Abstract
MicroRNAs (miRNAs) play a vital role in regulating tumor progression. Dysregulated miR-136 expression was linked to the development of various human cancers. In the present study, we investigated the expression and relationship of miR-136 and COX2 in hepatocellular carcinoma (HCC) using relevant experiments, involving CCK-8, Transwell assay, and luciferase reporter assay. We demonstrated that miR-136 expression is obviously decreased in HCC tissues and cells, and negatively correlated with the expression of COX2 mRNA. In vitro assay revealed that overexpression of miR-136 significantly changed the expression of proliferation- and metastasis-related proteins and inhibited the proliferation, migration, and invasion of HepG2 and Hep3B cells. Dual-luciferase reporter assay validated that the 3′-untranslated region (3′-UTR) of COX2 is a direct target of miR-136. Furthermore, COX2 siRNA partially enhanced the miR-136 overexpression-induced inhibitory effects. In conclusion, miR-136 was vital in the regulation of HCC cell proliferation and metastasis by targeting COX2. Thus, our findings provided novel evidence that miR-136 might be recommended as a potential target for the diagnosis and treatment of HCC patients.
Key words: miR-136, Cyclooxygenase 2 (COX2), Hepatocellular carcinoma (HCC)
INTRODUCTION
Hepatocellular carcinoma (HCC) is the most prevalent type among primary liver cancers and is the third leading cause of death in cancer-related mortalities1,2. In recent years, surgical resection and radio-/chemotherapy are considered the widely used treatments3. However, because of incomplete resection and resistance, the majority of HCC patients still suffer from recurrence and death4,5. Some reports identified that HCC-related oncogenes and signaling molecules were involved in tumorigenesis. However, the specific signaling pathways that regulate the expression of HCC-related oncogenes are still not elucidated. Therefore, it is essential to hunt for the molecular mechanisms and potential targets for new therapies.
MicroRNAs (miRNAs) play important roles during tumorigenesis, and dysregulation of miRNAs is often involved in the progression and development of human cancers6. In recent decades, it has been widely reported that miRNAs were involved in most types of human cancers7. It has been documented that the expression of miRNA is closely associated with cancer proliferation and metastasis8,9. For example, miR-103 promotes proliferation and metastasis by targeting Krüppel-like factor 4 (KLF4) in gastric cancer; however, miR-21 promotes tumor proliferation and invasion in gastric cancer by targeting phosphatase and tensin homolog (PTEN)10,11. In addition, miR-136 suppresses tumor invasion and metastasis by targeting Ras protein activator like 2 (RASAL2) in triple-negative breast cancer12. However, the specific role and regulatory network of miR-136 in the progression of hepatocellular cancer are rarely explored.
In the present study, miR-136 and its potential target cyclooxygenase 2 (COX2) were obviously downregulated in HCC specimens. Bioinformatics analysis revealed that binding sites of the tumor suppressor miR-136 were located in the 3′-untranslated region (3′-UTR) of COX2. Therefore, we assumed that miR-136 might regulate cell migration and invasion of HCC cancer cells by regulating the COX2 pathway.
MATERIALS AND METHODS
Tissue Specimens
A total of 60 tissue specimens, including 30 tumor tissues and 30 adjacent noncancerous tissues (at least 2.5 cm away from the tumor), were obtained from Shandong Provincial Third Hospital (Jinan Shandong, P.R. China). All tissues were collected from hepatocellular cancer patients at the time of radical cystectomy, snap frozen in liquid nitrogen, and stored at −70°C until use. All patients gave informed consent prior to collection of their specimens according to institutional guidelines. The study was approved by the Institute Research Ethics Committee at Shandong Provincial Third Hospital (Jinan Shandong, P.R. China).
Cell Culture
The human HCC cell lines (HepG2, Hep3B, and MHCC-97H) and an immortalized hepatic epithelial cell line, LO2, were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China) and routinely maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Life Technologies, Grand Island, NY, USA) in a humidified incubator at 37°C under 5% CO2. For luciferase assays, human embryonic kidney 293 (HEK293) cells were cultured with DMEM (Invitrogen–Life Technologies, Carlsbad, CA, USA) containing 10% FBS, with 100 U/ml of penicillin, and 100 μg/ml of streptomycin (Sigma-Aldrich, St. Louis, MO, USA).
Quantitative Real-Time Reverse Transcriptase Polymerase Chain Reaction (qRT-PCR) Assay
Total RNA was extracted from the samples and cells by TRIzol reagent (Invitrogen–Life Technologies) and purified with the RNeasy Maxi Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. The RNA concentration was measured using a NanoDrop 2000/2000c (Thermo Scientific, Waltham, MA, USA). One microgram of RNA from each sample was used to obtain cDNA by performing reverse transcription PCR with a miScript reverse transcription kit (Qiagen). Then qPCR was performed with a miScript SYBR Green PCR Kit (Qiagen) on the Roche Lightcycler 480 Real-Time PCR System (Roche Diagnostics, Basel, Switzerland). The thermocycling conditions were as follows: 95°C for 1 min, then 40 cycles of 95°C for 10 s, 55°C for 30 s, and 70°C for 30 s. The expression levels of miR-136 in tissues and cells were analyzed with the 2−ΔΔCq method.
The PCR primer sets (Shanghai Sangon Biotech, Shanghai, P.R. China) used here for miR-136 were designed as follows: miR-136, 5′-ACACAGCTGGGACTCCATAAGAATTG-3′ (forward) and 5′-CTCTGAAGTGTCGAATGTCTTGGCTTGAGTCCT-3′ (reverse). U6 was used as an internal control and amplified with forward primer 5′-CAAGCTTGGCGGCAGAAA-3′ and reverse primer 5′-ATTGGDTTCATTTACGC-3′.
Cell Transfection
Synthesized miR-136 mimic (GenePharma, Suzhou, P.R. China) or mimic negative control (NC) were transfected into cells with Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA), which was mixed in Opti-MEM® I reduced serum (Gibco, Grand Island, NY, USA) according to the manufacturer’s protocol. qPCR was performed to measure the expression of miR-136 in cells following transfection. All miRNA oligonucleotides were purchased from GenePharma (Shanghai, P.R. China).
Western Blotting
The concentration of protein extracted from tissues and cells was measured using the BCA Kit (Wuhan Boster Biological Technology, Ltd., Wuhan, Hubei, P.R. China). The extracted protein was then added to wells with loading buffer (30 μg/well) and boiled at 95°C for 10 min. Proteins were then separated on 10% polyacrylamide gels (Wuhan Boster Biological Technology, Ltd.) by electrophoresis. Proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes, blocked with 5% bovine serum albumin (BSA; Thermo Fisher Scientific, Rockford, IL, USA), and incubated with primary antibodies overnight at 4°C. Membranes were then rinsed with Tris-buffered saline/Triton X-100 (TBST) (Sigma-Aldrich) and incubated with corresponding secondary antibodies at room temperature for 1 h. The membranes were washed again, and chemiluminescence reagent was added for image development. Images were developed using a Gel Doc EZ imager (Bio-Rad, Hercules, CA, USA) with β-actin used as an internal reference. The gray value of the target band was analyzed using Western blotting image analysis software (Alpha Innotech, Hayward, CA, USA).
Cell Counting Kit-8 (CCK-8) Assay
Cell proliferation was measured using the CCK-8 according to the manufacturer’s instructions (Dojindo, Kumamoto, Japan). Cells (2 × 103 cells/well) were incubated in 96-well plates for 12, 24, 48, and 72 h, respectively. CCK-8 solution (10 μl) was added to each well, the plates were incubated for 1 h at 37°C, and absorbance at 450 nm wavelengths was measured in a microplate reader (Bio-Rad).
Transwell Assays
Cell migration and invasion were analyzed using a Transwell chamber assay (Corning Inc., Cambridge, MA, USA). Cells (1 × 105) were seeded into the upper chamber of the coated Transwell with 8-μm pore size, and the Transwell chamber was placed into 24-well culture plates. After 24 h, the cells that migrated or invaded into the lower chamber were stained with crystal violet (Sinopharm Chemical Reagent Co., Shanghai, P.R. China) and counted by microscopy.
Luciferase Assay
Site-directed mutagenesis was introduced into the miR-136 binding site of COX2 mRNA using QuikChange Lightning Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The 3′-UTR fragment of COX2 mRNA was then subcloned into the pGL3 luciferase vector (Promega, Madison, WI, USA) by the PCR method and cotransfected with miR-136 mimics into cells for 36 h in 96-well plates using Lipofectamine 2000 (Invitrogen). Dual-Luciferase Reporter Assay (Promega) was then performed to analyze the luciferase assays. Renilla luciferase (Promega) activity was used as the internal control.
Statistical Analysis
Statistical analysis was performed using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) and are shown as the mean ± standard deviation (SD). A t-test was used to analyze the difference between two groups. A value of p < 0.05 was considered to indicate a statistically significant difference.
RESULTS
The Expression and Correlation of miR-136 and COX2 in HCC Tissues
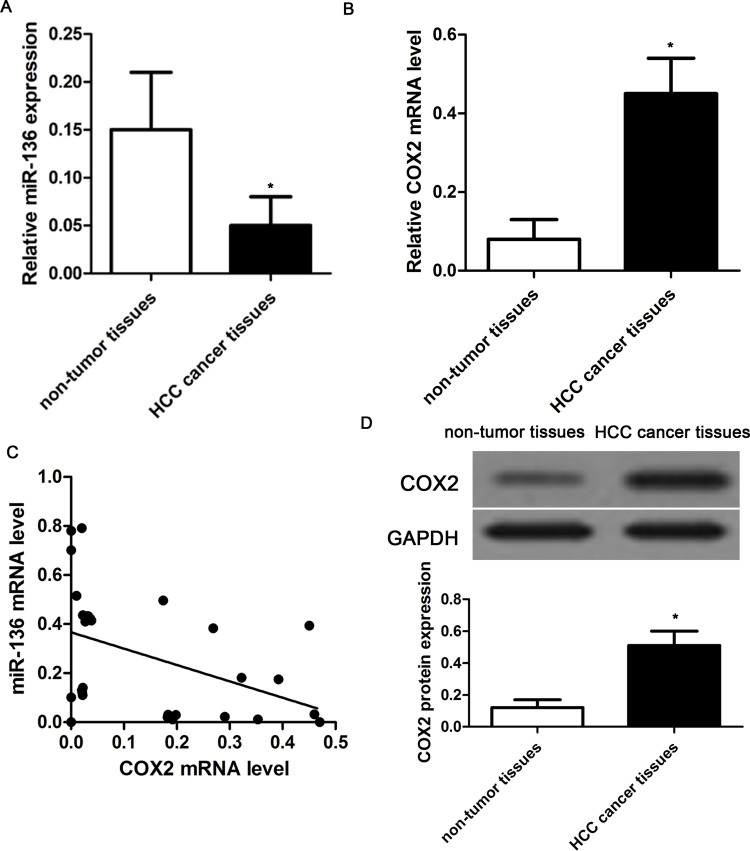
To investigate the expression and correlation of miR-136 and COX2 in HCC tissues, we analyzed the expression of miR-136 and COX2 (mRNA and proteins) in 30 pairs of human HCC cancer tissues and related normal tissues using qRT-PCR. Compared with normal tissues, miR-136 expression was significantly reduced in HCC cancer tissues (p < 0.01) (Fig. 1A). However, the expression of COX2 mRNA was obviously increased in tumor tissues compared with that in normal tissues (p < 0.01) (Fig. 1B). Meanwhile, we found that miR-136 expression was negatively associated with COX2 mRNA (r = −0.788, p = 0.005) (Fig. 1C). In addition, Western blot analysis also revealed that the expression of COX2 protein was significantly higher in HCC tumors than that in nontumor tissues (p < 0.01) (Fig. 1D).
Figure 1.
The expression of microRNA-136 (miR-136) and cyclooxygenase 2 (COX2) in hepatocellular carcinoma (HCC) tissues. (A) The relative expression of miR-136 was determined in 30 pairs of HCC tissues and the corresponding adjacent nontumor tissues by qualitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR). (B) The relative expression of COX2 mRNA in 30 pairs of HCC tissues and the corresponding adjacent nontumor tissues by qRT-PCR. The expression of miR-136 was normalized to U6. (C) According to Pearson’s correlation analysis, the expression of COX2 mRNA in 30 cases of HCC cancer tissues was inversely associated with the expression of miR-136 (p = 0.005). (D) The relative expression of COX2 protein was determined in 30 pairs of HCC tissues and the corresponding adjacent nontumor tissues by Western blot. *p < 0.05 compared to nontumor tissue.
The Expression of miR-136 and COX2 in HCC Cell Lines
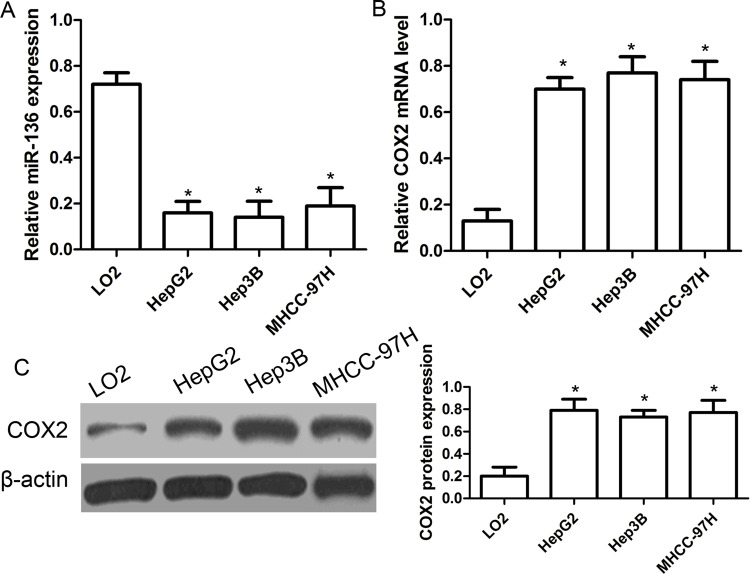
Based on the results mentioned above, we further analyzed the expression of miR-136 and COX2 in three human HCC cell lines (HepG2, Hep3B, and MHCC-97H) and an immortalized hepatic epithelial cell line, LO2. As shown in Figure 2A, the expression of miR-136 was significantly reduced in all three kinds of HCC cell lines compared with that in normal LO2 cells (p < 0.01). On the other hand, the expressions of COX2 mRNA and protein were significantly increased in HepG2, Hep3B, and MHCC-97H cells compared with that in LO2 cells (p < 0.01) (Fig. 2B and C).
Figure 2.
The expression of miR-136 and COX2 in HCC cell lines. (A) The relative expression of miR-136 was determined in HCC cell lines (HepG2, Hep3B, and MHCC-97H) and an immortalized hepatic epithelial cell line, LO2, by qRT-PCR. (B) The relative expression of COX2 mRNA was determined in HCC cell lines (HepG2, Hep3B, and MHCC-97H) and an immortalized hepatic epithelial cell line, LO2, by qRT-PCR. The expression of miR-136 was normalized to U6. (C) The relative expression of COX2 protein was determined in HCC cell lines (HepG2, Hep3B, and MHCC-97H) and an immortalized hepatic epithelial cell line, LO2, by Western blot. *p < 0.05 compared to LO2 cells.
miR-136 Suppresses Cell Proliferation In Vitro
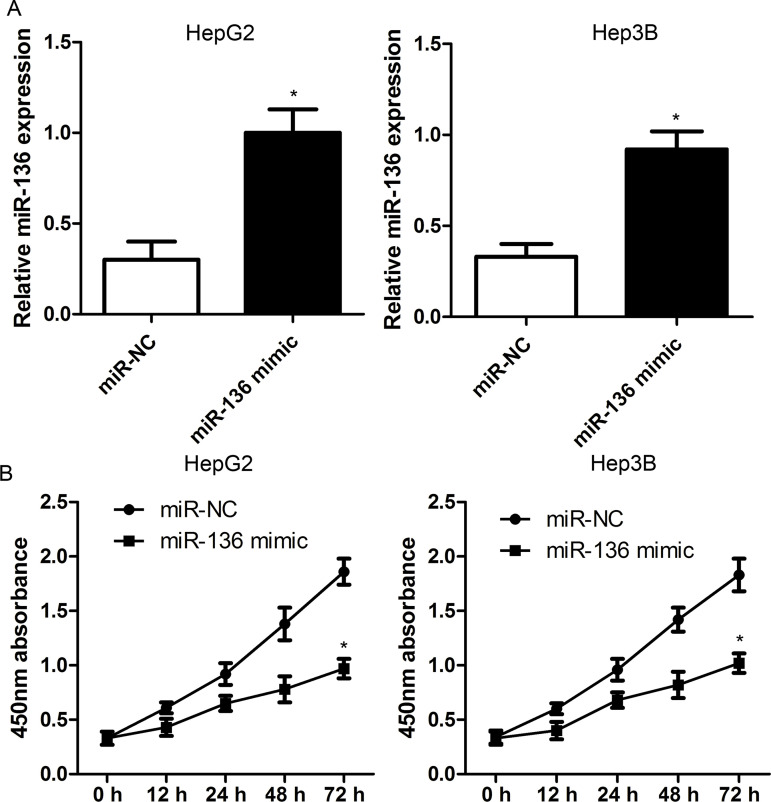
To figure out whether miR-136 inhibits the malignant phenotype of HCC cells, miR-136 mimics were transfected into HepG2 and Hep3B cell lines to overexpress miR-136 (Fig. 3A). The CCK-8 assay revealed that the overexpression of miR-136 significantly repressed the proliferation capacity of HepG2 and Hep3B cells compared with the miR-NC group (Fig. 3B), suggesting that miR-136 may be a tumor suppressor.
Figure 3.
miR-136 inhibits cell proliferation in HCC cells. (A) miR-136 mimics were transfected into HepG2 and Hep3B cell lines to overexpress miR-136. (B) Cell viability was determined in HepG2 and Hep3B cells by cell counting kit-8 (CCK-8) assay. Cells were transfected with miR-136 mimics or negative control (NC) duplex. The data are represented as mean ± standard deviation (SD). *p < 0.01 compared to miR-NC.
miR-136 Suppresses Cell Migration and Invasion In Vitro
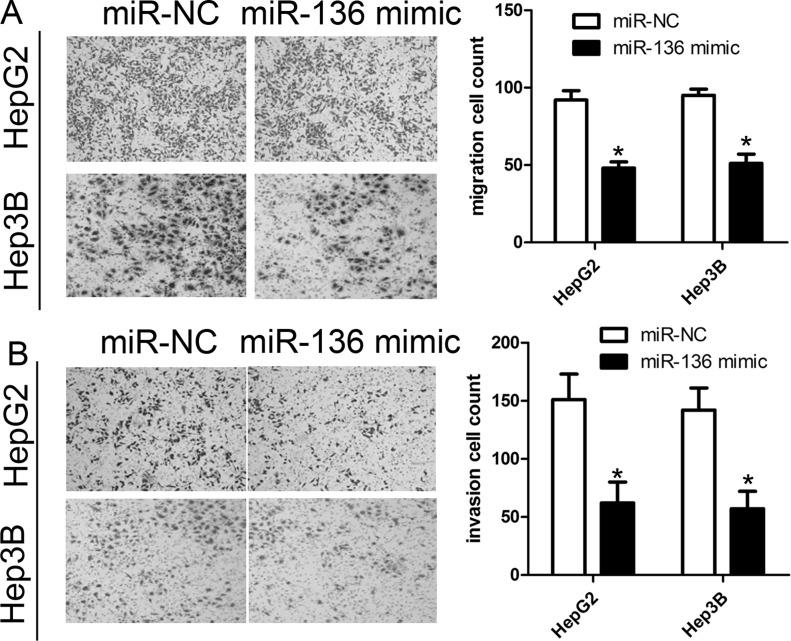
In view of the negative correlation of miR-136 with the metastatic property of HCC, we predicted that miR-136 would repress the migration and invasion of HCC cells. Transwell assays indicated that miR-136 overexpression robustly repressed the migration of HepG2 and Hep3B cells compared with the miR-NC group (Fig. 4A). The Transwell assay also revealed that miR-136 mimics could affect HepG2 and Hep3B cell invasion compared with the miR-NC group (p < 0.01) (Fig. 4B).
Figure 4.
miR-136 inhibits cell migration and invasion in HCC cells. (A) Cell migration assay was determined in HepG2 and Hep3B cells by Transwell assay. (B) Cell invasion assay was determined in HepG2 and Hep3B cells by Transwell assay. Cells were transfected with miR-136 mimics or miR-NC. The data are represented as mean ± SD. *p < 0.01 compared to miR-NC.
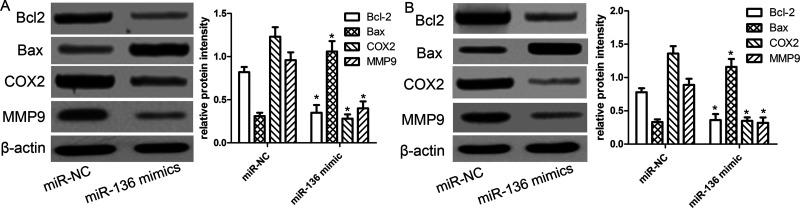
miR-136 Suppresses Proliferation and Metastasis-Related Proteins In Vitro
As known to all, proliferation- and metastasis-related proteins regulate the cancer cell’s biological properties. Furthermore, we used Western blot analysis to measure cell proliferation- and metastasis-related molecules, including B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax), matrix metalloproteinase 9 (MMP9), and COX2. We found that the expression of Bcl-2, MMP9, and COX2 protein was significantly decreased in HepG2 and Hep3B cells transfected with miR-136 mimics compared with the miR-NC group (Fig. 5A). Inversely, the expression of Bax protein was increased in HepG2 and Hep3B cells transfected with miR-136 mimics compared with the miR-NC group (Fig. 5B).
Figure 5.
miR-136 suppresses proliferation- and metastasis-related proteins in vitro. The relative expressions of B-cell lymphoma 2 (Bcl-2), Bcl-2-associated X protein (Bax), COX2, and matrix metalloproteinase 9 (MMP9) proteins were determined in HepG2 (A) and Hep3B (B) cells transfected with miR-NC or miR-136 mimics by Western blot. Levels of proteins were normalized to β-actin in each sample. The data are represented as mean ± SD. *p < 0.01 compared to miR-NC.
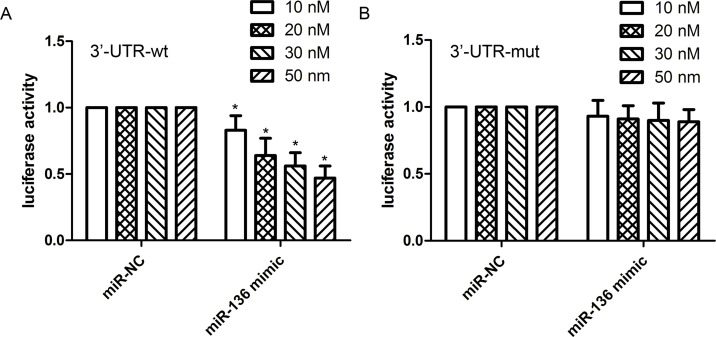
The 3′-UTR of COX2 Is a Direct Target of miR-136
According to the MiRanda software, we predicted that the COX2 gene may be a potential downstream target of miR-136. We carried out a dual-luciferase reporter assay to identify whether COX2 is a direct target of miR-136 in the HCC cellular conditions. Here miR-136 mimics or miR-NC with plasmids containing the 3′-UTR of wt-COX2 or mut-COX2 were cotransfected into HEK293 cell lines, and then the luciferase activity was measured. Our findings revealed that the overexpression of miR-136 significantly reduced the luciferase activity of HEK293 cell lines with wt-COX2-3′UTR and miR-136 mimics in a concentration-dependent manner (Fig. 6A). However, no difference in luciferase activity was observed in HEK293 cells with mut-COX2-3′UTR and miR-136 mimics (Fig. 6B).
Figure 6.
miR-136 directly targets COX2. The predicted miR-136 binding site within the COX2 3′-untranslated region (3′-UTR) and a mutated version generated by site-directed mutagenesis. Luciferase reporter assay illustrated direct binding of miR-136 to the wild-type (WT)-3′UTR of COX2 (A) in a concentration-dependent manner, but not to a mutant sequence (mut) within the 3′-UTR of COX2 (B). The data are represented as mean ± SD. *p < 0.01 compared to miR-NC.
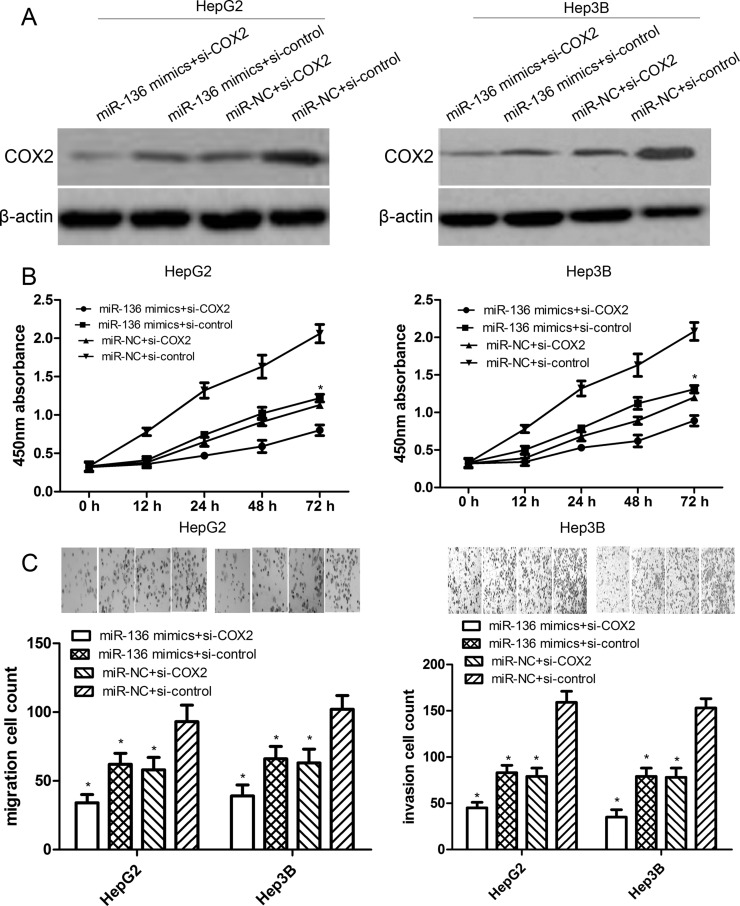
COX2 siRNA Partially Promotes miR-136 Overexpression-Induced Inhibition Effects
To observe the effect of COX2 small interfering RNA (siRNA) on miR-136-inhibited cell proliferation, migration, and invasion, we transfected COX2 siRNA or control siRNA into miR-136-overexpressing HepG2 or Hep3B cells. The expression level of COX2 protein following transfection of HepG2 or Hep3B cells with miR-136 mimics or COX2 siRNA was confirmed by Western blotting (Fig. 7A). We found that either COX2 siRNA or miR-136 mimics reduced the proliferation, migration, and invasion of HepG2 or Hep3B cells compared with their individual control. Notably, the cotransfection of COX2 siRNA and miR-136 mimics robustly inhibited the proliferation, migration, and invasion of HepG2 or Hep3B cells compared with the three other groups (both p < 0.01) (Fig. 7B and C). However, the cotransfection of COX2 siRNA and miR-136 mimics did not completely affect the tumor progression of HepG2 or Hep3B cells, suggesting that COX2 siRNA partially promotes the miR-136 overexpression-induced inhibition effects.
Figure 7.
COX2 small interfering RNA (siRNA) partially promotes miR-136 overexpression-induced inhibition effects. (A) The expression level of COX2 protein following transfection of HepG2 or Hep3B cells with miR-136 mimics and/or COX2 siRNA was confirmed by Western blotting. (B) The proliferation capacity of miR-136-overexpressing HepG2 or Hep3B cells cotransfected with COX2 siRNA (si-COX2) or control siRNA (si-control) was measured by CCK-8 assay. *p < 0.01 compared to miR-136 + si-COX2. (C) The migration or invasion capacity of miR-136-overexpressing HepG2 or Hep3B cells with si-COX2 or si-control was measured by Transwell assays. The data are represented by mean ± SD. *p < 0.01 compared to miR-NC + si-control.
DISCUSSION
To date, miRNAs have been demonstrated to play a key role in tumorigenesis. Some miRNAs can target oncogenes to suppress tumor development, while the rest of the miRNAs can target anti-oncogenes to promote tumor development13–17. Yuan et al. reported that miR-136 serves as a negative regulator in the progression of colon cancer by targeting liver receptor homolog 1 (LRH-1), and downregulated miR-136 expression contributes to high LRH-1 expression and aberrant activation of wingless-related integration site (Wnt) signaling18. Zheng et al. also showed that ectopic homeobox protein C10 (HOXC10) expression could reverse overexpressed miR-136-inhibited gastric cancer cell metastasis19. These findings provide new insights into the involvement of miR-136 into cancer-specific metastasis and are suggestive of the potential application of miR-136 in tumor treatment. However, the specific function and mechanism of miR-136 in hepatocellular cancer progression are not fully investigated.
In the present study, we investigate the miR-136 expression in HCC tissues and cells, and then elucidate the effects of miR-136 expression on the malignant features of HCC cells such as proliferation, migration, and invasion. We noticed that miR-136 expression was significantly decreased in HCC tumor tissues compared with the corresponding adjacent nontumorous tissues. Besides, we further applied HCC cells and human normal L02 liver cells to demonstrate that miR-136 expression was also downregulated in the HCC cells compared with normal L02 cells. These findings suggested that the aberrant expression of miR-136 may be recommended as an early detection of HCC patients. Additionally, the gain or loss assays of miR-136 expression were used to explore whether miR-136 could affect the HCC biology. These findings suggested that miR-136 suppressed the HCC proliferation, migration, and invasion in vitro. We also found that the inhibitory effects of miR-136 were associated with changes in proliferation- and metastasis-related proteins, indicating that miR-136 was a potential tumor suppressor in the development of HCC.
Next, we focused on the precise mechanisms underlying miR-136-induced inhibitory effects using the bioinformatics prediction programs. Previous studies have reported that COX2 was a leading gene relating to HCC progression. Li et al. reported that the overexpression of COX2 in pancreatic cancer cells promoted cell proliferation and migration, while the knockdown of the expression of COX2 inhibited tumorigenesis of pancreatic cancer cells. Mechanistically, COX2 regulated the expression of multiple genes involved in cell growth, migration, and cell apoptosis20. According to the MiRanda software, we predicted that the 3′-UTR of COX2 contained a conserved putative target site for miR-136. Dual-luciferase reporter and Western blot assays validated that overexpression of miR-136 inhibited the expression of COX2 protein by directly targeting its 3′-UTR. Furthermore, COX2 siRNA partially promotes miR-136 overexpression-induced inhibition effects, suggesting that the 3′-UTR of COX2 is a direct target of miR-136.
Besides, we noticed that miR-136 mimics indeed decreased the expression of Bcl-2 and MMP9 proteins and increased the expression of Bax proteins, suggesting that miR-136 indeed reduced HCC cell proliferation and invasion. These findings suggested that miR-136 may target more factors, such as transcription factors, to regulate Bcl-2 and MMP9 expression. We will investigate the more detailed mechanisms of the suppression of miR-136 in human HCCs in future studies.
In conclusion, we demonstrated that miR-136 acts as a tumor suppressor in HCC tumorigenesis and progression. Mechanically, miR-136 inhibits HCC cell proliferation, migration, and invasion partially by directly targeting the 3′-UTR of COX2. These findings enable us to explore more novel prognosis markers and potential targets for HCC treatment in the near future.
ACKNOWLEDGMENT
The authors are thankful to other members in their lab for their suggestions.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Cidon EU. Systemic treatment of hepatocellular carcinoma: Past, present and future. World J Hepatol. 2017;9:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Della Corte CM, Viscardi G, Papaccio F, Esposito G, Martini G, Ciardiello D, Martinelli E, Ciardiello F, Morgillo F. Implication of the hedgehog pathway in hepatocellular carcinoma. World J Gastroenterol. 2017;23:4330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nosratabadi R, Alavian SM, Zare-Bidaki M, Shahrokhi VM, Arababadi MK. Innate immunity related pathogen recognition receptors and chronic hepatitis B infection. Mol Immunol. 2017;90:64–73. [DOI] [PubMed] [Google Scholar]
- 4. Ghouri YA, Mian I, Rowe JH. Review of hepatocellular carcinoma: Epidemiology, etiology, and carcinogenesis. J Carcinog. 2017;16:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hao QQ, Chen GY, Zhang JH, Sheng JH, Gao Y. Diagnostic value of long noncoding RNAs for hepatocellular carcinoma: A PRISMA-compliant meta-analysis. Medicine 2017;96:e7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fan X, Chen W, Fu Z, Zeng L, Yin Y, Yuan H. MicroRNAs, a subpopulation of regulators, are involved in breast cancer progression through regulating breast cancer stem cells. Oncol Lett. 2017;14:5069–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S, Zhang JY, Lu LJ, Wang CH, Wang LH. MiR-630 promotes epithelial ovarian cancer proliferation and invasion via targeting KLF6. Eur Rev Med Pharmacol Sci. 2017;21:4542–7. [PubMed] [Google Scholar]
- 8. Zhang L, Ye Y, Tu H, Hildebrandt MA, Zhao L, Heymach JV, Roth JA, Wu X. MicroRNA-related genetic variants in iron regulatory genes, dietary iron intake, microRNAs and lung cancer risk. Ann Oncol. 2017;28:1124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Romano G, Veneziano D, Acunzo M, Croce CM. Small non-coding RNA and cancer. Carcinogenesis 2017;38:485–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng J, Liu Y, Qiao Y, Zhang L, Lu S. miR-103 promotes proliferation and metastasis by targeting KLF4 in gastric cancer. Int J Mol Sci. 2017;18:E910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY, Yan M. microRNA-21 promotes tumor proliferation and invasion in gastric cancer by targeting PTEN. Oncol Rep. 2012;27:1019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan M, Li X, Tong D, Han C, Zhao R, He Y, Jin X. miR-136 suppresses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep. 2016;36:65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Callegari E, Gramantieri L, Domenicali M, D’Abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: A model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gayral M, Jo S, Hanoun N, Vignolle-Vidoni A, Lulka H, Delpu Y, Meulle A, Dufresne M, Humeau M, Chalret du Rieu M, Bournet B, Sèlves J, Guimbaud R, Carrère N, Buscail L, Torrisani J, Cordelier P. MicroRNAs as emerging biomarkers and therapeutic targets for pancreatic cancer. World J Gastroenterol. 2014;20:11199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mei Q, Li X, Guo M, Fu X, Han W. The miRNA network: Micro-regulator of cell signaling in cancer. Expert Rev Anticancer Ther. 2014;14:1515–27. [DOI] [PubMed] [Google Scholar]
- 16. Zhang L, Fan XM. The pathological role of microRNAs and inflammation in colon carcinogenesis. Clin Res Hepatol Gastroenterol. 2015;39:174–9. [DOI] [PubMed] [Google Scholar]
- 17. Gandellini P, Rancati T, Valdagni R, Zaffaroni N. miRNAs in tumor radiation response: Bystanders or participants? Trends Mol Med. 2014;20:529–39. [DOI] [PubMed] [Google Scholar]
- 18. Yuan Q, Cao G, Li J, Zhang Y, Yang W. MicroRNA-136 inhibits colon cancer cell proliferation and invasion through targeting liver receptor homolog-1/Wnt signaling. Gene 2017;628:48–55. [DOI] [PubMed] [Google Scholar]
- 19. Zheng J, Ge P, Liu X, Wei J, Wu G, Li X. MiR-136 inhibits gastric cancer-specific peritoneal metastasis by targeting HOXC10. Tumour Biol. 2017;39:1010428317706207. [DOI] [PubMed] [Google Scholar]
- 20. Li W, Mao Z, Fan X, Cui L, Wang X. Cyclooxygenase 2 promoted the tumorigenicity of pancreatic cancer cells. Tumour Biol. 2014;35:2271–8. [DOI] [PubMed] [Google Scholar]