Abstract
It is increasingly evident that various long noncoding RNAs (lncRNAs) participate in the tumorigenesis of multiple tumors, including melanoma. lncRNAs have been validated as oncogenic factors in various tumors; however, the potential regulatory mechanism of CCAT1 in melanoma is still unclear. The purpose of this study was to investigate the regulation of CCAT1 on melanoma genesis. The expression of CCAT1 in melanoma tissue and cell lines was measured using qRT-PCR. Interference oligonucleotide or mimic sequences were applied to up- or downregulate RNA expression. CCK-8 and colony formation assays were performed to detect the proliferation capability. Transwell assay was used to assess the migration and invasion capacities. Bioinformatics analysis was performed to predict the target miRNAs of CCAT1. Expression of CCAT1 was significantly upregulated in melanoma tissue and cell lines. CCAT1 knockdown observably suppressed the proliferation, migration, and invasion abilities. Bioinformatics analysis predicted that miR-33a acted as a target of CCAT1, which was confirmed by dual-luciferase reporter assay. CCAT1 knockdown reversed the tumor-promoting ability of the miR-33a inhibitor. CCAT1 acts as an oncogenic factor in the genesis of melanoma and exerts tumor-promoting roles via sponging miR-33a, providing a novel insight for competing endogenous RNA (ceRNA) in the tumorigenesis of melanoma.
Key words: Melanoma, CCAT1, miR-33a, Competing endogenous RNA (ceRNA)
INTRODUCTION
Melanoma is the most aggressive subclass of malignant skin tumor1. The incidence of melanoma has increased all over the world yearly in recent decades2. Unlike any other neoplastic diseases, there is still no effective therapy for primary or metastatic melanoma3. Although melanoma results in an extremely high mortality, the authentic pathogenesis at the molecular level is still poorly understood4. In order to establish an effective and specific therapeutic method, in-depth pathogenesis needs to be probed from disparate molecular levels.
Long noncoding RNAs (lncRNAs) are a novel subclass of noncoding RNAs (ncRNA) having over 200 nucleotides in length and without protein-coding ability. lncRNAs have been reported to play a pivotal role in mediating tumorigenesis and the progression of various tumors, including melanoma. For instance, lncRNA HOTAIR is highly expressed in lymph node metastasis melanoma, indicating the association with motility, invasion, and metastatic potential5. lncRNA MHENCR overexpression induces poor survival in melanoma patients, and MHENCR knockdown significantly suppresses melanoma cell proliferation and induces cell cycle arrest and apoptosis in vitro through activating the miR-425/489-mediated PI3K–Akt pathway6.
Colon cancer-associated transcript-1 (CCAT1) has been validated as an oncogenic lncRNA in different types of malignancy, such as glioma, laryngeal squamous cell carcinoma, and acute myeloid leukemia7–9. In addition, Kaplan–Meier analysis with the log-rank test manifested that high expression of CCAT1 was associated with lower overall survival and progression-free survival in breast cancer and hepatocellular carcinoma10,11. However, the physiological function and potential molecular mechanisms of melanoma in the genesis and progression of melanoma remain to be further explored.
In this study, we aimed to detect the expression and abundance of CCAT1 in melanoma tissue and investigate the probable regulation. Furthermore, we analyzed the regulatory pathway of competing endogenous RNA (ceRNA) associated with CCAT1 and miR-33a in vitro, providing novel insight for the tumorigenesis of melanoma.
MATERIALS AND METHODS
Patient Samples and RNA Extraction
A total of 30 malignant melanoma patients who had not had any chemotherapy or radiotherapy were enrolled in our study. Tumor tissue samples and corresponding matched adjacent noncancerous tissue samples were obtained from the surgical resection tissue of osteosarcoma patients of the Department of Dermatology of Tianjin Hospital. All patients signed informed consent. The study obtained approval and supervision from the ethics committee of Tianjin Hospital.
All tumor tissue samples were confirmed by histological diagnosis. Tissues were frozen in liquid nitrogen immediately and stored at −80°C until further use. Total RNA was extracted from samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and then quantified using a NanoDrop ND-1000. Finally, RNA integrity was assessed using standard denaturing agarose gel electrophoresis.
Cell Lines and Culture
Human epidermal melanocytes neonatal (HEMn) cells and melanoma cell lines (M21, B16F10, melanoma200, MEL-RM, A375, and A2058) were purchased from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen) in a suitable environment at 37°C with 5% CO2.
Cell Transfection
Cells were seeded into six-well plates at a density of 4 × 104 cells/wells with 60–80% confluence. All oligonucleotides, including mimics, siRNAs, and scramble sequence negative control, were synthesized by GenePharma (Shanghai, P.R. China). The transfection was performed using Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions.
Quantitative Real-Time PCR
For quantitative real-time (qRT)-PCR, isolation of total cellular RNA from cells and tissues and reverse transcription were performed as previously described12. The cDNA was synthesized from primers and corresponding total RNA using RevertAid First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). qRT-PCR was then performed using LightCycler technology (Roche, Mannheim, Germany). The following pairs of primers were used in our study: CCAT1, 5′-AACTCTTCAGCCAGGGTCCAC-3′ (forward) and 5′-CACAGTGAAGATGATGAAGAC-3′ (reverse); miR-33a, 5′-GCTGGGAGCGCGATGGATACC-3′ (forward) and 5′-GGACAGAAGCCGTACGCCATCC-3′ (reverse).
Luciferase Assays
For the luciferase assay, the full-length 3′-UTR of CCAT1 containing miR-33a binding sites was cloned into the downstream of the firefly luciferase gene in pGL3 (Invitrogen) to establish pGL3-luc-CCAT1. Cells were seeded into 96-well plates at a density of 1 × 104 cells/well and then transfected with luciferase vector. Cells were transiently cotransfected with luciferase reporter plasmids (Promega, Madison, WI, USA) with Lipofectamine 2000 according to the manufacturer’s instructions. After 24 h of transfection, luciferase activity was measured using the dual-luciferase reporter gene assay kit (Promega) according to the manufacturer’s instructions. The relative firefly luciferase activities were normalized with the Renilla luciferase activities as a control for transfection efficiency.
Cell Proliferation Assay
Cell proliferation viability was measured using the cell counting kit 8 (CCK-8; Dojindo, Japan) according to the manufacturer’s instructions. Cells were seeded into 96-well plates at a density of 5 × 104 cells/well at 37°C, and cell viability was measured at the appointed time points using a microplate reader (Bio-Rad Laboratories Inc., Hertfordshire, UK) by spectrophotometry at 450 nm.
Colony Formation Assay
Melanoma cell lines (B16 and A375) were seeded into six-well plates at a density of 500/well for 2 weeks. Subsequently, the colonies were fixed with 4% paraformaldehyde for 5 min and stained with 1% crystal violet for 10 min. Colonies were examined and counted under a microscope. The assays were performed in triplicate.
Cell Cycle Analysis Assay
After transfection, cell cycle analysis was performed using a cell cycle analysis kit (Lianke, Shanghai, P.R. China). Briefly, cells (4 × 105 per well) were seeded into six-well plates and starved in FBS-free medium for 12 h. Cells were then washed with PBS, digested by trypsin, and collected by centrifugation at 1,000 rpm for 5 min. The cells (106 cells per sample) were fixed in 4 ml of 75% cold ethanol at −20°C overnight. After centrifugation, DNA staining was performed with 10 mg propidium iodide (PI)/ml PBS and 2.5 mg Ag DNase-free RNase/ml PBS for 30 min. Cell cycle profiles were generated with Modifit software (BD Biosciences, Franklin Lake, NJ, USA).
Apoptosis Assay
After being washed with cold PBS and resuspended in binding buffer, apoptotic cells were measured using an Annexin-V/Dead Cell Apoptosis Kit (Invitrogen). Briefly, cells (1 × 106 per well) were seeded into six-well plates and starved in FBS-free medium for 12 h. Cells were then stained with annexin V–FITC and PI. Finally, cells were calculated using a FACScan flow cytometer (Becton Dickinson, San Jose, CA, USA) equipped with cell quest software (Becton Dickinson).
Statistical Analysis
All quantitative data are represented as means ± SEM. GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA) statistical software was used to determine p values for the proliferation graphs by performing two-way ANOVA analysis and t-tests as indicated.
RESULTS
Expression of CCAT1 Was Upregulated in Human Melanoma Tissue and Closely Related to Poor Prognosis
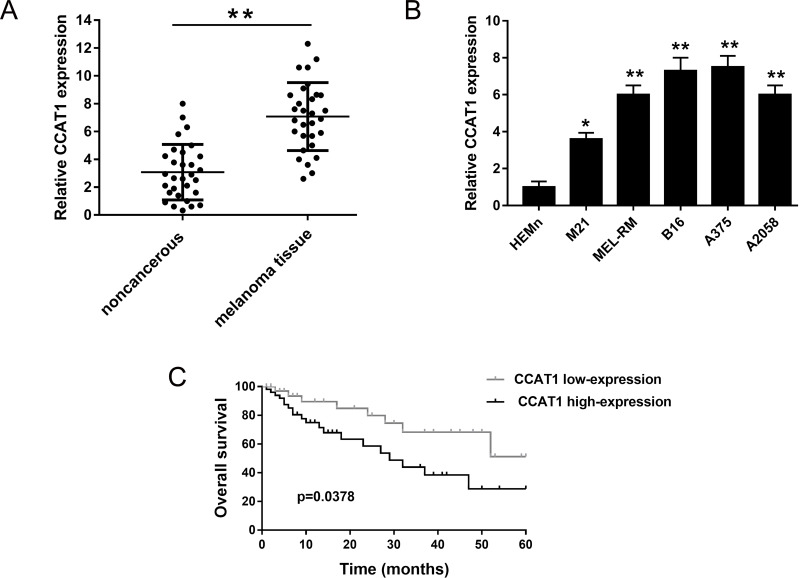
To assess the expression of CCAT1, qRT-PCR was performed on 30 melanoma tissue samples and their paired nontumor tissues. Results showed that CCAT1 was significantly upregulated in melanoma tissue compared to noncancerous tissues (Fig. 1A). In the meantime, the expression of CCAT1 was similarly overexpressed in melanoma cell lines (Fig. 1B). This obviously indicates that CCAT1 might act in an oncogenic role in melanoma tumorigenesis. We hypothesized that CCAT1 has a tumor-promoting role in melanoma genesis.
Figure 1.
Expression of colon cancer-associated transcript-1 (CCAT1) was upregulated in human melanoma tissue and cell lines, indicating poor prognosis. (A) Relative expression of CCAT1 in melanoma tissue compared to adjacent nontumor tissue examined using quantitative real-time (qRT)-PCR. (B) CCAT1 expression was significantly higher in melanoma cell lines than in normal human epidermal melanocytes neonatal (HEMn). (C) Overall survival between high and low CCAT1 expression demonstrated by Kaplan–Meier analysis. Data are presented as the mean ± SD. Statistical analysis was performed with Student’s t-test. *p < 0.05, **p < 0.01 compared to the control group.
To verify the hypothesis, we performed Kaplan–Meier analysis and log-rank test to evaluate the interrelation within CCAT1 expression and melanoma patients’ prognosis. Results showed that the overall survival of patients with high CCAT1 expression level had a poorer prognosis than those with low expression (Fig. 1C). Distinctly, the above results indicated that CCAT1 overexpression played an important oncogenic role in the occurrence and progression of melanoma.
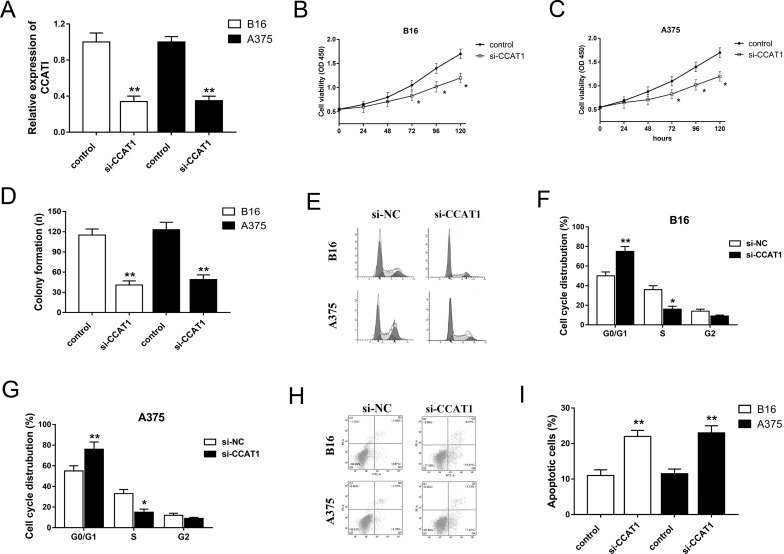
CCAT1 Knockdown Suppressed the Proliferation and Accelerated the Apoptosis of Melanoma Cell Lines
Owing to the hypothetical oncogenic role of CCAT1 in melanoma tumorigenesis, our study performed loss-of-function experiments on melanoma cell lines to validate the physiological effect of CCAT1. The expression of CCAT1 was significantly decreased in B16 and A375 cell lines transfected with interference sequence (Fig. 2A). The CCK-8 and colony formation assays showed that CCAT1 knockdown suppressed the proliferation and colony formation capacities of melanoma cell lines (Fig. 2B–D). Moreover, CCAT1 knockdown induced cell cycle arrest at the G0/G1 stage (Fig. 2E–G). In addition, CCAT1 knockdown promoted apoptosis of melanoma cells (Fig. 2H and I). Overall, the above results indicate that CCAT1 knockdown significantly resists melanoma progression, suggesting an oncogenic role in the oncogenesis of melanoma.
Figure 2.
CCAT1 knockdown suppressed the proliferation and accelerated the apoptosis of melanoma cell lines (B16 and A375). (A) CCAT1 knockdown in melanoma cells transfected with si-CCAT1. (B, C) Cell viability was measured at indicated time points using the cell counting kit 8 (CCK-8) assay. (D) Clone number of colony formation assay. (E–G) CCAT1 induced cell cycle arrest at the G0/G1 phase. (H, I) Apoptosis of B16 and A375 cells detected by flow cytometry. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01 compared to the si-NC group.
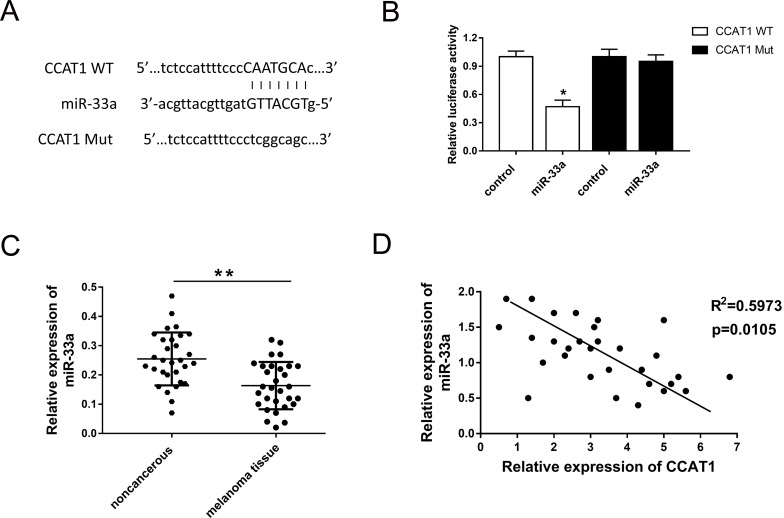
CCAT1 Directly Targeted miR-33a and Acted as an miR-33a Sponge
Our previous study validated that CCAT1 knockdown suppressed the malignant progression of melanoma cells and exerted a carcinogenic role in the physiological process. Because many studies had reported that lncRNAs function as miRNA sponges to regulate neoplastic processes, we hypothesized that CCAT1 might also act as an miRNA sponge in the oncogenesis of melanoma. With the aid of bioinformatics analysis (starBase, www.starbase.sysu.edu.cn), we discovered the target miR-33a, which had been verified to be a tumor suppressor in the oncogenesis of melanoma13. The complementary binding sites of miR-33a and 3′-UTR of CCAT1 are shown and verified by dual-luciferase reporter assay (Fig. 3A and B). The expression level of miR-33a was measured by qRT-PCR, indicating the lower expression of miR-33a in 30 melanoma sample tissues (Fig. 3C). Pearson’s correlation analysis showed a negative association within the expression of CCAT1 and miR-33a (Fig. 3D). In conclusion, we discovered the potential target miRNA of CCAT1 using bioinformatics analysis.
Figure 3.
CCAT1 directly targeted miR-33a and acted as an miR-33a sponge. (A) Putative complementary sites of miR-33a and 3′-UTR of CCAT1 predicted using bioinformatics analysis (starBase V2.0, http://starbase.sysu.edu.cn). (B) Dual-luciferase reporter assay validated the binding of miR-33a with CCAT1. (C) The expression level of miR-33a in 30 melanoma sample tissues. (D) Pearson’s correlation analysis revealed the correlation between CCAT1 and miR-33a expression in melanoma tissues. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01 compared to the control group.
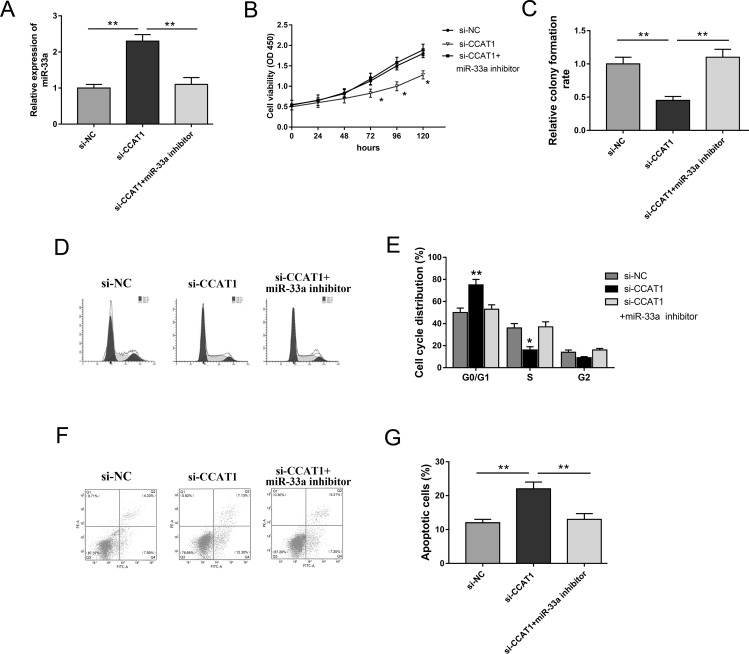
miR-33a Inhibitor Rescued the Inhibition of CCAT1 Knockdown on Melanoma Tumorigenesis
Our study validated that CCAT1 acts as an miR-33a sponge; thus, we performed rescue experiments in the A375 cell line to investigate the regulation of CCAT1 and miR-33a on proliferation, cell cycle, and apoptosis. Cotransfected with si-CCAT1, miR-33a expression was observably higher than in the si-NC group, which was reversed by the miR-33a inhibitor (Fig. 4A). The CCK-8 and colony formation assays showed that the miR-33a inhibitor rescued the inhibition of si-CCAT1 on cell proliferation and colony formation abilities (Fig. 4B and C). Moreover, the cotransfection of the miR-33a inhibitor significantly reversed cell cycle arrest at the G0/G1 phase and apoptosis promotion induced by si-CCAT1 (Fig. 4D–G). The above rescue experiments revealed that CCAT1 knockdown suppressed melanoma cell proliferation, induced cell cycle arrest, and accelerated apoptosis. However, miR-33a inhibitor reversed the inhibition of si-CCAT1, indicating that CCAT1 exerted a carcinogenic role by targeting miR-33a.
Figure 4.
The miR-33a inhibitor rescued the inhibition of CCAT1 knockdown on melanoma cell proliferation, cell cycle arrest, and apoptosis. (A) Expression of miR-33a was measured in the groups transfected with si-NC, si-CCAT1, and miR-33a inhibitor. (B) Cell proliferation viability of A375 cells detected by the CCK-8 assay. (C) Cell colony formation assay. (D, E) The miR-33a inhibitor reversed cell cycle arrest at the G0/G1 phase induced by CCAT1 knockdown. (F, G) Apoptosis of melanoma cells were detected by flow cytometry. Data are presented as the mean ± SD. *p < 0.05, **p < 0.01 compared to the si-NC group.
DISCUSSION
Melanoma is an extremely aggressive and malignant skin tumor with a high mortality. However, authentic pathogenesis at the molecular level is still poorly understood14. Recently, emerging evidence has demonstrated that lncRNAs exert an enormous crucial function in the physiological pathological process, especially multiple tumor oncogenesis15. Furthermore, various lncRNAs have been reported to be associated with the oncogenesis of melanoma16. In the present study, we aimed to investigate the undetermined moderating effect of CCAT1, known as an oncogenic lncRNA in multiple malignancies, and discover the endogenous competing mechanism involved in the genesis of melanoma.
lncRNA CCAT1 has been verified to be overexpressed in various malignant tumor tissues7–9. In the early stage, we measured the expression level of CCAT1 and found that it was upregulated in melanoma tissues and cell lines. In addition, a high CCAT1 expression level indicated lower overall survival and poor prognosis. Moreover, CCAT1 knockdown suppressed the proliferation, induced cell cycle arrest at the G0/G1 stage, and promoted apoptosis. Similarly, our study validated the oncogenic role of CCAT1 in the progression of melanoma. Aberrant expression of CCAT1 is involved in several processes associated with tumorigenesis, including cell proliferation, apoptosis, migration, and invasion through modulating several target genes or pathways17. In 70% of laryngeal squamous cell carcinoma (LSCC) patients, CCAT1 expression was higher in tumor tissues than in adjacent normal tissues, and the ectopic expression of CCAT1 accelerated the epithelial–mesenchymal transition (EMT) and tumorigenesis in LSCC via the let-7/HMGA2/Myc pathway9.
In general, lncRNAs may act as oncogenes or tumor suppressors to regulate tumorigenesis, proliferation, invasion, and metastasis18,19. lncRNAs play a critical role in many biological processes by silencing miRNAs, which is a canonical ceRNA regulating pattern. Aftab et al. reviewed the miRNAs and lncRNAs implicated in melanomagenesis, such as SPRY4-IT1, BANCR, and HOTAIR, indicating the pivotal interaction of lncRNA/miRNA on proliferation, cell cycle, migration, invasion, and immune evasion20. Our study indicates that CCAT1 acts as an miR-33a sponge and exerts a carcinogenic role by targeting miR-33a.
It is widely known that early diagnosis and therapy can significantly increase the survival rate of melanoma patients21,22. Thus, effective and specific biomarkers for early detection and diagnosis of melanoma are urgently required. In breast cancer, CCAT1 overexpression is significantly higher than those in adjacent normal tissues and associated with differentiation grades, TNM stages, and lymph node metastases, suggesting that CCAT1 could act as a novel biomarker in the prognosis of breast cancer patients11. In melanoma, lncRNA PVT1 is upregulated in melanoma tissues and the serum compared to that in nonneoplastic nevi23. Our study revealed that CCAT1 was specifically overexpressed in melanoma tissue, suggesting an efficient and valuable diagnostic marker.
In summary, our study uncovers the tumor-promoting role of CCAT1 in melanoma genesis via sponging miR-33a, providing insights into the mechanisms of the lncRNAs/miRNAs pathway and useful biomarkers or future therapeutic targets for melanoma oncogenesis.
ACKNOWLEDGMENTS
This work was supported by The Basic Medical Research Center of Tianjin Hospital. The authors want to thank Dr. Yang for her technical help and for correcting the manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. McNally RJ, Basta NO, Errington S, James PW, Norman PD, Craft AW. Socioeconomic patterning in the incidence and survival of children and young people diagnosed with malignant melanoma in northern England. J Invest Dermatol. 2014;134(11):2703–8. [DOI] [PubMed] [Google Scholar]
- 2. Erdmann F, Lortet-Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, Bray F. International trends in the incidence of malignant melanoma 1953–2008—Are recent generations at higher or lower risk? Int J Cancer 2013;132(2):385–400. [DOI] [PubMed] [Google Scholar]
- 3. Sharma KS, Lim P, Brotherston MT. Excision versus incision biopsy in the management of malignant melanoma. J Dermatolog Treat. 2016;27(1):88–90. [DOI] [PubMed] [Google Scholar]
- 4. Abildgaard C, Guldberg P. Molecular drivers of cellular metabolic reprogramming in melanoma. Trends Mol Med. 2015;21(3):164–71. [DOI] [PubMed] [Google Scholar]
- 5. Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen X, Dong H, Liu S, Yu L, Yan D, Yao X, Sun W, Han D, Gao G. Long noncoding RNA MHENCR promotes melanoma progression via regulating miR-425/489-mediated PI3K-Akt pathway. Am J Transl Res. 2017;9(1):90–102. [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L, Wang W, Cao L, Li Z, Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol Cells 2016;39(4):330–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang ZH, Guo XQ, Zhang QS, Zhang JL, Duan YL, Li GF, Zheng DL. Long non-coding RNA CCAT1 promotes glioma cell proliferation via inhibiting microRNA-410. Biochem Biophys Res Commun. 2016;480(4):715–20. [DOI] [PubMed] [Google Scholar]
- 9. Zhuang K, Wu Q, Jiang S, Yuan H, Huang S, Li H. CCAT1 promotes laryngeal squamous cell carcinoma cell proliferation and invasion. Am J Transl Res. 2016;8(10):4338–45. [PMC free article] [PubMed] [Google Scholar]
- 10. Deng L, Yang SB, Xu FF, Zhang JH. Long noncoding RNA CCAT1 promotes hepatocellular carcinoma progression by functioning as let-7 sponge. J Exp Clin Cancer Res. 2015;34:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang XF, Liu T, Li Y, Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int J Clin Exp Pathol. 2015;8(8):9440–5. [PMC free article] [PubMed] [Google Scholar]
- 12. Hellerbrand C, Muhlbauer M, Wallner S, Schuierer M, Behrmann I, Bataille F, Weiss T, Scholmerich J, Bosserhoff AK. Promoter-hypermethylation is causing functional relevant downregulation of methylthioadenosine phosphorylase (MTAP) expression in hepatocellular carcinoma. Carcinogenesis 2006;27(1):64–72. [DOI] [PubMed] [Google Scholar]
- 13. Zhou J, Xu D, Xie H, Tang J, Liu R, Li J, Wang S, Chen X, Su J, Zhou X, Xia K, He Q, Chen J, Xiong W, Cao P, Cao K. miR-33a functions as a tumor suppressor in melanoma by targeting HIF-1alpha. Cancer Biol Ther. 2015;16(6):846–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spencer KR, Mehnert JM. Mucosal melanoma: Epidemiology, biology and treatment. Cancer Treat Res. 2016;167:295–320. [DOI] [PubMed] [Google Scholar]
- 15. Rastrelli M, Tropea S, Rossi CR, Alaibac M. Melanoma: Epidemiology, risk factors, pathogenesis, diagnosis and classification. In Vivo 2014;28(6):1005–11. [PubMed] [Google Scholar]
- 16. Guo L, Yao L, Jiang Y. A novel integrative approach to identify lncRNAs associated with the survival of melanoma patients. Gene 2016;585(2):216–20. [DOI] [PubMed] [Google Scholar]
- 17. Guo X, Hua Y. CCAT1: An oncogenic long noncoding RNA in human cancers. J Cancer Res Clin Oncol. 2017;143(4):555–62. [DOI] [PubMed] [Google Scholar]
- 18. Sonkin D, Hassan M, Murphy DJ, Tatarinova TV. Tumor suppressors status in cancer cell line Encyclopedia. Mol Oncol. 2013;7(4):791–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pavel AB, Vasile CI. Identifying cancer type specific oncogenes and tumor suppressors using limited size data. J Bioinform Comput Biol. 2016;14(6):1650031. [DOI] [PubMed] [Google Scholar]
- 20. Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014;563:60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peng W, Jiang A. Long noncoding RNA CCDC26 as a potential predictor biomarker contributes to tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016;83:712–7. [DOI] [PubMed] [Google Scholar]
- 22. Zhao JH, Sun JX, Song YX, Chen XW, Yang YC, Ma B, Wang J, Gao P, Wang ZN. A novel long noncoding RNA-LOWEG is low expressed in gastric cancer and acts as a tumor suppressor by inhibiting cell invasion. J Cancer Res Clin Oncol. 2016;142(3):601–9. [DOI] [PubMed] [Google Scholar]
- 23. Chen X, Gao G. Long noncoding RNA PVT1 as a novel diagnostic biomarker and therapeutic target for melanoma. Biomed Res Int. 2017;2017:7038579. [DOI] [PMC free article] [PubMed] [Google Scholar]