Abstract
Colorectal cancer (CRC) is one of the most common oncological conditions worldwide, to date. MicroRNA-1290 (miR-1290) has been demonstrated to regulate its progression. We studied the role of miR-1290 in CRC progression. The gene was upregulated in CRC tissues and cells. Its overexpression promoted CRC cell proliferation analyzed by MTT assay, colony formation assay, and soft agar growth assay. In addition, miR-1290 knockdown inhibited CRC cell proliferation. We also found that miR-1290 overexpression reduced the p27 level and increased cyclin D1 at both the mRNA and protein levels, whereas miR-1290 knockdown increased p27 and reduced cyclin D1, confirming miR-1290 promoted CRC cell proliferation. Inositol polyphosphate 4-phosphatase B (INPP4B) was the target of miR-1290. Luciferase reporter assay revealed that miR-1290 directly bound to the 3′-UTR of INPP4B; the mutated seed sites in miR-1290 abrogated this effect. Double knockdown of INPP4B and miR-1290 promoted CRC cell proliferation, suggesting miR-1290 promoted CRC cell proliferation by targeting INPP4B.
Key words: miR-1290, INPP4B, Colorectal cancer (CRC), Cell proliferation
INTRODUCTION
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers worldwide. In 2012, about 1.4 million cases were diagnosed, and 693,900 patients died1. Although many genes and noncoding RNAs have been found to regulate CRC progression, new therapeutic approaches still need to be developed.
MicroRNAs (miRNAs) are small noncoding RNAs (about 19 nt in length) that regulate expression of many genes. This involves development and disease by binding to the 3′-untranslated region (3′-UTR) of a target mRNA and degrading mRNA (while inhibiting tmRNA translation). miRNAs have been demonstrated to regulate CRC progression; for example, patients with high miR-135b have poor clinical outcomes, induced by adenomatous polyposis coli (APC) loss, PTEN/PI3K pathway deregulation, and SRC overexpression, which promotes CRC transformation, proliferation, and invasion. miR-135b inhibition in CRC mouse models suppresses tumor growth2.
miR-1290 regulates tumor progression; for example, it is a potential prognostic factor for lung cancer and is upregulated in CD133+ marked lung cancer stem cells; it promoted the proliferation, clonogenicity, invasion, and migration of CD133+ cells by targeting fyn-related Src family tyrosine kinase3,4. It also promotes sphere formation and induces epithelial–mesenchymal transition (EMT), suggesting that miR-1290 promotes lung cancer initiation and progression5,6. Exosomal miR-1290 is a prognostic factor for castration-resistant prostate cancer7. miR-1290 is a new oncomiR in laryngeal carcinoma by targeting MAF and ITPR28. miR-1290 promotes pancreatic cancer cell proliferation and invasion and is a serum biomarker for low-stage pancreatic cancer9. miR-1290 is also upregulated in esophageal squamous cell carcinoma (ESCC) and promotes ESCC cell proliferation, migration, and invasion by targeting NFIX10,11. miR-1290 is upregulated in CRC tissues; it postpones cytokinesis to form multinucleated cells by targeting KIF13B12. In this study, we found that miR-1290 promoted CRC cell proliferation by targeting inositol polyphosphate 4-phosphatase B (INPP4B).
MATERIALS AND METHODS
Cell Culture and Clinical Specimens
Eight pairs of CRC tissues and their adjacent healthy tissues were collected from patients undergoing surgery at Shandong Provincial Hospital (affiliated with Shandong University) between September 2007 and May 2010 according to an institutional review board-approved protocol. Written informed consent was obtained from all patients involved. Eight CRC tissues including two clinical stage I, two clinical stage II, three clinical stage III, and one clinical stage VI, and the matched adjacent noncancerous tissues were examined by routine histopathological analysis. All tissues were immediately dissected and stored in liquid nitrogen until analyzed. The normal colorectal cell line FHC, and CRC cells SW480, HT-29, COLO205, SW403, KM202L, and SW620 were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (Gibco; Hyclone, Logan, UT, USA) in a humidified cell incubator with an atmosphere of 5% CO2 at 37°C.
Vector Construction and Transfection
The sequence of the miR-1290 precursor was cloned into pMSCV vector, pMSCV vector with miR-1290 overexpression, or empty vector, and packaging plasmid was cotransfected into 293T cells using PEI (Cat. No. 23966; Polysciences Asia Pacific, Inc., Taipei, Taiwan). The viruses were harvested after 48 h posttransfection and infected CRC to construct stable cell lines using puromycin (Cat. No. P8833; Sigma-Aldrich, St. Louis, MO, USA; Merck KGaA, Darmstadt, German). miR-1290 mimic, mutated miR-1290 mimic, miR-1290 inhibitor, small interfering RNAs for INPP4B (INPP4B siRNAs), and their negative control were purchased from Guangzhou BiboBio and transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time (qRT)-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). cDNA was synthesized using M-MLV Reverse Transcriptase (Cat. No. R011; Vazyme Biotech Co., Ltd., Nanjing, P.R. China); for miR-1290, the specific primer used was 5’-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCCCTG-3’; and for genes, Oligo dT18 was used. miR-1290, p27, and cyclin D1 levels were examined by AceQ qPCR SYBR Green Master Mix (Cat. No. Q111-02; Vazyme Biotech Co., Ltd.) on a CFX-96 system (Bioneer, Daejeon, South Korea). The primers used were as follows: miR-1290, 5′-GCCCGCGCCCGCTGGATTTTTGG-3′ (forward) and 5′-GTGCAGGGTCCGAGGT-3′ (reverse); p27, 5′-TAATTGGGGCTCCGGCTAACT-3′ (forward) and 5′-TGCAGGTCGCTTCCTTATTCC-3′ (reverse); cyclin D1, 5′-CAATGACCCCGCACGATTTC-3′ (forward) and 5′-CATGGAGGGCGGATTGGAA-3′ (reverse). U6 was used as the internal control for miR-1290, and GAPDH was used as the internal control for genes.
Western Blot
The proteins were separated by sodium dodecyl sulfate (SDS)-8% polyacrylamide gel electrophoresis and blotted to the PVDF membrane (Bio-Rad, Hercules, CA, USA). Anti-p27 (1:1,000 dilution; Cat. No. sc-1641; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-cyclin D1 (1:1,000 dilution; Cat. No. sc-753; Santa Cruz Biotechnology) antibodies were used. Anti-α-tubulin antibody (1:1,000 dilution; Cat. No. sc-53646; Santa Cruz Biotechnology) was used as the loading control. Goat anti-mouse immunoglobulin G was used as a secondary antibody, and protein signals were detected using an enhanced chemiluminescence system (National Institutes of Health, Bethesda, MD, USA).
Cell Proliferation Assay
MTT assay, colony formation assay, and soft agar growth assay determined the role of miR-1290 in CRC cell proliferation. For the MTT assay, 0.5 × 103 cells were seeded in 96-well plates; 25 μl of MTT (5 mg/ml) was added to each well at indicated time points and incubated for 2 h. Then the supernatant was discarded from each well, the colored formazan crystal was dissolved in 150 μl of isopropanol, and the optical density was measured at 590 nm. For the colony formation assay, cells were seeded in plates and cultured for 7 days. The colonies were fixed with 10% formaldehyde for 5 min and stained with 1.0% crystal violet for 30 s. For the soft agar growth assay, 0.5 × 103 cells were mixed with 2 ml of complete medium plus 0.3 agar (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA); the mixture was plated atop a bottom layer consisting of 1% agar in complete medium. After 10 days, the number of colonies whose diameter was larger than 0.1 mm was counted.
Luciferase Reporter Assay
The fragment of the 3′-UTR of INPP4B containing the binding site for miR-1290 was cloned into the pGL3 vector (BD Biosciences, Franklin Lakes, NJ, USA) and cotransfected with the miR-1290 mimic, mutated miR-1290 mimic, or miR-1290 inhibitor into the cells using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s instructions. Luciferase reporter assays were performed after 48 h of posttransfection according to the Dual-Luciferase Reporter Assay System (Promega).
Statistical Analysis
All statistical analyses were performed using SPSS 20.0 (SPSS, Inc., Chicago, IL, USA). Data are presented as the mean ± SD for three independent experiments. The differences between groups were analyzed using Student’s t-test, and a value of p < 0.05 was defined as significant.
RESULTS
miR-1290 Is Overexpressed in CRC
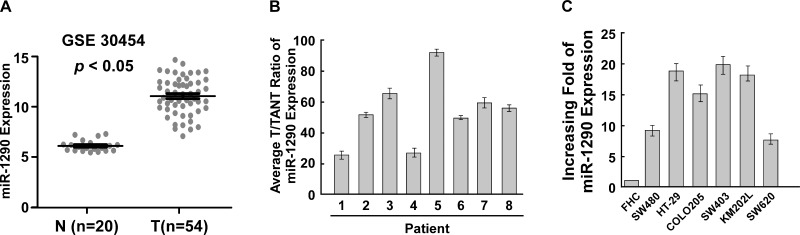
We used genome-wide miRNA expression profiles of 20 normal colonic tissues and 50 CRC tissues determined by miRNA microarray (GSE30454) to investigate the miR-1290 level13 and found that miR-1290 was significantly upregulated in CRC tissues (Fig. 1A). We also used eight pairs of CRC tissues and adjacent colonic tissues to examine the miR-1290 level and found that miR-1290 was upregulated in CRC tissues compared to paired normal colonic tissues (Fig. 1B). We also found miR-1290 was upregulated in CRC tissues compared to normal colonic cells (Fig. 1C). These findings suggested that miR-1290 was upregulated in CRC tissues and cells.
Figure 1.
MicroRNA-1290 (miR-1290) is upregulated to colorectal cancer (CRC) tissues and cells. (A) miR-1290 was upregulated in CRC tissues compared to the normal tissues; data from GSE3045. (B) miR-1290 was upregulated in CRC tissues (T) compared to the adjacent normal colorectal tissues (TANT). (C) miR-1290 was upregulated in CRC cells. Data are presented as mean ± SD for three independent experiments.
miR-1290 Contributes to the Cell Proliferation of CRC
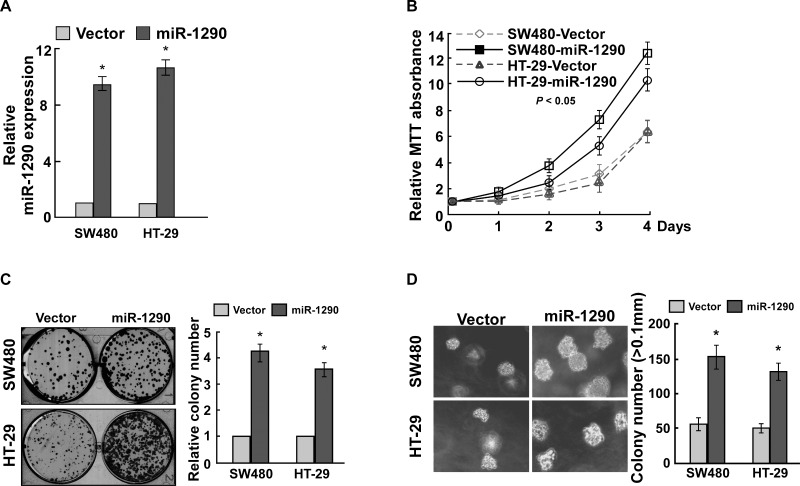
To elucidate the effect of miR-1290 on cell proliferation of CRC, miR-1290 was significantly upregulated in CRC cells SW480 and HT-29 after miR-1290 overexpression vector was transfected (Fig. 2A). MTT assay found that miR-1290 overexpression promoted cell proliferation (Fig. 2B). Colony formation assay suggested that miR-1290 overexpression significantly promoted cell proliferation (Fig. 2C). Anchorage-independent growth ability assay revealed that miR-1290 overexpression promoted cell proliferation (Fig. 2D). These findings suggested that miR-1290 overexpression promoted CRC cell proliferation.
Figure 2.
miR-1290 overexpression promotes CRC cell proliferation. (A) Quantitative real-time (qRT)-PCR assay demonstrated that miR-1290 was overexpressed in CRC cells infected with virus containing miR-1290 overexpression vector. (B) MTT assay suggested that miR-1290 overexpression promoted CRC cell proliferation. (C) Colony formation assay revealed that miR-1290 overexpression promoted CRC cell proliferation. (D) Soft agar growth assay suggested that miR-1290 overexpression promoted tumorigenesis in vitro. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
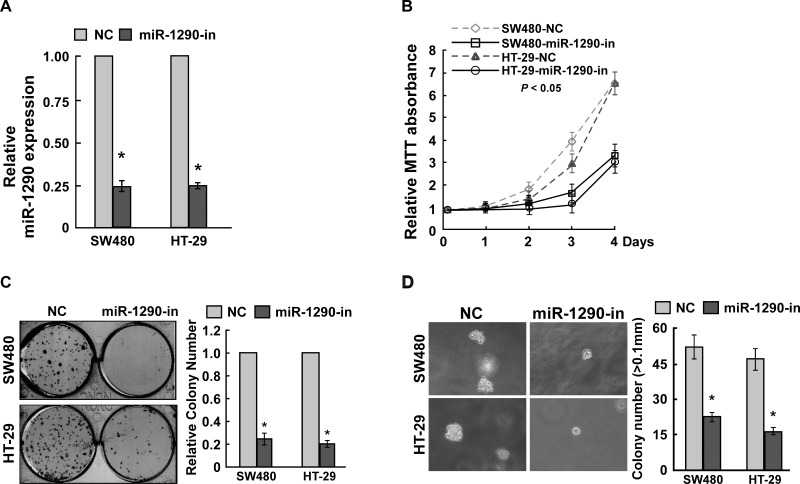
To confirm these findings, miR-1290 was downregulated in CRC cells SW480 and HT-29 using the miR-1290 inhibitor (Fig. 3A). MTT assay found that miR-1290 knockdown inhibited cell proliferation (Fig. 3B). Colony formation assay suggested that miR-1290 knockdown significantly suppressed cell proliferation (Fig. 3C). Anchorage-independent growth ability assay revealed that miR-1290 knockdown inhibited cell proliferation (Fig. 3D). These findings suggested that miR-1290 knockdown inhibited CRC cell proliferation. Taken together, miR-1290 contributed to CRC cell proliferation.
Figure 3.
miR-1290 knockdown inhibits CRC cell proliferation. (A) qRT-PCR assay suggested that miR-1290 was downregulated in CRC cells transfected with the miR-1290 inhibitor. (B) MTT assay suggested that miR-1290 knockdown inhibited CRC cell proliferation. (C) Colony formation assay suggested that miR-1290 knockdown inhibited CRC cell proliferation. (D) Soft agar growth assay revealed that miR-1290 knockdown inhibited tumorigenesis in vitro. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
miR-1290 Contributes to CRC Cell Proliferation by Targeting INPP4B
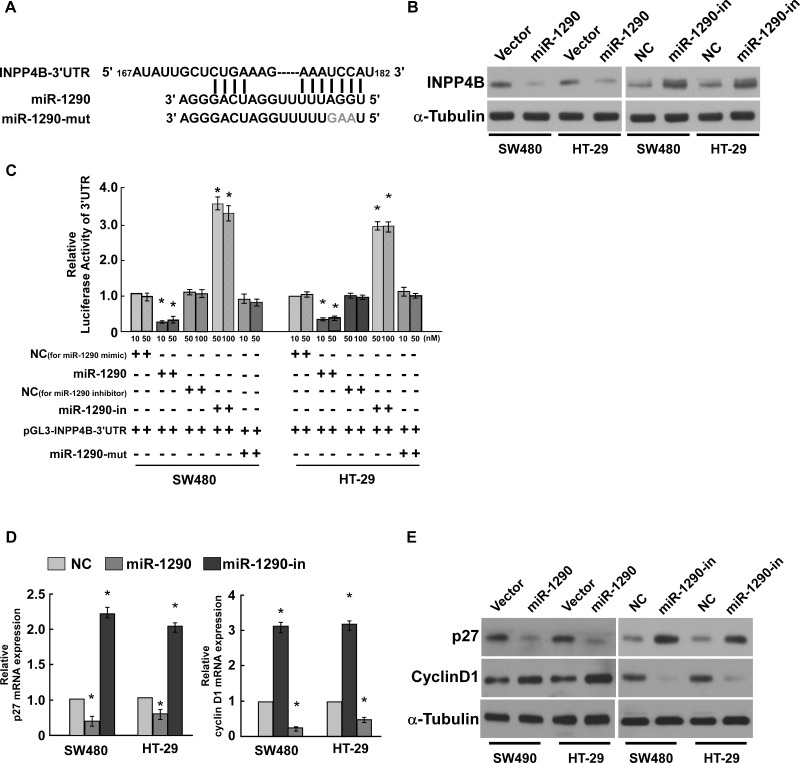
In our study we used public bioinformatics algorithms (including TargetScan and miRNase) to predict targets of miR-1290 and found that INPP4B might be a target of miR-1290 (Fig. 4A). Western blot assay suggested that miR-1290 overexpression inhibited INPP4B expression, whereas miR-1290 knockdown promoted INPP4B expression (Fig. 4B). Luciferase reporter system assay revealed that luciferase activity was significantly reduced in a dose-dependent manner when miR-1290 mimic was cotransfected with luciferase reporter vector pGL3-INPP4B-3′-UTR. Luciferase activity was significantly increased in a dose-dependent manner when miR-1290 inhibitor was cotransfected with luciferase reporter vector pGL3-INPP4B-3′-UTR. Luciferase activity was not changed when mutation miR-1290 mimic was cotransfected with luciferase reporter vector pGL3-INPP4B-3′-UTR (Fig. 4C), suggesting that miR-1290 directly bound to the 3′-UTR of INPP4B.
Figure 4.
Inositol polyphosphate 4-phosphatase B (INPP4B) is the target gene of miR-1290. (A) miR-1290 could bind to the 3′-untranslated region (3′-UTR) of INPP4B predicted using public software. The wild and mutated seed sites are shown. (B) Western blot assay revealed that miR-1290 had an opposite expression level in CRC cells. GAPDH was used as the loading control. (C) Luciferase reporter assay suggested that miR-1290 directly bound to the 3′-UTR of INPP4B, and the mutated seed sites abrogated the binding. (D, E) miR-1290 promoted cyclin D1 level and inhibited p27 level in CRC cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
We further confirmed that miR-1290 contributed to CRC cell proliferation by analyzing cell proliferation regulatory proteins p27 and cyclin D1. qRT-PCR and Western blot assay suggested that miR-1290 overexpression inhibited p27 expression and promoted cyclin D1 expression at both the mRNA and protein levels. miR-1290 knockdown promoted p27 expression and inhibited cyclin D1 expression (Fig. 4D and E), suggesting that miR-1290 contributed to CRC cell proliferation.
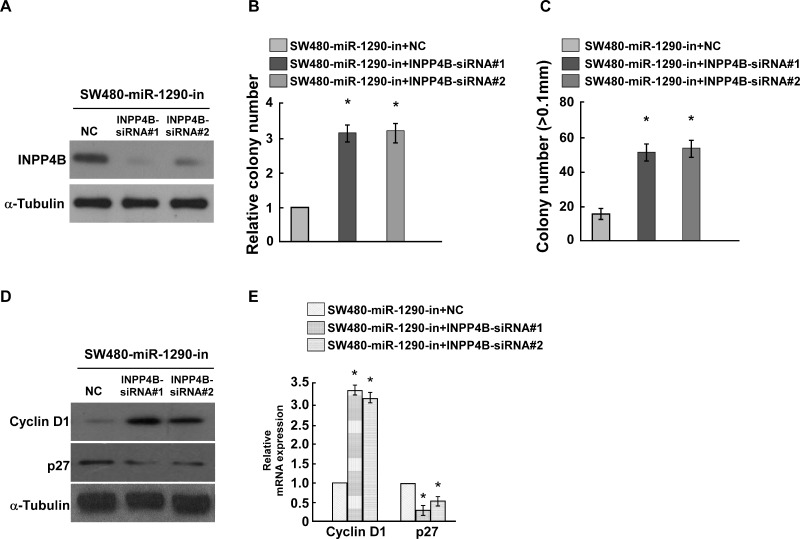
To demonstrate whether miR-1290 contributes to CRC cell proliferation by targeting INPP4B, INPP4B and miR-1290 were knocked down at the same time. Western blot assay suggested that siRNAs for INPP4B could effectively downregulate INPP4B expression (Fig. 5A). Double knockdown of INPP4B and miR-1290 could promote CRC cell proliferation analyzed by colony formation assay and anchorage-independent growth ability assay (Fig. 5B and C), copying the effects of miR-1290 overexpression. We also analyzed whether double knockdown of miR-1290 and INPP4B could regulate cyclin D1 and p27 and found that double knockdown of miR-1290 and INPP4B increased cyclin D1 expression and reduced p27 expression at both the protein and mRNA levels (Fig. 5D and E). These findings suggested that miR-1290 contributes to CRC cell proliferation by targeting INPP4B.
Figure 5.
miR-1290 promotes CRC proliferation through inhibiting INPP4B. (A) Western blot demonstrated that INPP4B small interfering RNAs (siRNAs) could inhibit the INPP4B level. α-Tubulin was used as the loading control. (B) Colony formation assay suggested that knockdown of miR-1290 and INPP4B promoted CRC cell proliferation. (C) Soft agar growth assay suggested that knockdown of miR-1290 and INPP4B promoted tumorigenesis in vitro. (D, E). Double knockdown of miR-1290 and INPP4B promoted cyclin D1 level and inhibited p27 level in CRC cells. Data are presented as mean ± SD for three independent experiments. *p < 0.05.
DISCUSSION
In this study, we studied the role of miR-1290 in CRC cell proliferation and found that miR-1290 was upregulated in human CRC tissues and cells. In addition, miR-1290 overexpression promoted CRC cell proliferation, and its downregulation inhibited CRC cell proliferation. Knockdown of INPP4B and miR-1290 promoted CRC cell proliferation, suggesting that miR-1290 promoted CRC cell proliferation by targeting INPP4B.
INPP4B, which catalyzes PI(3,4)P2 to PI(3)P, has dual roles in the progression of tumors. It acts as a tumor suppressor in prostate cancer14,15, melanocytic neoplasms16, thyroid cancer17, bladder cancer18, and breast cancers19,20, and it acts as an oncogene in breast cancers harboring PIK3CA mutation21 and melanomas22. This suggested that the role of INPP4B in tumors is dependent on genetic context. A previous report suggested that INPP4B is upregulated in CRC cells and tissues and promotes CRC cell proliferation by regulating Akt and SGK3 pathways and downregulating PTEN23. However, our study suggested that INPP4B inhibited CRC cell proliferation. This might be caused by a different cellular context. The role of INPP4B in CRC progression should be examined further.
p27 and cyclin D1 are important cell proliferation regulators; p27 inhibits cell cycle progression, and cyclin D1 accelerates G1/S phase transition24. We found that miR-1290 overexpression inhibited p27 expression and promoted cyclin D1 expression, whereas its knockdown promoted p27 expression and inhibited cyclin D1 expression, suggesting that miR-1290 promoted CRC cell proliferation, consistent with the results determined using MTT assay, colony formation assay, and soft agar growth assay. In summary, miR-1290 promoted CRC cell proliferation by inhibiting INPP4B.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2. Valeri N, Braconi C, Gasparini P, Murgia C, Lampis A, Paulus-Hock V, Hart JR, Ueno L, Grivennikov SI, Lovat F, Paone A, Cascione L, Sumani KM, Veronese A, Fabbri M, Carasi S, Alder H, Lanza G, Gafa R, Moyer MP, Ridgway RA, Cordero J, Nuovo GJ, Frankel WL, Rugge M, Fassan M, Groden J, Vogt PK, Karin M, Sansom OJ, Croce CM. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell 2014;25:469–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sun B, Yang N, Jiang Y, Zhang HF, Hou CY, Ji C, Liu YY, Zuo PP. Antagomir-1290 suppresses CD133(+) cells in non-small cell lung cancer by targeting fyn-related Src family tyrosine kinase. Tumor Biol. 2015;36:6223–30. [DOI] [PubMed] [Google Scholar]
- 4. Mo DP, Gu B, Gong X, Wu L, Wang H, Jiang Y, Zhang BF, Zhang MJ, Zhang Y, Xu J, Pan SY. miR-1290 is a potential prognostic biomarker in non-small cell lung cancer. J Thorac Dis. 2015;7:1570–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim G, An HJ, Lee MJ, Song JY, Jeong JY, Lee JH, Jeong HC. Hsa-miR-1246 and hsa-miR-1290 are associated with stemness and invasiveness of non-small cell lung cancer. Lung Cancer 2016;91:15–22. [DOI] [PubMed] [Google Scholar]
- 6. Kim KB, Kim K, Bae S, Choi Y, Cha HJ, Kim SY, Lee JH, Jeon SH, Jung HJ, Ahn KJ, An IS, An S. MicroRNA-1290 promotes asiatic acid-induced apoptosis by decreasing BCL2 protein level in A549 non-small cell lung carcinoma cells. Oncol Rep. 2014;32:1029–36. [DOI] [PubMed] [Google Scholar]
- 7. Huang XY, Yuan TZ, Liang MH, Du MJ, Xia S, Dittmar R, Wang D, See W, Costello BA, Quevedo F, Tan W, Nandy D, Bevan GH, Longenbach S, Sun ZF, Lu Y, Wang T, Thibodeau SN, Boardman L, Kohli M, Wang L. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015;67:33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Janiszewska J, Szaumkessel M, Kostrzewska-Poczekaj M, Bednarek K, Paczkowska J, Jackowska J, Grenman R, Szyfter K, Wierzbicka M, Giefing M, Jarmuz-Szymczak M. Global miRNA expression profiling identifies miR-1290 as novel potential oncomiR in laryngeal carcinoma. PLoS One 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li AG, Yu J, Kim H, Wolfgang CL, Canto MI, Hruban RH, Goggins M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19:3600–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH. MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis. World J Gastroenterol. 2015;21:3245–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mao Y, Liu J, Zhang DK, Li BS. MiR-1290 promotes cancer progression by targeting nuclear factor I/X (NFIX) in esophageal squamous cell carcinoma (ESCC). Biomed Pharmacother. 2015;76:82–93. [DOI] [PubMed] [Google Scholar]
- 12. Wu J, Ji XW, Zhu LL, Jiang QL, Wen ZZ, Xu S, Shao W, Cai JT, Du Q, Zhu YL, Mao JS. Up-regulation of microRNA-1290 impairs cytokinesis and affects the reprogramming of colon cancer cells. Cancer Lett. 2013;329:155–63. [DOI] [PubMed] [Google Scholar]
- 13. Balaguer F, Moreira L, Lozano JJ, Link A, Ramirez G, Shen Y, Cuatrecasas M, Arnold M, Meltzer SJ, Syngal S, Stoffel E, Jover R, Llor X, Castells A, Boland CR, Gironella M, Goel A. Colorectal cancers with microsatellite instability display unique miRNA profiles. Clin Cancer Res. 2011;17:6239–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, Ayala G, Ittmann MM, Weigel NL, Agoulnik IU. Decreased expression and androgen regulation of the tumor suppressor gene INPP4B in prostate cancer. Cancer Res. 2011;71:572–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rynkiewicz NK, Fedele CG, Chiam K, Gupta R, Kench JG, Ooms LM, McLean CA, Giles GG, Horvath LG, Mitchell CA. INPP4B is highly expressed in prostate intermediate cells and its loss of expression in prostate carcinoma predicts for recurrence and poor long term survival. Prostate 2015;75:92–102. [DOI] [PubMed] [Google Scholar]
- 16. Perez-Lorenzo R, Gill KZ, Shen CH, Zhao FX, Zheng B, Schulze HJ, Silvers DN, Brunner G, Horst BA. A tumor suppressor function for the lipid phosphatase INPP4B in melanocytic neoplasms. J Invest Dermatol. 2014;134:1359–68. [DOI] [PubMed] [Google Scholar]
- 17. Vo TTT, Fruman DA. INPP4B is a tumor suppressor in the context of PTEN deficiency. Cancer Discov. 2015;5:697–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsu IW, Yeh CR, Slavin S, Miyamoto H, Netto GJ, Tsai YC, Muyan M, Wu XR, Messing EM, Guancial EA, Yeh SY. Estrogen receptor alpha prevents bladder cancer development via INPP4B inhibited Akt pathway in vitro and in vivo. Oncotarget 2014;5:7917–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fedele CG, Ooms LM, Ho M, Vieusseux J, O’Toole SA, Millar EK, Lopez-Knowles E, Sriratana A, Gurung R, Baglietto L, Giles GG, Bailey CG, Rasko JEJ, Shields BJ, Price JT, Majerus PW, Sutherland RL, Tiganis T, McLean CA, Mitchell CA. Inositol polyphosphate 4-phosphatase II regulates PI3K/Akt signaling and is lost in human basal-like breast cancers. Proc Natl Acad Sci USA 2010;107:22231–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun Y, Ding H, Liu XG, Li XQ, Li L. INPP4B overexpression enhances the antitumor efficacy of PARP inhibitor AG014699 in MDA-MB-231 triple-negative breast cancer cells. Tumor Biol. 2014;35:4469–77. [DOI] [PubMed] [Google Scholar]
- 21. Gasser JA, Inuzuka H, Lau AW, Wei WY, Beroukhim R, Toker A. SGK3 mediates INPP4B-dependent PI3K signaling in breast cancer. Mol Cell 2014;56:595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chi MN, Guo ST, Wilmott JS, Guo XY, Yan XG, Wang CY, Liu XY, Jin L, Tseng HY, Liu T, Croft A, Hondermarck H, Scolyer RA, Jiang CC, Zhang XD. INPP4B is upregulated and functions as an oncogenic driver through SGK3 in a subset of melanomas. Oncotarget 2015;6:39891–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo ST, Chi MN, Yang RH, Guo XY, Zan LK, Wang CY, Xi YF, Jin L, Croft A, Tseng HY, Yan XG, Farrelly M, Wang FH, Lai F, Wang JF, Li YP, Ackland S, Scott R, Agoulnik IU, Hondermarck H, Thorne RF, Liu T, Zhang XD, Jiang CC. INPP4B is an oncogenic regulator in human colon cancer. Oncogene 2016;35:3049–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Agarwal R. Cell signaling and regulators of cell cycle as molecular targets for prostate cancer prevention by dietary agents. Biochem Pharmacol. 2000;60:1051–9. [DOI] [PubMed] [Google Scholar]