Abstract
Increasing evidence indicates that the dysregulation of microRNAs is associated with the development and progression of various cancers. MicroRNA-139-5p (miR-139-5p) has been reported to have a tumor suppressive role in many types of cancers. The role of miR-139-5p in ovarian cancer (OC) is poorly understood. The purpose of the present study was to explore the expression of miR-139-5p and its function in OC. The results showed that miR-139-5p expression was markedly downregulated in OC tissues and cell lines. In addition, underexpression of miR-139-5p was significantly associated with FIGO stage, lymph mode metastasis, and poor overall survival of OC patients. Functional analyses indicated that overexpression of miR-139-5p significantly inhibited proliferation, colony formation, migration, and invasion of OC cells. Rho-associated coiled-coil-containing protein kinase 2 (ROCK2) was identified as a direct target of miR-139-5p using luciferase reporter assays, qualitative real-time reverse transcriptase PCR (qRT-PCR), and Western blot. In addition, ROCK2 expression was upregulated and was inversely correlated with miR-139-5p levels in OC tissues. Rescue experiments showed that overexpression of ROCK2 effectively reversed the inhibitory effect of OC cells induced by miR-139-5p. Most interestingly, in vivo studies indicated that miR-139-5p markedly suppressed the growth of tumors by repressing ROCK2 expression in nude mice. Taken together, these findings demonstrated that miR-139-5p plays an important tumor suppressor role in OC by directly binding to ROCK2, providing a novel target for the molecular treatment of OC.
Key words: MicroRNAs, miR-139-5p, Ovarian cancer (OC), ROCK2, Proliferation, Invasion
INTRODUCTION
Ovarian cancer (OC) is a gynecological malignancy with high mortality rates worldwide1. Although great efforts have been made in the treatment of OC, the 5-year survival rate for all stages is 35%–38%, mainly due to late diagnosis, easy spreading, and rapid development of chemoresistance2,3. To improve the survival of patients with OC, there is an urgent need to explore the key molecular mechanisms of OC initiation and development to find new strategies and therapeutics.
MicroRNAs (miRNAs), a conserved class of small noncoding RNAs consisting of 18–25 nucleotides in length, have been the active focus as an anticancer therapy in recent years4,5. By binding to the 3′-untranslated region (3′-UTR) of mRNA sequences of their target genes, miRNAs have been reported to participate in a variety of biological processes, such as cell proliferation, cycle, differentiation, apoptosis, fat metabolism, oncogenesis, and drug resistance6,7. miRNAs are aberrantly expressed in various cancers and function as oncogenes or tumor suppressors due to their ability to regulate numerous other oncogenes or tumor suppressors8,9. Although a number of miRNAs have been found to regulate the procession and development of OCs10,11, effective and clinically applicable therapy targets against OC are yet to be developed.
Several laboratories have explored the suppressor roles of miR-139-5p in multiple types of cancers, such as colorectal cancer12, breast cancer13, hepatocellular carcinoma14, oral tongue squamous cell carcinoma15, bladder cancer16, esophageal squamous cell carcinoma17, and non-small cell lung cancer18. However, the role of miR-139-5p in OC is still unknown. Therefore, the purposes of the present study were to investigate the expression of miR-139-5p status in OC tissues and cell lines and to test its role in OC growth and metastasis, as well as to determine its underlying molecular mechanism in OC.
MATERIALS AND METHODS
Patients and Tissue Samples
Biopsy samples of OC tissues and paracancerous tissues were obtained from 46 patients who underwent surgery at the First Affiliated Hospital of Jilin University (Changchun, Jilin, P.R. China). The paracancerous tissues were 3 cm from the edge of the tumor, and there were no obvious tumor cells, as evaluated by an experienced pathologist. After surgery, all samples were immediately frozen in liquid nitrogen and stored at −80°C until use. Written informed consent was obtained from all subjects prior to the study. This study was approved by the Research Ethics Committee of the First Hospital, Jilin University (Changchun, P.R. China).
Cell Culture and Transfection
All cell lines (human OC cell lines SKOV3, A2780, OVCAR, and HO-8910, and a human ovarian surface epithelial cell line HOSEpiC) were purchased from the Tumor Cell Bank of the Chinese Academy of Medical Science (Beijing, P.R. China). All cells were routinely cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco BRL, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS; HyClone, Logan, Utah, USA) and 1% penicillin/streptomycin in a humidified incubator at 37°C and supplemented with 5% CO2.
miR-139-5p mimics or corresponding negative control mimics (miR-NC) were obtained from GenePharma (Shanghai, P.R. China). The Rho-associated coiled-coil-containing protein kinase 2 (ROCK2) overexpression plasmid (pCDNA3.1-ROCK2, without 3′-UTR) were given by Dr. Gou Long (Jilin University, P.R. China). SKOV3 cells were seeded in 24-well plates at a density of 2 × 104 cells/well and incubated for 24 h. Then cells were transfected with miRNA mimics/NC (100 mM) or vectors (5 μg) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in serum-free medium according to the manufacturer’s protocol.
RNA Extraction and Quantitative Real-Time Reverse Transcriptase PCR (qRT-PCR) Analysis
Total RNA was extracted from cultured cells or tissues using TRIzol reagent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. RNA quantity was measured by a SmartSpec Plus spectrophotometer (Bio-Rad, Hercules, CA, USA). RNA purity was evaluated by the A260/A280 ratio. The relative expression of miR-139-5p was detected using a SYBR PrimeScript miRNA RT PCR Kit (Takara, Dalian, P.R. China) in accordance with the manufacturer’s instructions, and U6 small nuclear RNA (snRNA) was used as an internal control. The specific primers of miR-139-5p and U6 were brought from Applied Biosystems (Foster City, CA, USA). For quantitative ROCK2 mRNA level, 2 μg of total RNA was reverse transcribed into cDNA using PrimeScript RT Reagent Kit with oligodT primer (Takara). Quantitative real-time PCR analysis was performed with SYBR Green Real-Time Master Mix (Toyobo, Co., Ltd, Osaka, Japan). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. qRT-PCR and data collection were performed on an Applied Biosystems 7900 Sequence Detection System (Applied Biosystems). Primers of ROCK2 and GAPDH were used in this study as described previously19. The relative expression of miR-139-5p and ROCK2 mRNA was normalized to that of U6 or GAPDH using the 2−ΔΔCt method.
Cell Proliferation and Colony Formation Assays
The cell proliferation assay was performed using the Cell Counting Kit-8 (CCK-8; Dojindo, Japan) according to the manufacturer’s instructions. The optical density (OD) value of each well was measured at 450 nm using a microplate spectrophotometer (Bio-Tek Instruments Inc., Winooski, VT, USA). For the colony formation assay, the methods were as previously described20.
Cell Migration and Invasion Assays
Cell migration was determined by wound healing assay as previously described20. The invasion assay was performed in triplicate using the 24-well Boyden chamber with 8-μm pore size polycarbonate membrane (Corning, Corning, NY, USA). Briefly, transfected cells (5 × 104) were plated into the upper chamber of the Boyden chambers coated with Matrigel in 200 μl of serum-free medium. Complete medium (600 μl) containing 10% FBS was added to the bottom chamber as a chemoattractant. Twenty-four hours after incubation, the noninvading cells in the upper chamber were removed with cotton swabs, whereas invaded cells on the bottom surface of the chamber were fixed, stained with Giemsa, and counted using ImageJ software (NIH, Bethesda, MD, USA).
Dual-Luciferase Reporter Assay
ROCK2 was identified as a miR-139-5p target in TargetScan7.1 (http://www.targetscan.org/vert_71/). The wild-type (Wt) and mutant DNA sequences of ROCK2 were custom synthesized by GeneChem Co., Ltd (Shanghai, P.R. China) and cloned into the pGL3-control vector (Ambion, Austin, TX, USA) at the NheI and XhoI restriction sites. Then SKOV3 cells were cotransfected with 100 ng of the above-described luciferase-ROCK2 mRNA 3′-UTR constructs and with 50 nM of either miR-139-5p mimics or miR-NC together with the Renilla luciferase construct, respectively, using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, the cells were collected, and the luciferase and Renilla luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). Firefly luciferase activity was normalized to Renilla luciferase activity.
Western Blot Analysis
Total proteins were extracted from tissue samples or cultured cells using lysis buffer (Beyotime Institute of Biotechnology, Haimen, P.R. China) on ice for 20 min and were centrifuged 3,000 × g at 4°C. The protein concentrations were determined using bicinchoninic acid (BCA Protein Assay Kit; Pierce, Rockford, IL, USA). Equal quantities of denatured protein samples were separated by 10% sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE) gels and transferred onto nitrocellulose membrane (Amersham BioSciences, Buckinghamshire, UK). After blocking with 5% nonfat milk, the blots were incubated with the following primary antibodies: antiROCK2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-Ki-67, and anti-GAPDH (Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). GAPDH was used as a loading control. The protein bands were detected by SuperSignal West Pico Chemiluminescent Substrate Kit (Pierce). Protein expression levels were detected using Image Lab software (Bio-Rad).
Immunohistochemistry (IHC)
The tumor tissues were fixed in formalin, embedded in paraffin, sectioned, and then heat immobilized or pepsin immobilized in accordance with the manufacturer’s instructions. The slides were stained with hematoxylin and eosin (H&E) or incubated with antibodies against ROCK2 (Santa Cruz Biotechnology). Staining was then performed using an IHC detection kit (Mingrui Biotechnology, Shanghai, P.R. China).
Tumor Xenografts
Stable cell lines with high expressions of miR-139-5p and miR-NC were established by transfecting SKOV3 cells with miR-139-5p mimics or miR-NC. Up to 2 × 106 cells stably expressing miR-139-5p or miR-NC were mixed with Matrigel and injected into 6-week-old female BALB/c mice (18–25 g; five mice in each group; the Laboratory Animal Center of Jilin University, Changchun, P.R. China ), respectively. The mice were monitored, and the tumor size was determined by measuring the tumor width (W) and length (L) every 7 days using a vernier caliper and calculating with the formula: volume (mm3) = 1/2 × W 2 × L. Five weeks after inoculation, the mice were euthanized using a subcutaneous injection with sodium pentobarbital (50 mg/kg), and the tumor tissues were stripped and weighed. The mice used in the experiments were handled in accordance with the guidelines set forth by the Ethics Committee of the Jilin University. All animal experiments were approved by the Animal Care and Use Committee and by the local ethics committee of Jilin University.
Statistical Analysis
Unless specified otherwise, data were presented as the mean ± standard deviation (SD) from at least three separate experiments. Differences among groups were evaluated by two-tailed Student’s t-test or a one-way analysis of variance (ANOVA) using the SPSS Statistics v20.0 software package (SPSS Inc., Chicago, IL, USA). The Kaplan–Meier method was used for analyzing survival data. Associations of miR-139-5p expression and ROCK2 expression were estimated using Pearson’s correlation analysis. In all cases, a value of p < 0.05 was considered statistically significant.
RESULTS
miR-139-5p Is Upregulated in OC and Correlated With Poor Survival
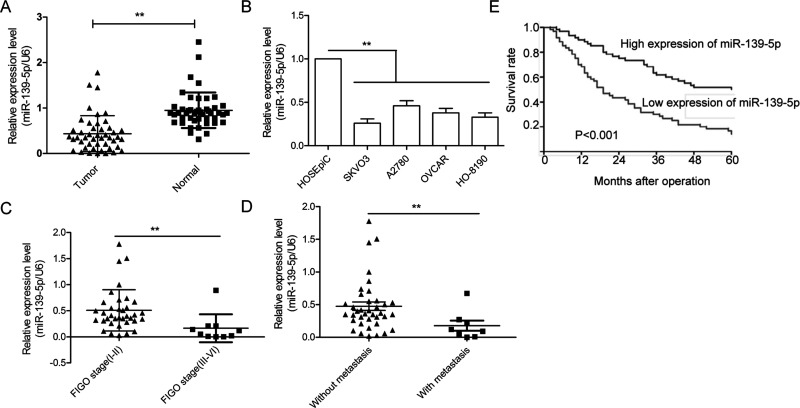
To analyze the expression of miR-139-5p, qRT-PCR was performed on 46 OC tissues paired with normal ovarian tissues. As shown in Figure 1A, miR-139-5p expression was significantly lower in cancer tissues than those of their paired normal tissues. In agreement with these observations, downregulation of miR-139-5p was also confirmed in four OC cell lines (SKOV3, A2780, OVCAR, and HO-8910) compared with a human ovarian surface epithelial cell line HOSEpiC (Fig. 1B). We also found that the expression levels of miR-139-5p in advanced International Federation of Gynecology and Obstetrics (FIGO) stage (III–IV) were significantly downregulated compared with those in low FIGO stage (I and II) (Fig. 1C). Consistent with the above results, miR-139-5p levels in tissues with lymph node metastases were markedly decreased compared to the tissues without lymph node metastases (Fig. 1D). Kaplan–Meier analysis indicated that the 5-year overall survival rates of OC patients with high miR-139-5p expression was significantly decreased compared to patients with low miR-139-5p expression (Fig. 1E). Taken together, these results strongly suggest that miR-139-5p is correlated with poor prognosis and is downregulated in OC.
Figure 1.
Identification of the expression of miR-139-5p in ovarian cancer (OC) tissues and cell lines. (A) The expression level of miR-139-5p was measured in 46 pairs of OC tissues and adjacent normal tissues by qualitative real-time reverse transcriptase PCR (qRT-PCR). (B) The expression of miR-139-5p was detected using qRT-PCR in four OC cell lines (SKOV3, A2780, OVCAR, and HO-8910) and a human ovarian surface epithelial cell line HOSEpiC. (C) Relative miR-139-5p expression in human OC tissues with different International Federation of Gynecology and Obstetrics (FIGO) stages. (D) Relative miR-139-5p expression in human OC tissues with or without lymph node metastasis. (E) Kaplan–Meier survival curves for OC patients were plotted based on high or low miR-139-5p. **p < 0.01.
miR-139-5p Inhibits OC Cell Proliferation, Migration, and Invasion In Vitro
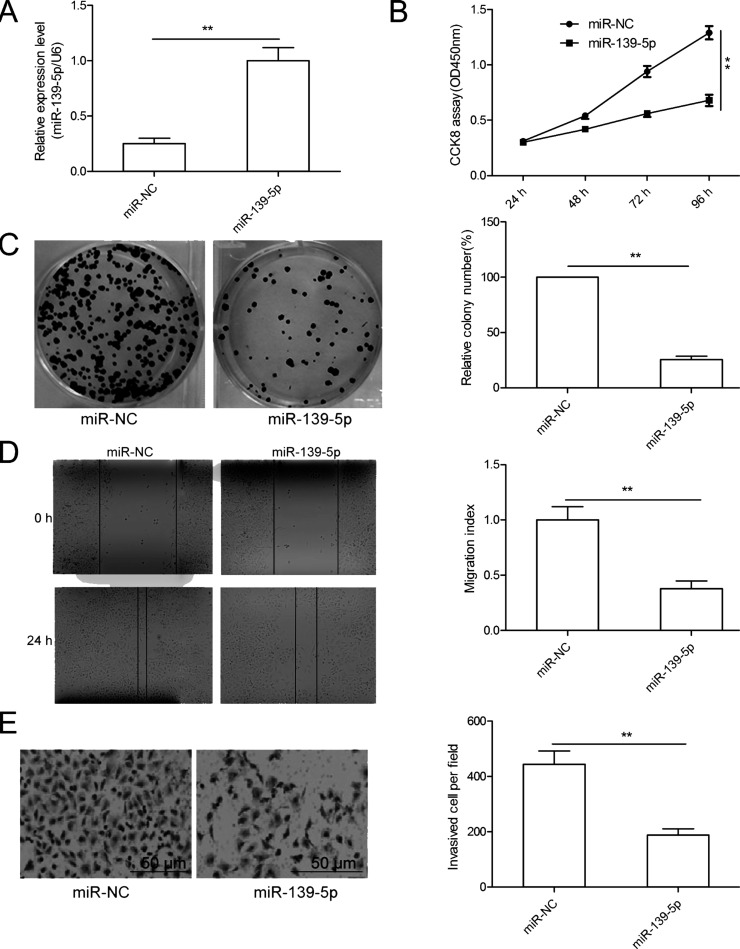
To evaluate miR-139-5p’s biological function in OC, the SKOV3 cells were transfected with miR-139-5p mimics or miR-NC, and then cell proliferation, colony formation, migration, and invasion were determined. qRT-PCR results showed that SKOV3 cells transfected with miR-139-5p mimics significantly enhanced miR-139-5p expression compared with cells transfected with miR-NC (Fig. 2A). We then used the CCK-8 assay to assess the effects of miR-139-5p on cell proliferation. The result showed that miR-139-5p overexpression significantly decreased cell proliferation of SKOV3 cells (Fig. 2B). This result was further confirmed by the colony formation assay (Fig. 2C). Next, we investigated the potential effect of miR-139-5p on cell motility and invasiveness. We found that overexpression of miR-139-5p significantly decreased the migration and invasion of SKOV3 cells (Fig. 2D and E). Collectively, these data clearly show that miR-139-5p can inhibit OC cell proliferation, migration, and invasion.
Figure 2.
miR-139-5p inhibits OC cell proliferation, migration, and invasion in vitro. (A) The expression level of miR-139-5p was measured in SKOV3 cells transfected with miR-139-5p mimic or its negative control (miR-NC) by qRT-PCR. (B–E) Cell proliferation, colony formation, migration, and invasion were determined in SKOV3 cells transfected with miR-139-5p mimic or miR-NC. **p < 0.01.
ROCK2 Is a Direct Target of miR-139-5p in OC Cells
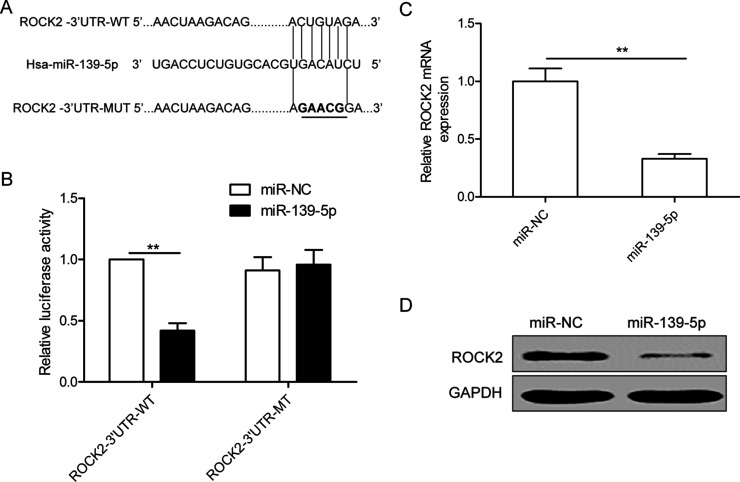
To identify a direct target gene of miR-139-5p, we used a bioinformatic prediction software (TargetScan) and selected ROCK2 as a potential downstream target gene, as shown in Figure 3A. To further confirm targeting of ROCK2 by miR-139-5p, luciferase activity assay was performed. Our results demonstrate that miR-139-5p obviously inhibited the luciferase activity of the Wt of ROCK2-3′UTR, but not that of the mutant-type (Mut) of ROCK2-3′UTR (Fig. 3B). Subsequently, we examined the ROCK2 expression by both mRNA and protein levels in SKOV3 cells transfected with miR-139-5p or miR-NC and found that ROCK2 expression at both mRNA and protein levels was downregulated in SKOV3 cells transfected with the miR-139-5p mimics compared to cells transfected with the miR-NC (Fig. 3C and D). These results suggested that ROCK2 might be a target of miR-139-5p in OC cells.
Figure 3.
Rho-associated coiled-coil-containing protein kinase 2 (ROCK2) is a direct target of miR-139-5p in OC cells. (A) The putative miR-139-5p binding sites and mutant (Mut) 3′-untranslated region (3′-UTR) ROCK2 sites are shown. The replaced site is underlined. WT, wild type; MT, mutant type. (B) Relative luciferase activity was measured in SKOV3 cells cotransfected with WT/MT-ROCK2-3′UTR reporter plasmid and miR-139-5p mimic or miR-NC. (C, D) The ROCK2 expression on mRNA and protein levels was measured in SKOV3 cells transfected with miR-139-5p mimic and miR-NC. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control. **p < 0.01.
ROCK2 Was Upregulated and Negatively Correlated With miR-139-5p in Ovarian Cancer Tissues
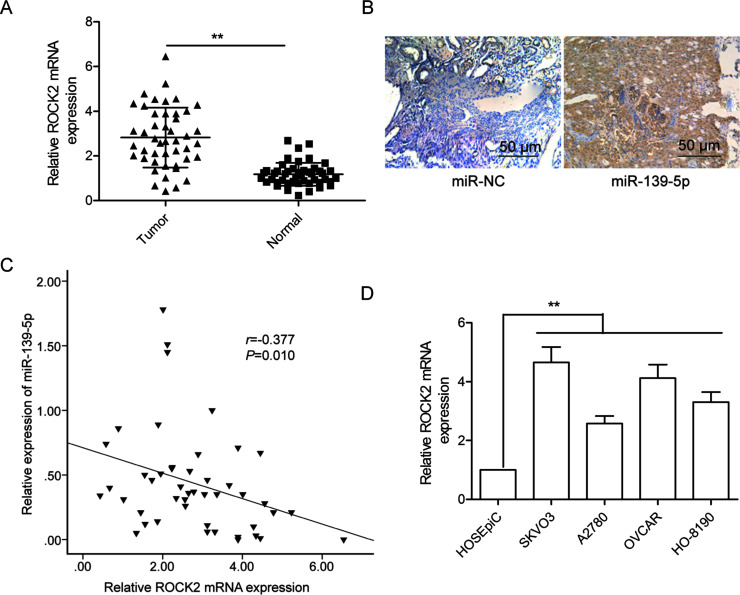
To further explore the relationship between miR-139-5p and ROCK2, we examined the mRNA expression of ROCK2 in OC tissues, and tissues adjacent to OC, by qRT-PCR. As shown in Figure 4A, ROCK2 mRNA expression was higher in OC tissues than in adjacent normal tissues. IHC further confirmed that ROCK2 protein expression was increased in OC tissues compared to adjacent normal tissues (Fig. 4B). Pearson’s correlation analysis demonstrated that ROCK2 mRNA expression was inversely correlated with miR-139-5p levels in OC tissues (r = −0.377, p = 0.01) (Fig. 4C). In addition, we also found that ROCK2 mRNA was significantly upregulated in four OC cell lines compared to the normal ovary cells (Fig. 4D).
Figure 4.
ROCK2 was upregulated and negatively correlated with miR-139-5p in OC tissues. (A) The ROCK2 mRNA expression level was measured in 46 pairs of OC tissues and adjacent normal tissues by qRT-PCR. GAPDH was used as an internal control. (B) The ROCK2 protein expression was determined in OC tissues and adjacent normal tissues by immunohistochemistry. (C) The correlation of the expression levels of ROCK2 and miR-139-5p was analyzed in OC tissues by Pearson’s correlation assay (n = 46). (D) The ROCK2 mRNA expression was determined in four OC cell lines (SKOV3, A2780, OVCAR, and HO-8910) and a human ovarian surface epithelial cell line (HOSEpiC) by qRT-PCR. GAPDH was used as an internal control. **p < 0.01.
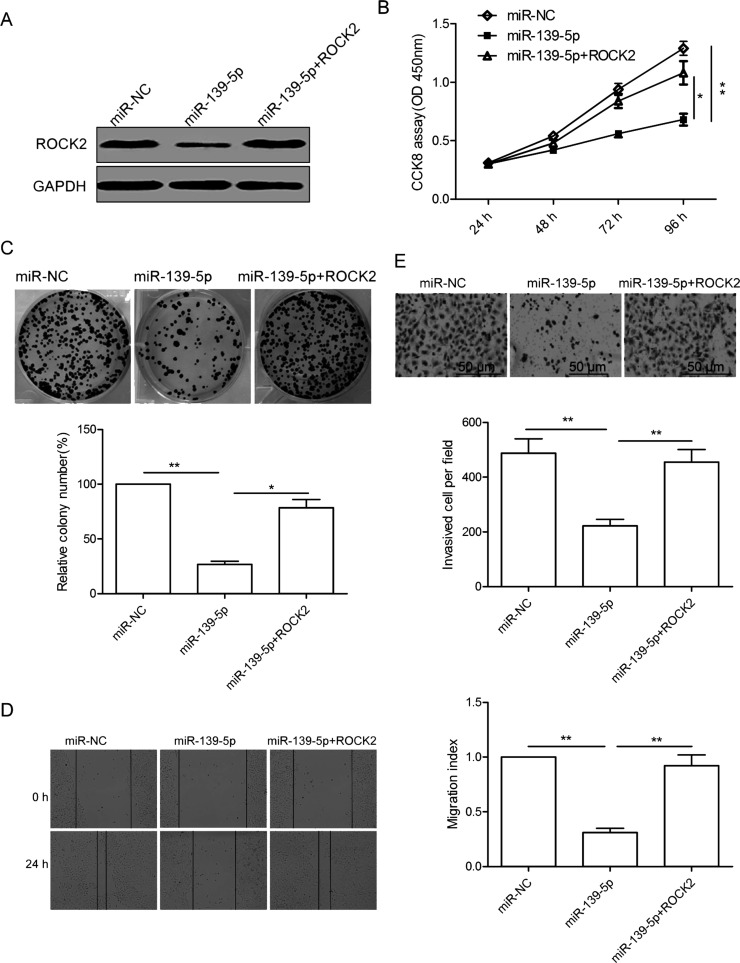
Overexpression of ROCK2 Ablates the Inhibitory Effects of miR-139-5p in Ovarian Cancer Cells
To validate whether miR-139-5p affects OS growth and invasion via targeting ROCK2, SKOV3 cells with miR-139-5p mimics or miR-NC were transfected with pcDNA3.1-ROCK2 plasmid (without the 3′-UTR). The forced expression of the ROCK2 vector could recover ROCK2 protein expression in SKOV3 cells transfected with miR-139-5p (Fig. 5A). In addition, overexpression of ROCK2 reversed the inhibitory effects of miR-139-5p on SKOV3 cell proliferation, colony formation, migration, and invasion (Fig. 5B–E). These results suggested that miR-139-5p exerts a suppressive role in OC by repressing ROCK2.
Figure 5.
Overexpression of ROCK2 ablates the inhibitory effects of miR-139-5p in OC cells. (A) Western blot analysis of ROCK2 levels in SKOV3 cells after transfection with miR-139-5p mimic or miR-NC, along with (or without) ROCK2 cDNA vector lacking the 3′-UTR region. GAPDH was used as an internal control. (B–E) Cell proliferation, colony formation, migration, and invasion were determined in SKOV3 cells after transfection with miR-139-5p mimic or miR-NC, along with (or without) ROCK2 cDNA vector lacking the 3′-UTR region. *p < 0.05, **p < 0.01.
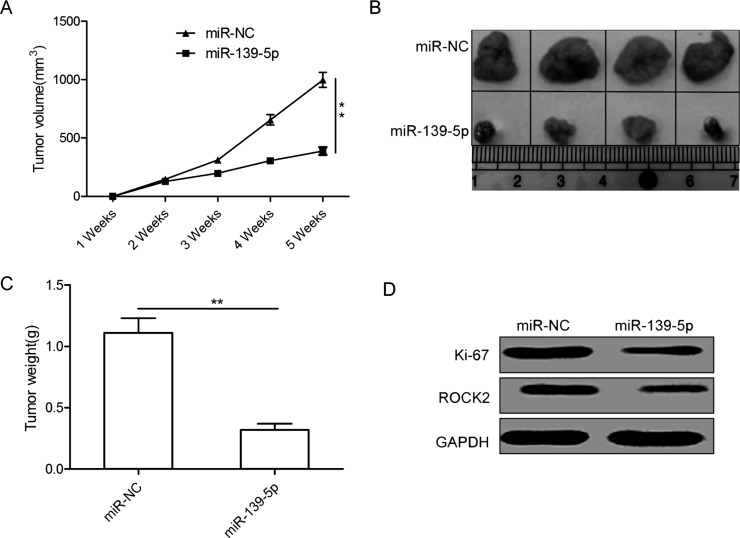
miR-139-5p Inhibits Ovarian Cancer Growth in a Mouse Model
To evaluate the effects of miR-139-5p on tumor growth in vivo, we manipulated the expression levels of miR-139-5p in SKOV3 cells, and then injected cells into the flanks of nude mice to establish subcutaneous OC xenografts. It was found that tumor growth was slower in the SKOV3/miR-139-5p group than in the SKOV3/miR-NC group (Fig. 6A). Consistent with the tumor growth curve, the size and weight of the tumor from the SKOV3/miR-139-5p group were significantly decreased compared to the tumor from the SKOV3/miR-NC group 35 days after implantation (Fig. 6B and C). In addition, overexpression of miR-139-5p downregulated the protein expression of Ki-67, a cell proliferation marker (Fig. 6D). We also found that ROCK2 protein expression was downregulated in the SKOV3/miR-139-5p tumors compared to the tumors from the SKOV3/miR-NC group. These data suggest that increasing miR-139-5p levels can suppress the growth of OC in vivo.
Figure 6.
miR-139-5p inhibits tumor growth in a mouse model. (A) The curve of tumor growth in different groups. (B) Photographs of xenograft tumors in different groups. (C) Average tumor weight in different groups. (D) Western blot analysis of ROCK2 and Ki-67 expression in xenograft tumors. GAPDH was used as an internal control. *p < 0.05, **p < 0.01.
DISCUSSION
Recent advances in the understanding of the molecular mechanism of OC have revealed that numerous miRNAs play an important role in the progression and development of tumors by binding to specific sites in the target mRNA10,11. For example, Wu et al. reported that downregulation of miR-221-3p in OC cell lines suppressed cell proliferation and migration and promoted tumor cell apoptosis by inducing apoptosis protease-activating factor 1 (APAF1) protein21. Li et al. found that miR-494 overexpression in OC cells remarkably inhibits cell proliferation, colony formation, migration, and invasion; induces cell apoptosis and G0/G1 phase arrest in vitro; and suppresses tumor growth in a nude mouse xenograft model system by targeting insulin-like growth factor 1 receptor (IGF1R)22. Lin et al. found that ectopic overexpression of miR-215 could significantly inhibit OC cell proliferation, colony formation, migration, and invasion in vitro, as well as suppress tumor growth in vivo by repressing ribosome assembly factor nuclear integrity/regulatoy particle non-ATPase 12 (NIN/RPN12)-binding protein (NOB1)23. Our present study revealed that the expressions of miR-139-5p were markedly downregulated in OC tissues and cell lines. Additionally, ectopic expression of miR-139-5p inhibited OC proliferation, colony formation, migration, and invasion in vitro and decreased tumor growth in vivo. Collectively, these findings indicated that miR-139-5p may have an essential role to play in the tumorigenesis and progression of OC.
Two precursors of miR-139, miR-139-5p and miR-139-3p, are located within the second intron of the phosphodiesterase 2A (PDE2A) gene on chromosome 11q13.424. The miRNA miR-139-5p has been reported to be downregulated and play a suppressive role in multiple cancers12–18. However, the expression state and function of miR-139-5p remain unclear. In this study, we assessed miR-139-5p expression levels in both OC cell lines and human OC tissue samples and compared them to the expression of miR-139-5p in a normal ovarian cell line and matched normal ovarian tissues and found that miR-139-5p expression was lower in OC tissues and OC cell lines compared to normal ovarian tissues and cell lines. We also showed that decreased miR-139-5p was significantly associated with lymph node metastasis, FIGO stage, and poor overall survival of OC patients. Moreover, we investigated the biological function of miR-139-5p in OC by CCK-8, colony formation, wound healing, Transwell chamber assays in vitro, and tumorigenesis of mouse OC in vivo. It was found that overexpression of miR-139-5p in SKOV3 cells inhibited proliferation, colony formation, migration, and invasion in vitro, as well as suppressed tumor growth in vivo. These results implied that miR-139-5p might function as a tumor suppressor in OC.
It was well known that miRNAs behave as oncogenes or tumor suppressor genes depending on the cellular function of their targets9. In this study, we used TargetScan to predict targets of miR-139-5p and found that ROCK2 contains a highly conserved miR-139-5p binding site on the 3′-UTR. ROCK2 is a serine/threonine kinase that was involved in diverse biological processes, such as regulation of cytokinesis, smooth muscle contraction, the formation of actin stress fibers and focal adhesions25,26. ROCK2, as a key downstream effector of RhoA small guanosine triphosphatase (GTPase), has been reported to play a crucial role in proliferation, differentiation, apoptosis, and oncogenic transformation in many types of cancer including OC27,28. It has been shown that downregulation of ROCK2 inhibits OC growth and invasion29 and sensitizes the cell to cisplatin30. In the present study, through the luciferase assay, qRT-PCR, and Western blot assays, ROCK2 was confirmed as a direct target of miR-139-5p in OC, which was consistent with the findings of previous studies in that ROCK2 was a direct target of hepatocellular carcinoma31 and colorectal cancer32. Furthermore, ROCK2 mRNA level was inversely correlated with miR-139-5p level in OC tissues. Exogenous overexpression of ROCK2 partially reversed the inhibitory effects of miR-139-5p on OC proliferation, colony formation, migration, and invasion. These results suggest that miR-139-5p exerts its biological role in OC partially by repressing ROCK2.
In summary, our results first describe that the miR-139-5p level in both tissue and cell lines was significantly lower in OC patients, and its expression level was significantly associated with lymph node metastasis, FIGO stage, and poor overall survival of OC patients. In addition, our results demonstrated that miR-139-5p significantly inhibited OC cell proliferation, colony formation, migration, and invasion in vitro and impaired tumor growth in vivo by targeting ROCK2. These findings suggested that miR-139-5p plays a suppressive role in OC. Although many of the mechanisms still remain unclear for miR-139-5p-based suppression of OC, our findings are encouraging and might provide a new therapeutic strategy for OC prevention and treatment.
ACKNOWLEDGMENTS
This work was supported by the Jilin Province Key Scientific and Technical Project (20150203016NY) and the General Project of the Department of Science and Technology of Jilin Province (20160203016NY and 20160519018JH).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. [DOI] [PubMed] [Google Scholar]
- 2. Permuth-Wey J, Sellers TA. Epidemiology of ovarian cancer. Methods Mol Biol. 2009;472:413–37. [DOI] [PubMed] [Google Scholar]
- 3. Dembo AJ, Davy M, Stenwig AE, Berle EJ, Bush RS, Kjorstad K. Prognostic factors in patients with stage I epithelial ovarian cancer. Obstet Gynecol. 1990;75(2):263–73. [PubMed] [Google Scholar]
- 4. He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. [DOI] [PubMed] [Google Scholar]
- 5. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. [DOI] [PubMed] [Google Scholar]
- 6. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 7. Jovanovic M, Hengartner MO. miRNAs and apoptosis: RNAs to die for. Oncogene 2006;25(46):6176–87. [DOI] [PubMed] [Google Scholar]
- 8. Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: A new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15(6):341–55. [DOI] [PubMed] [Google Scholar]
- 9. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6(11):857–66. [DOI] [PubMed] [Google Scholar]
- 10. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14(9):2690–5. [DOI] [PubMed] [Google Scholar]
- 11. Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–707. [DOI] [PubMed] [Google Scholar]
- 12. Miyoshi J, Toden S, Yoshida K, Toiyama Y, Alberts SR, Kusunoki M, Sinicrope FA, Goel A. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci Rep. 2017;7:43393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang HD, Sun DW, Mao L, Zhang J, Jiang LH, Li J, Wu Y, Ji H, Chen W, Wang J, Ma R, Cao HX, Wu JZ, Tang JH. MiR-139-5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Commun. 2015;465(4):702–13. [DOI] [PubMed] [Google Scholar]
- 14. Qiu G, Lin Y, Zhang H, Wu D. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;463(3):315–21. [DOI] [PubMed] [Google Scholar]
- 15. Chen Z, Yu T, Cabay RJ, Jin Y, Mahjabeen I, Luan X, Huang L, Dai Y, Zhou X. miR-486-3p, miR-139-5p, and miR-21 as biomarkers for the detection of oral tongue squamous cell carcinoma. Biomark Cancer 2017;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107(9):1233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu R, Yang M, Meng Y, Liao J, Sheng J, Pu Y, Yin L, Kim SJ. Tumor-suppressive function of miR-139-5p in esophageal squamous cell carcinoma. PLoS One 2013;8(10):e77068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu W, Hang M, Yuan CY, Wu FL, Chen SB, Xue K. MicroRNA-139-5p inhibits cell proliferation and invasion by targeting insulin-like growth factor 1 receptor in human non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(4):3864–70. [PMC free article] [PubMed] [Google Scholar]
- 19. Xu Z, Hong Z, Ma M, Liu X, Chen L, Zheng C, Xi X, Shao J. Rock2 promotes RCC proliferation by decreasing SCARA5 expression through beta-catenin/TCF4 signaling. Biochem Biophys Res Commun. 2016;480(4):586–593. [DOI] [PubMed] [Google Scholar]
- 20. Wang Y, Xu C, Wang Y, Zhang X. MicroRNA-365 inhibits ovarian cancer progression by targeting Wnt5a. Am J Cancer Res. 2017;7(5):1096–106. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21. Wu Q, Ren X, Zhang Y, Fu X, Li Y, Peng Y, Xiao Q, Li T, Ouyang C, Hu Y, Zhang Y, Zhou W, Yan W, Guo K, Li W, Hu Y, Yang X, Shu G, Xue H, Wei Z, Luo Y, Yin G. MiR-221-3p targets ARF4 and inhibits the proliferation and migration of epithelial ovarian cancer cells. Biochem Biophys Res Commun. 2017;497(4):1162–70. [DOI] [PubMed] [Google Scholar]
- 22. Li N, Zhao X, Wang L, Zhang S, Cui M, He J. miR-494 suppresses tumor growth of epithelial ovarian carcinoma by targeting IGF1R. Tumour Biol. 2016;37(6):7767–76. [DOI] [PubMed] [Google Scholar]
- 23. Lin Y, Jin Y, Xu T, Zhou S, Cui M. MicroRNA-215 targets NOB1 and inhibits growth and invasion of epithelial ovarian cancer. Am J Transl Res. 2017;9(2):466–77. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Zhang HD, Jiang LH, Sun DW, Li J, Tang JH. MiR-139-5p: Promising biomarker for cancer. Tumour Biol. 2015;36(3):1355–65. [DOI] [PubMed] [Google Scholar]
- 25. Denk-Lobnig M, Martin AC. Modular regulation of Rho family GTPases in development. Small GTPases 2017:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang P, Li W, Peng J, Qi S, Song L, Liu C, Li F. Clinicopathological significance of RhoA expression in digestive tract cancer: A systematic review and meta-analysis. Clin Lab. 2016;62(10):1955–64. [DOI] [PubMed] [Google Scholar]
- 27. Guo J, Chen L, Luo N, Yang W, Qu X, Cheng Z. Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep. 2015;33(6):3124–30. [DOI] [PubMed] [Google Scholar]
- 28. Wang HF, Takenaka K, Nakanishi A, Miki Y. BRCA2 and nucleophosmin coregulate centrosome amplification and form a complex with the Rho effector kinase ROCK2. Cancer Res. 2011;71(1):68–77. [DOI] [PubMed] [Google Scholar]
- 29. Sawada K, Morishige K, Tahara M, Ikebuchi Y, Kawagishi R, Tasaka K, Murata Y. Lysophosphatidic acid induces focal adhesion assembly through Rho/Rho-associated kinase pathway in human ovarian cancer cells. Gynecol Oncol. 2002;87(3):252–9. [DOI] [PubMed] [Google Scholar]
- 30. Ohta T, Takahashi T, Shibuya T, Amita M, Henmi N, Takahashi K, Kurachi H. Inhibition of the Rho/ROCK pathway enhances the efficacy of cisplatin through the blockage of hypoxia-inducible factor-1alpha in human ovarian cancer cells. Cancer Biol Ther. 2012;13(1):25–33. [DOI] [PubMed] [Google Scholar]
- 31. Wong CC, Wong CM, Tung EK, Au SL, Lee JM, Poon RT, Man K, Ng IO. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology 2011;140(1):322–31. [DOI] [PubMed] [Google Scholar]
- 32. Shen K, Mao R, Ma L, Li Y, Qiu Y, Cui D, Le V, Yin P, Ni L, Liu J. Post-transcriptional regulation of the tumor suppressor miR-139-5p and a network of miR-139-5p-mediated mRNA interactions in colorectal cancer. FEBS J. 2014;281(16):3609–24. [DOI] [PubMed] [Google Scholar]