Abstract
As a member of the miRNA family, let-7c has been identified as a tumor suppressor in many cancers. However, the molecular biological function of let-7c in glioma has not been elucidated. The aim of this study was to explore let-7c expression levels and evaluate its function in glioma cells. We first measured the expression of let-7c in four glioma cell lines and a normal cell line by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR), and the results showed that let-7c was downregulated in glioma cells. By applying gain-of-function and loss-of-function assays, the experiments suggested that dysregulation of let-7c could obviously affect cell proliferation, metastasis, and invasion. Based on online bioinformatics analysis and Dual-Luciferase Reporter assays, we found that E2F5 was a target gene of let-7c and contributed to the function of let-7c in glioma cells. Our investigations indicated that loss of let-7c contributed to the progression of glioma cells.
Key words: Glioma, let-7c, E2F transcription factor 5 (E2F5), Proliferation, Invasion, Migration
INTRODUCTION
Glioma is one of the most common and the most aggressive malignant tumors in the central nervous system (CNS), posing a large threat to human health1. Malignant glioma is characterized with unique features such as rapid growth and high migration. Because of the rapid growth, angiogenesis, and metastasis of glioma cells, it is extremely difficult to cure this disease2. Surgeries combined with radiotherapy, chemotherapy, and targeted therapy are currently the standard treatments3. However, surgical treatment to remove all malignant glioma cells is not possible. Therefore, there is an urgent need to develop new therapies for treating glioma.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that bind to the 3′-untranslated regions (3′-UTRs) of target mRNAs to induce transcript degradation and/or inhibit translation4. miRNAs play important roles in various pathophysiological processes5,6. let-7c was considered to be a tumor suppressor by regulating oncogenic mRNAs and was most frequently associated with clinical outcomes in patients with cancer. Previous research on let-7c has primarily focused on its functions and roles in cancer. let-7c has been shown to inhibit cancer cell survival by regulating cell proliferation7,8. Recent studies suggest that let-7c is an important regulator of macrophage polarization. let-7c was observed to be downregulated in most cancers, and it played crucial roles in carcinogenesis and cancer metastasis9–11. Ma et al. identified let-7c as being able to inhibit influenza virus replication12. However, its roles in glioma and the regulation pathways have not been previously investigated.
E2F transcription factor 5 (E2F5) is a key oncogenic protein and has been reported to be functional in various cancers13. For example, miR-34a enhances the sensitivity of gastric cancer cells to treatment with paclitaxel by targeting E2F513. miR-154 inhibits growth and invasion of breast cancer cells through targeting E2F514. The transcription factor forkhead box N3 (FOXN3) inhibits cell proliferation by downregulating E2F5 expression in hepatocellular carcinoma cells15. Here we investigated the function of E2F5 in glioma and found that E2F5 could enhance proliferation and neoplastic transformation, and its silencing resulted in inhibition of growth, migration, and invasion of glioma cells. E2F5 was revealed to be a potential target of let-7c. In this study, we determined that let-7c inhibited the proliferation, invasion, and migration of glioma cells via targeting E2F5. All in all, the let-7c/E2F5 axis seems to be a potential therapeutic target for better treatment of malignant glioma.
MATERIALS AND METHODS
Cell Lines
Four human glioma cell lines (U87, U251, SHG44, and A172) were used in this study. Human normal astrocytes were used as the negative control (NC). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) supplemented with 10% bovine calf serum (Gibco). Cells were maintained at 37°C in an atmosphere of humidified air with 5% CO2.
Bioinformatics Analysis
The 3′-UTR sequences for E2F5 that are complementary to let-7c were determined using TargetScan (http://www.targetscan.org) and miRDB (http://mirdb.org) to confirm that they are potential binding partners.
Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
Total RNA from tissues and cells was isolated with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse transcription was performed with PrimeScript RT Reagent Kit (Takara, Dalian, P.R. China) according to the manufacturer’s protocol. qRT-PCR was performed with SYBR Prime Script RT-PCR Kits (Takara) based on the manufacturer’s instructions. The expression levels of let-7c and E2F5 were calculated with the 2−ΔΔCt method, which were normalized to U6 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA, respectively. All assays were performed in triplicate. The expression levels were relative to the fold change of the corresponding controls, which were defined as 1.0.
Cell Viability
Cell viability was assessed via 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT; Dojindo Molecular Technologies, Inc., Kumamoto, Japan) assay. Cells (5 × 103/well) were seeded in a 96-well flat-bottomed plate for 24 h and then transfected with corresponding short hairpin RNA (sh-RNA) or pc-DNA3.1 and cultured in normal medium. At 0, 24, 48, 72, and 96 h after transfection, the MTT solution (5 mg/ml, 20 μl) was added to each well. Following incubation for 4 h, the medium was removed, and 100 μl of dimethyl sulfoxide (DMSO) was added to each well. The relative number of surviving cells was assessed by measuring the optical density (OD) of cell lysates at 560 nm. All assays were performed in triplicate.
Colony Formation Assay
Cells (500 cells/well) were plated in six-well plates and incubated in DMEM with 10% bovine calf serum at 37°C. Two weeks later, the cells were fixed and stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA). The number of visible colonies was counted manually.
Cell Migration and Invasion Assays
The cell invasive and migratory abilities were evaluated using Transwell assays. For the migration assay, 200 μl of treated U87 and SHG44 cells (5 × 105) with serum-free medium was seeded in the top 24-well Transwell chamber (8-μm pore; Corning, Painted Post, NY, USA) of the inserts, and 600 μl of complete medium was added to the lower chamber. After 24 h of incubation, the migratory cells were fixed, stained in 0.1% crystal violet solution, and counted under a confocal microscope. For the invasion assay, the invasive cells were detected by Transwell Matrigel (BD Biosciences, San Diego, CA, USA) invasion assay. Matrigel (BD Biosciences) was precoated onto the upper chambers. Other experimental procedures were similar to the migration assay. Three independent experiments were carried out.
Wound Healing Assay
Cells were seeded in 24-well plates and cultured. A germ-free plastic scriber was used to create 1.0-mm wounds. The cells were washed and incubated in serum-free medium for 48 h. The wound healing process was observed under a microscope (Leica, Wetzlar, Germany).
Cell Attachment and Detachment Assays
For the attachment assay, cells were seeded in 24-well plates at 5 × 104 cells/well. Unattached cells were removed after 1 h of incubation, and the attached cells were counted after trypsinization. The data are presented as a percentage of the attached cells compared to total cells. For the cell detachment assay, after 24 h of incubation, the cells were incubated with 0.05% trypsin for 3 min to detach the cells. The culture medium was added to inactivate the trypsin, and the detached cells were collected. The remaining cells were incubated with 0.25% trypsin to detach them, and the cells were then counted. The data are presented as a percentage of the detached cells to total cells.
Cell Cycle Assay
For the cell cycle assay, the treated U87 and SHG44 cells were obtained, washed with PBS three times, fixed with 80% ethanol, subsequently incubated with RNase A (0.25 mg/ml, Sigma-Aldrich) for 30 min at 37°C, and then incubated with 20 μg/ml propidium iodide (KeyGen, Nanjing, P.R. China) for 20 min at room temperature. The cell cycle was analyzed by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences). The experiments were performed in triplicate.
Western Blot Analysis
Total proteins in cells were obtained using RIPA buffer containing phenylmethylsulfonyl fluoride (PMSF; Beyotime Biotechnology, Beijing, P.R. China) on ice. Equal amounts of protein (50 μg of protein per lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% denaturing gel and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The immunoblots were blocked with 5% skim milk in Tris-buffered saline (TBS)-Tween 20 (0.05%, v/v) for 1 h at room temperature. TBST was used to wash these membranes. Then the membranes were probed with primary antibodies at 4°C overnight. Subsequently, the membranes were incubated with respective secondary antibody for 1 h at room temperature. The immunoblots were then exposed and detected using an enhanced chemiluminescence Western blotting detection system (Bio-Rad, Hercules, CA, USA).
Cell Transfection
Transfections with pcDNA3.1/E2F5, si-E2F5, let-7c mimic (10 nM), let-7c inhibitor, and appropriate NCs (all obtained from GenePharma, Shanghai, P.R. China) were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol.
Dual-Luciferase Reporter Assay
pmirGLO Dual-Luciferase Vector (Promega, Madison, WI, USA) was used for cloning. pmirGLO-E2F5-wt or pmirGLO-E2F5-mut was cotransfected with let-7c mimics or miR-NC into HEK293 cells by Lipofectamine-mediated gene transfer. The relative luciferase activity was normalized to Renilla luciferase activity 48 h after transfection. The data are relative to the fold change of the corresponding control groups defined as 1.0.
Statistical Analysis
Data are shown as the mean ± standard error of at least three independent experiments. SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for statistical analysis. Two group comparisons were performed with a Student’s t-test. Multiple group comparisons were analyzed with one-way ANOVA. All tests performed were two sided. A value of p < 0.05 was considered statistically significant.
RESULTS
let-7c Is Downregulated in Glioma Cells and Associated With Tumor Proliferation
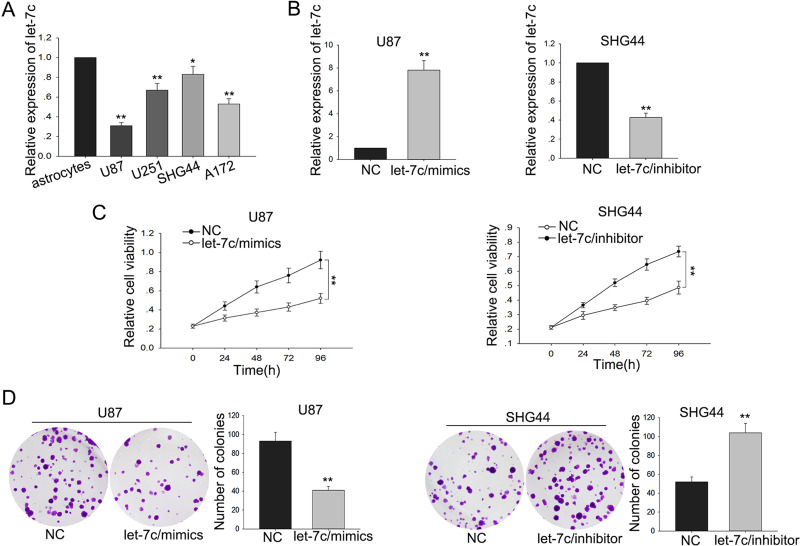
To explore the function of let-7c in glioma cells, we examined the let-7c level in four glioma cell lines (U87, U251, SHG44, A172) and one normal astrocyte cell line. Compared with the normal cell line, the levels of let-7c in the four glioma cell lines were significantly decreased (Fig. 1A). These results indicated that let-7c might play an important role in glioma. To further investigate the biological functions of let-7c in glioma, glioma cells were transfected with let-7c mimic or let-7c inhibitor using miR-NC or anti-miR-NC as NCs (Fig. 1B). MTT assay and colony formation assay showed decreased proliferation ability of glioma cells transfected with let-7c mimic compared with that in the NC-transfected cells, whereas deletion of let-7c obviously promoted cell proliferation ability (p < 0.01) (Fig. 1C and D). These data together revealed that let-7c was associated with glioma cell proliferation.
Figure 1.
let-7c is downregulated in glioma cells and associated with tumor proliferation. (A) let-7c expression levels between glioma cells (U87, U251, SHG44, and A172) and the normal astrocyte cell line were measured by quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR). (B) let-7c mimic and inhibitor were separately transfected into U87 and SHG44 cells. (C) MTT and (D) colony formation assays were performed to measure the proliferation of glioma cells transfected with let-7c mimic and inhibitor. Error bars represented the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01 versus control group.
Downregulation of let-7c Enhances the Migration and Invasion Abilities of Glioma Cells
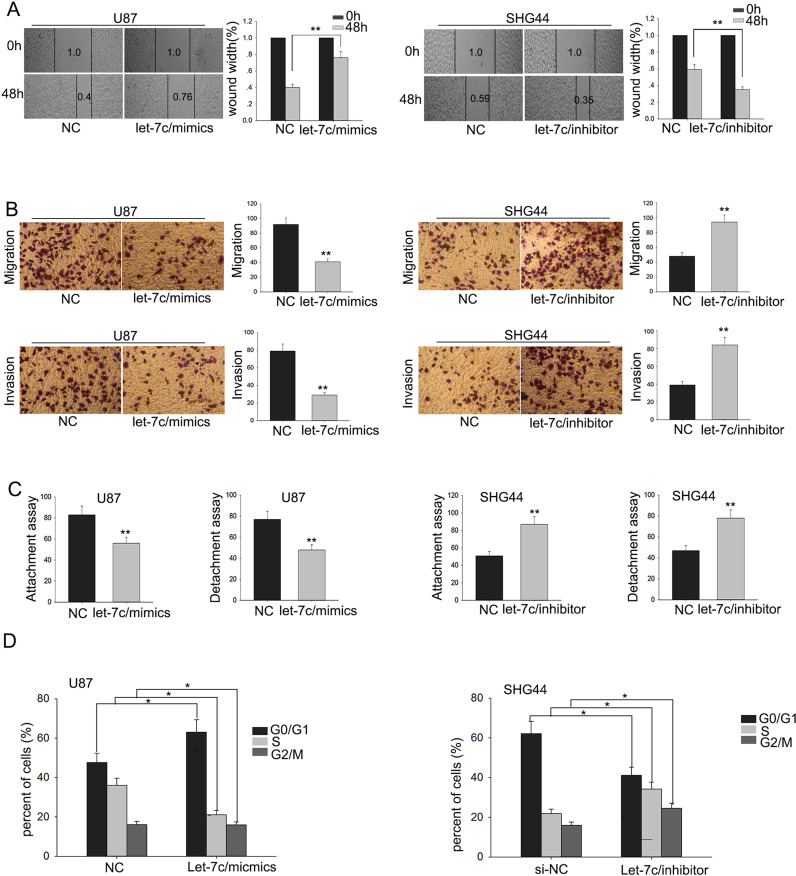
Metastasis is one of the most important characteristics of malignant tumors. Therefore, we examined the effect of let-7c on the migratory and invasive capabilities of glioma cells. The wound healing assay and invasion assay were utilized to detect the migratory capability of glioma cells. The results of the wound healing and migration assays revealed that overexpression of let-7c could inhibit the migratory capability of glioma cells (Fig. 2A and B). Transwell invasion assay revealed that the overexpression of let-7c impaired cell invasion, while downregulation of let-7c increased cell invasion (Fig. 2B). Additionally, consistent with the results of the migration and wound healing assays, attachment/detachment assays revealed that upregulated let-7c suppressed glioma cell attachment/detachment (Fig. 2C), which was contrary to that in the cells with deletion of let-7c. These results demonstrated that let-7c could effectively inhibit the migration and invasion of glioma cells.
Figure 2.
Downregulation of let-7c enhances the migration ability of glioma cells. (A) Wound healing, (B) metastasis (migration and invasion), and (C) attachment/detachment assays were utilized to detect the migratory capability of glioma cells. (D) Flow cytometry was used to check the effect of let-7c on the cell cycle. Error bars represented the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01 versus control (NC) group.
Downregulation of let-7c Induces the Cell Cycle Arrest of Glioma Cells
As shown in Figure 2D, cell numbers in the G0/G1 phase were greater than those in the S and G2/M phase in cells transfected with let-7c mimic, in contrast to cells transfected with let-7c inhibitor. The results revealed that knockdown of let-7c induced cell cycle arrest of glioma cells.
E2F5 Is Identified as a Target of let-7c
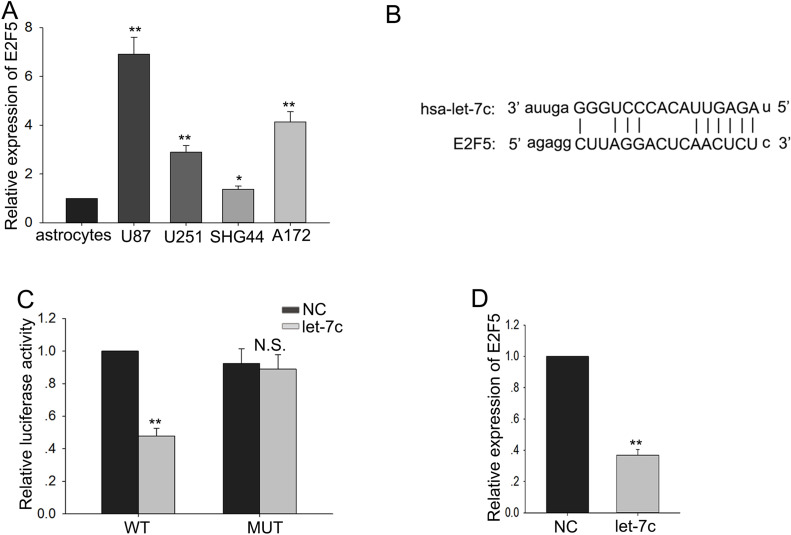
Overexpression of E2F5 had been found to be associated with tumor aggressiveness, metastasis, and poor prognosis in multiple cancers. To research whether E2F5 was involved in the function of let-7c, we first detected the level of E2F5 in four glioma cell lines and a normal cell line, and the trend of the E2F5 level was opposite to the level of let-7c (Fig. 3A). Then we utilized two bioinformatic analysis websites (http://www.targetscan.org/cgi-bin/targetscan/vert_71/view_gene.cgi?rs=ENST00000418930.2&taxid=9606&members=let-7-5p/98-5p&showcnc=0&shownc=0&subset=1 and http://mirdb.org/cgi-bin/target_detail.cgi?targetID=2060091) to forecast the binding sites between let-7c and E2F5 (Fig. 3B). Therefore, we supposed that the functions of let-7c in glioma cells might be mediated by E2F5. To further explore the regulatory relationship between let-7c and E2F5, we performed luciferase reporter assays. The let-7c mimic reduced the luciferase activity of wild-type (WT) E2F5 reporter vector (Fig. 3C). Additionally, we examined the expression of E2F5 in response to the high level of let-7c and found that overexpression of let-7c could inhibit the expression of E2F5 (Fig. 3D). These data suggested that E2F5 was a target of let-7c.
Figure 3.
E2F5 is identified as target of let-7c. (A) The level of E2F5 in four glioma cell lines and a normal astrocyte cell line was detected. (B) Bioinformatics analysis helped us obtain the binding sites between let-7c and E2F5. (C). Luciferase reporter assays further confirmed the regulating relationship between let-7c and E2F5 after wild-type (WT) or mutant (Mut) E2F5 transfection. (D) The expression of E2F5 in response to the levels of let-7c. Error bars represent the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01 versus control group (astrocytes or NC).
The Function of E2F5 on the Proliferation Ability and Migration as Well as Invasion Capacities of Glioma Cells
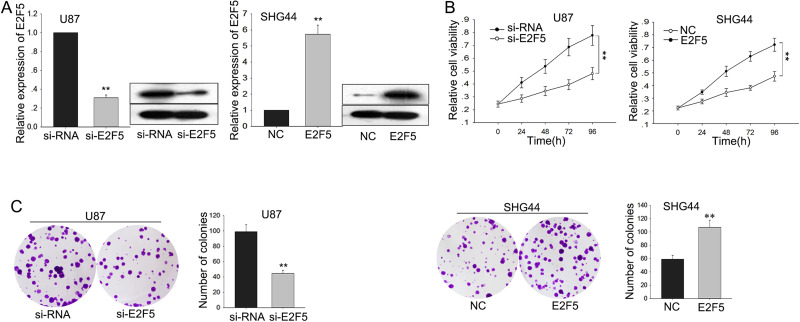
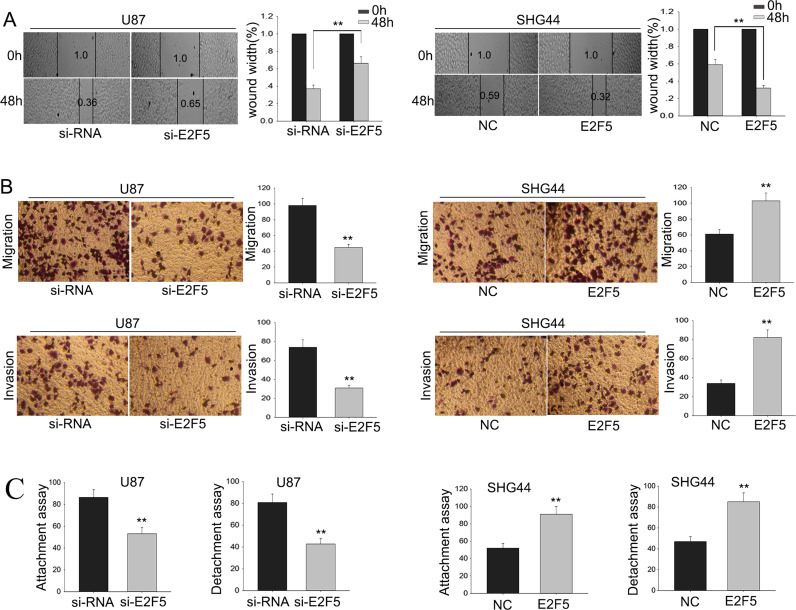
To determine the biological functions of E2F5 in glioma, glioma cells were stably transfected with short interfering RNA for E2F5 (si-E2F5) and E2F5 expression vector using siRNA and empty vector as NCs, respectively. Satisfactory transfection efficiency was acquired at 48 h posttransfection (Fig. 4A). Moreover, Western blot assay suggested that knockdown of E2F5 decreased the protein level of E2F5, whereas the ectopic overexpression of E2F5 increased its protein level (Fig. 4A). MTT assays revealed the weakened proliferation ability of glioma cells transfected with si-E2F5 compared with that in siRNA-transfected cells, whereas cells transfected with pcDNA-E2F5 increased proliferation (p < 0.01) (Fig. 4B). Additionally, colony formation assays revealed low proliferation of glioma cells transfected with si-E2F5 compared with that in siRNA-transfected cells, and an opposite result was obtained in cells transfected with pcDNA-E2F5 (p < 0.01) (Fig. 4C).
Figure 4.
The function of E2F5 on the proliferation ability of glioma cells. Glioma cells were transfected with short interfering RNA for E2F5 (si-E2F5), pcDNA-E2F5, or appropriate controls (si-RNA or NC). (A) The transfection efficiency was obtained after 48 h; Western blot assay was performed to determine the protein level of E2F5. (B) MTT and (C) colony formation assays were performed to detect the cell proliferation of transfected glioma cells. Error bars represented the mean ± SD of at least three independent experiments. **p < 0.01 versus control group (si-RNA or NC).
Metastasis and wound healing assays were then utilized to detect the function of E2F5 on the migratory capability of glioma cells. The results of the wound healing and migration assays indicated that si-E2F5 could inhibit the migratory ability of glioma cell (Fig. 5A and B). In addition, attachment/detachment assays revealed that si-E2F5 suppressed glioma cell attachment/detachment, and, on the contrary, cells transfected with pcDNA-E2F5 presented an increased metastasis capacity (Fig. 5C). The invasion assay showed that the invasive ability in cells transfected with si-E2F5 was obviously decreased, and invasion in cells transfected with pcDNA-E2F5 was enhanced (Fig. 5B). These data indicated that downregulated E2F5 could inhibit the glioma cell migration and invasion abilities.
Figure 5.
The function of dysregulated E2F5 on the migration and invasion of glioma cells. Glioma cells were transfected with si-E2F5, pcDNA-E2F5, or appropriate controls (si-RNA or NC). (A) Wound healing, (B) metastasis (migration and invasion), and (C) attachment/detachment assays were utilized to detect the migratory capability of glioma cells. Error bars represented the mean ± SD of at least three independent experiments. **p < 0.01 versus control group (si-RNA or NC).
The Effect of let-7c on Glioma Cells Is in an E2F5-Dependent Manner
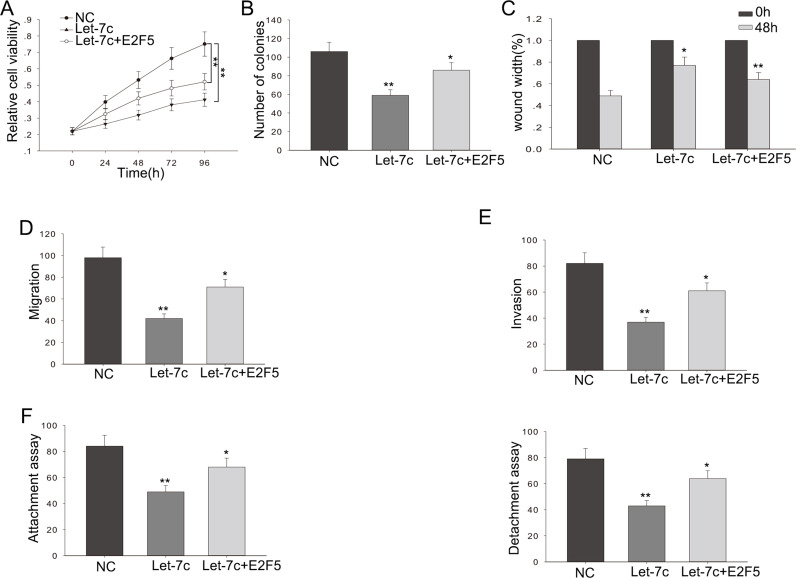
We performed rescue experiments to determine the influence of let-7c on glioma cell proliferation and migration by targeting E2F5. let-7c or NC was transfected into glioma cells cotransfected with E2F5. The weakened proliferation ability caused by let-7c in glioma cells was partially abolished by cotransfected E2F5 (p < 0.01) (Fig. 6A). The decreased colony formation capacity induced by let-7c in glioma cells was abrogated by the introduction of E2F5 (p < 0.01) (Fig. 6B). The anti-metastasis and anti-invasion effects of let-7c in glioma could be partly abolished by cotransfected E2F5 (p < 0.01) (Fig. 6C–F). These results showed that let-7c could influence cell proliferation, metastasis, and invasion in glioma cells in vitro, at least partially in an E2F5-dependent manner.
Figure 6.
The effect of let-7c on glioma cells is in an E2F5-dependent manner. Glioma cells were cotransfected with pcDNA-E2F5 and pcDNA-let-7c or appropriate controls (NC). (A) MTT and (B) colony formation assays were applied to detect the proliferation ability of cotransfected glioma cells. (C) Wound healing and (D) metastasis assays were utilized to detect the migratory capability of cotransfected glioma cells. (E) Transwell assay was employed to detect the invasive ability of cotransfected glioma cells (F) Attachment/detachment assay was used to detect the migratory capability of cotransfected glioma cells. Error bars represented the mean ± SD of at least three independent experiments. *p < 0.05, **p < 0.01 versus control group (NC).
DISCUSSION
An accumulating number of studies has shown that miRNAs are a type of small, highly conservative molecules that play a crucial role in regulating multiple cellular activities including cell proliferation, differentiation, and apoptosis16–18. let-7c has been reported in many cancers and functions as a tumor suppressor19–25. For instance, upregulation of miRNA let-7c by quercetin inhibits pancreatic cancer progression by activation of Numbl25; and detection of let-7c and miR-152 in plasma can serve as a noninvasive biomarker for non-small cell lung cancer26. In line with the above studies, the present study revealed important findings that let-7c could suppress the proliferation ability and metastasis capacity in glioma cells, which suggested a potential role of let-7c in the treatment of glioma. Even though let-7c was previously observed to be altered in many different cancer cells7–11, the underlying mechanisms have not been completely elucidated. To investigate the mechanistic function of let-7c in glioma, we measured the level of let-7c in glioma cells. Results from the qRT-PCR demonstrated that let-7c was significantly downregulated in glioma cells. We transfected glioma cell lines with a let-7c mimic or inhibitor and found that ectopic overexpression of let-7c could inhibit glioma cell proliferation and metastasis. Deletion of let-7c could significantly promote the proliferation ability and metastatic potential of glioma cells. Additionally, we found that the 3′-UTR of the E2F5 gene contained binding sites for let-7c by utilizing the bioinformatics TargetScan and miRDB. In glioma cells, let-7c interacted with the 3′-UTR of the oncogene E2F5, and the increased level of let-7c resulted in a low expression level of E2F5, which could inhibit the progression of glioma cells.
In summary, our study investigated the role of let-7c/E2F5 in glioma cells. Our results showed that let-7c was downregulated in glioma cells, which might provide a strong rationale for its potential use as a therapeutic target for treating glioma.
ACKNOWLEDGMENT
The authors thank the laboratory members.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Dolecek TA, Propp JM, Stroup NE, Kruchko C. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2005-2009. Neuro Oncol. 2012;14(Suppl 5):v1–49 [Erratum Neuro Oncol. 2013;15(5):646–7]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: Genetics, biology, and paths to treatment. Genes Dev. 2007;21(21):2683–710. [DOI] [PubMed] [Google Scholar]
- 3. Chen J, McKay RM, Parada LF. Malignant glioma: Lessons from genomics, mouse models, and stem cells. Cell 2012;149(1):36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tan KS, Armugam A, Sepramaniam S, Lim KY, Setyowati KD, Wang CW, Jeyaseelan K. Expression profile of microRNAs in young stroke patients. PLos One 2009;4(11):e7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eacker SM, Dawson TM, Dawson VL. Understanding microRNAs in neurodegeneration. Nat Rev Neurosci. 2009;10(12):837–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marchal JA, Lopez GJ, Peran M, Comino A, Delgado JR, Garcia-Garcia JA, Conde V, Aranda FM, Rivas C, Esteban M, Garcia MA. The impact of PKR activation: From neurodegeneration to cancer. FASEB J. 2014;28(5):1965–74. [DOI] [PubMed] [Google Scholar]
- 7. Bjorner S, Fitzpatrick PA, Li Y, Allred C, Howell A, Ringberg A, Olsson H, Miller CJ, Axelson H, Landberg G. Epithelial and stromal microRNA signatures of columnar cell hyperplasia linking let-7c to precancerous and cancerous breast cancer cell proliferation. PLoS One 2014;9(8):e105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhan M, Qu Q, Wang G, Zhou H. Let-7c sensitizes acquired cisplatin-resistant A549 cells by targeting ABCC2 and Bcl-XL. Pharmazie 2013;68(12):955–61. [PubMed] [Google Scholar]
- 9. Nadiminty N, Tummala R, Lou W, Zhu Y, Shi X-B, Zou JX, Chen H, Zhang J, Chen X, Luo J, White RWd, Kung H-J, Evans CP, Gao AC. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One 2012;7(3):e32832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, Zheng Q, Ma Y, Zhang J, Wu N, Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342(1):43–51. [DOI] [PubMed] [Google Scholar]
- 11. Lin KY, Ye H, Han BW, Wang WT, Wei PP, He B, Li XJ, Chen YQ. Genome-wide screen identified let-7c/miR-99a/miR-125b regulating tumor progression and stem-like properties in cholangiocarcinoma. Oncogene 2016;35(26):3376–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma Y-J, Yang J, Fan X-L, Zhao H-B, Hu W, Li Z-P, Yu G-C, Ding X-R, Wang J-Z, Bo X-C, Zheng X-F, Zhou Z, Wang S-Q. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influenza A virus in infected human lung epithelial cells. J Cell Mol Med. 2012;16(10):2539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li L, Wu C, Zhao Y. miRNA-34a enhances the sensitivity of gastric cancer cells to treatment with paclitaxel by targeting E2F5. Oncol Lett. 2017;13(6):4837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu H, Fei D, Zong S, Fan Z. MicroRNA-154 inhibits growth and invasion of breast cancer cells through targeting E2F5. Am J Transl Res. 2016;8(6):2620–30. [PMC free article] [PubMed] [Google Scholar]
- 15. Sun J, Li H, Huo Q, Cui M, Ge C, Zhao F, Tian H, Chen T, Yao M, Li J. The transcription factor FOXN3 inhibits cell proliferation by downregulating E2F5 expression in hepatocellular carcinoma cells. Oncotarget 2016;7(28):43534–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 17. Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, function and role in cancer. Curr Genomics 2010;11(7):537–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics Proteomics Bioinformatics 2015;13(1):17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen C, Hu Y, Li L. NRP1 is targeted by miR-130a and miR-130b, and is associated with multidrug resistance in epithelial ovarian cancer based on integrated gene network analysis. Mol Med Rep. 2016;13(1):188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sun J, Cheng L, Shi H, Zhang Z, Zhao H, Wang Z, Zhou M. A potential panel of six-long non-coding RNA signature to improve survival prediction of diffuse large-B-cell lymphoma. Sci Rep. 2016;6:27842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen C, Wang H-j, Yang L-Y, Jia X-b, Xu P, Chen J, Liu Y. Expression of miR-130a in serum samples of patients with epithelial ovarian cancer and its association with platinum resistance. Sichuan Da Xue Xue Bao 2016;47(1):60–3. [PubMed] [Google Scholar]
- 22. Cui Y-L, Wang B, Gao H-M, Xing Y-H, Li J, Li H-J, Lin Z, Wang Y-Q. Interleukin-18 and miR-130a in severe sepsis patients with thrombocytopenia. Patient Prefer Adherence 2016;10:313–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Han B, Lian L, Li X, Zhao C, Qu L, Liu C, Song J, Yang N. Chicken gga-miR-130a targets HOXA3 and MDFIC and inhibits Marek’s disease lymphoma cell proliferation and migration. Mol Biol Rep. 2016;43(7):667–76. [DOI] [PubMed] [Google Scholar]
- 24. Song C-L, Liu B, Shi Y-F, Liu N, Yan Y-Y, Zhang J-C, Xue X, Wang J-P, Zhao Z, Liu J-G, Li Y-X, Zhang X-H, Wu J-D. MicroRNA-130a alleviates human coronary artery endothelial cell injury and inflammatory responses by targeting PTEN via activating PI3K/Akt/eNOS signaling pathway. Oncotarget 2016;7(44):71922–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nwaeburu CC, Bauer N, Zhao Z, Abukiwan A, Gladkich J, Benner A, Herr I. Up-regulation of microRNA let-7c by quercetin inhibits pancreatic cancer progression by activation of Numbl. Oncotarget 2016;7(36):58367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dou H, Wang Y, Su G, Zhao S. Decreased plasma let-7c and miR-152 as noninvasive biomarker for non-small-cell lung cancer. Int J Clin Exp Med. 2015;8(6):9291–8. [PMC free article] [PubMed] [Google Scholar]