Abstract
Cervical cancer is the fourth most common malignancy among females worldwide. MicroRNA-379 (miR-379) is aberrantly expressed in multiple human cancer types. However, the expression pattern, roles, and detailed regulatory mechanisms of miR-379 in cervical cancer remain unknown. In this study, we found that miR-379 expression was downregulated in cervical cancer tissues and cell lines. Low miR-379 expression was correlated with International Federation of Gynecology and Obstetrics (FIGO) stage, lymph node metastasis, and distant metastasis. Additionally, miR-379 overexpression suppressed the proliferation and invasion of cervical cancer cells. Furthermore, V-crk avian sarcoma virus CT10 oncogene homolog-like (CRKL) was identified as a direct target of miR-379 in cervical cancer. CRKL was upregulated in cancer tissues and negatively correlated with miR-379 expression. Moreover, restored CRKL expression rescued the inhibitory effects of miR-379 overexpression on cell proliferation and invasion. In conclusion, miR-379 may serve as a tumor suppressor in cervical cancer by directly targeting CRKL. Restoring miR-379 expression may be an effective strategy for the treatment of cervical cancer.
Key words: Cervical cancer, MicroRNA-379 (miR-379), Proliferation, Invasion, V-crk avian sarcoma virus CT10 oncogene homolog-like (CRKL)
INTRODUCTION
Cervical cancer is the fourth most common malignancy among females worldwide1. Approximately 530,000 new cases of cervical cancer are diagnosed, and 270,000 cancer-related deaths are recorded annually2. Most cervical cancer cases are caused by infection with human papillomavirus (HPV), and HPV DNA is identified in approximately 95% of malignant cervical lesions3. The abnormal expression or activity of specific genes is responsible for the pathogenesis of cervical cancer4–6. Currently, the main treatments for patients with cervical cancer include surgery resection, radiotherapy, and chemotherapy7. Despite improvements in cervical cancer treatments, about 30% of patients develop cancer recurrence, lymph node metastasis, or distant metastasis and eventually obtain an unfavorable prognosis8. The 5-year overall survival rate is less than 40%, particularly for patients with cervical cancer at advanced stages9. Therefore, the molecular mechanisms underlying the occurrence and development of cervical cancer must be elucidated to develop novel therapeutic strategies.
MicroRNAs (miRNAs), a class of endogenous noncoding small RNAs, negatively regulate gene expression at the posttranscriptional level by either inducing mRNA degradation or inhibiting translation via interaction with the 3′-untranslated region (3′-UTR) of target mRNAs10,11. Computational estimations suggest that the human genome possesses more than 1,000 miRNAs, which regulate one third of human protein-encoding genes12. miRNAs participate in a wide range of physiological processes, including cell proliferation, cycle, apoptosis, differentiation, development, metastasis, and metabolism13–15. The dysregulation of miRNAs is involved in various diseases, particularly cancers16, such as cervical17, bladder18, gastric19, and lung cancers20. Aberrantly expressed miRNAs may serve as either tumor suppressors or oncogenes depending on the tumor type and biological roles of their target genes21. Hence, miRNAs exhibit potential as novel therapeutic targets for the diagnosis and therapy of human cancers.
miR-379, located at chromosome 14q32.31, is aberrantly expressed in multiple human cancer types22,23. However, the expression pattern, roles, and detailed regulatory mechanisms of miR-379 in cervical cancer remain unknown. Therefore, in the current study, the expression levels, biological roles, and direct target genes of miR-379 in cervical cancer were investigated.
MATERIALS AND METHODS
Tissue Samples and Cell Lines
This study was approved by the Ethics Committee of the Chinese PLA 101 Hospital. Written informed consent was also obtained from all participants involved in this study. Fifty-three paired cervical cancer tissues and corresponding adjacent normal tissues were obtained from patients who underwent surgery at the Department of Obstetrics and Gynecology, Chinese PLA 101 Hospital, between May 2014 and October 2016. None of the cervical cancer patients had received radiotherapy, chemotherapy, or other treatments prior to surgery. All tissues were immediately frozen in liquid nitrogen and stored at −80°C.
Cervical cancer cell lines (HeLa, Ca-Ski, SiHa, and C-33A) were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, P.R. China). A human normal cervical epithelial cell line (Ect1/E6E7) was acquired from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS) and 1% penicillin/streptomycin (all from Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Cells were grown at 37°C in a humidified atmosphere under 5% CO2.
Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. To detect miR-379 expression, total RNA was reverse transcribed into cDNA using a TaqMan® MicroRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was performed using TaqMan MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) following the manufacturer’s instructions. To quantify mRNA expression, the complementary DNA was synthesized with a Moloney murine leukemia virus reverse transcription system (Promega Corporation, Madison, WI, USA) and subjected to qPCR using a SYBR Premix Ex Taq™ kit (Takara Biotechnology Co., Ltd., Dalian, P.R. China). The expression levels of miR-379 and CRKL mRNA were normalized with reference to the expression levels of U6 small nuclear RNA and GAPDH, respectively. Each sample was analyzed in triplicate. The relative expression was calculated by the 2−ΔΔCt method24.
RNA Oligoribonucleotides and Plasmids
The miR-379 mimics and miRNA mimic negative control (miR-NC) used in this study were synthesized by Shanghai GenePharma Co. Ltd (Shanghai, P.R. China). CRKL overexpression plasmid without the 3′-UTR (pcDNA3.1-CRKL) and empty plasmid (pcDNA3.1) were acquired from Guangzhou RiboBio Co., Ltd. (Guangzhou, P.R. China). Cells were seeded into six-well culture plates at a density of 7 × 105 cells per well. When cell density reached 70%–80% confluence, transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Cell medium was replaced with fresh DMEM containing 10% FBS. Cell transfection efficiency was evaluated using RT-qPCR or Western blotting analysis.
Cell Counting Kit-8 (CCK-8) Assay
The CCK-8 assay was used to evaluate cervical cancer cell proliferation. Transfected cells were collected at 24 h posttransfection and suspended in DMEM containing 10% FBS. A total of 3,000 transfected cells were seeded in 96-well culture plates and incubated at 37°C with 5% CO2 for 0, 24, 48, or 72 h. At the indicated time points, the CCK-8 assay was performed according to the manufacturer’s protocols. Briefly, 10 μl of CCK-8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was added to each well and incubated for another 2 h at 37°C. The optical density (OD) was measured at 450-nm wavelength with a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Experiments were performed in triplicate and repeated at least three times independently.
In Vitro Invasion Assay
In vitro invasion assay was performed using Transwell chambers coated with Matrigel (Becton-Dickinson, Franklin Lakes, NJ, USA). After transfection for 48 h, cells were collected and suspended in FBS-free DMEM. A total of 5 × 104 cells were plated into the upper chamber. DMEM containing 20% FBS was added to the lower chamber. After incubation at 37°C with 5% CO2 for 24 h, the cells remaining on the upper surface of the filters were removed with a cotton swab. The cells that had invaded into the lower surface of the filters were fixed with 100% methanol, stained with 0.5% crystal violet, and dried in air. The invasive cells were photographed and counted under an inverted microscope (magnification: 200×; Olympus Corporation, Tokyo, Japan) in five random fields. All assays were performed in triplicate.
Bioinformatics Analysis and Luciferase Reporter Assay
TargetScan online software (www.targetscan.org/) and miRanda (www.microrna.org) were used to predict the potential targets of miR-379. From the candidates, CRKL was selected as it has previously been demonstrated to contribute to cervical cancer progression25. To confirm this hypothesis, luciferase reporter plasmid, pMIR-CRKL-3′-UTR wild type (Wt), and pMIR-CRKL-3′-UTR mutant (Mut) were synthesized and confirmed by Shanghai GenePharma Co., Ltd. Cells were seeded into 24-well plates at a density of 60%–70% confluence. After incubation overnight, cells were transfected with miR-379 mimics or miR-NC, along with pMIR-CRKL-3′-UTR Wt or pMIR-CRKL-3′-UTR Mut, using Lipofectamine 2000. The luciferase activities were determined at 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions. Renilla luciferase activities were normalized to firefly luciferase activities.
Western Blotting Analysis
Total protein was extracted from tissues or cells using 1× radioimmunoprecipitation assay lysis buffer (Santa Cruz Biotechnology, Santa Cruz, CA, USA) and subjected to quantification using a BCA Protein Assay kit (Bio-Rad). Equal amount of protein was separated in 10% sodium dodecyl sulfate-polyacrylamide (SDS-PAGE) gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). Membranes were then blocked for 2 h at room temperature with 5% skimmed milk in TBS containing 0.05% Tween 20 (TBST) and incubated at 4°C overnight with primary antibodies against human CRKL (1:1,000 dilution; Cat. No. sc-365471; Santa Cruz Biotechnology) and GAPDH (1:1,000 dilution; Cat. No. sc-69778; Santa Cruz Biotechnology). After washing three times with TBST, the membranes were incubated at 37°C for 1 h with corresponding horseradish peroxidase-conjugated secondary antibody (1:5,000 dilution; Cat. No. sc-2005; Santa Cruz Biotechnology). Advanced Western Blot Detection Kit (Thermo Fisher Scientific) was utilized to visualize the specific protein bands. GAPDH served as control. The experiment was repeated at least three times.
Statistics Analysis
All statistical analyses were carried out using the SPSS 22.0 software (SPSS, Inc., Chicago, IL, USA). Data are expressed as the means ± standard deviations (SD), and differences between groups were compared using Student’s t-test or ANOVA, as appropriate. Student–Newman–Keuls test was used as a post hoc test following ANOVA. Spearman’s correlation analysis was utilized to investigate the correlation between miR-379 and CRKL mRNA expression level in cervical cancer tissues. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-379 Is Downregulated in Cervical Cancer Tissues and Cell Lines
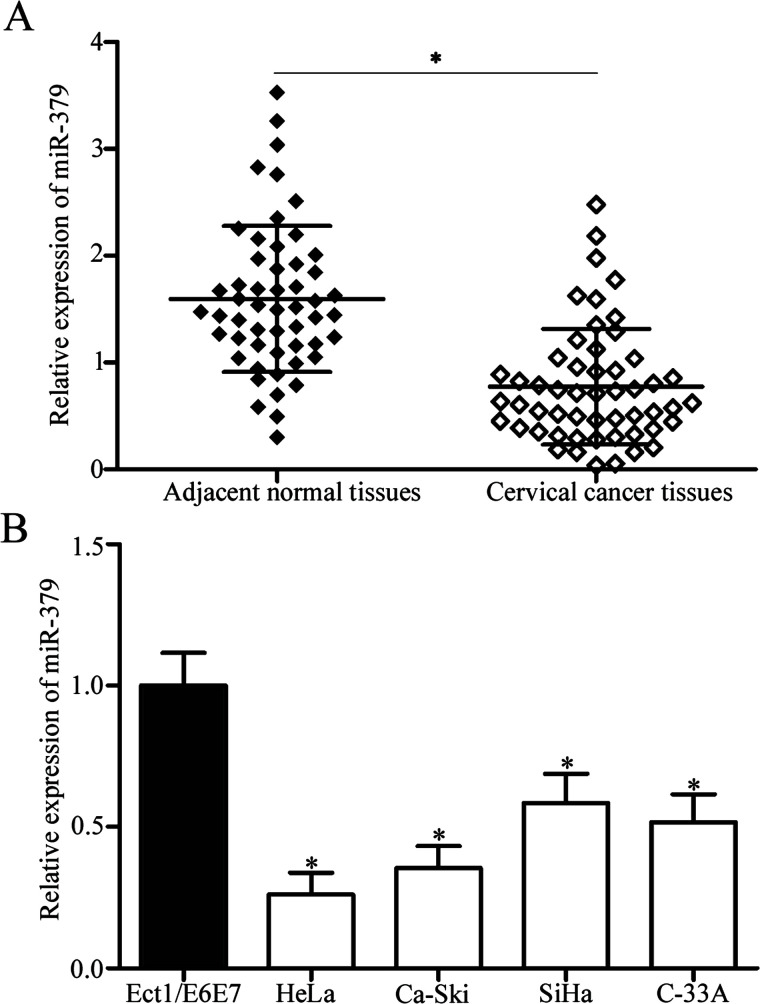
To characterize the role of miR-379 in cervical cancer, we determined miR-379 expression in 53 paired cervical cancer tissues and their corresponding adjacent normal tissues. RT-qPCR data showed that the miR-379 expression in cervical cancer tissues decreased compared with that in the corresponding adjacent normal tissues (p < 0.05) (Fig. 1A). We also analyzed the association between miR-379 and clinicopathological features of patients with cervical cancer, including age, histology, tumor size, International Federation of Gynecology and Obstetrics (FIGO) stage, lymph node metastasis, and distant metastasis. All patients with cervical cancer were categorized based on the median miR-379 expression into either miR-379 low-expression group (n = 27) or miR-379 high-expression group (n = 26). As shown in Table 1, low miR-379 expression was significantly correlated with FIGO stage (p = 0.003), lymph node metastasis (p = 0.001), and distant metastasis (p = 0.021). However, miR-379 expression was not significantly correlated with age (p = 0.498), histology (p = 0.465), and tumor size (p = 0.695).
Figure 1.
MicroRNA-379 (miR-379) is downregulated in cervical cancer tissues and cell lines. (A) miR-379 expression was determined in 53 paired cervical cancer tissues and their corresponding adjacent normal tissues through reverse transcription quantitative polymerase chain reaction (RT-qPCR) analysis. *p < 0.05 compared with adjacent normal tissues. (B) miR-379 expression levels in four human cervical cancer cell lines (HeLa, Ca-Ski, SiHa, and C-33A) and a human normal cervical epithelial cell line (Ect1/E6E7) were detected through RT-qPCR analysis. *p < 0.05 compared with Ect1/E6E7.
Table 1.
The Association Between MicroRNA-379 Expression and Clinicopathologic Features of Patients With Cervical Cancer
| Clinicopathologic Features | No. of Cases | miR-379 Expression | p Value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | 0.498 | |||
| <50 | 24 | 11 | 13 | |
| ≥50 | 29 | 16 | 13 | |
| Histology | 0.465 | |||
| Squamous cell carcinoma | 41 | 22 | 19 | |
| Adenocarcinoma | 12 | 5 | 7 | |
| Tumor size (cm) | 0.695 | |||
| <4 | 21 | 10 | 11 | |
| ≥4 | 32 | 17 | 15 | |
| FIGO stage | 0.003* | |||
| I–II | 18 | 4 | 14 | |
| III–IV | 35 | 23 | 12 | |
| Lymph node metastasis | 0.001* | |||
| Yes | 29 | 21 | 8 | |
| No | 24 | 6 | 18 | |
| Distant metastasis | 0.021* | |||
| Yes | 16 | 12 | 4 | |
| No | 37 | 15 | 22 | |
FIGO stage, International Federation of Gynecology and Obstetrics stage.
p < 0.05.
We also examined miR-379 expression in four human cervical cancer cell lines (HeLa, Ca-Ski, SiHa, and C-33A) and a human normal cervical epithelial cell line (Ect1/E6E7). As shown in Figure 1B, the miR-379 expression was downregulated in all four cervical cancer cell lines compared with that in Ect1/E6E7 (p < 0.05). Among the cervical cancer cell lines, HeLa and Ca-Ski cells expressed relatively lower miR-379 levels and were thus selected for further experiments. Hence, miR-379 may play important roles in cervical cancer initiation and progression.
miR-379 Overexpression Inhibits the Proliferation and Invasion of Cervical Cancer Cells
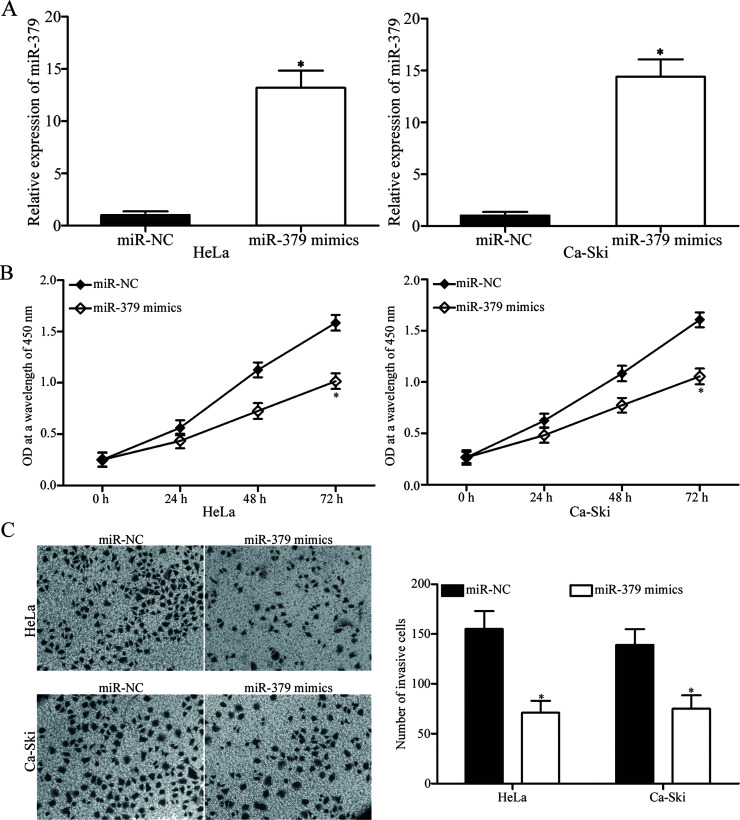
To explore the biological functions of miR-379 in cervical cancer, we transfected HeLa and Ca-Ski cells with miR-379 mimics or miR-NC. RT-qPCR analysis was conducted to evaluate transfection efficiency. As shown in Figure 2A, miR-379 expression was dramatically upregulated in HeLa and Ca-Ski cells following transfection with miR-379 mimics (p < 0.05). Afterward, CCK-8 assay and in vitro invasion assay were performed to investigate the effects of miR-379 on the proliferation and invasion of cervical cancer cells, respectively. Upregulation of miR-379 suppressed the proliferation (p < 0.05) (Fig. 2B) and invasion (p < 0.05) (Fig. 2C) of HeLa and Ca-Ski cells compared with that in miR-NC-transfected cells. Hence, miR-379 may function as a tumor suppressor in cervical cancer.
Figure 2.
Tumor-suppressive effects of miR-379 on the proliferation and invasion of cervical cancer cells. (A) HeLa and Ca-Ski cells were transfected with miR-379 mimics or miR-NC. After 48 h of transfection, RT-qPCR analysis was conducted to evaluate transfection efficiency. *p < 0.05 compared with miR-NC. (B) Effect of miR-379 overexpression on cervical cancer cell proliferation was assessed using cell counting kit-8 (CCK-8) assay. *p < 0.05 compared with miR-NC. (C) In vitro invasion assay was performed to examine the invasive ability of HeLa and Ca-Ski cells transfected with miR-379 mimics or miR-NC. *p < 0.05 compared with miR-NC.
CRKL Is a Direct Target of miR-379 in Cervical Cancer
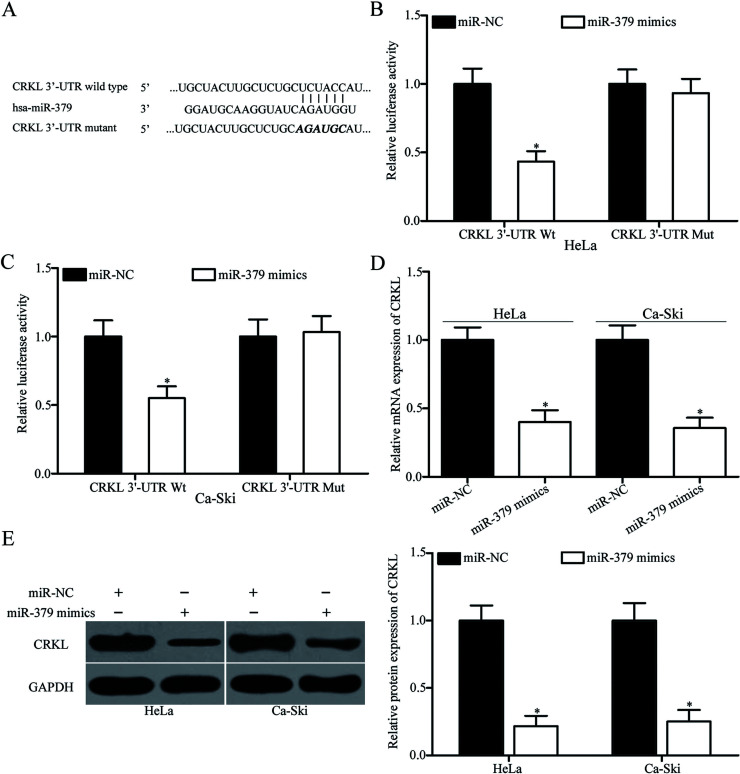
To determine the molecular mechanism underlying the tumor-suppressing roles of miR-379 in cervical cancer, we performed bioinformatics analysis for predicting the potential targets of miR-379. Among the candidate targets, CRKL (Fig. 3A) was selected for further confirmation because it contributes to cervical cancer progression25. To confirm whether miR-379 could directly target the 3′-UTR of CRKL, we conducted luciferase reporter assays. HeLa and Ca-Ski cells were transfected with miR-379 mimics or miR-NC, together with pMIR-CRKL-3′-UTR Wt or pMIR-CRKL-3′-UTR Mut. As shown in Figure 3B and C, restoration of miR-379 expression significantly repressed the luciferase activities of pMIR-CRKL-3′-UTR Wt (p < 0.05). However, the change in the luciferase activities of pMIR-CRKL-3′-UTR Mut in HeLa and Ca-Ski cells was insignificant. To confirm the potential role of miR-379 in CRKL regulation, we detected CRKL mRNA and protein expression levels in HeLa and Ca-Ski cells transfected with miR-379 mimic or miR-NC. Ectopic expression of miR-379 decreased the CRKL expression in HeLa and Ca-Ski cells at both the mRNA (p < 0.05) (Fig. 3D) and protein levels (p < 0.05) (Fig. 3E). Hence, CRKL is a direct target of miR-379 in cervical cancer.
Figure 3.
V-crk avian sarcoma virus CT10 oncogene homolog-like (CRKL) is a direct target of miR-379 in cervical cancer. (A) Predicted miR-379 binding sites in the 3′-untranslated region (3′-UTR) of CRKL and its mutant containing altered nucleotides in the 3′-UTR. (B, C) HeLa and Ca-Ski cells were cotransfected with luciferase reporter plasmid and miR-379 mimics or miR-NC. Luciferase activities were detected 48 h after the transfection. Firefly luciferase activity was normalized to Renilla luciferase activity. *p < 0.05 compared with miR-NC. RT-qPCR and Western blotting analysis were conducted to measure the mRNA (D) and protein (E) expression levels of CRKL in HeLa and Ca-Ski cells transfected with miR-379 mimics or miR-NC, respectively. *p < 0.05 compared with miR-NC.
CRKL Is Upregulated in Cervical Cancer and Inversely Correlated With miR-379 Expression
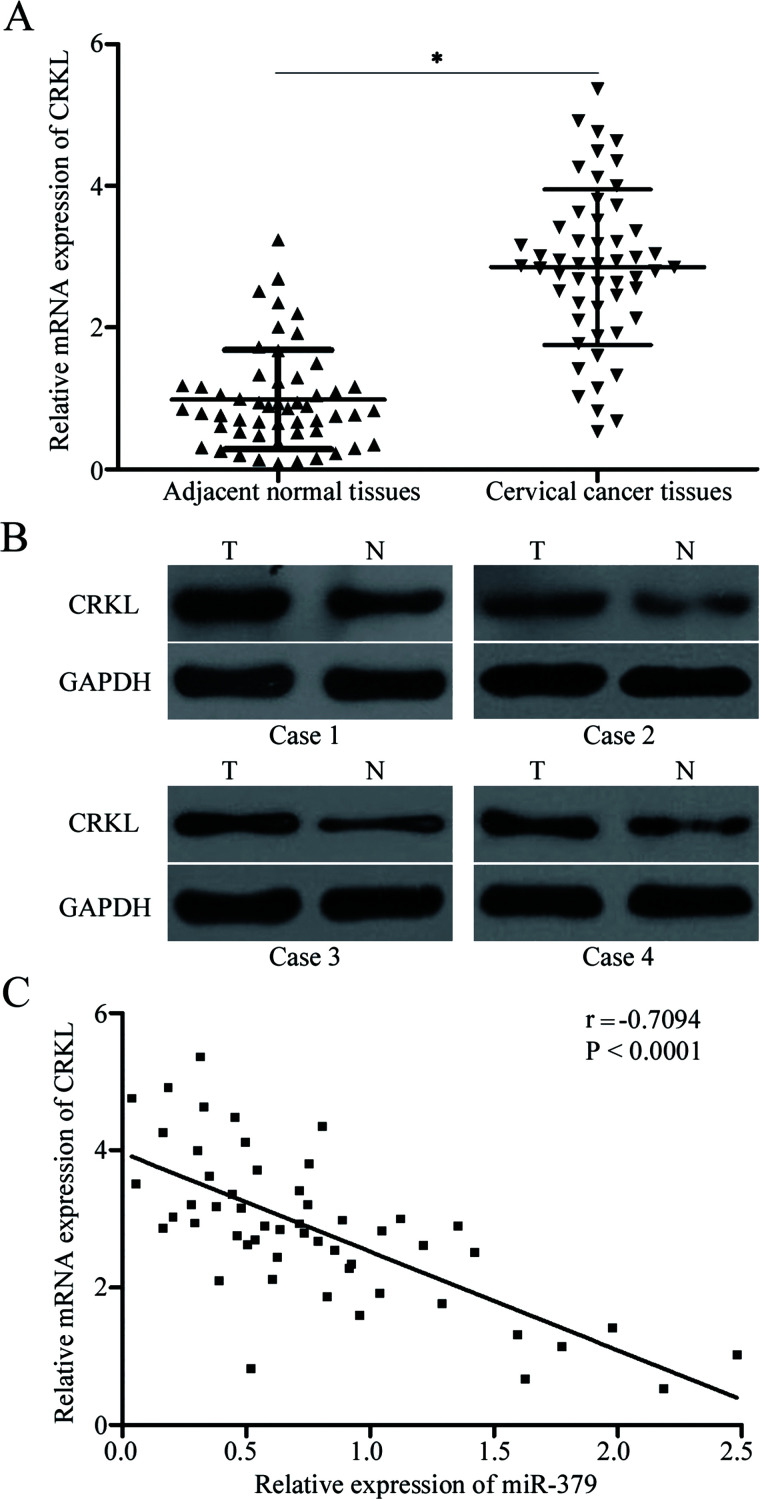
To further examine the association between miR-379 and CRKL, we measured CRKL mRNA expression in 53 paired cervical cancer tissues and their corresponding adjacent normal tissues. RT-qPCR data indicated that CRKL mRNA was highly expressed in cervical cancer tissues compared with that in adjacent normal tissues (p < 0.05) (Fig. 4A). Additionally, Western blotting analysis revealed that the CRKL protein was significantly upregulated in cervical cancer compared with that in adjacent normal tissues (Fig. 4B). Furthermore, Spearman’s correlation analysis demonstrated the negative correlation of miR-379 expression with the mRNA expression of CRKL in cervical cancer tissues (r = −0.7094, p < 0.0001) (Fig. 4C).
Figure 4.
CRKL upregulation in cervical cancer and its negative correlation with miR-379 expression. (A) RT-qPCR analysis of CRKL mRNA expression in 53 paired cervical cancer tissues and their corresponding adjacent normal tissues. *p < 0.05 compared with adjacent normal tissues. (B) Quantification of CRKL protein level in cervical cancer tissues and corresponding adjacent normal tissues through Western blotting analysis. (C) Association between CRKL mRNA and miR-379 expression in cervical cancer tissues was determined through Spearman’s correlation analysis.
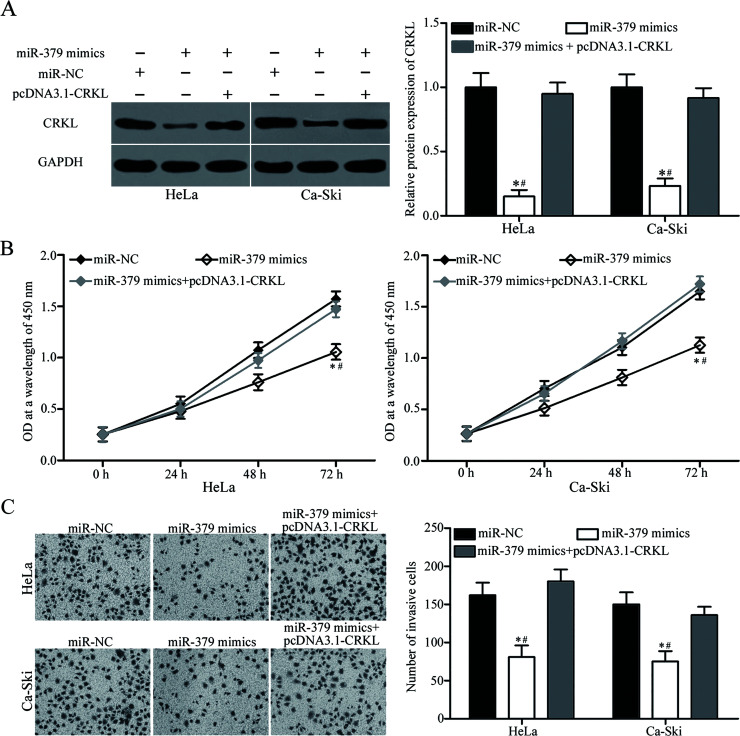
Restored CRKL Expression Abrogates miR-379-Induced Inhibitory Effects on Cervical Cancer Cells
Considering that CRKL is a direct target of miR-379, we employed the CRKL overexpression plasmid to determine whether CRKL overexpression can abolish the tumor-suppressing roles of miR-379 in cervical cancer cells. HeLa and Ca-Ski cells were transfected with miR-379 mimics with or without pcDNA3.1-CRKL. As shown in Figure 5A, the CRKL expression decreased in HeLa and Ca-Ski cells transfected with miR-379 mimics but was restored in the cells cotransfected with miR-379 mimics and pcDNA3.1-CRKL (p < 0.05). Functional experiments showed that the resumed CRKL expression effectively reversed the inhibitory effects of miR-379 overexpression on the proliferation (p < 0.05) (Fig. 5B) and invasion (p < 0.05) (Fig. 5C) of HeLa and Ca-Ski cells. These results suggest that miR-379 exerts tumor-suppressive roles in cervical cancer cells, at least in part, by inhibiting CRKL.
Figure 5.
CRKL overexpression rescued the tumor-suppressing effects of miR-379 on cervical cancer cells. HeLa and Ca-Ski cells were transfected with miR-379 mimics with or without pcDNA3.1-CRKL. (A) After 72 h of transfection, Western blot analysis was carried out to detect CRKL protein expression. *p < 0.05 compared with miR-NC. #p < 0.05 compared with miR-379 mimics + pcDNA3.1-CRKL. CCK-8 and in vitro invasion assays were performed to determine cell proliferation (B) and invasion (C). *p < 0.05 compared with miR-NC. #p < 0.05 compared with miR-379 mimics + pcDNA3.1-CRKL.
DISCUSSION
Over the past decades, miRNAs have been recognized as key regulatory molecules involved in tumorigenesis and tumor development26,27. Scholars have investigated novel miRNAs involved in cervical cancer formation and progression to improve the prognosis of patients affected with this malignancy. In the present study, the expression of miR-379 was downregulated in both cervical cancer tissues and cell lines. Low miR-379 expression was correlated with FIGO stage, lymph node metastasis, and distant metastasis of cervical cancer. Additionally, upregulation of miR-379 repressed the proliferation and invasion of cervical cancer cells. Furthermore, CRKL was identified as a direct target of miR-379 in cervical cancer. CRKL was highly expressed in cervical cancer tissues and negatively correlated with miR-379 expression. Moreover, CRKL overexpression rescued the miR-379-induced inhibitory effects on cervical cancer cells. These findings suggest that restoring miR-379 expression may be a novel strategy for the treatment of patients with cervical cancer.
miR-379 is aberrantly expressed in multiple human cancer types. For example, miR-379 is downregulated in gastric cancer tissues and cell lines. Low miR-379 expression is associated with lymph node metastasis and advanced TNM stage. Additionally, miR-379 is a validated novel independent prognostic marker for predicting the 5-year survival of patients with gastric cancer28. In breast cancer, the miR-379 expression was low in tumor tissues and correlated with tumor stage22. In hepatocellular carcinoma, the miR-379 expression decreases in tumor tissues29 and is associated with advanced TNM stage and metastasis29. Moreover, miR-379 is downregulated in bladder cancer30 and osteosarcoma31,32 but upregulated in prostate cancer tissues and cell lines. High miR-379 expression is correlated with progression-free survival of patients with prostate cancer23. These findings suggest that miR-379 expression is tissue specific and may be developed as a potential prognostic factor in several cancer types.
Numerous studies have provided sufficient evidence that miR-379 is a tumor suppressor in human cancer. For instance, Xu et al. reported that miR-379 overexpression decreased cell migration, invasion, and epithelial–mesenchymal transition in gastric cancer28. Wu et al. demonstrated that upregulation of miR-379 inhibited cell growth and metastasis in bladder cancer in vitro30. Xie et al. revealed that restoring miR-379 expression suppressed the growth and motility of osteosarcoma cells in vitro and reduced the growth of osteosarcoma xenografts in vivo31. Yamamoto et al. found that ectopic miR-379 expression attenuated the malignant pleural mesothelioma and cell invasive capacity and increased the cell chemosensitivity to vorinostat33. Chen et al. showed that miR-379 reexpression suppressed the proliferation, migration, invasion, and epithelial-to-mesenchymal transition of cancer cells both in vitro and in vivo29,34. Nevertheless, miR-379 is known as an oncogene in prostate cancer. Downregulation of miR-379 inhibited epithelial-to-mesenchymal transition in prostate cancer23. These conflicting findings suggest that miR-379 exhibits tissue-specific biological roles and may be a potential target in antitumor therapy.
Multiple target genes of miR-379 have been validated; these genes include FAK28 in gastric cancer, MDM230 in bladder cancer, EIF4G231, PDK1 in osteosarcoma, cyclin B122 in breast cancer, IL-1833 in malignant pleural mesothelioma, and FAK29 in hepatocellular carcinoma. In the present study, CRKL was identified as a direct and functional target of miR-379 in cervical cancer. CRKL, a member of the human Crk adapter protein family, is a key substrate and effector of BCR-ABL oncogenic tyrosine kinase in chronic myelogenous leukemia35,36. Studies have reported CRKL overexpression in many malignant tumors, such as pancreatic cancer37, breast cancer38, non-small cell lung cancer39, gastric cancer40, and colon cancer41. CRKL is involved in tumorigenesis and tumor development by regulating cell proliferation, cycle, apoptosis, metastasis, and epithelial-to-mesenchymal transition40,42,43. CRKL is also upregulated in cervical cancer. Overexpressing CRKL promoted the proliferation, invasion, and chemoresistance of cervical cancer cells25. Hence, CRKL may be an effective therapeutic target for the treatment of patients with cervical cancer.
In conclusion, the miR-379/CRKL pathway regulated the proliferation and invasion of cervical cancer cells. The abnormal miR-379 expression may be used as a therapeutic target for the treatment of patients with cervical cancer.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (31300624 and 81470684), the Natural Science Foundation of Jiangsu Province (BK20161168), the Clinical Special Fund of Jiangsu Province (b12014032), the Postdoctoral Science Foundation of China ( 2015M571818), the Six Major Categories Talent (2014-WSN-043 and 2011-WS-074), the Innovation and Entrepreneurship Training Program for College Students in Jiangsu Province (2015-10313003Z, 201510313003, KYLX14-1455, and 201610313002Z), and the Colleges and Universities Foundation in Jiangsu Province (16KJB320016).
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 3. Wardak S. Human papillomavirus (HPV) and cervical cancer. Med Dosw Mikrobiol. 2016;68:73–84. [PubMed] [Google Scholar]
- 4. Bertelsen BI, Steine SJ, Sandvei R, Molven A, Laerum OD. Molecular analysis of the PI3K-AKT pathway in uterine cervical neoplasia: Frequent PIK3CA amplification and AKT phosphorylation. Int J Cancer 2006;118:1877–83. [DOI] [PubMed] [Google Scholar]
- 5. Zhang P, Kong F, Deng X, Yu Y, Hou C, Liang T, Zhu L. MicroRNA-326 suppresses the proliferation, migration and invasion of cervical cancer cells by targeting ELK1. Oncol Lett. 2017;13:2949–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yang Y, Xie YJ, Xu Q, Chen JX, Shan NC, Zhang Y. Down-regulation of miR-1246 in cervical cancer tissues and its clinical significance. Gynecol Oncol. 2015;138:683–8. [DOI] [PubMed] [Google Scholar]
- 7. Printz C. Expert panel issues new global cervical cancer screening guideline. Cancer 2017;123:2387–8. [DOI] [PubMed] [Google Scholar]
- 8. de Freitas AC, Gomes Leitao Mda C, Coimbra EC. Prospects of molecularly-targeted therapies for cervical cancer treatment. Curr Drug Targets 2015;16:77–91. [DOI] [PubMed] [Google Scholar]
- 9. Dai S, Lu Y, Long Y, Lai Y, Du P, Ding N, Yao D. Prognostic value of microRNAs in cervical carcinoma: A systematic review and meta-analysis. Oncotarget 2016;7:35369–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kanekura K, Nishi H, Isaka K, Kuroda M. MicroRNA and gynecologic cancers. J Obstet Gynaecol Res. 2016;42:612–7. [DOI] [PubMed] [Google Scholar]
- 11. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–14. [DOI] [PubMed] [Google Scholar]
- 12. Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–21. [DOI] [PubMed] [Google Scholar]
- 13. Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57(Suppl 1):i106–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Z, Yu X, Shen J, Jiang Y. MicroRNA dysregulation in uveal melanoma: A new player enters the game. Oncotarget 2015;6:4562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li Z, Yu X, Shen J, Law PT, Chan MT, Wu WK. MicroRNA expression and its implications for diagnosis and therapy of gallbladder cancer. Oncotarget 2015;6:13914–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–4. [DOI] [PubMed] [Google Scholar]
- 17. Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun. 2017;485:556–62. [DOI] [PubMed] [Google Scholar]
- 18. Shin SS, Park SS, Hwang B, Moon B, Kim WT, Kim WJ, Moon SK. MicroRNA-892b influences proliferation, migration and invasion of bladder cancer cells by mediating the p19ARF/cyclin D1/CDK6 and Sp-1/MMP-9 pathways. Oncol Rep. 2016;36:2313–20. [DOI] [PubMed] [Google Scholar]
- 19. Lu Y, Tang L, Zhang Q, Zhang Z, Wei W. MicroRNA-613 inhibits the progression of gastric cancer by targeting CDK9. Artif Cells Nanomed Biotechnol. 2017. (epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 20. Ma Z, Cai H, Zhang Y, Chang L, Cui Y. MiR-129-5p inhibits non-small cell lung cancer cell stemness and chemoresistance through targeting DLK1. Biochem Biophys Res Commun. 2017;490:309–16. [DOI] [PubMed] [Google Scholar]
- 21. Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Khan S, Brougham CL, Ryan J, Sahrudin A, O’Neill G, Wall D, Curran C, Newell J, Kerin MJ, Dwyer RM. miR-379 regulates cyclin B1 expression and is decreased in breast cancer. PLoS One 2013;8:e68753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gururajan M, Josson S, Chu GC, Lu CL, Lu YT, Haga CL, Zhau HE, Liu C, Lichterman J, Duan P, Posadas EM, Chung LW. miR-154* and miR-379 in the DLK1-DIO3 microRNA mega-cluster regulate epithelial to mesenchymal transition and bone metastasis of prostate cancer. Clin Cancer Res. 2014;20:6559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 25. Ji H, Li B, Zhang S, He Z, Zhou Y, Ouyang L. Crk-like adapter protein is overexpressed in cervical carcinoma, facilitates proliferation, invasion and chemoresistance, and regulates Src and Akt signaling. Oncol Lett. 2016;12:3811–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng H, Liu JY, Song FJ, Chen KX. Advances in circulating microRNAs as diagnostic and prognostic markers for ovarian cancer. Cancer Biol Med. 2013;10:123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–5. [DOI] [PubMed] [Google Scholar]
- 28. Xu M, Qin S, Cao F, Ding S, Li M. MicroRNA-379 inhibits metastasis and epithelial-mesenchymal transition via targeting FAK/AKT signaling in gastric cancer. Int J Oncol. 2017;51:867–76. [DOI] [PubMed] [Google Scholar]
- 29. Chen JS, Li HS, Huang JQ, Dong SH, Huang ZJ, Yi W, Zhan GF, Feng JT, Sun JC, Huang XH. MicroRNA-379-5p inhibits tumor invasion and metastasis by targeting FAK/AKT signaling in hepatocellular carcinoma. Cancer Lett. 2016;375:73–83. [DOI] [PubMed] [Google Scholar]
- 30. Wu D, Niu X, Tao J, Li P, Lu Q, Xu A, Chen W, Wang Z. MicroRNA-379-5p plays a tumor-suppressive role in human bladder cancer growth and metastasis by directly targeting MDM2. Oncol Rep. 2017;37:3502–8. [DOI] [PubMed] [Google Scholar]
- 31. Xie X, Li YS, Xiao WF, Deng ZH, He HB, Liu Q, Luo W. MicroRNA-379 inhibits the proliferation, migration and invasion of human osteosarcoma cells by targetting EIF4G2. Biosci Rep. 2017;37(3). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32. Li Z, Shen J, Chan MT, Wu WK. MicroRNA-379 suppresses osteosarcoma progression by targeting PDK1. J Cell Mol Med. 2017;21:315–23. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Yamamoto K, Seike M, Takeuchi S, Soeno C, Miyanaga A, Noro R, Minegishi Y, Kubota K, Gemma A. MiR-379/411 cluster regulates IL-18 and contributes to drug resistance in malignant pleural mesothelioma. Oncol Rep. 2014;32:2365–72. [DOI] [PubMed] [Google Scholar]
- 34. Chen JS, Huang JQ, Dong SH, Huang XH. [Effects of microRNA-379-5p on proliferation, migration and invasion of hepatocellular carcinoma cell line]. Zhonghua Yi Xue Za Zhi 2016;96:1450–3. [DOI] [PubMed] [Google Scholar]
- 35. ten Hoeve J, Morris C, Heisterkamp N, Groffen J. Isolation and chromosomal localization of CRKL, a human crk-like gene. Oncogene 1993;8:2469–74. [PubMed] [Google Scholar]
- 36. Feller SM. Crk family adaptors-signalling complex formation and biological roles. Oncogene 2001;20:6348–71. [DOI] [PubMed] [Google Scholar]
- 37. Fu L, Dong Q, Xie C, Wang Y, Li Q. CRKL protein overexpression enhances cell proliferation and invasion in pancreatic cancer. Tumour Biol. 2015;36:1015–22. [DOI] [PubMed] [Google Scholar]
- 38. Zhao T, Miao Z, Wang Z, Xu Y, Wu J, Liu X, You Y, Li J. Overexpression of CRKL correlates with malignant cell proliferation in breast cancer. Tumour Biol. 2013;34:2891–7. [DOI] [PubMed] [Google Scholar]
- 39. Wang Y, Dong QZ, Fu L, Stoecker M, Wang E, Wang EH. Overexpression of CRKL correlates with poor prognosis and cell proliferation in non-small cell lung cancer. Mol Carcinog. 2013;52:890–9. [DOI] [PubMed] [Google Scholar]
- 40. Han G, Wu D, Yang Y, Li Z, Zhang J, Li C. CrkL meditates CL20/CCR6-induced EMT in gastric cancer. Cytokine 2015;76:163–9. [DOI] [PubMed] [Google Scholar]
- 41. Lan B, Zhang J, Shan J, Zhang P, Zhang W, Chen Y, Zhen W, Yan S. Downregulation of CRKL expression can inhibit tumorigenesis in colon cancer. Front Biosci. (Landmark Ed) 2014;19:528–34. [DOI] [PubMed] [Google Scholar]
- 42. Cai L, Wang H, Yang Q. CRKL overexpression promotes cell proliferation and inhibits apoptosis in endometrial carcinoma. Oncol Lett. 2017;13:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chafik A. The role of CRKL in breast cancer metastasis: Insights from systems biology. Syst Synth Biol. 2015;9:141–6. [DOI] [PMC free article] [PubMed] [Google Scholar]