Abstract
Recent studies have demonstrated that miR-202 is associated with several types of cancer; however, the expression and function of miR-202 have not been investigated in bladder cancer. We analyzed the expression of miR-202 in bladder cancer tissues and adjacent noncancerous tissues. The effect of miR-202 on the proliferation, migration, and invasion was evaluated by in vitro assays. The target gene of miR-202 was assessed by luciferase reporter assay. In this study, miR-202 was found to be significantly downregulated in bladder cancer cell lines and tissues and was highly correlated with the T classification, N classification, grade, and recurrence. Ectopic expression of miR-202 suppressed cell viability, colony formation, cell migration, and invasion in vitro and inhibited xenograft tumor growth in vivo. Inversely, downregulation of miR-202 had contradictory effects. The 3′-untranslated region (3′-UTR) of epidermal growth factor receptor (EGFR) was identified as a direct target of miR-202 using luciferase reporter assays, and knockdown of EGFR enhanced miR-202-inhibited cell proliferation, migration, and invasion. In conclusion, miR-202 suppresses bladder cancer carcinogenesis and progression by targeting EGFR, thereby representing a potential target for miRNA-based therapy for bladder cancer in the future.
Key words: miR-202, Epidermal growth factor receptor (EGFR), Bladder cancer
INTRODUCTION
Bladder cancer is one of the most frequent malignancies in the world. In spite of the great advances in the diagnosis and therapy in recent decades, patients with bladder cancer still have a poor 5-year survival rate, which is less than 60%1,2. Bladder cancer is responsible for the highest cost of therapy among all the malignancies, so it is essential to improve early diagnosis and prediction3,4. In addition, it is imperative to investigate the molecular mechanism and potential targets that regulate bladder cancer aggressiveness and chemoresistance3–5.
MicroRNAs (miRNAs) are a type of small noncoding RNAs that are 19–25 nucleotides in length. It has been reported that miRNAs control a wide range of biological and pathological processes by base pairing with the 3′-untranslated region (3′-UTR) of their target mRNA transcripts6–9. In addition, the expression of miRNAs is dysregulated in some cancer types and might serve as either tumor suppressors or oncogenes10–13. To date, several miRNAs have been demonstrated to affect cell growth, proliferation, and metastasis, such as miR-136, miR-23a, and miR-99a14,15. Notably, miR-202 also has an important role in tumorigenesis. Furthermore, understanding the effects and molecular mechanisms of miRNAs might contribute to the elucidation of the biological progression of bladder cancer. However, the specific function and regulatory mechanism of miR-202 in bladder cancer progression are rarely investigated.
In the present study, we assumed that the miR-202/epidermal growth factor receptor (EGFR) pathway may promote the invasion and metastasis of bladder cancer cells. We analyzed the expression levels of miR-202 in bladder cancer tissues in different clinical stages, in normal tissues, as well as in several bladder cancer cell lines to explore the potential role of miR-202 in bladder cancer progression.
MATERIALS AND METHODS
Patient Tissue Specimens
A total of 50 cases of resected specimens from bladder cancer patients were collected for this study. Bladder cancer specimens were compared with the paired normal bladder tissue (NT) from the same patient. All specimens had been histologically and clinically diagnosed at the General Hospital of Jinan Military Command from 2013 to 2016, independently by two experienced pathologists. For the use of these clinical materials for research purposes, prior patient consent was obtained, and the study was approved by the Institutional Research Ethics Committee.
Cell Culture
The human bladder cancer cell lines (EJ, T24, and BIU-87) were purchased from China Academia Sinica Cell Repository (Shanghai, P.R. China) and cultured in DMEM (Invitrogen, Carlsbad, CA, USA) or RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal calf serum at 37°C in a 5% CO2 incubator. Human urothelial SV-HUC-1 cells were purchased from the Institute of Cell Research, Chinese Academy of Sciences, Shanghai, P.R. China. The SV-HUC-1 cells were cultured in F-12K (Corning, Manassas, VA, USA) plus 10% fetal bovine serum (FBS). The other cells were cultured in DMEM plus 10% FBS. The medium was replaced every 3 days.
Real-Time PCR
The cells or spheroids were harvested after transfection, and RNA was isolated using TRI Reagent® (Sigma-Aldrich, St. Louis, MO, USA). Ten nanograms of RNA was used for reverse transcription using the TaqMan MicroRNA RT Kit (Applied Biosystems, Life Technologies, Madison, WI, USA). Briefly, 5 μl of the RNA was added to 10 μl of the master mix containing 0.15 μl of dNTP (100 nM), 1 μl of multiscribe enzyme (50 U/μl), 1.5 μl of 10× RT buffer, 0.19 μl of RNAse inhibitor (20 U/μl), 4.16 μl of RNAse-free H2O, and 3 μl of primers (miR-202). The conditions for the reverse transcription were 30 min at 16°C, 30 min at 42°C, and 5 min at 85°C. Quantitative real-time (RT)-PCR of the individual miRNAs was performed in a total volume of 10 μl containing 5 μl of TaqMan master mix, 3.17 μl of RNase-free H2O, 0.5 μl of TaqMan primer, and 1.33 μl of cDNA. The PCR was performed in quadruplicate in a RotorGene Corbett 6000 QPCR. The conditions for the reaction were 2 min at 50°C, 10 min at 95°C, cycling (50 repeats): step 1, 15 s at 95°C; step 2, 60 s at 60°C. The data were collected and analyzed using the quantitative RotorGene software. The miRNA levels were normalized to the stable internal control miRNA RNU6.
Transfection of siRNA and miRNA
Cells (1.5 × 105) were seeded per well in a 24-well plate; 5 nM siRNA and 50 nM miRNA inhibitor, mimics, and their respective controls were diluted into 100 μl of serum-free medium. Subsequently, 3 μl of HiPerFect Transfection Reagent (Qiagen, Valencia, CA, USA) was added to the mixture and incubated at room temperature for 10 min. The transfection complexes formed were then added dropwise onto cells and incubated under normal growth conditions. Cells were harvested 48 h after the transfections.
Western Blot
The protein was extracted from the specimens, or from the cultured cells, in RIPA lysis buffer (1% NP40, 0.1% SDS, 100 μg/ml phenylmethylsulfonyl fluoride, 0.5% sodium deoxycholate, in PBS) on ice. The supernatants were collected after centrifugation at 12,000 × g at 4°C for 20 min. Protein concentration was determined using a BCA protein assay kit (Bio-Rad, P.R. China), and whole lysates were mixed with 4× SDS loading buffer (125 mmol/L Tris-HCl, 4% SDS, 20% glycerol, 100 mmol/L DTT, and 0.2% bromophenol blue) at a ratio of 1:3. Samples were heated at 100°C for 5 min and were separated on SDS-polyacrylamide gels. The separated proteins were then transferred to a PVDF membrane. The membrane blots were first probed with a primary antibody. After incubation with horseradish peroxidase-conjugated second antibody, autoradiograms were prepared using the enhanced chemiluminescence system to visualize the protein antigen. The signals were recorded using X-ray film. Blotting images were representatives from five repeats.
Cell Proliferation and Colony Formation Assays
Cell proliferation was monitored using Cell Counting Kit-8 (CCK-8; Sigma-Aldrich). Cells (3,000 cells/well, 5 wells/group) were allowed to grow in 96-well plates. Cell proliferation was documented every 24 h following the manufacturer’s protocol. CCK-8 reagent was added to each well at 1.5 h before the endpoint of incubation. The optical density (OD) 450-nm values were determined by a microplate reader. All experiments were repeated at least three times. For the colony formation assay, cells (100/well) were allowed to grow in a culture dish (8 cm2) and maintained in media containing 10% FBS, replacing the medium every 4 days. After 14 days, cells were fixed with methanol and stained with 10% Giemsa (Solarbio, Beijing, P.R. China). Only positive colonies (diameter: >40 um) in the dishes were counted and compared. All experiments were performed in triplicate.
Wound Healing Assay
Cells were seeded in six-well culture plates and grown to confluence forming a monolayer covering the surface of the entire well. After cells were serum starved in serum-free RPMI for 18 h, the wound was created in the center of the cell monolayer by the gentle removal of the attached cells with a 10-μl pipette tip. Debris was removed by PBS wash, and the cells received fresh RPMI with 10% FBS and 10 mmol/L of hydroxyurea. Cells were photographed with a Zeiss Axiovert 200M inverted microscope at the intervals of 0, 24, and 72 h.
Transwell Migration Assay
A suspension of 1 × 105 stably transduced cells in 0.5% FBS medium was added to BD cell culture inserts with 8-μm porous membrane coated with 20% FBS (BD Biosciences, Bedford, MA, USA). Cells were incubated for 2–5 days at 37°C in a 5% CO2 incubator. To quantify migrating cells, cells remaining on the top side of the membrane were removed using a cotton-tipped swab, and cells that had migrated to the underside were visualized with a Zeiss Axiovert 200M inverted microscope and photographed.
Luciferase Assay
Fragments of the EGFR mRNA 3′-UTRs containing the putative or mutated miRNA binding sites for miR-202 were cloned into the GV306 luciferase reporter vector (GeneChem, P.R. China). The constructs were then cotransfected with miR-202 mimic or negative control oligos into HEK293 T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Luciferase activity was measured 48 h after transfection using the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA) according to the manufacturer’s protocol.
In Vivo Tumorigenesis Model
An in vivo model was established by subcutaneously injecting 5-week-old female BALB/c nude mice with 1 × 107 cells transfected with miR-202 (or mock transfected) suspended in PBS (10 mice per group). The weight of the mice and tumor volumes were determined every 3 days; tumor volume was assessed by measuring the length (L) and width (W) using calipers (tumor volume, mm3 = 0.5 × L × W 2). After 8 weeks, the mice were euthanized, and the tumors were excised, measured, and photographed. All mice were obtained from Shanghai SLAC Laboratory Animal, Co., Ltd. (Shanghai, P.R. China) and housed in a specific pathogen-free environment. This experiment was in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of the General Hospital of Jinan Military Command.
Statistical Analyses
The results were represented as mean ± SEM of independent experiments. The p value was calculated according to Student’s t-test when comparing two groups using the GraphPad p value calculator. Multiple groups were compared by one-way ANOVA, when necessary, followed by pairwise comparisons with post hoc test. The differences were considered significant with a value of p < 0.05.
RESULTS
The Expression of miR-202 and Clinicopathologic Characteristics
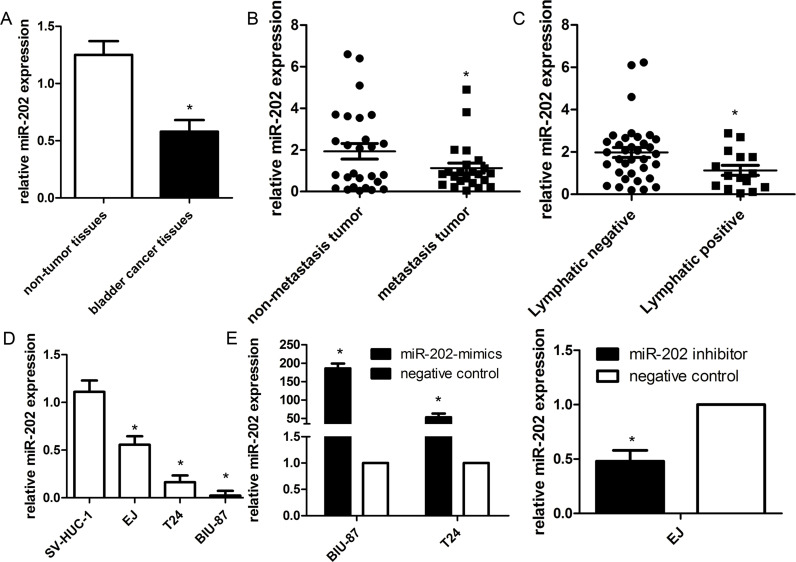
To investigate the expression of miR-202 in bladder cancer tissues, we evaluated the expression of miR-202 in 50 pairs of frozen bladder cancer tissues and the corresponding normal tissues by qRT-PCR. We found that miR-202 expression was downregulated in cancer tissues compared with the matching normal tissues (Fig. 1A). The clinical characteristics of bladder cancer patients included 27 cases of non-muscle-invasive and 23 cases of muscle-invasive bladder cancer. A strong correlation occurred between miR-202 expression and clinicopathologic characteristics of these patients, including T classification, N classification, recurrence, and mortality (all p < 0.001) (Fig. 1B and C). By contrast, miR-202 expression did not correlate with gender, age, grade, and surgery type (all p > 0.10). We also evaluated miR-202 expression in three bladder cancer cell lines (EJ, T24, and BIU-87) and urothelial SV-HUC-1 cells. The mean expression levels for miR-202 in the three bladder cancer cell lines were 0.023, 0.164, and 0.556, respectively, compared with that (1.125) of SV-HUC-1 cells (Fig. 1D). Notably, BIU-87 cells (highest aggressiveness in the three cell lines) expressed the lowest miR-202 level, indicating miR-202 was associated with bladder cancer metastasis.
Figure 1.
miR-202 is downregulated in bladder cancer and associated with clinical data. (A) Expression of mature miR-202 was determined by way of quantitative real-time (qRT)-PCR in 50 paired human bladder cancer and their corresponding nontumorous samples and normalized against an endogenous U6 RNA control. Low-level expression of miR-202 was associated with T classification (B) and N classification (C) of bladder cancer. (D) miR-202 expression was detected in bladder cancer cell lines and SV-HUC-1. (E) The level of miR-202 was significantly changed in bladder cancer cells after being infected with miR-202 lentivirus or miR-202 inhibitor. Data are represented as the mean ± SEM of three independent experiments. *p < 0.01.
miR-202 Inhibits Cell Proliferation, Migration, and Invasion In Vitro
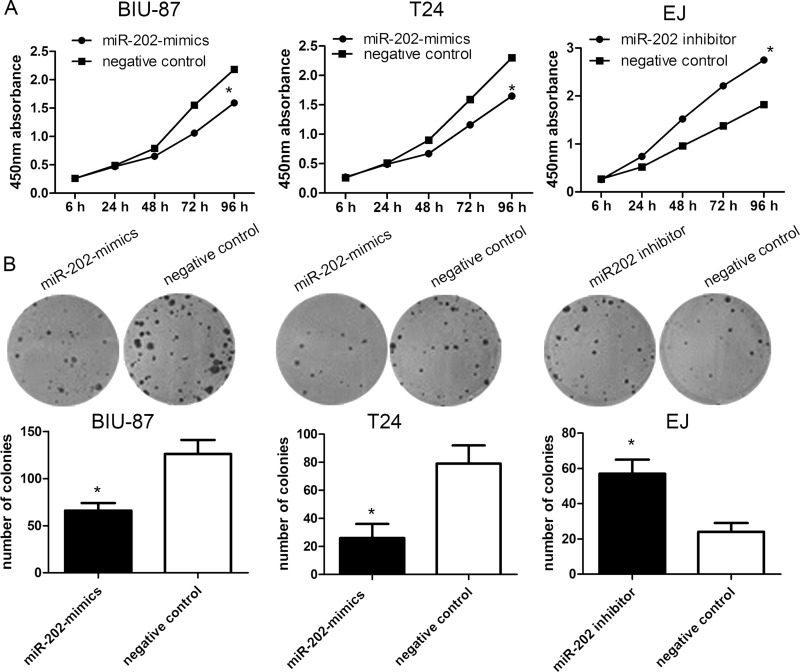
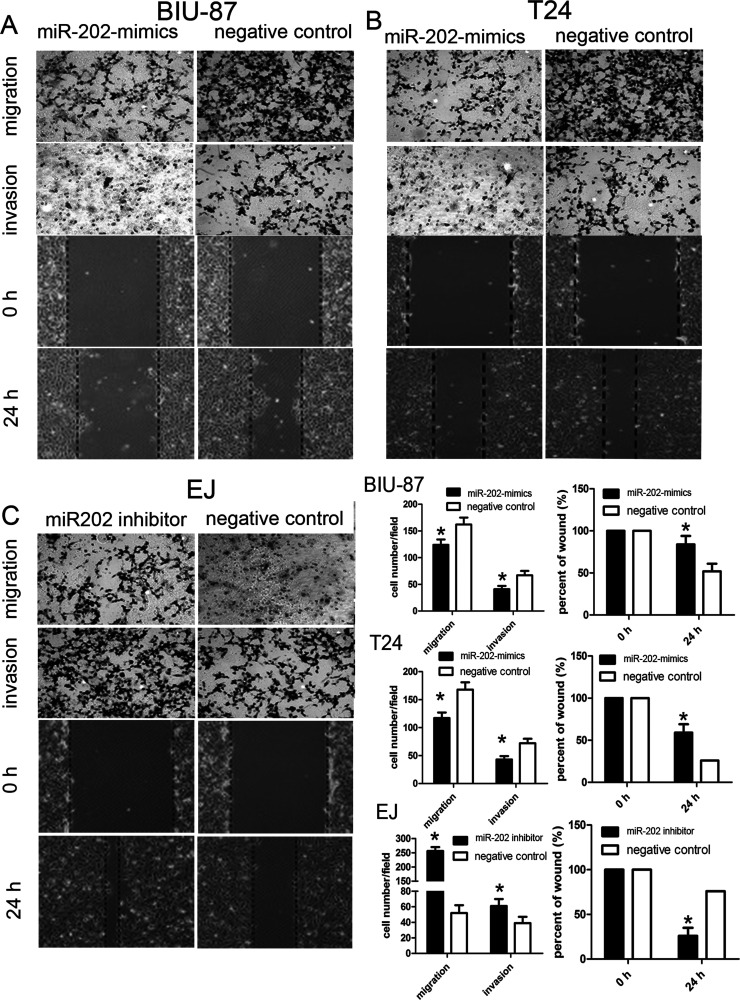
To functionally characterize miR-202 in the development of bladder cancer, we promoted the expression of miR-202 by ectopic expression of this miRNA in the relatively lowest expressing bladder cancer cell lines BIU-87 and T24, and downregulated the expression of miR-202 in the highest expressing EJ cell line. So BIU-87 miR-202 mimic and T24 miR-202 mimic cells, and EJ miR-202 inhibitor cells were generated (Fig. 1E). We then conducted CCK-8 and colony formation assays to evaluate the role of miR-202 in cell proliferation of bladder cancer. Compared with the control group, forced expression of miR-202 impaired cell proliferation in both BIU-87 miR-202 mimics and T24 miR-202 mimics (Fig. 2A and B). Inversely, the cell proliferation of EJ miR-202 inhibitor cells was significantly enhanced. Subsequently, we evaluated the role of miR-202 in the cell migration and invasion of bladder cancer. The wound healing assay revealed that wound closure was obviously impaired in both BIU-87 miR-202 mimics and T24 miR-202 mimics. Furthermore, Transwell assays showed that the migratory and invasive abilities were significantly decreased (Fig. 3A and B), whereas reduced miR-202 expression in EJ miR-202 inhibitor cells resulted in the opposite effects (Fig. 3C). These findings suggested that miR-202 inhibits bladder cancer cell proliferation and metastasis in vitro.
Figure 2.
The expression profile of miR-202 in bladder cell lines. (A) Cells were transfected with miR-202 mimics and identified by RT-PCR. Cell proliferation was measured using a cell counting kit-8 (CCK-8) assay. (B) miR-202 inhibited bladder cancer cell proliferation determined using colony formation. Data are represented as the mean ± SEM of three independent experiments. *p < 0.05.
Figure 3.
miR-202 inhibited bladder cancer cell migration and invasion in vitro. (A) Upregulated miR-202 suppressed migration and invasion in vitro in BIU-87 cells. (B) Upregulated miR-202 suppressed migration and invasion in vitro in T24 cells. (C) Downregulated miR-202 enhanced migration and invasion in vitro in EJ cells compared with controls. Data on the right side are represented as the mean ± SEM of three independent experiments. *p < 0.01.
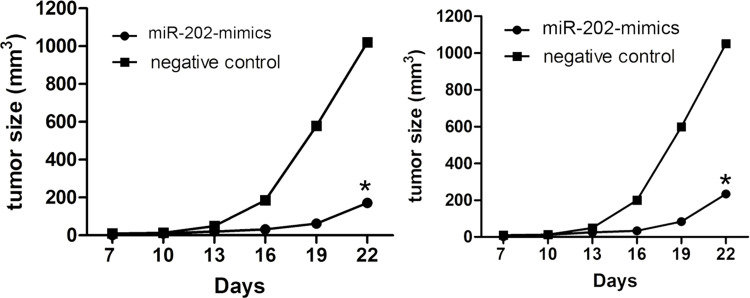
Overexpression of miR-202 Inhibits the Growth of Bladder Cancer Cells In Vivo
In view of the in vitro assays, we tested whether miR-202 could affect tumorigenicity and metastasis in vivo. BIU-87 and T24 cells stably expressing miR-202 and negative control vector were injected subcutaneously into nude mice, and the mice were sacrificed 4 weeks after tumor cell implantation. We found that the average tumor volume of T24 cells transfected with miR-202 mimics was significantly smaller than that in the control group. The tumor growth curve was made according to time, and a significant difference was shown between the two groups (Fig. 4).
Figure 4.
Overexpression of miR-202 inhibits bladder cancer in vivo. The mouse subcutaneous implantation model in mice was constructed using BIU-87 and T24 cells infected with negative control and miR-202 mimics. Tumor growth curves of subcutaneous implantation models of bladder cancer. Data are represented as the mean ± SEM of three independent experiments. *p < 0.01.
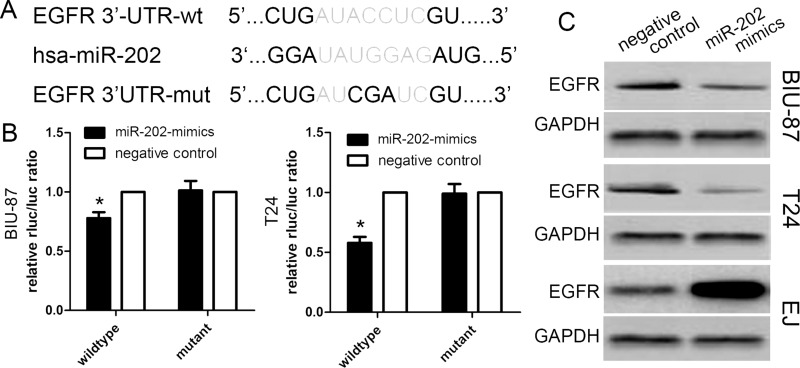
EGFR Is a Direct Downstream Target of miR-202
In this study, we determined potential target genes of miR-202 using available databases. Of various candidates, EGFR showed the most complementary sequences with miR-202 (Fig. 5A). The overexpression of the EGFR protein has been identified in various types of cancers, and high EGFR expression is correlated with aggressive phenotypes of malignancies. We carried out a luciferase-based assay to validate whether the EGFR gene was regulated by miR-202. Luciferase vectors containing the 3′-UTR of EGFR were generated and transfected along with or without the miR-202 mimics into three bladder cancer cells. The luciferase activity assay revealed that miR-202 expression markedly reduced the activity of the wild-type EGFR 3′-UTR of BIU-87 and T24 cells. However, miR-202 expression did not reduce the activity of the mutant EGFR 3′-UTR of BIU-87 cells and T24 cells (Fig. 5B). Based on the protein level, Western blot analysis showed that EGFR protein expression was obviously decreased in BIU-87 cells and T24 cells transfected with miR-202 mimics, but increased in EJ cells transfected with the miR-202 inhibitor (Fig. 5C).
Figure 5.
Epidermal growth factor receptor (EGFR) is a candidate target of miR-202. (A) Schematic of the construction of wild-type or mutant EGFR 3′-untranslated region (3′-UTR) vectors. (B) Relative luciferase activity was analyzed in BIU-87 and T24 cells. Firefly luciferase vector was used as an internal control. (C) Western blot results of EGFR protein in BIU-87, T24, and EJ cells infected with miR-202 mimics lentivirus or miR-202 inhibitor. Data are represented as the mean ± SEM of three independent experiments. *p < 0.01.
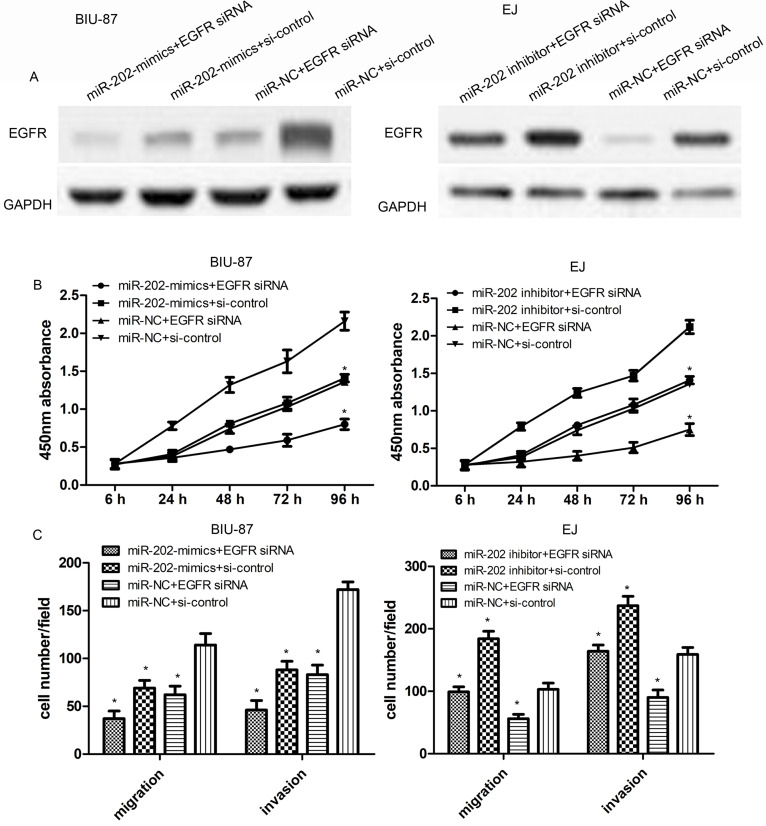
Knockdown of EGFR Inhibits Cell Proliferation, Migration, and Invasion
To further validate the effect of EGFR knockdown on cell proliferation, migration, and invasion, EGFR siRNA or control siRNA was transfected into miR-202-overexpressed BIU-87 cells or miR-202-inhibited EJ cells. Then the protein levels of EGFR were determined using Western blot assays. As shown in Figure 6A, EGFR siRNA significantly reduced miR-202-suppressed EGFR protein level in BIU-87 and EJ cells. Then miR-202 mimics or EGFR siRNA significantly reduced cell proliferation, compared with the negative control (p < 0.01) (Fig. 6B). Results of migration and invasion assays showed that the cotransfection of EGFR siRNA and miR-202 mimics strongly inhibited the migration and invasion of BIU-87 cells compared with the three other groups. On the other hand, EGFR siRNA also reduced the effect of the miR-202 inhibitor in EJ cells (both p < 0.01) (Fig. 6C).
Figure 6.
Knockdown of EGFR inhibits cell proliferation, migration, and invasion. (A) The expression level of EGFR protein in BIU-87 cells with miR-202 mimics and/or EGFR siRNA or EJ cells with miR-202 inhibitor and/or EGFR siRNA was confirmed by Western blotting. (B) The proliferation of miR-202-overexpressing BIU-87 cells with EGFR siRNA or miR-202 inhibitor EJ cells with EGFR siRNA was measured by CCK-8 assay. (C) The migration or invasion capacity of miR-202-overexpressing BIU-87 cells with EGFR siRNA or miR-202 inhibitor EJ cells with EGFR siRNA was measured by Transwell assays. The data are represented as the mean ± SEM of three independent experiments. *p < 0. 01.
DISCUSSION
Recently, accumulating evidence has suggested that miRNAs play important roles in regulating diverse cellular processes, including proliferation, apoptosis, migration, and invasion16–19. Zhao et al. indicated that miR-202 functions as a tumor suppressor in non-small cell lung cancer by targeting STAT320. However, to date, the role of miR-202 in the cell biology of human bladder cancer has remained unclear. In the present study, we demonstrated that miR-202 was significantly downregulated in three bladder cancer cell lines and tissue compared with normal cells and tissues. We further analyzed the correlation of miR-202 expression with clinical characteristics. A marked correlation between low miR-202 expression with the T classification, N classification, grade, and recurrence was confirmed in 50 patients with bladder cancer. These findings suggested that miR-202 plays a crucial role in the initiation and development of bladder cancer. Moreover, Jiang et al. showed that the expression of miR-202 is aberrantly downregulated in lung cancer and serves as a tumor-suppressing gene via targeting cyclin D121. Meng et al. reported that the expression of miR-202 is constantly decreased in ESCC tissues and cell lines22. Together with these findings, we assumed that miR-202 acts as a tumor suppressor in bladder cancer and may be used as a prognostic biomarker.
miRNAs may exert diverse biological functions by regulating the expression of downstream target genes in different tissues or cancers. miRNAs can negatively regulate many target genes through binding their 3′-UTR in order to induce cleavage of the message or inhibit translation. In our study, prediction of the candidate target genes for miR-202 was performed by bioinformatics tools (Targetscan, PicTar, and MiRanda). Then EGFR, one of putative target genes, was verified by luciferase reporter assay and Western blot assay. The ErbB family contains EGFR (also known as ErbB1/HER-1), ErbB2/Neu/HER-2, ErbB3/HER-3, and ErbB4/HER-4. EGFR is a kind of receptor tyrosine kinase located in the cell membrane, which activates homo- or heterodimerization and receptor activation, subsequently inducing downstream signaling pathways (the extracellular signal-regulated kinase and phosphatidylinositol 3-kinase/protein kinase B pathways). These pathways further modulate the proliferation, apoptosis, migration, and angiogenesis23,24. EGFR overexpression has been identified in many types of cancers, including head and neck cancer, pancreatic cancer, and colorectal cancer25–27. Importantly, EGFR is highly overexpressed in most bladder cancer patients, and EGFR expression plays a pivotal role in the prognosis or survival of bladder cancer patients28. In this work, miR-202 could directly bind the 3′-UTR of EGFR, and overexpression of miR-202 downregulated EGFR protein expression, suggesting a strong negative regulation between miR-202 and EGFR in bladder cancer.
In conclusion, the present study showed that miR-202 is downregulated in bladder cancer cells and tissues, and its inhibitory effects on cell proliferation, migration, invasion, and tumor growth are mediated by downregulating EGFR. The present results indicate that miR-202 could be a useful marker and potential therapeutic target in bladder cancer.
ACKNOWLEDGMENT
The authors are thankful to other members in their lab for their suggestions.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Manikandan R, Rodriguez O, Parada R, Palou Redorta J. Nonmuscle-invasive bladder cancer: What’s changing and what has changed. Urologia. 2017;84:1–8. [DOI] [PubMed] [Google Scholar]
- 2. Olkhov-Mitsel E, Savio AJ, Kron KJ, Pethe VV, Hermanns T, Fleshner NE, van Rhijn BW, van der Kwast TH, Zlotta AR, Bapat B. Epigenome-wide DNA methylation profiling identifies differential methylation biomarkers in high-grade bladder cancer. Transl Oncol. 2017;10:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu B, Qi L, Liu S, Liu W, Ou Z, Chen M, Liu L, Zu X, Wang J, Li Y. CLASP2 is involved in the EMT and early progression after transurethral resection of the bladder tumor. BMC Cancer. 2017;17:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kang M, Lee KH, Lee HS, Jeong CW, Kwak C, Kim HH, Ku JH. Concurrent autophagy inhibition overcomes the resistance of epidermal growth factor receptor tyrosine kinase inhibitors in human bladder cancer cells. Int J Mol Sci. 2017;18(2)pii: E321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cho YM, Hasumura M, Imai T, Takami S, Nishikawa A, Ogawa K. Horseradish extract promotes urinary bladder carcinogenesis when administered to F344 rats in drinking water. J Appl Toxicol. 2017;37:853–62. [DOI] [PubMed] [Google Scholar]
- 6. He B, Yin B, Wang B, Xia Z, Chen C, Tang J. MicroRNAs in esophageal cancer (review). Mol Med Rep. 2012;6:459–65. [DOI] [PubMed] [Google Scholar]
- 7. Feifei N, Mingzhi Z, Yanyun Z, Huanle Z, Fang R, Mingzhu H, Mingzhi C, Yafei S, Fengchun Z. MicroRNA expression analysis of mammospheres cultured from human breast cancers. J Cancer Res Clin Oncol. 2012;138:1937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vira D, Basak SK, Veena MS, Wang MB, Batra RK, Srivatsan ES. Cancer stem cells, microRNAs, and therapeutic strategies including natural products. Cancer Metastasis Rev. 2012;31(3–4):733–51. [DOI] [PubMed] [Google Scholar]
- 9. Ling H, Zhang W, Calin GA. Principles of microRNA involvement in human cancers. Chin J Cancer 2011;30:739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sempere LF. Integrating contextual miRNA and protein signatures for diagnostic and treatment decisions in cancer. Expert Rev Mol Diagn. 2011;11:813–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim K, Chadalapaka G, Pathi SS, Jin UH, Lee JS, Park YY, Cho SG, Chintharlapalli S, Safe S. Induction of the transcriptional repressor ZBTB4 in prostate cancer cells by drug-induced targeting of microRNA-17-92/106b-25 clusters. Mol Cancer Ther. 2012;11:1852–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu J, Zheng M, Tang YL, Liang XH, Yang Q. MicroRNAs, an active and versatile group in cancers. Int J Oral Sci. 2011;3:165–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu S, Howell PM, Riker AI. Up-regulation of miR-182 expression after epigenetic modulation of human melanoma cells. Ann Surg Oncol. 2013;20:1745–52. [DOI] [PubMed] [Google Scholar]
- 14. Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, Noro R, Yoshimura A, Cai L, Gemma A. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int J Oncol. 2012;41:869–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yan B, Fu Q, Lai L, Tao X, Fei Y, Shen J, Chen Z, Wang Q. Downregulation of microRNA 99a in oral squamous cell carcinomas contributes to the growth and survival of oral cancer cells. Mol Med Rep. 2012;6:675–81. [DOI] [PubMed] [Google Scholar]
- 16. Yin S, Bleul T, Zhu Y, Isayev O, Werner J, Bazhin AV. MiRNAs are unlikely to be involved in retinoid receptor gene regulation in pancreatic cancer cells. Cell Physiol Biochem. 2017;44:644–56. [DOI] [PubMed] [Google Scholar]
- 17. Guo J, Xiao Z, Yu X, Cao R. miR-20b promotes cellular proliferation and migration by directly regulating phosphatase and tensin homolog in prostate cancer. Oncol Lett. 2017;14:6895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. López-Aguilar JE, Velázquez-Flores MA, Simón-Martínez LA, Ávila-Miranda R, Rodríguez-Florido MA, Ruiz-Esparza Garrido R. Circulating microRNAs as biomarkers for pediatric astrocytomas. Arch Med Res. 2017;48:323–32. [DOI] [PubMed] [Google Scholar]
- 19. Xiong Y, Wang R, Peng L, You W, Wei J, Zhang S, Wu X, Guo J, Xu J, Lv Z, Fu Z. An integrated lncRNA, microRNA and mRNA signature to improve prognosis prediction of colorectal cancer. Oncotarget 2017;8:85463–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao Z, Lv B, Zhang L, Zhao N, Lv Y. miR-202 functions as a tumor suppressor in non-small cell lung cancer by targeting STAT3. Mol Med Rep. 2017;16:2281–9. [DOI] [PubMed] [Google Scholar]
- 21. Jiang J, Huang J, Wang XR, Quan YH. MicroRNA-202 induces cell cycle arrest and apoptosis in lung cancer cells through targeting cyclin D1. Eur Rev Med Pharmacol Sci. 2016;20:2278–84. [PubMed] [Google Scholar]
- 22. Meng X, Chen X, Lu P, Ma W, Yue D, Song L, Fan Q. miR-202 promotes cell apoptosis in esophageal squamous cell carcinoma by targeting HSF2. Oncol Res. 2017;25:215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Citri A, Yarden Y. EGF-ERBB signalling: Towards the systems level. Nat Rev Mol Cell Biol. 2006;7:505–16. [DOI] [PubMed] [Google Scholar]
- 24. Wells A. EGF receptor. Int J Biochem Cell Biol. 1999;31:637–43. [DOI] [PubMed] [Google Scholar]
- 25. Salomon DS, Brandt R, Ciardiello F, Normanno N. Epidermal growth factor-related peptides and their receptors in human malignancies. Crit Rev Oncol Hematol. 1995;19:183–232. [DOI] [PubMed] [Google Scholar]
- 26. Nicholson RI, Gee JMW, Harper ME. EGFR and cancer prognosis. Eur J Cancer 2001;37:9–15. [DOI] [PubMed] [Google Scholar]
- 27. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–81. [DOI] [PubMed] [Google Scholar]
- 28. Arfaoui AT, Mejri S, Belhaj R, Karkni W, Chebil M, Rammeh S. Prognostic value of immunohistochemical expression profile of epidermal growth factor receptor in urothelial bladder cancer. J Immunoassay Immunochem. 2016;37:359–67. [DOI] [PubMed] [Google Scholar]