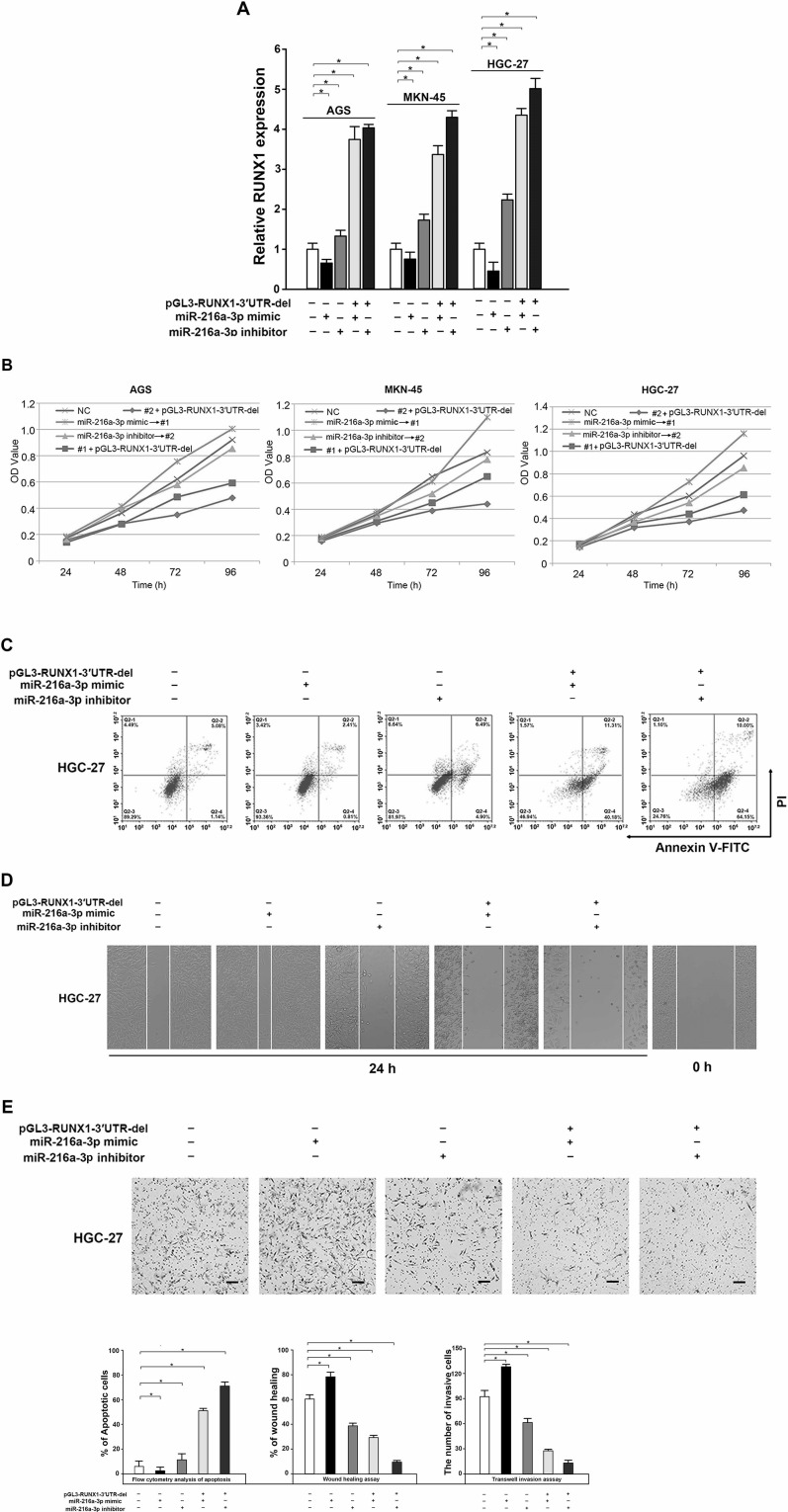
We found the regulatory effect of miR-216a-3p on GC proliferation, migration, and invasion as well as RUNX1 expression. To further demonstrate the tumor-suppressive activity of miR-216a-3p by downregulating RUNX1, the RUNX1 3′-UTR defect plasmid (pGL3-RUNX1-3′UTR-del) and the miR-216a-3p mimic or inhibitor were both transfected into HGC-27 GC cells, and the transfection efficiency as well as the cell viability, migration, and invasion were analyzed. Our results showed that the mRNA expression of RUXN1 in cotransfection of the miR-216a-3p inhibitor and the RUNX1 overexpression plasmid group was higher than that of the other groups (p < 0.05) (Fig. 5A). As expected, decreased cell proliferation, accompanied by decreased cell migration and invasion, was also detected in HGC-27 cells transfected with the miR-216a-3p inhibitor following treatment of pGL3-RUNX1-3′UTR-del construct (p < 0.05) (Fig. 5B and E). In addition, overexpression of RUNX1 significantly reversed the miR-216a-3p-induced promotion of proliferation, migration, and invasion in GC cells (p < 0.05) (Fig. 5B and E). These results illustrate that the promoting effect of miR-216a-3p on the proliferation, migration, and invasion of GC cells is realized by targeting inhibition of the expression of RUNX1.
Figure 5.
RUNX1 suppressed the activity of miR-216a-3p on the proliferation, migration, and invasion of GC cells. (A) Expression of RUNX1 was increased when cotransfected with the miR-216a-3p mimic or inhibitor and the pGL3-RUNX1-3′UTR-del compared to a single transfection of the miR-216a-3p mimic or inhibitor, determined by qRT-PCR. (B–E) The stimulative effect of miR-216a-3p on the proliferation, migration, and invasion of GC cells was reversed by the upregulation of RUNX1, detected by the MTS cell proliferation assay (B), analysis of apoptosis by flow cytometry (C), wound healing assay (D), and Transwell invasion assay (E). All experiments were conducted in triplicate. GAPDH served as the internal control. *p < 0.05, one-way ANOVA.
miR-216a-3p Promotes the NF-κB Signaling Pathway Through Targeting RUXN1
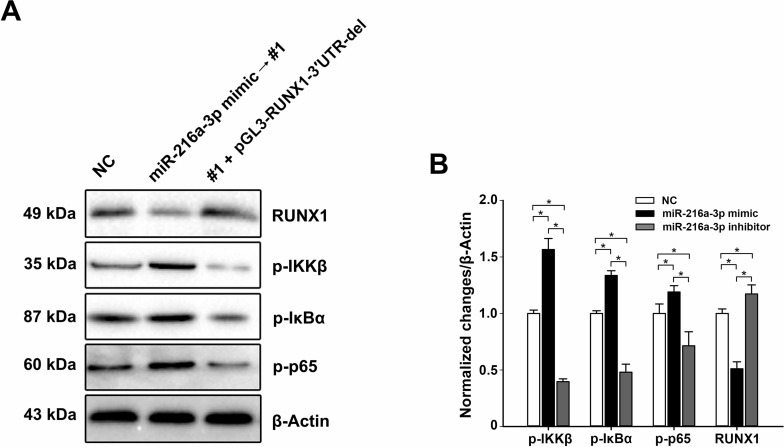
Previous studies have revealed that RUNX1 negatively regulates the NF-κB signaling pathway via interacting with cytoplasm IKK complex, which is a key element in this signaling pathway31. Additionally, the ectopic expression of miR-216a-3p in HGC-27 cells significantly decreased the expression of RUNX1 (Fig. 3C and D). In the present study, Western blot was used to investigate whether miR-216a-3p regulates the NF-κB signaling pathway through targeting RUXN1. Compared to the negative control, miR-216a-3p-overexpressing cells exhibited higher expression levels of p-IKKβ, p-IκBα, and p-p65, while they were downregulated when cotransfected with the RUXN1-overexpressing plasmid (Fig. 6). These observations indicate that miR-216a-3p positively regulates the activities of GC cells by inhibiting the NF-κB signaling pathway.
Figure 6.
miR-216a-3p activates the NF-κB signaling pathway. (A) The expression levels of p-IKKβ, p-IκBα, p-p65, and RUNX1 were detected using Western blot in HGC-27 cells transfected with miR-216a-3p alone or in combination with the RUNX1 overexpression plasmid. (B) Corresponding statistical treatment of the Western blot data. Grayscale intensities of immunoreactive bands were assessed using ImageJ software (National Institutes of Health). The blots shown are representatives of three independent replicates. β-Actin was used as the internal control. *p < 0.05.
DISCUSSION
miRNAs are a family of naturally existing, small, noncoding RNAs that can suppress translation or regulate transcription via binding with the 3′-UTR of target gene mRNA, which is consequently characterized by the decrease in the corresponding target gene expression5–7. Undoubtedly, miRNAs participate in many biological functions, including regulation of metabolism, stress resistance, immunity, cellular differentiation, proliferation, and apoptosis8–10. An increasing body of evidence has shown that the aberrant miRNA expression was correlated with the development and progression of a variety of cancers, including GC13,14, suggesting its potential importance for both disease-specific biomarkers and therapeutic targets.
miR-216a (ENSG00000207798), a classic multifunctional miRNA, has been found to be aberrantly expressed in several human malignancies and identified as being involved in tumorigenesis. For instance, Xia et al.15 reported that miR-216a was found to cause EMT and adjust the drug resistance in liver cancer via activating the PI3K/Akt and TGF-β pathways. Also, the study by Chen et al.16 discovered that miR-216a was apparently upregulated in liver cancer tissues and stimulated transcriptionally by the androgen pathway, which would be an innovative mechanism for the role that the androgen pathway played in early hepatocarcinogenesis. Recently, Liu et al.17 showed for the first time that miR-216a was greatly increased in ovarian cancer tissues as well as in cell lines and supported metastasis and EMT by repressing the PTEN/Akt signaling pathway. Contrarily, other reports indicated that miR-216a was a tumor suppressor in pancreatic carcinoma18, lung carcinoma19, and glioma20. These findings thus indicate that miR-216a may play dual roles in tumorigenicity, determined by tissue type and specific targets. However, how miR-216a-3p is involved in the development and progression of GC has been largely unexplained. In this study, we focused our research on the biological functions of miR-216a-3p on GC. Our findings showed that the expression of miR-216a-3p was significantly elevated in GC tissues and cell lines and was firmly correlated with the prognosis and clinicopathological characteristics of GC patients. Additionally, miR-216a-3p positively regulates GC cell proliferation, migration, and invasion, which was consistent with the previous studies15–17, suggesting that miR-216a-3p might be an auspicious marker for GC prognosis and targeted therapy.
Proliferation, migration, and invasion are characteristics of highly malignant cancer cells, which are regulated by proliferation-related genes such as cyclin D1 and Bcl-2, as well as migration- and invasion-related genes such as MMP2 and MMP9. Cyclin D1 is a protein needed for the progression of the cell cycle through the G1 phase, which is encoded by the CCND1 gene32 in humans. It has been previously demonstrated that the continuous presence of cyclin D1 is essentially required to maintain tumor cell proliferation33,34. Additionally, cyclin D1 overexpression can also downregulate Fas expression, resulting in enhanced chemotherapeutic resistance and protection from apoptosis35. Cyclin D1, during the G1 phase of the cell cycle, binds and activates cdk4 and cdk6, two catalytic partners, which catalyze phosphorylation of the retinoblastoma protein, resulting in the release of E2F and the stimulation of proliferation-related E2F target genes36. Also, Bcl-2 family members are regulatory proteins that are of significance in regulating cell apoptosis via the intrinsic pathway. There are two major categories in this family, proapoptotic proteins (like Bid, Bax, and Bad) and antiapoptotic proteins (such as Bcl-2, Bcl-xL, and Bcl-w)37. Bcl-2 includes four conserved BH domains which heterodimerize with Bax, and its overexpression prevents cells from apoptosis caused by different stimuli38. MMP2 and MMP9 are representatives of the MMP gene family, which is involved in the proteolytic processing of structural proteins in the extracellular matrix. A number of studies have reported that both MMP2 and MMP9 promote cancer cell migration and invasion39–41 and are related to the prognosis of cancer patients42,43. Results in this study displayed that the protein levels of cyclinD1, Bcl-2, MMP2, and MMP9 were significantly upregulated in miR-216a-3p mimic-transfected cells and downregulated in miR-216a-3p inhibitor-transfected cells. These data thus provide a molecular mechanism underlying the ability of miR-216a-3p to promote proliferation, migration, and invasion of GC cells, which might also be employed to describe the clinicopathological characteristics of GC patients in Table 1.
The luciferase reporter assay in the present study authenticated that miR-216a-3p could target the regulation of RUNX1, although the participation of miR-mediated posttranscription in the regulation of RUNX1 was investigated, such as miR-9-144, miR-21527, and miR-13945. Additionally, overexpression of RUNX1 significantly reversed the miR-216a-3p-induced promotion of proliferation, migration, and invasion in GC cells, which might be mediated through the inhibitory effect of RUNX1 on the NF-κB signaling pathway. RUNX1, also known as AML1 protein, is a transcription factor that consists of 453 amino acids and modulates the differentiation of hematopoietic stem cells into functional blood cells21. As a transcription factor, RUNX1 collaborates with another protein, CBFβ, composing one version of a complex acknowledged as CBF, which binds to the core section, 5′-PYGPYGGT-3′, of many enhancers and promoters. In fact, the RUNX1 gene is frequently mutated in sporadic myeloid and lymphoid leukemia via point mutation22, translocation23, or amplification24, suggesting its role in the carcinogenesis of blood cancers. Contrarily, RUNX1 was also regarded as a tumor suppressor in other solid cancer types including GC25–27, with lower expression in corresponding tumor tissues.
NF-κB signaling is known to be involved in the regulation of a large number of normal cellular and organismal activities, such as immune and inflammatory responses, cellular growth, developmental processes, and apoptosis. A number of proteins are included in NF-κB signaling, such as the IKK complex, IκBα, p65, and NF-κB, which are phosphorylated upon activation of this signaling pathway, thus resulting in the nuclear translocation of phosphorylated NF-κB to transfer series target genes. Aberrant NF-κB signaling activation has been identified in various human malignancies and plays a key role in cancer development and progression46,47. For instance, several studies have reported that NF-κB signaling promotes cell proliferation by regulating the expression of several genes that participate in the cell cycle process, such as cyclin D1, D2, D3, and cyclin E48,49. In addition, NF-κB signaling-motivated cyclin D1 expression appears to be an essential element in breast carcinogenesis30. NF-κB signaling also promotes the expression of some members of the antiapoptotic Bcl-2 family28,50, which antagonize the activity of proapoptotic members containing Bcl-XL and A1/BFL1, subsequently resulting in the suppression of apoptosis. Li et al.29 reported that NF-κB signaling activation is required for MMP2 and MMP9 gene expression, which participates in cell mobility, suggesting a role that the NF-κB signaling might play in the migration and invasion of cancer cells. Interestingly, it was also reported that NF-κB signaling was suppressed by Wt RUNX1 through interaction with the IκB kinase complex, thus indicating its kinase activity, which adequately blocked the growth and progression of leukemia31. On the contrary, Mut RUNX1 forms fail to inhibit IKK, leading to the anomalous activation of NF-κB signaling in AML cases with RUNX1 anomalies31. In this study, the expression of RUNX1 was decreased compared to that in the noncancerous tissues and correlated significantly with the histologic grade of GC. In addition, the NF-κB signaling of the miR-216a-3p group was significantly activated compared with that of the NC group, while it apparently was inhibited after transfection of RUNX1. All these results confirmed that miR-216a-3p acted on promotive phenotypes in GC cells through RUNX1 and its downstream NF-κB signaling pathway, which may also be used as a mechanism to describe the effect of miR-216a-3p on the upregulation of cyclin D1, Bcl-2, MMP2, and MMP9 as shown in Figure 2.
Our study presents the first in vitro evidence that miR-216a-3p could promote proliferation, migration, and invasion of GC cells through RUNX1 and its downstream NF-κB signaling pathway. However, the potential role of miR-216a-3p in GC tumorigenesis in vivo has not yet been investigated. Thus, more in vivo experiments should be conducted in our future work, which would superinduce more depth to the theory that miR-216a-3p might be critically involved in GC tumorigenesis. Additionally, identification of other targets of miR-216a-3p or other miRNAs that target RUNX1 in GC is also critical.
In summary, our work showed that miR-216a-3p promotes GC cell proliferation, migration, and invasion via targeting RUNX1 and activating the NF-κB signaling pathway. Therefore, miR-216a-3p/RUNX1 could serve as a conceivable treating molecular target for innovative therapeutic agents for GC.
ACKNOWLEDGMENTS
This article was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LY14H160041) and the Zhejiang Medical and Health Science and Technology Plan Project (Nos. 2013KYB007 and 2014PYA001).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60(5):277–300. [DOI] [PubMed] [Google Scholar]
- 2. Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010;127(12):2893–917. [DOI] [PubMed] [Google Scholar]
- 3. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mackenzie M, Spithoff K, Jonker D. Systemic therapy for advanced gastric cancer: A clinical practice guideline. Curr Oncol. 2011;18(4):e202–e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cha YJ, Lee JH, Han HH, Kim BG, Kang S, Choi YD, Cho NH. MicroRNA alteration and putative target genes in high-grade prostatic intraepithelial neoplasia and prostate cancer: STAT3 and ZEB1 are upregulated during prostate carcinogenesis. Prostate 2016;76(10):937–47. [DOI] [PubMed] [Google Scholar]
- 6. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. [DOI] [PubMed] [Google Scholar]
- 7. Braun J, Misiak D, Busch B, Krohn K, Hüttelmaier S. Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification). Nucleic Acids Res. 2014;42(8):e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambros V. The functions of animal microRNAs. Nature 2004;431(7006):350–5. [DOI] [PubMed] [Google Scholar]
- 9. Hoffman Y, Pilpel Y. Gene expression. MicroRNAs silence the noisy genome. Science 2015;348(6230):41–2. [DOI] [PubMed] [Google Scholar]
- 10. Li S, Castillo-Gonzalez C, Yu B, Zhang X. The functions of plant small RNAs in development and in stress responses. Plant J. 2017;90(4):654–70. [DOI] [PubMed] [Google Scholar]
- 11. Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27(34):5848–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cartier F, Indersie E, Lesjean S, Charpentier J, Hooks KB, Ghousein A, Desplat A, Dugot-Senant N, Trézéguet V, Sagliocco F. New tumor suppressor microRNAs target glypican-3 in human liver cancer. Oncotarget 2017;8(25):41211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zheng Q, Chen C, Guan H, Kang W, Yu C. Prognostic role of microRNAs in human gastrointestinal cancer: A systematic review and meta-analysis. Oncotarget 2017;8(28):46611–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li C, Dong J, Han Z, Zhang K. MicroRNA-219-5p represses the proliferation, migration, and invasion of gastric cancer cells by targeting the LRH-1/Wnt/β-catenin signaling pathway. Oncol Res. 2017;25(4):617–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xia H, Ooi LLP, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology 2013;58(2):629–41. [DOI] [PubMed] [Google Scholar]
- 16. Chen PJ, Yeh SH, Liu WH, Lin CC, Huang HC, Chen CL, Chen DS, Chen PJ. Androgen pathway stimulates microRNA-216a transcription to suppress the tumor suppressor in lung cancer-1 gene in early hepatocarcinogenesis. Hepatology 2012;56(2):632–43. [DOI] [PubMed] [Google Scholar]
- 17. Liu H, Pan Y, Han X, Liu J, Li R. MicroRNA-216a promotes the metastasis and epithelial-mesenchymal transition of ovarian cancer by suppressing the PTEN/AKT pathway. Onco Targets Ther. 2017;10:2701–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou B-h, Jian Z-x, Cui P, Li S-j, Tian R-q, Ou J-r. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589(17):2224–32. [DOI] [PubMed] [Google Scholar]
- 19. Wang R-T, Xu M, Xu C-X, Song Z-G, Jin H. Decreased expression of miR216a contributes to non-small-cell lung cancer progression. Clin Cancer Res. 2014;20(17):4705–16. [DOI] [PubMed] [Google Scholar]
- 20. Zhang J, Xu K, Shi L, Zhang L, Zhao Z, Xu H, Liang F, Li H, Zhao Y, Xu X. Overexpression of microRNA-216a suppresses proliferation, migration, and invasion of glioma cells by targeting leucine-rich repeat-containing G protein-coupled receptor 5. Oncol Res. 2017;25(8):1317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ichikawa M, Yoshimi A, Nakagawa M, Nishimoto N, Watanabe-Okochi N, Kurokawa M. A role for RUNX1 in hematopoiesis and myeloid leukemia. Int J Hematol. 2013;97(6):726–34. [DOI] [PubMed] [Google Scholar]
- 22. Osato M. Point mutations in the RUNX1/AML1 gene: Another actor in RUNX leukemia. Oncogene 2004;23(24):4284–96. [DOI] [PubMed] [Google Scholar]
- 23. Lam K, Zhang D-E. RUNX1 and RUNX1-ETO: Roles in hematopoiesis and leukemogenesis. Front Biosci. 2012;17:1120–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moosavi SA, Sanchez J, Adeyinka A. Marker chromosomes are a significant mechanism of high-level RUNX1 gene amplification in hematologic malignancies. Cancer Genet Cytogenet. 2009;189(1):24–8. [DOI] [PubMed] [Google Scholar]
- 25. Supernat A, Łapińska-Szumczyk S, Sawicki S, Wydra D, Biernat W, Żaczek AJ. Deregulation of RAD21 and RUNX1 expression in endometrial cancer. Oncol Lett. 2012;4(4):727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. [DOI] [PubMed] [Google Scholar]
- 27. Li N, Zhang Q-Y, Zou J-L, Li Z-W, Tian T-T, Dong B, Liu X-J, Ge S, Zhu Y, Gao J. miR-215 promotes malignant progression of gastric cancer by targeting RUNX1. Oncotarget 2016;7(4):4817–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Catz SD, Johnson JL. Transcriptional regulation of bcl-2 by nuclear factor [kappa] B and its significance in prostate cancer. Oncogene 2001;20(50):7342–51. [DOI] [PubMed] [Google Scholar]
- 29. Li J, Lau GK-K, Chen L, Dong S-s, Lan H-Y, Huang X-R, Li Y, Luk JM, Yuan Y-F, Guan X-y. Interleukin 17A promotes hepatocellular carcinoma metastasis via NF-kB induced matrix metalloproteinases 2 and 9 expression. PLoS One 2011;6(7):e21816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hassanzadeh P. Colorectal cancer and NF-κB signaling pathway. Gastroenterol Hepatol Bed Bench 2011;4(3):127–32. [PMC free article] [PubMed] [Google Scholar]
- 31. Nakagawa M, Shimabe M, Watanabe-Okochi N, Arai S, Yoshimi A, Shinohara A, Nishimoto N, Kataoka K, Sato T, Kumano K. AML1/RUNX1 functions as a cytoplasmic attenuator of NF-κB signaling in the repression of myeloid tumors. Blood 2011;118(25):6626–37. [DOI] [PubMed] [Google Scholar]
- 32. Motokura T, Bloom T. A novel cyclin encoded by a bc/1-linked candidate oncogene. Nature 1991;350(6318):512–5. [DOI] [PubMed] [Google Scholar]
- 33. Yu Q, Sicinska E, Geng Y, Ahnström M, Zagozdzon A, Kong Y, Gardner H, Kiyokawa H, Harris LN, Stål O. Requirement for CDK4 kinase function in breast cancer. Cancer Cell 2006;9(1):23–32. [DOI] [PubMed] [Google Scholar]
- 34. Karim S, Al-Maghrabi JA, Farsi HM, Al-Sayyad AJ, Schulten H-J, Buhmeida A, Mirza Z, Al-boogmi AA, Ashgan FT, Shabaad MM. Cyclin D1 as a therapeutic target of renal cell carcinoma—A combined transcriptomics, tissue microarray and molecular docking study from the Kingdom of Saudi Arabia. BMC Cancer 2016;16(Suppl 2):741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shintani M, Okazaki A, Masuda T, Kawada M, Ishizuka M, Doki Y, Weinstein IB, Imoto M. Overexpression of cyclin DI contributes to malignant properties of esophageal tumor cells by increasing VEGF production and decreasing Fas expression. Anticancer Res. 2001;22(2A):639–47. [PubMed] [Google Scholar]
- 36. Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5(3):a008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cory S, Adams JM. The Bcl2 family: Regulators of the cellular life-or-death switch. Nat Rev Cancer 2002;2(9):647–56. [DOI] [PubMed] [Google Scholar]
- 38. Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993;74(4):609–19. [DOI] [PubMed] [Google Scholar]
- 39. Wang X, Tong X, Gao H, Yan X, Xu X, Sun S, Wang Q, Wang J. Silencing HIWI suppresses the growth, invasion and migration of glioma cells. Int J Oncol. 2014;45(6):2385–92. [DOI] [PubMed] [Google Scholar]
- 40. Theodore LN, Hagedorn EJ, Cortes M, Natsuhara K, Liu SY, Perlin JR, Yang S, Daily ML, Zon LI, North TE. Distinct roles for matrix metalloproteinases 2 and 9 in embryonic hematopoietic stem cell emergence, migration, and niche colonization. Stem Cell Reports 2017;8(5):1226–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao Y, Zhou F-L, Li WP, Wang J, Wang LJ. Slit2-Robo1 signaling promotes the adhesion, invasion and migration of tongue carcinoma cells via upregulating matrix metalloproteinases 2 and 9, and downregulating E-cadherin. Mol Med Rep. 2016;14(3):1901–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Daniele A, Zito A, Giannelli G, Divella R, Asselti M, Mazzocca A, Paradiso A, Quaranta M. Expression of metalloproteinases MMP-2 and MMP-9 in sentinel lymph node and serum of patients with metastatic and non-metastatic breast cancer. Anticancer Res. 2010;30(9):3521–7. [PubMed] [Google Scholar]
- 43. Fan H-X, Li H-X, Chen D, Gao Z-X, Zheng J-H. Changes in the expression of MMP2, MMP9, and ColIV in stromal cells in oral squamous tongue cell carcinoma: Relationships and prognostic implications. J Exp Clin Cancer Res. 2012;31:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fu L, Shi J, Liu A, Zhou L, Jiang M, Fu H, Xu K, Li D, Deng A, Zhang Q. A minicircuitry of microRNA-9-1 and RUNX1-RUNX1T1 contributes to leukemogenesis in t(8;21) acute myeloid leukemia. Int J Cancer 2017;140(3):653–61. [DOI] [PubMed] [Google Scholar]
- 45. Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H, Xi Z, Li Z, Liu Y. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. 2016; 24(10):1806–22. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46. Karin M, Cao Y, Greten FR, Li Z-W. NF-κB in cancer: From innocent bystander to major culprit. Nat Rev Cancer 2002;2(4):301–10. [DOI] [PubMed] [Google Scholar]
- 47. Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-κB: A blossoming of relevance to human pathobiology. Cell 2017;168(1):37–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS. NF-κB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol Cell Biol. 1999;19(8):5785–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rubio MF, Fernandez PNL, Alvarado CV, Panelo L, Grecco MR, Colo GP, Martínez-Noel GA, Micenmacher SM, Costas MA. Cyclin D1 is a NF-κB corepressor. Biochim Biophys Acta 2012;1823(6):1119–31. [DOI] [PubMed] [Google Scholar]
- 50. Chen C, Edelstein LC, Gélinas C. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol Cell Biol. 2000;20(8):2687–95. [DOI] [PMC free article] [PubMed] [Google Scholar]