Abstract
Accumulating evidence has indicated that long noncoding RNA (lncRNA) PlncRNA-1 plays an important regulatory role in cancers. However, the expression and biological functions of PlncRNA-1 in colorectal cancer (CRC) are still unclear. In the present study, we determined the expression of PlncRNA-1 in CRC and explored the function of PlncRNA-1 on CRC cell progression. The results showed that PlncRNA-1 was significantly increased in CRC tissues and cell lines; high PlncRNA-1 expression was associated with depth of invasion, lymph node metastasis, and TNM stage of CRC patients. Kaplan–Meier curve analysis showed that patients with high PlncRNA-1 expression had a poor overall survival. PlncRNA-1 knockdown remarkably reduced cell proliferation, migration, and invasion and promoted cell apoptosis in vitro. In vivo xenograft experiments showed that PlncRNA-1 inhibition significantly suppressed tumor growth. Finally, we used an agonist (740Y-P) of the PI3K/Akt signaling pathway; function assays showed that PlncRNA-1 exerted its effects by targeting the PI3K/Akt signaling pathway in CRC. Taken together, our data suggested that PlncRNA-1 might act as an oncogene in CRC progression and serve as a potential biomarker and therapeutic target for the treatment of CRC.
Key words: Long noncoding RNAs (lncRNAs), PlncRNA-1, Colorectal cancer (CRC), Progression, PI3K/Akt
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignancies and a leading cause of cancer-related deaths worldwide1. It is also a cancer type closely related to genetic and epigenetic alterations2. Despite the great advances achieved in molecularly targeted agents and chemotherapy for CRC, many patients do not clinically benefit from treatment because of different tumor mutation types, chemotherapy resistance, and adverse reactions3,4. Therefore, there is an urgent need to identify and develop a novel promising prognostic marker and therapeutic target.
Long noncoding RNAs (lncRNAs) are a group of RNA transcripts greater than 200 nucleotides in length and without protein-coding capacity5. lncRNAs account for more than 80% of the entire genome transcripts and were perceived as transcriptions of “noise” or clonal artifacts in the past decades6. But now, increasing evidence has revealed that lncRNAs were involved in diverse biological processes, such as cellular development, proliferation, differentiation, and apoptosis7. The dysregulation of lncRNAs has been confirmed to be related to tumor initiation, progression, invasion, and metastasis in various kinds of cancers. For example, Chen et al. showed that upregulation of lncRNA HOTTIP promoted metastasis of esophageal squamous cell carcinoma via induction of epithelial–mesenchymal transition (EMT)8. Peng and Fan found that lncRNA CCHE1 indicated a poor prognosis of hepatocellular carcinoma and promoted carcinogenesis via activation of the ERK/MAPK pathway9. Wang et al. indicated that lncRNA TUG1 promoted migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells10. However, the roles and clinical values of lncRNAs in tumors are still unknown.
In the present study, we showed that lncRNA PlncRNA-1 (prostate cancer upregulated long noncoding RNA 1, also known as CBR3-AS1) expression was increased in CRC tissues and cell lines; high PlncRNA-1 expression was associated with advanced clinical features and poor overall survival of CRC patients. Reduced expression of lncRNA PlncRNA-1 significantly suppressed cell proliferation, migration, and invasion and promoted cell apoptosis. In vivo xenograft experiments revealed that PlncRNA-1 inhibition suppressed tumor growth. In addition, we found that PlncRNA-1 exerted its effects by targeting the PI3K/Akt signaling pathway in CRC. Thus, these results suggested that PlncRNA-1 might play a crucial role in the progression of CRC.
MATERIALS AND METHODS
Human Tissue Samples
A total of 77 pairs of CRC tissues and adjacent normal tissues were collected from CRC patients with informed consent at The First Affiliated Hospital of Zhengzhou University during 2011–2012. All specimens were frozen immediately in liquid nitrogen and stored at −80°C until RNA extraction. Patients underwent surgical resection without preoperative chemotherapy or radiotherapy. Informed consents were obtained from all patients, and this study was approved by the Clinical Research Ethics Committee at The First Affiliated Hospital of Zhengzhou University. The results for the clinical information are presented in Table 1.
Table 1.
Association Between PlncRNA-1 Expression and Clinicopathological Features of Colorectal Cancer Patients
| Parameters | Total | PlncRNA-1 Expression | p Value | |
|---|---|---|---|---|
| High | Low | |||
| Gender | 0.576 | |||
| Male | 45 | 24 | 21 | |
| Female | 32 | 15 | 17 | |
| Age (years) | 0.427 | |||
| <60 | 37 | 17 | 20 | |
| ≥60 | 40 | 22 | 18 | |
| Tumor size (cm) | 0.570 | |||
| <5 | 39 | 21 | 18 | |
| ≥5 | 38 | 18 | 20 | |
| Differentiation status | 0.119 | |||
| Well and moderately | 48 | 21 | 27 | |
| Poorly | 29 | 18 | 11 | |
| Depth of invasion | 0.014 | |||
| T1–T2 | 28 | 9 | 19 | |
| T3–T4 | 49 | 30 | 19 | |
| Lymph nodes metastasis | 0.004 | |||
| Negative | 53 | 21 | 32 | |
| Positive | 24 | 18 | 6 | |
| TNM stage | 0.030 | |||
| I–II | 33 | 12 | 21 | |
| III–IV | 44 | 27 | 17 | |
TNM, tumor, node, and metastasis.
RNA Extraction and Quantitative Real-Time PCR
Total RNA was isolated from cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. cDNA was synthesized from no more than 5 μg of total RNA by the RecertAid™ First-Strand cDNA Synthesis Kit (Applied Biosystems, Carlsbad, CA, USA). Real-time PCR was performed with SYBR Premix Ex Taq™ (Takara, Dalian, P.R. China) in a 25-μl reaction volume on an MJ Opticon Monitor Chromo4™ instrument (Bio-Rad, Hercules, CA, USA). GAPDH was used as an internal control. The PCR primers were as follows: PlncRNA-1, 5′-AGTAGTTGCTTGTCCTAT-3′ (forward) and 5′-AAGTCAGTAAGTCCTAAG-3′ (reverse); GAPDH, 5′-CGCTCTCTGCTCCTCCTGTTC-3′ (forward) and 5′-ATCCGTTGACTCCGACCTTCAC-3′ (reverse). The 2−ΔΔCT method was used to determine relative gene expression.
Cell Culture and Transfection
Human CRC cell lines (SW480, DLD-1, HCT116, and SW620) and human colonic epithelial cells (HcoEpiC), purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, P.R. China), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) in humidified air with 5% CO2 at 37°C. The siRNAs targeting PlncRNA-1 (si-PlncRNA-1) and negative control siRNA (si-NC) were adopted and synthesized by GeneChem (Shanghai, P.R. China). Cells were transfected with si-PlncRNA-1 or si-NC using Lipofectamine 2000 reagents according to the manufacturer’s instruction (Invitrogen).
Cell Proliferation Assay
Cell proliferation was detected with CCK-8 assay (Dojindo, Tokyo, Japan). Briefly, after cell transfection for 24 h, cells were seeded in 96-well plates at a density of 100 μl/well (1 × 104 cells/well), 5 repeated wells for each group cells, and the cells were cultured in an incubator at 37°C, 5% CO2. A 10-μl CCK-8 solution was added to each well at the 24-, 48-, 72-, and 96-h time points followed by 4 h of incubation at 37°C. The absorbance OD value was measured by a microplate reader at 450 nm (ThermoFisher Scientific, Waltham, MA, USA).
Cell Apoptosis Assay
After 48 h of transfection, Annexin-V-FITC Kit (BD Biosciences, San Jose, CA, USA) was applied to detect cell apoptosis. Briefly, the cells were digested with trypsin and resuspended in PBS. Cells (1 × 105) were centrifuged (Beckman Coulter, Brea, CA, USA), and 195 μl of annexin V-FITC-conjugated solution was added to suspend the cells. Afterward, 5 μl of Annexin V-FITC was added and mixed evenly, and the cells were incubated at room temperature shielded from light for 10 min. The cells were centrifuged, and then 190 μl of Annexin V-FITC conjugate was added to suspend the cells; 10 μl of PI staining solution was added, and the cells were put on ice shielded from light. Apoptosis was detected with flow cytometry (Beckman) within 1 h.
Transwell Invasion Assay
The 24-well Transwell chamber with 8-μm pores was purchased from BD Biosciences. The transfected cells (1 × 105 cells per well) were suspended in 100 μl of serum-free medium and then seeded into the upper chamber of the 24-well plate, which was precoated with 30 μl of Matrigel (Sigma-Aldrich, St. Louis, MO, USA). The lower chamber was filled with 600 μl of medium with 10% FBS. After incubating for 48 h at 37°C, the cells that migrated into the lower membrane surface were fixed with methanol, stained with 0.5% crystal violet, and counted under a light microscope at 100× magnification in five randomly selected fields across the center and the periphery of the membrane.
In Vivo Tumorigenesis Assays
Five-week-old athymic BALB/c mice were maintained under specific pathogen-free conditions and manipulated according to protocols approved by the Shanghai Medical Experimental Animal Care Commission. HCT116 cells were transfected with Scramble (sh-NC) or sh-PlncRNA-1. After 48 h, cells were collected and injected into either side of the posterior flank of the nude mouse. Tumor volumes were examined every 7 days when the implantations started to grow. Tumor volumes (length × width2 × 0.5) and weights were measured every 7 days in mice from the control or sh-PlncRNA-1 groups. Six weeks after injection, the mice were killed, and the tumor weights were measured.
Western Blotting
Total proteins were extracted using RIPA protein extraction reagent (Millipore, Boston, MA, USA). Proteins were quantified using the BCA Protein Assay Kit. Equal amounts (20 μg) of protein extracts were separated by 10% SDS gel electrophoresis and then transferred to polyvinylidene fluoride membrane (Bio-Rad). After blocking with 5% skim milk, the membranes were incubated with primary antibodies at 4°C overnight (Abcam, Cambridge, UK). Membranes were washed in TBST and incubated with corresponding secondary antibodies for 2 h at 37°C. An enhanced chemiluminescence system (ECL; Pierce, Rockford, IL, USA) was used to view the bands. GAPDH was used as the loading control.
Statistical Analysis
All statistical data were analyzed by the SPSS18.0 software (SPSS Inc., Chicago, IL, USA). Experimental data are shown as the means ± standard deviation (SD) and were assessed using Student’s t-test, chi-square test, or ANOVA as appropriate. Statistically significant differences were defined as a value of p < 0.05.
RESULTS
lncRNA PlncRNA-1 Is Overexpressed in Colorectal Cancer
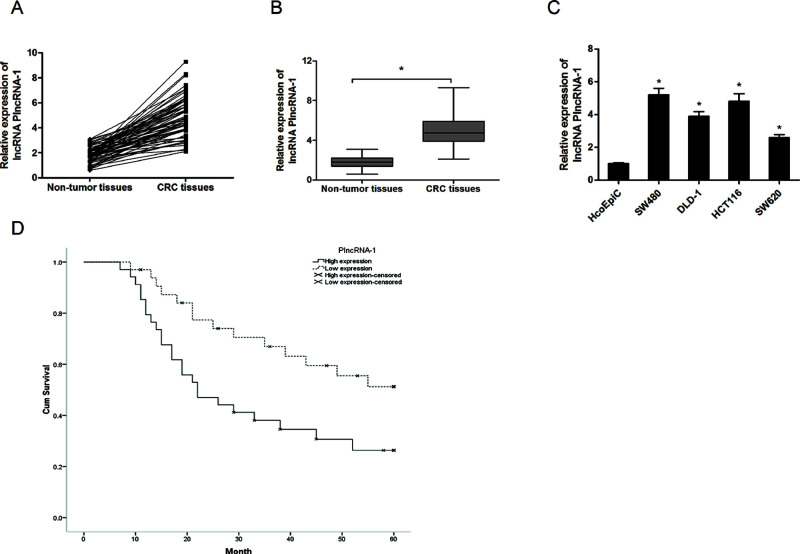
In the present study, we explored the expression of PlncRNA-1 in CRC by qRT-PCR. Our data showed that PlncRNA-1 expression was remarkably increased in CRC tissues compared with adjacent nontumor tissues (p < 0.05) (Fig. 1A and B). Similarly, the expression of PlncRNA-1 in the CRC cell lines (SW480, DLD-1, HCT116, and SW620) was higher than in the human colonic epithelial cells (HcoEpiC) (p < 0.05) (Fig. 1C). Next we determined the association between PlncRNA-1 expression and clinicopathological features in CRC patients. The results showed that PlncRNA-1 expression was significantly correlated with depth of invasion, lymph node metastasis, and TNM stage. However, no significant associations were identified between PlncRNA-1 expression and other clinicopathological features (Table 1). In addition, we evaluated the value of PlncRNA-1 on the prognosis of CRC patients. Kaplan–Meier survival analysis revealed that patients with high PlncRNA-1 expression had a shorter overall survival than patients with low PlncRNA-1 expression (p < 0.05) (Fig. 1D). These findings indicated that PlncRNA-1 might play critical roles in the development of CRC.
Figure 1.
Long noncoding RNA (lncRNA) PlncRNA-1 (prostate cancer upregulated long noncoding RNA 1) expression was increased in colorectal cancer (CRC). (A, B) The relative lncRNA-1 expression in CRC tissues and adjacent nontumor tissues. (C) The relative lncRNA-1 expression in CRC cell lines (SW480, DLD-1, HCT116, and SW620) and human colonic epithelial cells (HcoEpiC). (D) Kaplan–Meier survival analysis revealed that patients with high PlncRNA-1 expression had a poor overall survival compared with patients with low PlncRNA-1 expression. *p < 0.05.
PlncRNA-1 Inhibition Suppressed CRC Cell Proliferation
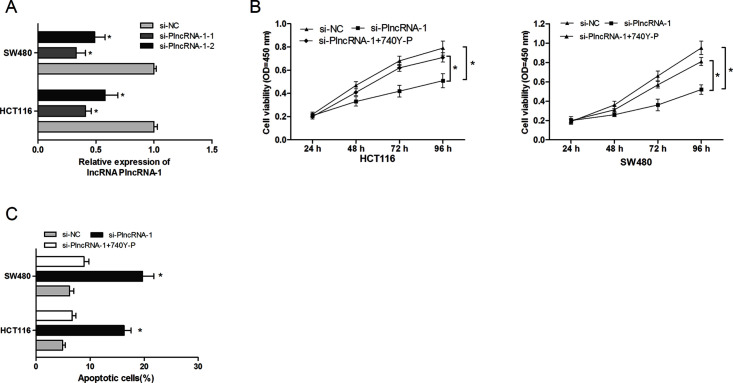
To explore the biological functions of PlncRNA-1 in CRC, we knocked down PlncRNA-1 expression in HCT116 and SW480 cells by transfecting si-PlncRNA-1 (p < 0.05) (Fig. 2A). The CCK-8 assays revealed that PlncRNA-1 inhibition significantly suppressed the proliferation rate of HCT116 and SW480 cells compared to the si-NC group (p < 0.05) (Fig. 2B). In addition, flow cytometry was used to explore the role of PlncRNA-1 on cell apoptosis. The results indicated that PlncRNA-1 inhibition dramatically induced apoptosis of CRC cells compared with the si-NC group (p < 0.05) (Fig. 2C). Those results suggested that PlncRNA-1 inhibition might suppress CRC cell proliferation by inducing the cell apoptosis process.
Figure 2.
Effects of the PI3K/Akt signaling pathway agonist (740Y-P) on the role of PlncRNA-1 in CRC cell proliferation and apoptosis. (A) Relative expression of PlncRNA-1 in HCT116 and SW480 cells was detected through qRT-PCR. (B) The CCK-8 assay showed that PlncRNA-1 inhibition suppressed proliferation of HCT116 and SW480 cells, whereas 740Y-P reversed the effect of PlncRNA-1 inhibition on CRC cell proliferation. (C) The cell apoptosis assay revealed that PlncRNA-1 suppression induced apoptosis of HCT116 and SW480 cells, whereas 740Y-P reversed the effect of PlncRNA-1 inhibition on CRC cell apoptosis. *p < 0.05.
PlncRNA-1 Inhibition Suppressed CRC Cell Migration and Invasion
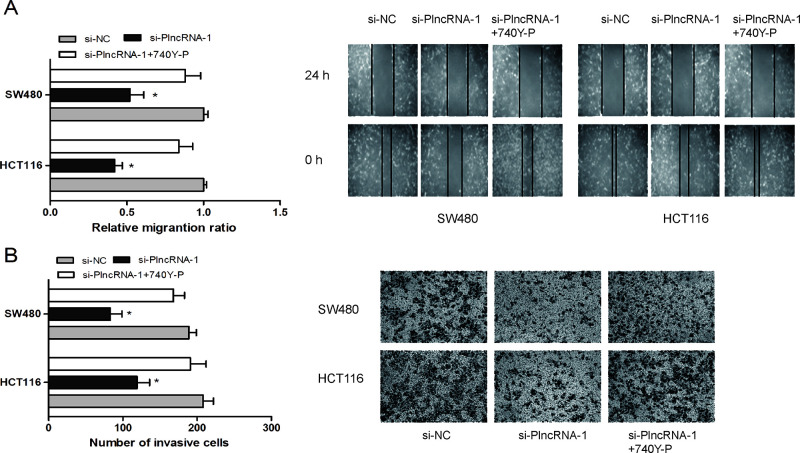
We further determined the influence of PlncRNA-1 on CRC cell migration and invasion. The wound healing assay showed that PlncRNA-1 depletion significantly reduced cell migration ability in HCT116 and SW480 cells (p < 0.05) (Fig. 3A). A Transwell invasion assay revealed that PlncRNA-1 inhibition suppressed HCT116 and SW480 cell invasion ability compared with the si-NC group (p < 0.05) (Fig. 3B). Those data indicated that PlncRNA-1 suppression might inhibit CRC cell metastasis in vitro.
Figure 3.
Effects of the PI3K/Akt signaling pathway agonist (740Y-P) on the role of PlncRNA-1 in CRC cell migration and invasion. (A) The wound healing assay showed that PlncRNA-1 knockdown reduced the cell migration ability of HCT116 and SW480 cells, whereas 740Y-P reversed the effect of PlncRNA-1 inhibition on CRC cell migration. (B) The Transwell invasion assay found that PlncRNA-1 suppression inhibited the cell invasion ability of HCT116 and SW480 cells, whereas 740Y-P reversed the effect of PlncRNA-1 inhibition on CRC cell invasion. *p < 0.05.
PlncRNA-1 Suppression Reduced Tumor Growth In Vivo
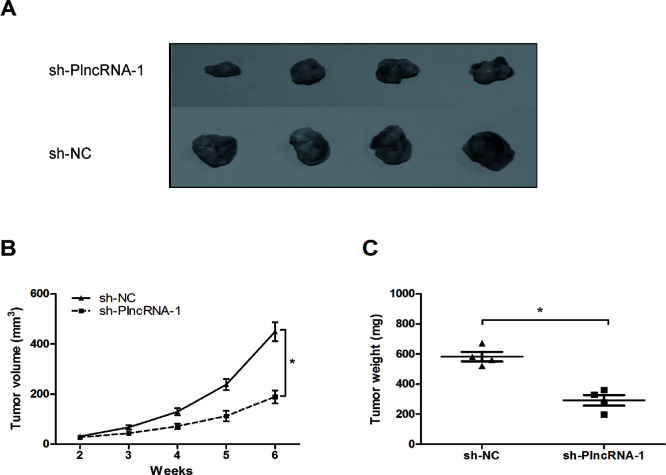
To explore the effect of PlncRNA-1 on tumor growth in vivo, we injected sh-PlncRNA-1- or sh-NC-transfected HCT116 cells into nude mice. The results showed that the growth in the sh-PlncRNA-1 group was slower compared to the sh-NC group (p < 0.05) (Fig. 4A and B). At 6 weeks after injection, the tumors were weighed and measured. We found that the average tumor weight in the sh-PlncRNA-1 group was obviously lower than that in the sh-NC group (p < 0.05) (Fig. 4C). Those data suggested that PlncRNA-1 suppression depresses tumor growth in vivo.
Figure 4.
The effect of PlncRNA-1 inhibition on tumorigenesis in vivo. (A) Photographs of tumor xenografts 6 weeks after inoculation. (B) Sh-PlncRNA-1 or sh-NC was transfected into HCT116 cells, which were injected into nude mice. The tumor volumes were calculated every 7 days from 2 to 6 weeks. (C) Tumor weight in the sh-PlncRNA-1 group was lower than that in the sh-NC group. *p < 0.05.
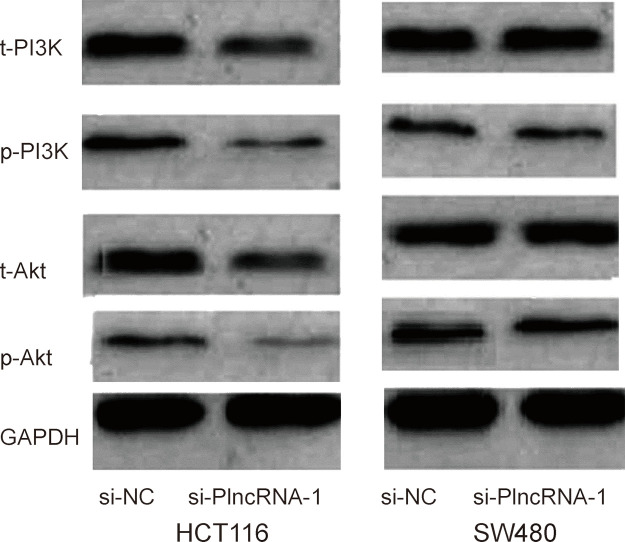
PlncRNA-1 Suppression Inhibited PI3K/Akt Signaling Pathway in CRC Cells
In order to determine the possible mechanism by which PlncRNA-1 regulated proliferation and metastasis of CRC cells, we used Western blot analysis to explore the effects of PlncRNA-1 knockdown on the PI3K/Akt signaling pathway. The Western blot showed that PlncRNA-1 silencing markedly decreased the phosphorylation of PI3K and Akt protein expression compared to the si-NC group (Fig. 5). Furthermore, to confirm whether the PI3K/Akt signaling pathway contributed to the function of lncRNA PlncRNA-1, an agonist (740Y-P) of the PI3K/Akt signaling pathway was added to the PlncRNA-1 knockdown CRC cells. When compared with the PlncRNA-1 knockdown cells, 740Y-P increased CRC cell proliferation and inhibited CRC cell apoptosis (Fig. 2C and D). Furthermore, the in vitro assay showed that cell migration and invasion abilities were significantly increased in CRC cells transfected with 740Y-P + si-PlncRNA-1 when compared with the PlncRNA-1 knockdown group (Fig. 3A and B). Thus, these results further confirmed that the downregulation of PlncRNA-1 had an effect on the biological behavior of CRC cells by regulating the PI3K/Akt signaling pathway.
Figure 5.
Effect of PlncRNA-1 on the PI3K/Akt signaling pathway in CRC cells. The Western blot showed that PlncRNA-1 inhibition suppressed the phosphorylation of PI3K and Akt protein expression compared to the si-NC group.
DISCUSSION
CRC is a group of diverse heterogeneous diseases arising through various molecular pathways. This heterogeneity determines tumor prognosis and response to therapy and brings great challenges not only in studying the molecular basis of the disease but also in clinical patient management11. Increasing evidence showed that lncRNAs might act as oncogenes or cancer suppressors, contributing to CRC pathogenesis and progression. For example, Liu et al. showed that overexpression of lncRNA DANCR was associated with advanced tumor progression and poor prognosis in patients with CRC12. Zhou et al. suggested that knockdown of lncRNA GHET1 could inhibit cell proliferation and invasion of CRC13. Han et al. found that lncRNA CRNDE could promote CRC cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling14. These studies indicated that lncRNAs play important roles in CRC progression.
PlncRNA-1, a new lncRNA that locates in the antisense region of carbonyl reductase 3 (CBR3), was first found to be generally overexpressed in prostate cancer cell lines and tissues15. Wang et al. found that upregulation of the PlncRNA-1 promoted esophageal squamous carcinoma cell proliferation and correlated with advanced clinical stage16. Dong et al. revealed that PlncRNA-1 could promote metastasis and induce EMT in hepatocellular carcinoma17. However, the role of PlncRNA-1 in the progression of CRC is still unclear.
In the current study, our data showed that the expression of PlncRNA-1 was increased in CRC tissues and cell lines. High PlncRNA-1 expression was correlated with depth of invasion, lymph node metastasis, TNM stage, and poor overall survival of CRC patients. In addition, we explored the function of PlncRNA-1 in CRC cells by applying loss-of-function approaches. The CCK-8 assay revealed that PlncRNA-1 suppression reduced CRC cell proliferation in vitro. Flow cytometry indicated that PlncRNA-1 inhibition induced cell apoptosis in vitro. The wound healing and Transwell invasion assays suggested that PlncRNA-1 inhibition suppressed CRC cell migration and invasion in vitro. Additionally, the xenograft tumor growth assay showed that PlncRNA-1 inhibition reduced CRC cell growth in vivo. Therefore, those results illuminated that PlncRNA-1 was one of the critical lncRNAs contributing to CRC carcinogenesis and progression.
The PI3K/Akt signaling pathway regulates various biological events in cells, such as gene expression, cell growth, metabolism, and metastasis18. Recent studies showed that lncRNAs could regulate tumor progression via the PI3K/Akt signaling pathway. For example, Lu et al. found that lncRNA HULC promoted cell proliferation by regulating the PI3K/Akt signaling pathway in chronic myeloid leukemia19. Zhang et al. found that lncRNA ANRIL indicated a poor prognosis of cervical cancer and promoted carcinogenesis via the PI3K/Akt pathways20. In the present study, the Western blot showed that PlncRNA-1 knockdown decreased the phosphorylation of PI3K and Akt protein expression. Furthermore, 740Y-P, an agonist of PlncRNA-1, increased proliferation, migration, and invasion of CRC cells transfected with si-PlncRNA-1. These data indicated that PlncRNA-1 might contribute to malignant CRC cell phenotypes via the activation of the PI3K/Akt signaling pathway.
In conclusion, our findings indicated that lncRNA PlncRNA-1 plays critical roles in CRC development and progression. PlncRNA-1 is a potential diagnostic and therapeutic agent for the treatment of CRC patients.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–17. [DOI] [PubMed] [Google Scholar]
- 3. Dolatkhah R, Somi MH, Bonyadi MJ, Asvadi Kermani I, Farassati F, Dastgiri S. Colorectal cancer in Iran: Molecular epidemiology and screening strategies. J Cancer Epidemiol. 2015;2015:643020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stegeman I, de Wijkerslooth TR, Stoop EM, van Leerdam ME, Dekker E, van Ballegooijen M, Kuipers EJ, Fockens P, Kraaijenhagen RA, Bossuyt PM. Colorectal cancer risk factors in the detection of advanced adenoma and colorectal cancer. Cancer Epidemiol. 2013;37:278–83. [DOI] [PubMed] [Google Scholar]
- 5. Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer 2011;10:38–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. [DOI] [PubMed] [Google Scholar]
- 7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–9. [DOI] [PubMed] [Google Scholar]
- 8. Chen X, Han H, Li Y, Zhang Q, Mo K, Chen S. Upregulation of long noncoding RNA HOTTIP promotes metastasis of esophageal squamous cell carcinoma via induction of EMT. Oncotarget 2016;7:84480–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peng W, Fan H. Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother. 2016;83:450–5. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Yang T, Zhang Z, Lu M, Zhao W, Zeng X, Zhang W. Long noncoding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108:859–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, Di Narzo AF, Yan P, Hodgson JG, Weinrich S, Bosman F, Roth A, Delorenzi M. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231:63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu Y, Zhang M, Liang L, Li J, Chen Y-X. Over-expression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8:11480–4. [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res. 2016;23:303–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Han P, Li J-w, Zhang B-m, Lv J-c, Li Y-m, Gu X-y, Yu Z-w, Jia Y-h, Bai X-f, Li L. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer 2017;16:9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cui Z, Ren S, Lu J, Wang F, Xu W, Sun Y, Wei M. The prostate cancer-up-regulated long noncoding RNA PlncRNA-1 modulates apoptosis and proliferation through reciprocal regulation of androgen receptor. Urol Oncol. 2013;31:1117–23. [DOI] [PubMed] [Google Scholar]
- 16. Wang C-M, Wu Q-Q, Li S-Q, Chen F-J, Tuo L, Xie H-W, Tong Y-S, Ji L, Zhou G-Z, Cao G. Upregulation of the long non-coding RNA PlncRNA-1 promotes esophageal squamous carcinoma cell proliferation and correlates with advanced clinical stage. Dig Dis Sci. 2014;59:591–7. [DOI] [PubMed] [Google Scholar]
- 17. Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of long non-coding RNA PlncRNA-1 promotes metastasis and induces epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38:836–46. [DOI] [PubMed] [Google Scholar]
- 18. Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, Gonzalez-Baron M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193–204. [DOI] [PubMed] [Google Scholar]
- 19. Lu Y, Li Y, Chai X, Kang Q, Zhao P, Xiong J, Wang J. Long noncoding RNA HULC promotes cell proliferation by regulating PI3K/AKT signaling pathway in chronic myeloid leukemia. Gene 2017;607:41–6. [DOI] [PubMed] [Google Scholar]
- 20. Zhang D, Sun G, Zhang H, Tian J, Li Y. Long non-coding RNA ANRIL indicates a poor prognosis of cervical cancer and promotes carcinogenesis via PI3K/Akt pathways. Biomed Pharmacother. 2017;85:511–6. [DOI] [PubMed] [Google Scholar]