Abstract
The direct roles of miR-139-3p on hepatocellular carcinoma (HCC) cell growth and metastasis remain poorly understood. We attempted to demonstrate the regulatory role of miR-139-3p in HCC progression and its underlying mechanisms. Here we showed that miR-139-3p expression was significantly reduced in the HCC tissues compared to paratumor tissues. Exogenous overexpression of miR-139-3p inhibited the migration and invasion of HCC cells, whereas downregulation of miR-139-3p was able to induce HCC HepG2 and SNU-449 cell migration and invasion. In addition, miR-139-3p inhibited HCC growth and lung metastasis in an in vivo mouse model, which is mainly regulated by annexin A2 receptor (ANXA2R). Finally, we identified that the expression of miR-139-3p was inversely correlated with ANXA2R expression in human HCC tissue. All these results demonstrated that miR-139-3p inhibited the metastasis process in HCC by downregulating ANXA2R expression.
Key words: MicroRNA 139-3p (miR-139-3p), Hepatocellular carcinoma (HCC), Annexin A2 receptor (ANXA2R), Metastasis
INTRODUCTION
Hepatocellular carcinoma (HCC) is one of the most prevalent cancers and the second most frequent cause of cancer-related deaths worldwide1. Although the cure rate is 20%–30% in HCC patients who undergo surgical resection, the 5-year survival rate is very poor, with a high incidence of postoperative recurrence rate2. For reducing patient survival rate, postoperative recurrence is a major problem, and it often occurs because of intrahepatic metastasis from the original primary tumor tissues or from de novo carcinogenesis. The precise molecular mechanisms involved in HCC metastasis are still not known, but sufficient evidence demonstrates that the epithelial–mesenchymal transition (EMT) process plays a major role in HCC cell metastasis3. The EMT process is a reversible and transient switch from a polarized and epithelial cell phenotype to a mesenchymal cellular phenotype, with high migration and invasion properties. In principle, the EMT process is a fundamental event in the early stages of embryo development and pathophysiological situations, including chronic inflammation, wound healing, and carcinoma progression. A previous study identified that downexpression or functional loss of E-cadherin is one of the most important hallmarks of EMT, and transcriptional control of E-cadherin during EMT seems to be required4. Although EMT entails the downregulation of other epithelial-specific genes, such as those for components of tight and gap junctions or desmosomes, E-cadherin downregulation is thought to be inherent to EMT.
MicroRNAs (miRNAs) are an evolutionarily conserved family of small (approximately 22 nucleotides) noncoding RNAs, known to be potent modifiers of gene expression at the posttranscriptional level5. They control multiple cellular processes including tumorigenesis and metastasis by suppressing gene expression in various malignancies, including breast cancer, non-small cell lung cancer (NSCLC), and HCC6. miR-139-3p, a member of the miR-139 family, is considered to be a potential tumor suppressor. miR-139-3p has been reported to suppress tumor growth and metastasis of cervical cancer through the Nin one-binding protein (Nob1p) signaling pathway7, and downregulation of miR-139-3p in tumor tissue was associated with poor overall survival for patients who underwent curative surgery for colon cancer8. The result of quantitative real-time (qRT)-PCR showed that miR-139-3p was downregulated in CC tissues and cell lines. In the present study, we found that miR-139-3p suppresses tumor progression in HCC by inhibiting tumor growth and suppressing cancer cell migration and metastasis. Overexpression of miR-139-3p significantly suppressed HCC cell migration and invasion and induced cell growth in vivo. Furthermore, overexpressing miR-139-3p significantly promoted the tumorigenicity of HCC cells in vivo. Bioinformatics analysis and luciferase reporter gene assay confirmed that annexin A2 receptor (ANXA2R) was targeted by miR-139-3p at the 3′-untranslated region (3′-UTR) of its mRNA sequence. Furthermore, the level of miR-139-3p was inversely correlated with ANXA2R expression in HCC. Collectively, miR-139-3p has strong potential as a promising metastasis promoter in HCC progression.
MATERIALS AND METHODS
Patients, Specimens, and Follow-Up
In this study, HCC specimens used in the immunohistochemical assay were obtained randomly from patients who underwent radical resection of pathologically confirmed tumors from 2015 to 2016 at The Third Affiliated Hospital of SunYat-sen University (Guangzhou, P.R. China). No patients received any preoperative anticancer treatment. The present research was approved by the research ethics committee of The Third Affiliated Hospital of SunYat-sen University. A total of 31 cases were used in this study to examine ANXA2R expression, with 31 HCC patients expressing miR-139-3p. All patients provided written informed consent to participate in this study.
Cell Line Culture and Morphological Observation
The human HCC cell lines HepG2, Bel-7402, SNU-449, SNU-423, SNU-387, and PLC/PRF/5 (obtained from the Cell Bank of the Chinese Academy of Sciences, Shanghai, P.R. China) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS) and 100 U ml−1 penicillin and 50 mg ml−1 streptomycin. All cells were incubated at 37°C with a humidified atmosphere of 5% CO2.
miRNA and Transfection
To modify miR-139-3p expression levels in HCC cell lines, we obtained recombinant lentivirus vectors from Genechem (Shanghai, P.R. China) that included genes such as pre-miR-139-3p, the negative control precursor miRNA; anti-miRNA-locked nucleic acids (LNAs) against miR-139-3p; and the negative control of anti-miRNA-LNAs. These vectors, with their packaging vectors, were transfected into 293 T cells using Lipofectamine 2000 (Invitrogen). HepG2 cells or SNU-449 cells were then transfected with virus following the manufacturer’s instructions. To knock down ANXA2R, a siRNA sequence targeting human ANXA2R (5′-GCCUCAACCCUGGCUUGUATT-3′) was cloned into pSuper-retro-puro (Promega, Madison, WI, USA) to generate pSuper-retro-ANXA2R-RNAi (referred to as siANXA2R). The esophageal HCC cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions9.
Identifying Target Gene of miR-139-3p
We predicted target genes of miR-139-3p based on TargetScan using predicted context score <−0.210. TargetScan Human Release 7.1 was used.
Western Blot
Cells were lysed using cell lysis buffer [150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100] containing protease and phosphatase inhibitors. Equivalent amounts of protein were subjected to SDS-polyacrylamide electrophoresis (PAGE) and transferred to nitrocellulose membranes. The membranes were blocked with 5% nonfat milk for 2 h and then incubated with the respective primary antibody overnight at 4°C, followed by the incubation of the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody for 2 h at room temperature. Blots were visualized with an enhanced chemiluminescence (ECL) detection kit (Millipore, Braunschweig, Germany) and analyzed using Quantity One 1-D Analysis Software (Bio-Rad, San Francisco, CA, USA)11.
Quantitative Real-Time RT-PCR
Total cellular RNA was extracted using TRIzol reagent (Invitrogen), and 2 μg of RNA was subjected to complementary DNA (cDNA) synthesis using random hexamers. RT-qPCRwas performed using an Applied Biosystems 7500 Sequence Detection system with an initial denaturation step at 95°C for 10 min, followed by 28 cycles of denaturation at 95°C for 60 s, primer annealing at 58°C for 30 s, and primer extension at 72°C for 30 s, with a final extension step at 72°C for 5 min. Target gene expression was calculated using the threshold cycle (Ct) values and the formula 2 − [(Ct of Genes) − (Ct of UBC)] relative to the internal control gene ubiquitin C (UBC)12. PCR primers were designed using Primer Express version 2.0 (Applied Biosystems, Foster City, CA, USA) and were as follows: E-cadherin, 5′-CGAGAGCTACACGTTCACGG-3′ (forward) and 5′-GGGTGTCGAGGGAAAAATAGG-3′ (reverse); N-cadherin, 5′-TTTGATGGAGGTCTCCTAACACC-3′ (forward) and 5′-ACGTTTAACACGTTGGAAATGTG-3′ (reverse); vimentin, 5′-GACGCCATCAACACCGAGTT-3′ (forward) and 5′-CTTTGTCGTTGGTTAGCTGGT-3′ (reverse); UBC, 5′-GACTCATGACCACAGTCCATGC-3′ (forward) and 5′-AGAGGCAGGGATGATGTTCTG-3′ (reverse).
Wound Healing Assay
We examined the migration of HCC cells using a wound healing assay. Briefly, cells were each grown on 3.5-cm plates. After the growing cell layers had reached confluence, we inflicted a uniform wound in each plate using a pipette tip and washed the wounded layers with phosphate-buffered saline (PBS) to remove all cell debris. Then cells were cultured with culture supernatants from the indicated cells. We evaluated the closure at 24 h using bright-field microscopy13.
Transwell Invasion Assay
Quantitative cell invasion assays were performed using a chamber (Corning, Tewksbury, MA, USA) with 8.0-μm polycarbonate filter inserts in 24-well plates as described previously14. About 1 × 105 cells per well were added to the upper chamber, and the lower chamber was filled with complete medium (containing 20% FBS). Cells were allowed to migrate for 24 h at 37°C. Noninvaded cells were removed from the upper chamber using a cotton swab, and the migrated cells were fixed with methanol, stained with crystal violet, and photographed under an inverted microscope. Invasion was assessed by counting the number of stained cells from five random fields15.
Luciferase Activity Assay
HEK293T cells were seeded in a 96-well plate at 50% to 60% confluence. After 24 h, cells were transfected with 50 ng of miR-139-3p expression vector, miR-139-3p inhibitor, control vector, or negative control. Cells were cotransfected with 10 ng of the wild-type (WT) or mutant (MUT) 3′-UTR of the target gene. Transfections were performed using 0.45 μl of FuGENE (Promega). Forty-eight hours after transfection, cells were collected, and Renilla luciferase activity was measured using a dual-luciferase reporter assay (Promega) and detected using an Orion II microplate luminometer (Berthold, Bad Wildbad, Germany). Luciferase reporter assays were performed in quadruplicate and repeated in three independent experiments16.
Xenograft Model of HCC in Nude Mice
BALB/c nude mice (5 weeks old, weighing 18–20 g) were purchased from the Shanghai Silaike Experimental Animals Co., Ltd. and housed under specific pathogen-free conditions. The experimental protocol was approved by The Third Affiliated Hospital of SunYat-sen University. The various cells (5 × 106 cells) in saline were implanted by subcutaneous injection (100 μl per mouse) to obtain subcutaneous tumors (n = 6 per group). The mice were euthanized after 25 days to obtain the tumor tissues. After their volume was measured, the tumors (removed from the liver) were placed in a 4% paraformaldehyde solution. The tumor volume was calculated according to the formula: tumor volume = (largest diameter × perpendicular height2)/217.
In Vivo Lung Metastasis Assay
To evaluate the metastatic potential of HCC cells, 2 × 106 cells in 100 μl of serum-free medium were injected into the tail veins of 6-week-old female nude mice (n = 6 per group). After 12 weeks, mice were sacrificed, and quantitation of metastatic colonies was performed on representative hematoxylin and eosin (H&E)-stained sections of formalin-fixed and paraffin-embedded lungs.
Statistical Analysis
Data analysis was performed using SPSS 19.0 for Windows. Differences in the results of two groups were evaluated using either two-tailed Student’s t-test or one-way ANOVA followed by post hoc Dunnett’s test. Pearson’s correlation test was used to compare quantitative variables. A value of p < 0.05 was considered statistically significant.
RESULTS
Downregulation of miR-139-3p in HCC
First, the online database YM500 (http://ngs.ym.edu.tw/ym500/) was analyzed to compare the miRNA expression in HCC and normal solid tissues18. This database compared the miRNA expression profiles between 50 normal solid tissues and 147 primary HCCs. For these miRNAs, we found that miR-139-3p was significantly downregulated in HCC, compared to normal tissues (Fig. 1A). We then examined the expression pattern of miR-139-3p in human HCC tissues. In the 31 CRC tissues and 18 normal adjacent tissues, we found that miR-139-3p levels were significantly decreased in HCC tissues compared to normal adjacent tissues (Fig. 1B). Finally, the reduced miR-139-3p expression was also confirmed in a panel of HCC cell lines and normal liver cell line LO2. As shown in Figure 1C, miR-139-3p was markedly reduced in all six cancer lines examined (SNU-387, HepG2, SNU-423, Bel-7402, SNU-449, and PLC/PRF/5).
Figure 1.
MicroRNA-139-3p (miR-139-3p) was downregulated in hepatocellular carcinoma (HCC) tissues compared to normal tissues. (A) miRNA meta-analysis in YM500v2 (http://ngs.ym.edu.tw/ym500v2/index.php) was performed to detect differently expressed miRNA of HCC and control noncancerous solid tissues. The hierarchical clustering of miRNAs that were differentially expressed among HCC patients (n = 147) and normal controls (n = 50). (B) Quantitative real-time (qRT)-PCR analysis showing relative miR-139-3p expression levels in HCC tissues (n = 31) and in normal adjacent tissues (n = 18). **p < 0.01 compared with normal. (C) qRT-PCR analysis of miR-139-3p mRNA levels in the HCC cell lines and normal liver cells LO2. **p < 0.01 compared with LO2 cells.
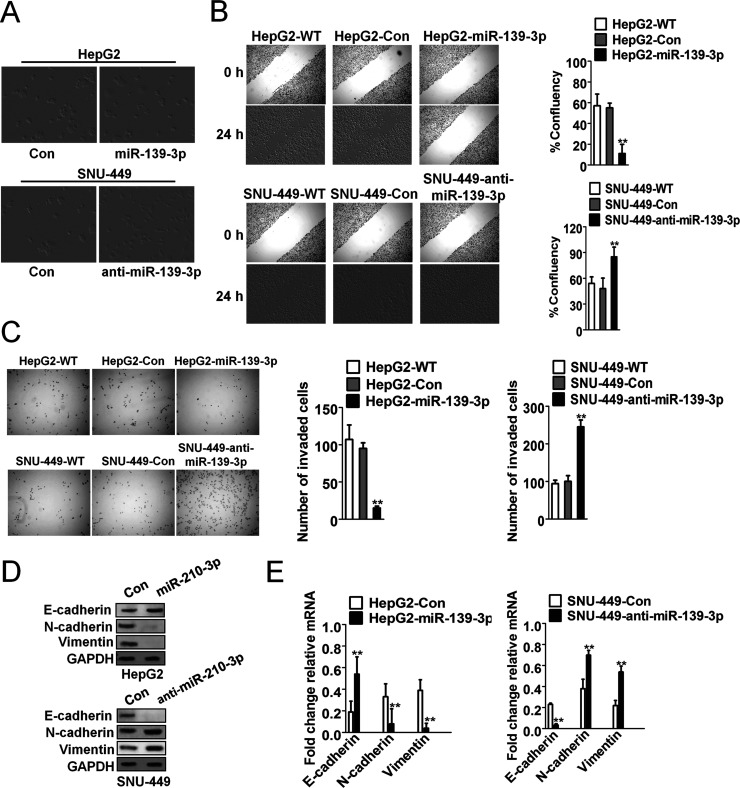
Downregulation of miR-139-3p Is Associated With EMT in HCC Cells
To investigate the potential roles of miR-139-3p in human HCC, HepG2 and SNU-449 cells were transfected with recombinant lentivirus vector containing pre-miR-139-3p to increase the level of miR-139-3p. These subclones were HepG2-WT (wild type of HepG2), HepG2-control (HepG2 transfected with the negative control of precursor miRNA), and HepG2-miR-139-3p (HepG2 transfected with pre-miR-139-3p), as well as SNU-449-WT (wild type of SNU-449), SNU-449-control (SNU-449 transfected with the negative control of anti-miRNA-LNAs), and SNU-449-anti-miR-139-3p (SNU-449 transfected with anti-miRNA-LNAs against miR-139-3p). The SNU-449-anti-miR-139-3p cells gained an EMT-like phenotype, compared to SNU-449-control cells, showing an irregular fibroblast-like shape instead of a typical epithelial cobblestone appearance, whereas HepG2-miR-139-3p cells had no morphological changes compared to HepG2-control cells (Fig. 2A). In the cell wound healing migration and Transwell invasion assays, HepG2-control cells exhibited markedly higher migration ability than HepG2-miR-139-3p cells, while SNU-449-anti-miR-139-3p cells exhibited significantly higher migration potential than control cells (Fig. 2B and C). Western blot (Fig. 2D) and qRT-PCR assays (Fig. 2E) demonstrated that miR-139-3p upregulated the expression of the epithelial cell marker E-cadherin and downregulated the expression of the mesenchymal cell markers N-cadherin and vimentin, whereas anti-miR-139-3p caused an opposite outcome. These results indicate that downregulation of miR-139-3p induced EMT in HCC cells.
Figure 2.
Downregulation of miR-139-3p induced epithelial–mesenchymal transition (EMT) in HCC cells. (A) SNU-449-anti-miR-139-3p cells exhibited morphologic changes consistent with EMT, compared to control cells. HepG2 cells did not show morphological changes. (B) Cell migration through wound healing analysis was compared between HCC cells transfected with miR-139-3p or anti-miR-139-3p. The percentage of wound closure was evaluated. **p < 0.01 compared with HepG2-wild type (WT) or SNU-449-WT cells. (C) HepG2 cells were transfected with miR-139-3p, and SNU-449 cells were transfected with anti-miR-139-3p, and then the invasion ability of both cell lines was investigated by Transwell invasion assay. Quantitative analysis of the total invasive cells from three independent experiments is shown on the right. **p < 0.01 compared with HepG2-WT or SNU-449-WT cells. (D) Western blot assays were performed to evaluate the expression of E-cadherin, N-cadherin, and vimentin in SNU-449-anti-miR-139-3p and HepG2-miR-139-3p cells. (E) The mRNA of E-cadherin, N-cadherin, and vimentin in SNU-449-anti-miR-139-3p and HepG2-miR-139-3p cells was assayed by qRT-PCR assay. **p < 0.01 compared with HepG2-Con or SNU-449-Con cells.
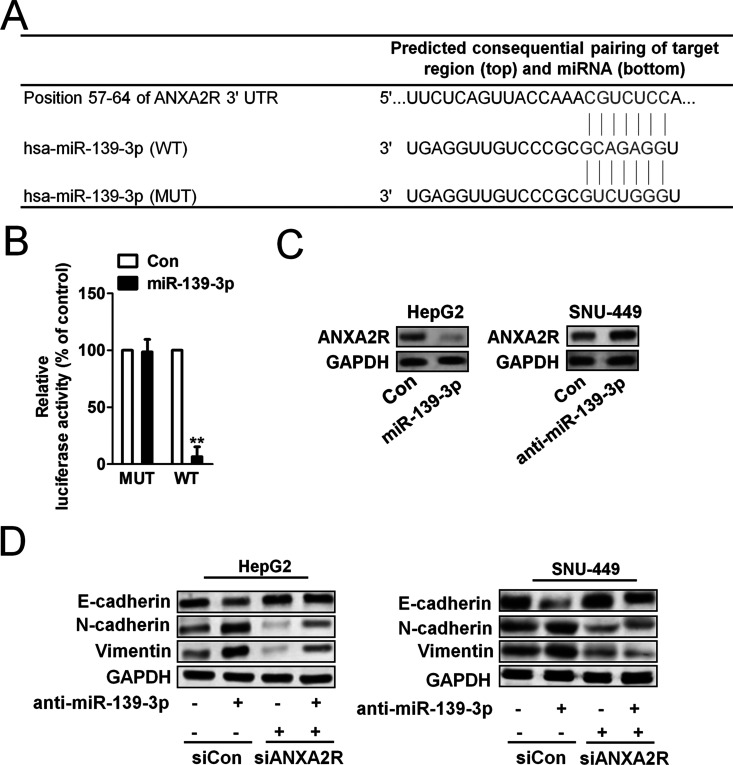
miR-139-3p Regulated EMT by Targeting ANXA2R
miRNAs generally regulate target gene expression by directly binding to its 3′-UTR region. TargetScan (http://www.targetscan.org) was used to predict the target genes of miR-139-3p and inspired us that ANXA2R might be the target of miR-139-3p (Fig. 3A). Overexpression of ANXA2R has been demonstrated to correlate with tumor metastasis and poor prognosis in various cancer types, including breast cancer, pancreatic cancer, and multiple myeloma. In order to verify the 3′-UTR of ANXA2R as a direct target of miR-139-3p, luciferase reporter assay was performed to determine whether miR-139-3p directly regulated ANXA2R expression in HepG2 cells. The target sequence of ANXA2R 3′-UTR (WT 3′-UTR), which contained a miR-139-3p putative binding site, could be mutated, so a mutant sequence (MUT 3′-UTR) messenger RNA (mRNA) was cloned downstream of the luciferase reporter gene vector19. A significant decrease in luciferase activity was observed when the cells were transfected with miR-139-3p compared to the control. The activity of the MUT 3′-UTR vector was unaffected by a simultaneous transfection with miR-139-3p (Fig. 3B). To explore the mechanisms of miR-139-3p in moderating tumor growth and metastasis of HCC tumors, several assays were performed. Western blot analysis showed that ANXA2R expression was decreased after transfection with miR-139-3p in HepG2 cells and increased after downregulation of miR-139-3p in SNU-449 cells compared to respective controls (Fig. 3C). To investigate the relationship of ANXA2R and E-cadherin expression, we transfected our two HCC cell lines with ANXA2R short hairpin RNA (shRNA) to explore whether miR-139-3p could regulate E-cadherin expression by directly targeting ANXA2R expression. The results show that ANXA2R shRNA reduced E-cadherin expression and eliminated the difference in E-cadherin expression between HepG2-control and HepG2-miR-139-3p (Fig. 3D, left) and between SNU-449-control and SNU-449-anti-miR-139-3p (Fig. 3D, right). These results suggest that miR-139-3p directly interacted with ANXA2R mRNA and inhibited the expression of ANXA2R.
Figure 3.
Annexin A2 receptor (ANXA2R) played a critical role in miR-139-3p modulating EMT process. (A) Diagram of the putative binding sequence of miR-139-3p in the 3′-untranslated region (3′-UTR) containing reporter constructs of ANXA2R is shown. (B) Luciferase reporter assays showed that miR-139-3p significantly decreased the luciferase activity of WT ANXA2R but not mutant (MUT) ANXA2R. **p < 0.05 compared with control. (C) Western blot assay showed that miR-139-3p downregulated ANXA2R expression in HepG2 cells, and anti-miR-139-3p upregulated the expression of ANXA2R in SNU-449 cells. (D) ANXA2R shRNA transfection in HepG2 cells eliminated the expression of E-cadherin in miR-139-3p overexpression cells. ANXA2R shRNA transfection in SNU-449 cells rescued the expression of E-cadherin in cells transfected with anti-miR-139-3p.
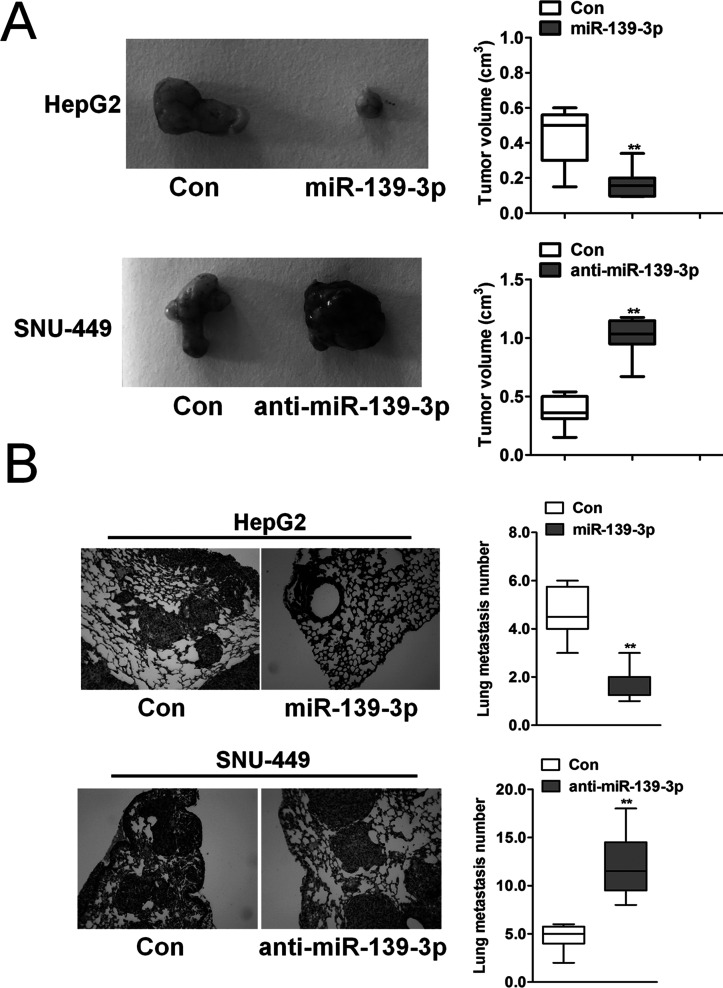
miR-139-3p Inhibited Tumor Growth and Metastasis In Vivo
To explore the role of miR-139-3p in tumor growth and metastasis, orthotopic nude mouse models were established with HepG2 and SNU-449 cell lines. Mice with SNU-449-anti-miR-139-3p tumors had larger tumors (Fig. 4A, top) than those with SNU-449-control tumors. Mice with HepG2-miR-139-3p tumors had smaller tumors (Fig. 4A, bottom) compared to those with HepG2-control tumors. Finally, more pulmonary metastatic foci occurred in the SNU-449-anti-miR-139-3p group than in the SNU-449-control group (Fig. 4B). Nevertheless, only few pulmonary metastases were detected in the mouse models established with HepG2-miR-139-3p cells (Fig. 4B). These results indicate that miR-139-3p expression inhibited tumor progression in vivo, and targeting the ANXA2R-related EMT process may have been one of the main mechanisms.
Figure 4.
miR-139-3p inhibited metastasis process in vivo. (A) HCC subcutaneous tumor tissues (SNU-449-control, SNU-449-anti-miR-139-3p, HepG2-control, and HepG2-miR-139-3p) were implanted into nude mouse to establish the xenograft HCC models. Twenty-five days after implantation, the mice were euthanized, and tumor volumes were assessed. **p < 0.01 compared with control cells. (B) Quantification of lung metastatic foci showed that downregulation of miR-139-3p significantly accelerated pulmonary metastasis of SNU-449-anti-miR-139-3p compared to SNU-449-control tumors. Few pulmonary metastases occurred in HepG2-miR-139-3p xenografts based on quantification of metastatic foci. **p < 0.01 compared with control cells.
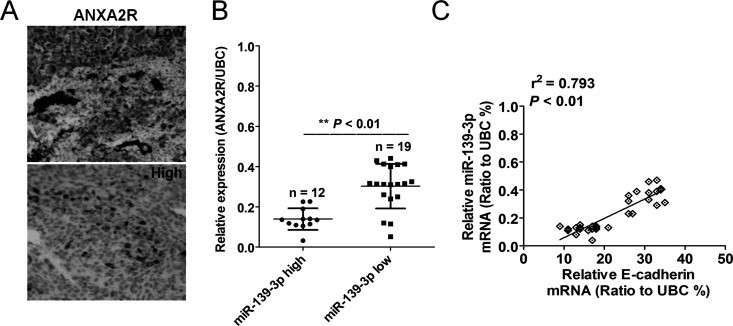
miR-139-3p Expression Was Inversely Correlated With ANXA2R in Human HCC Tissue
To investigate the relationships between miR-139-3p expression and ANXA2R expression, we examined miR-139-3p expression in 31 HCC patients with expression of ANXA2R. Representative images of immunostaining for ANXA2R are shown in Figure 5A. Using a median value of miR-139-3p expression as a cutoff point, we divided the 31 cases of HCC patients into two groups. Patients with high miR-139-3p expression had significantly lower ANXA2R expression than those with low miR-139-3p expression (Fig. 5B). We then checked the relevance of the ANXA2R protein and miR-139-3p mRNA levels using Pearson’s correlation scatter plots. The results showed that there was negative correlation between the two levels (Fig. 5C). The results further support an inverse relationship between miR-139-3p and ANXA2R expression in human HCC tissue.
Figure 5.
The relationship between miR-139-3p expression and the expression of ANXA2R in human HCC tissue. (A) Representative images of ANXA2R staining are shown. (B) An inverse association between miR-139-3p and ANXA2R expression was found. **p < 0.01 compared with miR-139-3p high group. (C) Patients with high miR-139-3p expression had significantly lower ANXA2R expression compared with those with low miR-139-3p expression.
DISCUSSION
miR-139-3p plays a critical role in various types of human cancer. Although a previous study has shown that it may function as an oncogene in malignant glioma, lung cancer, cholangiocarcinoma, and chronic lymphocytic leukemia, emerging evidence demonstrates that miR-139-3p may serve as a potential tumor suppressor in other malignant tumors20. A previous study showed that miR-139-3p was downregulated in colon cancer samples with a poor prognosis, while high levels of miR-139-3p expression were associated with a good prognosis for patients8. Moreover, another study found that miR-139-3p inhibited cervical cancer cell proliferation, migration, and invasion, and induced cell apoptosis through downregulation of ANXA2R expression7. However, the underlying mechanisms associated with miR-139-3p in HCC growth and metastasis are still not fully understood. Here we demonstrated that miR-139-3p inhibits the migration and invasion processes by regulating ANXA2R expression in HCC through a cell autonomous mechanism.
Metastasis and tumorigenesis are multistep processes, and a considerable amount of evidence indicates that EMT is responsible for converting noninvasive tumor cells into cells with metastatic potential, enabling invasion into the basement membrane and the vascular system, survival in the circulation, and extravasion at a distant secondary site21. In addition, EMT has been demonstrated to significantly contribute to liver fibrosis, which provides a premetastatic niche or metastatic-supportive microenvironments. A previous study has demonstrated the critical role of miRNAs in EMT through regulation of E-cadherin expression22. Moreover, some reports have found that miR-139-3p strongly reduces the expression of ANXA2R in some tumors8,23. In the present study, miR-139-3p was found to be inversely correlated with ANXA2R expression in the HCC cells and HCC tissues, and overexpression of miR-139-3p decreased the luciferase reporter activity of the WT 3′-UTR of ANXA2R but not the MUT 3′-UTR. More importantly, the effects of miR-139-3p modulation on EMT in HCC were affected by the ANXA2R shRNA. The data support that miR-139-3p could inhibit EMT by repressing the expression of ANXA2R.
This study has some potential limitations. Our results showed that miR-139-3p inhibited EMT through targeting ANXA2R expression. However, they did not exclude other signal pathways that may modulate EMT and could be mediated by miR-139-3p. Since EMT has been critically discussed as the key process in tumor aggressiveness and metastasis, our findings in the present study demonstrate that miR-139-3p as a suppressive miRNA could inhibit tumor metastasis and invasion partly by impeding EMT through repression of ANXA2R. The data also suggest that miR-139-3p could be a marker and potential therapeutic target in HCC patients in the future. In conclusion, in view of the anti-invasive effects of miR-139-3p on tumor cells, therapeutic miR-139-3p delivery in the treatment of HCC deserves further investigation.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Lo CH, Yang JF, Liu MY, Jen YM, Lin CS, Chao HL, Huang WY. Survival and prognostic factors for patients with advanced hepatocellular carcinoma after stereotactic ablative radiotherapy. PLoS One 2017;12(5):e0177793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu SH, Huang JZ, Xu ML, Yu G, Yin XF, Chen D, Yan GR. ACK1 promotes gastric cancer epithelial-mesenchymal transition and metastasis through AKT-POU2F1-ECD signalling. J Pathol. 2015;236(2):175–85. [DOI] [PubMed] [Google Scholar]
- 3. Xie CH, Cao YM, Huang Y, Shi QW, Guo JH, Fan ZW, Li JG, Chen BW, Wu BY. Long non-coding RNA TUG1 contributes to tumorigenesis of human osteosarcoma by sponging miR-9-5p and regulating POU2F1 expression. Tumour Biol. 2016;37(11):15031–41. [DOI] [PubMed] [Google Scholar]
- 4. Hu X, Zhai Y, Kong P, Cui H, Yan T, Yang J, Qian Y, Ma Y, Wang F, Li H, Cheng C, Zhang L, Jia Z, Li Y, Yang B, Xu E, Wang J, Yang J, Bi Y, Chang L, Wang Y, Zhang Y, Song B, Li G, Shi R, Liu J, Zhang M, Cheng X, Cui Y. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93. [DOI] [PubMed] [Google Scholar]
- 5. Liu Y, Wang Y, Sun X, Mei C, Wang L, Li Z, Zha X. miR-449a promotes liver cancer cell apoptosis by downregulation of Calpain 6 and POU2F1. Oncotarget 2016;7(12):13491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Geng L, Chaudhuri A, Talmon G, Wisecarver JL, Are C, Brattain M, Wang J. MicroRNA-192 suppresses liver metastasis of colon cancer. Oncogene 2014;33(46):5332–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang P, Xi J, Liu S. MiR-139-3p induces cell apoptosis and inhibits metastasis of cervical cancer by targeting NOB1. Biomed Pharmacother. 2016;83:850–6. [DOI] [PubMed] [Google Scholar]
- 8. Liu X, Duan B, Dong Y, He C, Zhou H, Sheng H, Gao H, Zhang X. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7(11):8046–52. [PMC free article] [PubMed] [Google Scholar]
- 9. Wang L, Yao J, Sun H, He K, Tong D, Song T, Huang C. MicroRNA-101 suppresses progression of lung cancer through the PTEN/AKT signaling pathway by targeting DNA methyltransferase 3A. Oncol Lett. 2017;13(1):329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol Cell 2007;27(1):91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noel A, Pouyssegur J, Barbry P, Mazure NM, Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013;4:e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshino H, Yonemori M, Miyamoto K, Tatarano S, Kofuji S, Nohata N, Nakagawa M, Enokida H. MicroRNA-210-3p depletion by CRISPR/Cas9 promoted tumorigenesis through revival of TWIST1 in renal cell carcinoma. Oncotarget 2017;8(13):20881–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding L, Zhao L, Chen W, Liu T, Li Z, Li X. miR-210, a modulator of hypoxia-induced epithelial-mesenchymal transition in ovarian cancer cell. Int J Clin Exp Med. 2015;8(2):2299–307. [PMC free article] [PubMed] [Google Scholar]
- 14. Han F, Wang C, Wang Y, Zhang L. Long noncoding RNA ATB promotes osteosarcoma cell proliferation, migration and invasion by suppressing miR-200s. Am J Cancer Res. 2017;7(4):770–83. [PMC free article] [PubMed] [Google Scholar]
- 15. Khotskaya YB, Goverdhan A, Shen J, Ponz-Sarvise M, Chang SS, Hsu MC, Wei Y, Xia W, Yu D, Hung MC. S6K1 promotes invasiveness of breast cancer cells in a model of metastasis of triple-negative breast cancer. Am J Transl Res. 2014;6(4):361–76. [PMC free article] [PubMed] [Google Scholar]
- 16. Li P, Xue WJ, Feng Y, Mao QS. MicroRNA-205 functions as a tumor suppressor in colorectal cancer by targeting cAMP responsive element binding protein 1 (CREB1). Am J Transl Res. 2015;7(10):2053–9. [PMC free article] [PubMed] [Google Scholar]
- 17. Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-kappaB signaling pathway. Oncotarget 2016;7(9):9939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng WC, Chung IF, Tsai CF, Huang TS, Chen CY, Wang SC, Chang TY, Sun HJ, Chao JY, Cheng CC, Wu CW, Wang HW. YM500v2: A small RNA sequencing (smRNA-seq) database for human cancer miRNome research. Nucleic Acids Res. 2015;43(Database issue):D862–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tsuchiya S, Fujiwara T, Sato F, Shimada Y, Tanaka E, Sakai Y, Shimizu K, Tsujimoto G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1). J Biol Chem. 2011;286(1):420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu X, Li J, Yu Z, Li J, Sun R, Kan Q. MiR-935 promotes liver cancer cell proliferation and migration by targeting SOX7. Oncol Res. 2017;25(3):427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miao Y, Zheng W, Li N, Su Z, Zhao L, Zhou H, Jia L. MicroRNA-130b targets PTEN to mediate drug resistance and proliferation of breast cancer cells via the PI3K/Akt signaling pathway. Sci Rep. 2017;7:41942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang D, Shi W, Tang Y, Liu Y, He K, Hu Y, Li J, Yang Y, Song J. Prefoldin 1 promotes EMT and lung cancer progression by suppressing cyclin A expression. Oncogene 2017;36(7):885–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107(9):1233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]