Abstract
Maternally expressed gene 3 (MEG3), a long noncoding RNA, has been reported to be associated with the pathogenesis of multiple malignancies. However, little is known regarding the role of MEG3 in leukemia. In this study, we found that the expression of MEG3 was decreased in leukemia patients and cell lines and has potential to be considered as a biomarker for leukemia. In addition, overexpression of MEG3 inhibited cell proliferation and invasion in vitro and in vivo. Moreover, a potential bonding site between miR-184 and MEG3 was predicted, and low expression of miR-184 was found in leukemia patients and cell lines. In vitro loss and gain of function showed that overexpression of MEG3 significantly decreased the expression of miR-184, and MEG3 knockdown markedly increased it. Furthermore, our results showed that MEG3 interacted with miR-184 and subsequently mitigated the proliferation and invasion of leukemia cells by downregulating related proteins. In conclusion, our study has identified a novel pathway through which MEG3 acts as a tumor suppressor in leukemia at the level of miRNAs and provided a molecular basis for potential applications of MEG3 in the prognosis and treatment of leukemia.
Key words: Long noncoding RNAs (lncRNAs), Maternally expressed gene 3 (MEG3), miR-184, Leukemia
INTRODUCTION
Chronic myeloid leukemia (CML) is a major myeloproliferative neoplasm, which accounts for around 15% of newly diagnosed leukemia in adults1. Currently, CML treatment is based on the use of the tyrosine kinase inhibitors (TKIs), such as imatinib, which inhibit the tyrosine kinase activity of the breakpoint cluster region (BCR)-Abelson murine leukemia (ABL) protein2–4. However, TKIs can effectively target proliferating mature cells, but do not eradicate leukemic stem cells5. Patients can develop resistance to TKIs, which limits the long-term benefits of the drug6. Thus, a deeper understanding of the molecular and genetic mechanisms that induce the initiation and progression of CML is crucial.
Long noncoding RNAs (lncRNAs) are a type of RNAs that are longer than 200 nucleotides and are implicated in multiple biological processes. Recently, lncRNAs were recognized to be diagnostic and prognostic biomarkers for malignant tumors. Maternally expressed gene 3 (MEG3) is one of the most important lncRNAs, which is located on chromosome 14q32 and expressed in many normal tissues7. Loss of MEG3 has been shown in various human cancers, including bladder, bone marrow, breast, and liver cancers, and exogenous expression of MEG3 significantly suppresses the proliferation of human cancer cells8,9. However, the biological significance of MEG3 in leukemia is not well explored.
MicroRNAs (miRNAs) are a conserved family of small noncoding RNA molecules that posttranscriptionally regulate gene expression. It was estimated that about 60% of genes can be regulated by miRNAs. miR-184 is a recently identified cancer-related miRNA. It was reported to be highly expressed in many cancers, such as hepatocellular carcinoma, glioblastoma, and pancreatic ductal adenocarcinoma10–12. In addition, the upregulation of miR-184 may facilitate cell proliferation and invasion10–12. However, the exact expression of miR-184 and the molecular mechanism underlying its role in leukemia have not been reported.
In this study, we examined the expression of MEG3 and miR-184 in leukemia patients, cell lines, and mouse models in an attempt to provide a new basis for the development of the mechanism of leukemia.
MATERIALS AND METHODS
Patients and Samples
Peripheral blood samples were obtained from 57 patients with acute myeloid leukemia (AML) (diagnosed from January 2015 to December 2016 at the Department of Hematology in the First Affiliated Hospital of Xinxiang Medical University) and 57 healthy volunteers. Written informed consent was signed by all participants. This study was approved by the ethics committee of the First Affiliated Hospital of Xinxiang Medical University.
Cell Culture
THP-1, HL-60, CCL-240, and CRL-1582 cells (ATCC, Manassas, VA, USA) were grown in RPMI-1640 medium complemented with 10% FBS (v/v; Life Technologies, Grand Island, NY, USA). All cells were cultured at 37°C in a 5% CO2 incubator.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Equal amounts of RNA were reverse transcribed to cDNA with the SuperScript Reverse Transcriptase Kit (Thermo Fisher Scientific, Waltham, MA, USA). Then the total cDNA was amplified and analyzed by SYBR Green PCR Master Mix (Thermo Fisher Scientific) in a Fast Real-Time PCR 7500 System (Applied Biosystems, Foster City, CA, USA). The following primers were used12,13: MEG3, 5′-CTGCCCATCTACACCTCACG-3′ (forward) and 5′-CTCTCCGCCGTCTGCGCTAGGGGCT-3′ (reverse); miR-184, 5′-TACGACTATGTGACCTGCCTG-3′ (forward) and 5′-TGGTTCAACT CTCCT TTCCA-3′ (reverse); GAPDH, 5′-GGCCTTCCGTGTTCCTAC-3′ (forward) and 5′-TGTCATCATATCTGGCAGGTT-3′ (reverse). The original Ct values were adjusted to GAPDH.
Construction of the Recombinant Lentiviral Vector
The MEG3 gene was designed and synthesized by PCR according to the GenBank of human MEG3 (NR_002766) gene sequence14. The LV-MG3 was constructed essentially as previously described15.
Cell Viability Analysis
HL-60 cells were cultured on a 96-well plate and transfected with LV-GFP or LV-MEG3 for various times. Cell viability was then measured by the CCK-8 kit (Beyotime Biotechnology, Shanghai, P.R. China) according to the manufacturer’s instructions.
Western Blot
Total protein samples from cells and tissues were prepared with standard protocol. Western blot was performed essentially as previously described16.
Cell Invasion Analyses
The invasion capacity of the HL-60 cells was examined using Transwell invasion assay. Briefly, cells were plated in the upper chamber in serum-free medium, and 20% FBS was added to the medium in the lower chamber. After incubating for 24 h, noninvading cells were removed from the top well with a cotton swab; the bottom cells were fixed in 95% ethanol and stained with hematoxylin. The cell numbers were determined by counting of the penetrating cells under a microscope at 200× magnification on 10 random fields in each well.
Bioinformatics Data Set
Prediction of the interaction between MEG3 and miR-184 was performed using DIANA TOOLS (http://diana.imis.athena-innovation.gr/DianaTools) as previously described17.
Luciferase Reporter Assays
The 3′-UTR of MEG3 mRNA containing miR-184 binding sites was PCR amplified and inserted to downstream of a Renilla luciferase reporter gene in the pGL3 vector. In addition, a mutant construct containing mutations within the binding sites was generated using the TaKaRa MutanBEST Kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. HL-60 cells were cotransfected with miR-184 mimics and wild-type or mutant luciferase reporter plasmid by Lipofectamine 2000 reagent (Invitrogen). At 24 h after transfection, the luciferase activities were measured with a dual-luciferase reporter assay system (Promega, Madison, WI, USA) according to the manufacturer’s instructions.
Animal Work and Experimental Protocols
Fifty athymic BALB/c mice (Laboratory Animal Center of Xinxiang Medical University, Xinxiang, P.R. China) were used in the present study. Mice were housed under controlled conditions (25 ± 2°C, 70% humidity, and 12-h light/dark periods) and fed a regular sterile chow diet and water ad libitum. All mice were randomly divided into two groups. One group was subcutaneously inoculated in the back with 5 × 106 HL-60 cells transfected with LV-MEG3, and another group was treated with scrambled HL-60 cells. Animals were monitored for signs of tumor growth. Tumor volumes were calculated at 6–30 days after injection. Upon termination, the tumor tissues were collected. All the animal experiments were performed according to relevant national and international guidelines and were approved by the Animal Experimental Ethical Committee.
Statistical Analysis
All results were presented as mean ± SD. The statistical significance of the studies was analyzed using Student’s t-test. The difference was considered statistically significant with a value of p < 0.05.
RESULTS
Expression of MEG3 Is Decreased in Leukemia Patients and Cell Lines
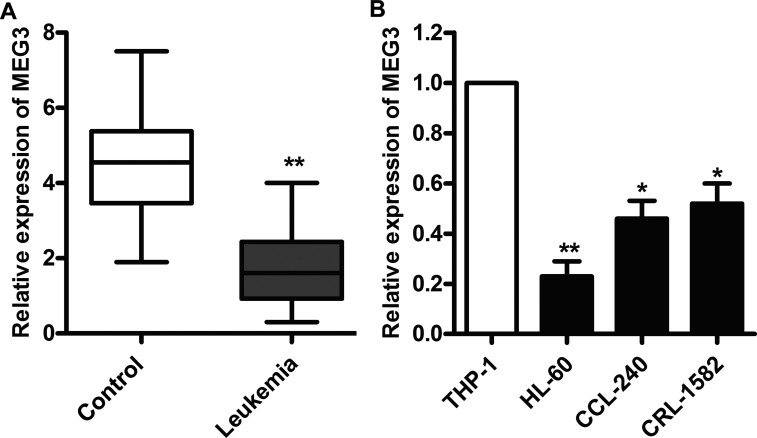
To understand the biological significance of lncRNA MEG3 in leukemia, the mRNA levels of MEG3 were examined in serum from 57 patients. The results showed that the expressions of MEG3 in patients with leukemia were significantly higher than those in healthy controls (p < 0.01) (Fig. 1A). The expressions of MEG3 were subsequently measured in THP-1 cells and three leukemia cell lines (HL-60, CCL-240, and CRL-1582). As shown in Figure 1B, the mRNA expression levels of MEG3 were significantly increased in leukemia cells compared with HL-60 cells (p < 0.01). These results indicate that leukemia cells display a low expression of MEG3.
Figure 1.
The expression of maternally expressed gene 3 (MEG3) is decreased in leukemia patients and cell lines. (A) The mRNA levels of MEG3 in serum from 57 leukemia patients were assayed by quantitative real-time polymerase chain reaction (qRT-PCR). (B) The mRNA levels of MEG3 in THP-1, HL-60, CCL-240, and CRL-1582 cells were assayed by qRT-PCR. All the experiments were repeated at least three times. *p < 0.05, **p < 0.01 versus normal cells or serum.
MEG3 Overexpression Suppresses Proliferation and Invasion of Leukemia Cells
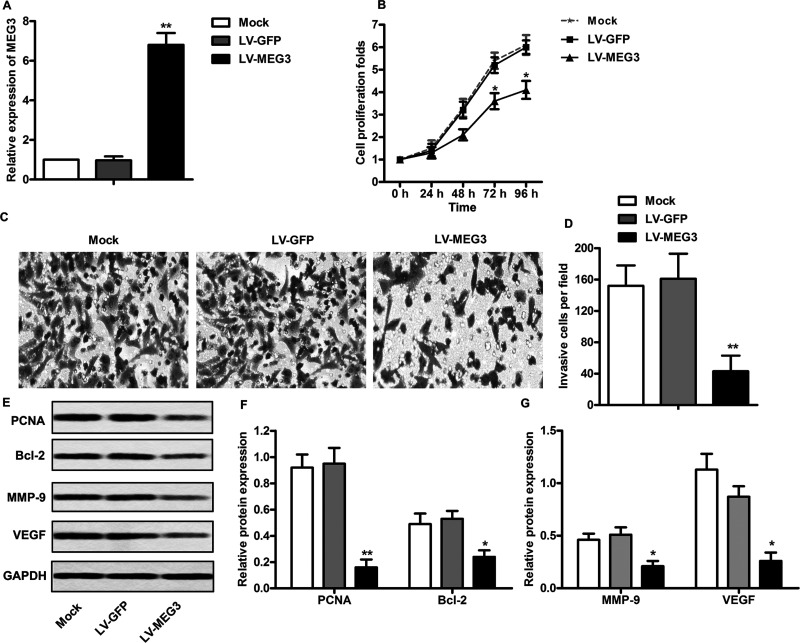
To investigate the effect of MEG3 on cell proliferation and invasion, HL-60 cells were transfected with LV-MEG3 to overexpress MEG3. According to the qRT-PCR analysis, the relative mRNA level of MEG3 was significantly increased after the transfection of LV-MEG3 (p < 0.01) (Fig. 2A). This revealed that the LV-MEG3 used in the present study successfully overexpressed MEG3 in HL-60 cells. The cell viability analysis showed that MEG3 overexpression significantly decreased cell proliferation after 72 or 96 h posttransfection in HL-60 cells (p < 0.05) (Fig. 2B). Then a Matrigel Transwell assay was carried out. As shown in Figure 2C and D, the capacity of cell invasion was obviously decreased in HL-60 cells after MEG3 overexpression (p < 0.01). In addition, the expression levels of proliferation-associated proteins were measured in LV-MEG3-treated cells. As shown in Figure 2E and F, the expression levels of proliferating cell nuclear antigen (PCNA) and B-cell lymphoma 2 (BCL-2) were significantly decreased in LV-MEG3-treated HL-60 cells (p < 0.01). Because vascular endothelial growth factor (VEGF) and matrix metalloproteinases (MMPs) play an important role in tumor progression by promoting migration and invasion, the effect of MEG3 overexpression on the expression levels of these proteins was determined in HL-60 cells. As anticipated, the expressions of MMP-9 and VEGF were significantly decreased after transfecting with LV-MEG3 (p < 0.05) (Fig. 3E and G). Taken together, these findings indicate that MEG3 overexpression may inhibit the proliferation and invasion of leukemia cells through the downregulation of proliferation- and invasion-related proteins.
Figure 2.
MEG3 overexpression suppresses proliferation and invasion of leukemia cells. (A) HL-60 cells were transfected with LV-GFP or LV-MEG3 for 24 h, and the mRNA levels of MEG3 were measured by qRT-PCR. (B) HL-60 cells were transfected with LV-GFP or LV-MEG3 for 24, 48, 72, and 96 h, and the cell viability was assayed by the CCK-8 kit. (C) Matrigel invasion assay. (D) Quantifications of (C). (E) Protein levels of proliferating cell nuclear antigen (PCNA), B-cell lymphoma 2 (BCL-2), matrix metalloproteinase 9 (MMP9), and vascular endothelial growth factor (VEGF) were assayed by Western blot. (F, G) Quantifications of (E). All the experiments were repeated at least three times, and the representative data are shown. GAPDH was used as loading control. *p < 0.05, **p < 0.01 versus mock.
Figure 3.
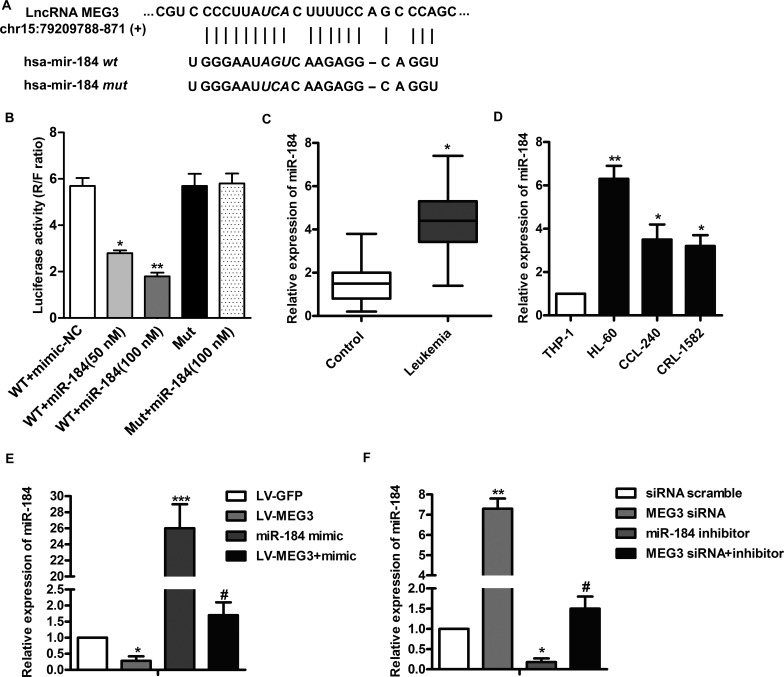
MEG3 downregulates miR-184 expression in leukemia. (A) Bioinformatics analysis of MEG3 and miR-184. (B) HL-60 cells were transfected with luciferase reporter plasmid that contained the wild-type or mutant binding site of miR-184 and cotransfected with different levels of the miR-184 mimic for 24 h. Cell lysates were assayed for luciferase activity. (C) The mRNA levels of miR-184 in serum from 57 leukemia patients were assayed by qRT-PCR. (D) The mRNA levels of miR-184 in THP-1, HL-60, CCL-240, and CRL-1582 cells were assayed by qRT-PCR. (E) HL-60 cells were transfected with the LV-MEG3 and/or miR-184 mimic for 24 h, and the mRNA levels of miR-184 were assayed by qRT-PCR. (F) HL-60 cells were transfected with the MEG3 siRNA and/or miR-184 inhibitor for 24 h, and the mRNA levels of miR-184 were assayed by qRT-PCR. All the experiments were repeated at least three times. *p < 0.05, **p < 0.01, ***p < 0.001 versus normal cells or scramble. #p < 0.05 versus miR-184 mimic or inhibitor treated alone.
MEG3 Downregulates miR-184 Expression in Leukemia
Many studies have reported that miR-184 is highly expressed in various tumors10,12, and our bioinformatics analysis indicated that MEG3 and miR-184 have a targeted correlation (Fig. 3A). Based on this, we established a luciferase reporter containing exact or mutant miR-184 binding sites. The results showed that the miR-184 mimic dose-dependently decreased the luciferase activity of wild-type MEG3 reporter plasmid (p < 0.01) (Fig. 3B). However, the reducing effect was abolished on mutant MEG3 reporter plasmid. We next investigated the expressions of miR-184 in serum from 57 patients. The results showed that the expressions of miR-184 in patients with leukemia were significantly higher than those in healthy control (p < 0.05) (Fig. 3C). As before, we subsequently compared the expressions of miR-184 in THP-1 cells and leukemia cell lines (HL-60, CCL-240, and CRL-1582). As shown in Figure 3D, the mRNA expression levels of miR-184 were significantly elevated in leukemia cells compared with THP-1 cells (p < 0.05). To further determine the relationship between MEG3 and miR-184, the expression levels of miR-184 in cells transfected with LV-MEG3 and/or the miR-184 mimic were detected. As shown in Figure 3E, the expression levels of miR-184 were significantly decreased in HL-60 cells transfected with LV-MEG3 (p < 0.05). LV-MEG30 also partially downregulated miR-184 mimic-induced overexpression of miR-184 (p < 0.05). Then inhibition of MEG3 and miR-184 was carried out. The results showed that MEG3 siRNA markedly increased the expression of miR-184 (p < 0.01) (Fig. 3F), but the promotion was reduced in the presence of the miR-184 inhibitor (p < 0.05) (Fig. 3F). These results suggest that MEG3 may downregulate the expression of miR-184.
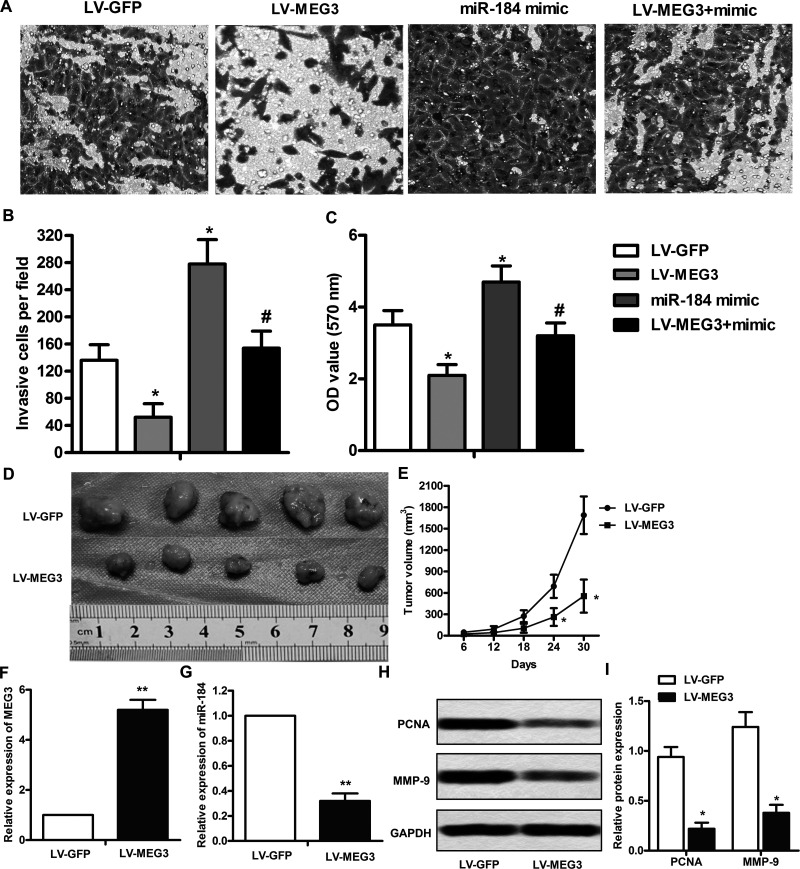
MEG3 Downregulates miR-184 and Mitigates the Progression of Leukemia
To further determine the biological significance of miR-184 in the progression of leukemia, cell invasion and proliferation assays were carried out. As shown in Figure 4A and B, LV-MEG3 significantly reduced miR-184 overexpression-induced cell invasion (p < 0.05). Similar results were found in the cell proliferation assay (p < 0.05) (Fig. 4C). We next assessed the effect of MEG3 overexpression on tumor growth and metastasis in mice injected with HL-60 cells. As shown in Figure 4D and E, the tumor volumes in the MEG3 overexpression mouse model were markedly smaller than those in the scrambled model at 24–30 days (p < 0.05). In addition, the expression of MEG3 was significantly increased in tumor tissues (p < 0.01) (Fig. 4G), whereas the expression of miR-184 was decreased (p < 0.01) (Fig. 4H). Furthermore, Western blot analysis showed that the expressions of PCNA and MMP9 were significantly decreased in the MEG3 overexpression model (Fig. 4I and J). Taken together, these results suggest that the interaction between MEG3 and miR-184 may contribute to the progression of leukemia.
Figure 4.
MEG3 downregulates miR-184 and mitigates the progression of leukemia. HL-60 cells were transfected with the LV-MEG3 and/or miR-184 mimic for 24 h. (A) Matrigel invasion assay. (B) Quantifications of (A). (C) Cell viability was assayed by the CCK-8 kit. (D) Mice were injected intraperitoneally with MEG3 overexpression or scramble HL-60 cells, and the tumor size was measured by ruler at 30 days. (E) Calculation of tumor volume at 6, 12, 18, 24, and 30 days after injection. (G, H) The expressions of MEG3 and miR-184 in tumor tissues were assayed by qRT-PCR. (I) Western blot analysis of PCNA and MMP9 in the tumor tissues. (J) Quantitation of (I). All the experiments were repeated at least three times. *p < 0.05, **p < 0.01 versus scramble. #p < 0.05 versus miR-184 mimic treated alone.
DISCUSSION
In this study, we have investigated the role of MEG3 in leukemia and made several novel observations. MEG3 has a low expression in leukemia cells. miR-184 is highly expressed in leukemia and closely related to tumor progression. MEG3 can target and downregulate the expression of miR-184.
CML is a malignant hematopoietic stem cell disorder whose most important characterization is Philadelphia chromosome translocation. The translocation results in a tyrosine kinase driving expansion of myeloid progenitors18. At least one third of patients will eventually fail imatinib treatment, resulting in rapid disease relapse, and a significant proportion of these will progress toward blast crisis (BC), which is usually rapidly disastrous19. Increased proliferation and apoptosis resistance are among the mechanisms of myeloproliferative disorder20. Thus, there has been increasing interest in finding new strategies to override apoptosis resistance and consequently improve treatment outcome in CML patients.
Accumulating evidence suggests that lncRNAs play diverse roles in human carcinoma. Many lncRNAs have been linked to tumorigenesis, either as oncogenes or tumor suppressors. While the underlying mechanism of many of these lncRNAs remains to be elucidated, it is clear that lncRNAs contribute to dysregulation of gene expression in leukemia, which then results in cancer initiation, development, and progression. For example, lncRNA HULC is frequently overexpressed in CML cells and positively correlated with malignancy of patients16. Another lncRNA, MIAT, is upregulated in established leukemia/lymphoma cell lines and functions to protect malignant mature B cells from cell apoptosis21. Xing et al. found that HOTAIR knockdown inhibits cell growth and colony formation and also induces the apoptosis of leukemia cells22.
Unlike the majority of lncRNAs, MEG3 has been verified in several tumors to function as a tumor suppressor. MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis23. Upregulation of MEG3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma24. Recently, MEG3 was found downregulated in AML. Moreover, there was a hypermethylation of the MEG3 promoter in AML patients, which may result from the decreased Tet methylcytosine dioxygenase 2 (TET2) activity25,26. In the present study, the role of MEG3 in leukemia was investigated in more depth. The results showed that MEG3 expression was lower in CML patients and leukemia cell lines than in the controls. In addition, overexpression of MEG3 notably decreased cellular proliferation and invasion in HL-60 cells, which may be attributed to the downregulation of proliferation- and invasion-related proteins. This provides strong evidence that MEG3 acts as a tumor suppressor in human leukemia.
Although much remains to be learned, emerging evidence suggests that miRNAs play an important role in the pathogenesis of tumor development by regulating target mRNA. As one newly discovered miRNA family member, miR-184 has been found to be abnormally expressed in various tumor cells, either as an oncogene or a tumor suppressor. Gao et al. showed that the expression of miR-184 was upregulated in hepatocellular carcinoma tissues10. Inhibition of miR-184 suppressed HepG2 cell proliferation and induced HepG2 apoptosis by regulating INPPL1 expression10. Su et al. demonstrated that overexpression of miR-184 repressed renal cancer cell migration, inhibited cell proliferation, and induced cell apoptosis27. In this study, the bioinformatics analysis revealed that MEG3 contains a potential miRNA response element (MRE) for miR-184 in its 3′-UTR. Moreover, we validated miR-184 as a target of MEG3 using overexpression, knockdown, and reporter plasmid techniques. Furthermore, miR-184 was found overexpressed in leukemia patients and cell lines. These observations strongly indicate that MEG3 acts as an oncogene and downregulates by MEG3 in human leukemia.
MMPs are a proteolytic enzyme metal ion-dependent Zn superfamily, and the main function is to degrade a variety of extracellular matrix (ECM), especially the basement membrane in ECM tumor invasion. MMP2 and MMP9 are two important factors in the MMP family and are closely related with tumor invasion and metastasis28–31. A recent study showed that overexpression of MEG3 can inhibit the invasion and migration of the pancreatic cancer cell line PANC-1 by inhibiting the expression of MMP2 and MMP932. In our study, we found that MEG3 overexpression effectively inhibited the expression of miR-184 and subsequently mitigated the proliferation and invasion of leukemia cells by downregulating PCNA and MMP9 in vitro and in vivo. These findings not only indicated the regulatory role but also presented a potential downstream target of MEG3 in leukemia.
In summary, we found that MEG3 is downregulated in leukemia and proposed a new mechanism through which MEG3 exerts its antitumor role at the level of miRNAs. Our results revealed that MEG3 may competitively bind miR-184 and regulate the progression of leukemia. Understanding the precise molecular mechanism is vital for exploring new potential strategies for early diagnosis and therapy. Our experimental data also suggested that targeting the MEG3-miR-184 axis may represent a novel therapeutic application in leukemia.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jabbour E, Kantarjian H. Chronic myeloid leukemia: 2014 update on diagnosis, monitoring, and management. Am J Hematol. 2014;89(5):547–56. [DOI] [PubMed] [Google Scholar]
- 2. Hughes TP, Saglio G, Quintas-Cardama A, Mauro MJ, Kim DW, Lipton JH, Bradley-Garelik MB, Ukropec J, Hochhaus A. BCR-ABL1 mutation development during first-line treatment with dasatinib or imatinib for chronic myeloid leukemia in chronic phase. Leukemia 2015;29(9):1832–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arora R, Sharma M, Monif T, Iyer S. A multi-centric bioequivalence trial in Ph+ chronic myeloid leukemia patients to assess bioequivalence and safety evaluation of generic imatinib mesylate 400 mg tablets. Cancer Res Treat. 2016;48(3):1120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Soverini S, De Benedittis C, Castagnetti F, Gugliotta G, Mancini M, Specchia G, Russo D, Iurlo A, Bonifacio M, Salvucci M. BCR-ABL mutations in chronic myeloid leukemia (CML) patients (pts) with failure and warning to first-and second-line tyrosine kinase inhibitor (TKI) therapy: What is the advantage of next-generation sequencing (NGS) over conventional sequencing? Blood 2015;126(23):346.26048910 [Google Scholar]
- 5. Wu L, Yu J, Chen R, Liu Y, Lou L, Wu Y, Huang L, Fan Y, Gao P, Huang M, Wu Y, Chen Y, Xu J. Dual inhibition of Bcr-Abl and Hsp90 by C086 potently inhibits the proliferation of imatinib-resistant CML cells. Clin Cancer Res. 2015;21(4):833–43. [DOI] [PubMed] [Google Scholar]
- 6. O’Hare T, Eide CA, Tyner JW, Wong MJ, Smith CA, Corbin AS, Buchanan S, Jessen KA, Tang C, Holme K. Inhibition of T315I Bcr-Abl and other imatinib-resistant Bcr-Abl mutants by the selective Abl kinase inhibitor SGX70393. Blood 2006;108(11). [Google Scholar]
- 7. Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000;5(3):211–20. [DOI] [PubMed] [Google Scholar]
- 8. Zhang X, Rice K, Wang Y, Chen W, Zhong Y, Nakayama Y, Zhou Y, Klibanski A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010;151(3):939–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang X, Zhou Y, Mehta KR, Danila DC, Scolavino S, Johnson SR, Klibanski A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J Clin Endocrinol Metab. 2003;88(11):5119–26. [DOI] [PubMed] [Google Scholar]
- 10. Gao B, Gao K, Li L, Huang Z, Lin L. miR-184 functions as an oncogenic regulator in hepatocellular carcinoma (HCC). Biomed Pharmacother. 2014;68(2):143–8. [DOI] [PubMed] [Google Scholar]
- 11. Zhang X, Ding H, Han Y, Sun D, Wang H, Zhai XU. The significance of microRNA-184 on JAK2/STAT3 signaling pathway in the formation mechanism of glioblastoma. Oncol Lett. 2015;10(6):3510–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Xiang H, Ge W, Wang H, Wang T, Xiong M. Expression and functional perspectives of miR-184 in pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol. 2015;8(10):12313–8. [PMC free article] [PubMed] [Google Scholar]
- 13. Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012;113(6):1868–74. [DOI] [PubMed] [Google Scholar]
- 14. Liu LX, Deng W, Zhou XT, Chen RP, Xiang MQ, Guo YT, Pu ZJ, Li R, Wang GF, Wu LF. The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells. Int J Oncol. 2016;48(1):421–9. [DOI] [PubMed] [Google Scholar]
- 15. Chen RP, Huang ZL, Liu LX, Xiang MQ, Li GP, Feng JL, Liu B, Wu LF. Involvement of endoplasmic reticulum stress and p53 in lncRNA MEG3-induced human hepatoma HepG2 cell apoptosis. Oncol Rep. 2016;36(3):1649–57. [DOI] [PubMed] [Google Scholar]
- 16. Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene 2017;607:41–6. [DOI] [PubMed] [Google Scholar]
- 17. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Reczko M, Maragkakis M, Dalamagas TM, Hatzigeorgiou AG. DIANA-LncBase: Experimentally verified and computationally predicted microRNA targets on long non-coding RNAs. Nucleic Acids Res. 2013;41(Database issue):D239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eiring AM, Khorashad JS, Anderson DJ, Yu F, Redwine HM, Mason CC, Reynolds KR, Clair PM, Gantz KC, Zhang TY, Pomicter AD, Kraft IL, Bowler AD, Johnson K, Partin MM, O’Hare T, Deininger MW. β-Catenin is required for intrinsic but not extrinsic BCR-ABL1 kinase-independent resistance to tyrosine kinase inhibitors in chronic myeloid leukemia. Leukemia 2015;29(12):2328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lucas CM, Harris RJ, Holcroft AK, Scott LJ, Carmell N, McDonald E, Polydoros F, Clark RE. Second generation tyrosine kinase inhibitors prevent disease progression in high-risk (high CIP2A) chronic myeloid leukaemia patients. Leukemia 2015;29(7):1514–23. [DOI] [PubMed] [Google Scholar]
- 20. Perekhrestenko T, Diachenko M, Sviezhentseva I, Gordienko A, Bilko D. Mechanisms of resistance in patients with chronic myeloid leukemia treated with tyrosine kinase inhibitors. Georgian Med News 2015(240):43–50. [PubMed] [Google Scholar]
- 21. Sattari A, Siddiqui H, Moshiri F, Ngankeu A, Nakamura T, Kipps TJ, Croce CM. Upregulation of long noncoding RNA MIAT in aggressive form of chronic lymphocytic leukemias. Oncotarget 2016;7(34):54174–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xing CY, Hu XQ, Xie FY, Yu ZJ, Li HY, Bin Z, Wu JB, Tang LY, Gao SM. Long non-coding RNA HOTAIR modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589(15):1981–7. [DOI] [PubMed] [Google Scholar]
- 23. Qin R, Chen Z, Ding Y, Hao J, Hu J, Guo F. Long non-coding RNA MEG3 inhibits the proliferation of cervical carcinoma cells through the induction of cell cycle arrest and apoptosis. Neoplasma 2013;60(5):486–92. [DOI] [PubMed] [Google Scholar]
- 24. Xiu YL, Sun KX, Chen X, Chen S, Zhao Y, Guo QG, Zong ZH. Upregulation of the lncRNA Meg3 induces autophagy to inhibit tumorigenesis and progression of epithelial ovarian carcinoma by regulating activity of ATG3. Oncotarget 2017;8(19):31714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lyu Y, Lou J, Yang Y, Feng J, Hao Y, Huang S, Yin L, Xu J, Huang D, Ma B and others. Dysfunction of the WT1-MEG3 signaling promotes AML leukemogenesis via p53-dependent and -independent pathways. Leukemia 2017;89:1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yao H, Duan M, Lin L, Wu C, Fu X, Wang H, Guo L, Chen W, Huang L, Liu D and others. TET2 and MEG3 promoter methylation is associated with acute myeloid leukemia in a Hainan population. Oncotarget 2017;8(11):18337–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Su Z, Chen D, Li Y, Zhang E, Yu Z, Chen T, Jiang Z, Ni L, Yang S, Gui Y and others. microRNA-184 functions as tumor suppressor in renal cell carcinoma. Exp Ther Med. 2015;9(3):961–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yan Y, Liang H, Li T, Li M, Li R, Qin X, Li S. The MMP-1, MMP-2, and MMP-9 gene polymorphisms and susceptibility to bladder cancer: A meta-analysis. Tumour Biol. 2014;35(4):3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao H, Yuan X, Jiang J, Wang P, Sun X, Wang D, Zheng Q. Antimetastatic effects of licochalcone B on human bladder carcinoma T24 by inhibition of matrix metalloproteinases-9 and NF-small ka, CyrillicB activity. Basic Clin Pharmacol Toxicol. 2014;115(6):527–33. [DOI] [PubMed] [Google Scholar]
- 30. Magee PJ, Allsopp P, Samaletdin A, Rowland IR. Daidzein, R-(+)equol and S-(-)equol inhibit the invasion of MDA-MB-231 breast cancer cells potentially via the down-regulation of matrix metalloproteinase-2. Eur J Nutr. 2014;53(1):345–50. [DOI] [PubMed] [Google Scholar]
- 31. Kuo CL, Lai KC, Ma YS, Weng SW, Lin JP, Chung JG. Gallic acid inhibits migration and invasion of SCC-4 human oral cancer cells through actions of NF-kappaB, Ras and matrix metalloproteinase-2 and -9. Oncol Rep. 2014;32(1):355–61. [DOI] [PubMed] [Google Scholar]
- 32. Gu L, Zhang J, Shi M, Zhan Q, Shen B, Peng C. lncRNA MEG3 had anti-cancer effects to suppress pancreatic cancer activity. Biomed Pharmacother. 2017;89:1269–76. [DOI] [PubMed] [Google Scholar]